ABSTRACT
Background: Rapid identification of gram-positive bacteria and resistance determinants from blood cultures can reduce the time to optimal antibiotic therapy.
Objective: This study evaluates the use of technology to rapidly identify gram-positive bacteria in combination with a pharmacist-directed antimicrobial stewardship protocol in a tertiary-care facility.
Methods: Rapid diagnostic testing was performed on gram-positive blood cultures. Pharmacists were instructed to notify prescribers of results and recommend appropriate antimicrobial therapy based on targeted treatment chart. The primary outcomes were mean time to optimal antibiotic therapy, mean time antibiotics were avoided before traditional culture results, and percent of patients with time to optimal antibiotic therapy reached in less than or equal to 2 hours.
Results: Inclusion criteria were met for 297 patients. Mean time to identify bacteria was 26.8 hours with nucleic acid assay versus 75.3 hours with traditional culture (difference = 48.5 hours, p < .0001). The rapid identification of gram-positive bacteria combined with accepted pharmacist intervention improved time to optimal antibiotic therapy (8.4 vs 15.4 hours, p = .0095). When contaminants were identified, antibiotics were avoided for 39.5 hours before traditional culture with pharmacist intervention versus 37.2 hours (p > .05). Antibiotic change occurred in less than or equal to 2 hours in more patients in the pharmacist intervention group (28% vs 10.5%, p = .0002).
Conclusions: Rapid identification combined with pharmacist intervention significantly improved time to optimal antibiotic therapy and significantly increased the number of patients receiving optimal antibiotic therapy in less than or equal to 2 hours over rapid identification alone. A pharmacist-directed protocol combined with rapid identification enhanced antimicrobial stewardship.
Keywords: antimicrobial stewardship, automated microarray nucleic acid test, gram-positive bacteremia, pharmacist intervention
Gram-positive bacteremia is associated with increased rates of morbidity and mortality and has remained as such in the face of new antibiotic therapies, optimized dosing regimens, and increased collaborative decision-making across disciplines.1 Delays in administration of appropriate antibiotics is an independent predictor of mortality in patients with bactermia.2,3 Recent studies have shown that rapid identification of bacteria and genetic resistance determinants from blood cultures can improve patient outcomes and reduce hospital expenditures.4–6
Hospital microbiology laboratories typically require 24 to 72 hours to identify and determine susceptibility of bacteria isolated from a blood culture. The benefits of rapid identification include reduced time to optimal antibiotic therapy, time to discontinuation of unnecessary antibiotics, length of stay, mortality, risk of antibiotic resistance, and hospital costs.4–6 A previous study showed that a pharmacist-managed antimicrobial stewardship program (ASP) is independently associated with decreased time to administration of antibiotics in addition to the use of rapid identification testing systems.7
The automated nanoparticle probe microarray-based nucleic acid test (Verigene; Nanosphere, Illinois) is an array that identifies common gram-positive bacteria and genetic resistance determinants in 2.5 hours from the time of positive blood culture.8 The grampositive bacteria identified by the microarray-based test include Staphylococcus spp., S. aureus, S. epidermidis, S. lugdudensis, Streptococcus spp., S. pyogenes, S. agalactiae, S. anginosus group, S. pneumoniae, Enterococcus faecalis, E. faecium, and Listeria spp. Additionally, the test can identify the presence of mecA gene for S. aureus or S. epidermidis, which confers resistance to methicillin, and the presence of vanA and vanB genes for E. faecalis or E. faecium, which confers resistance to vancomycin.
Previous published studies have focused on identification of only a few species, have been conducted at large academic hospitals, have been run primarily by infectious disease pharmacists, or have only investigated a small sample of patients. Our study investigates the impact of a pharmacist-directed ASP with rapid identification in a tertiary-care community hospital.
METHODS
Study Design
This single-center, retrospective cohort study was conducted at a 396-bed tertiary care hospital. The hospital's institutional review board approved the study protocol prior to initiation. The hospital's clinical microbiology laboratory implemented the nucleic acid gram-positive array in December 2013 following internal validation. Adult patients with positive blood cultures that were tested with the nucleic acid gram-positive array between March 1, 2014 and November 30, 2014 were included. Patients with a length of stay (LOS) of less than 2 days were excluded. Patients were retrospectively divided into 2 groups. The intervention group comprised those patients with an accepted, documented pharmacist intervention. The control group included all other patients without an accepted pharmacist intervention. An accepted, documented pharmacist intervention was defined as a recommendation provided to the prescriber involving the patient's antibiotic therapy that was acted upon. This includes recommendations to continue current therapy, escalate, de-escalate, or discontinue antibiotic therapy based on array results. The control group was all other patients for whom only the result was reported or the intervention was not accepted by the physician (Figure 1).
Figure 1.
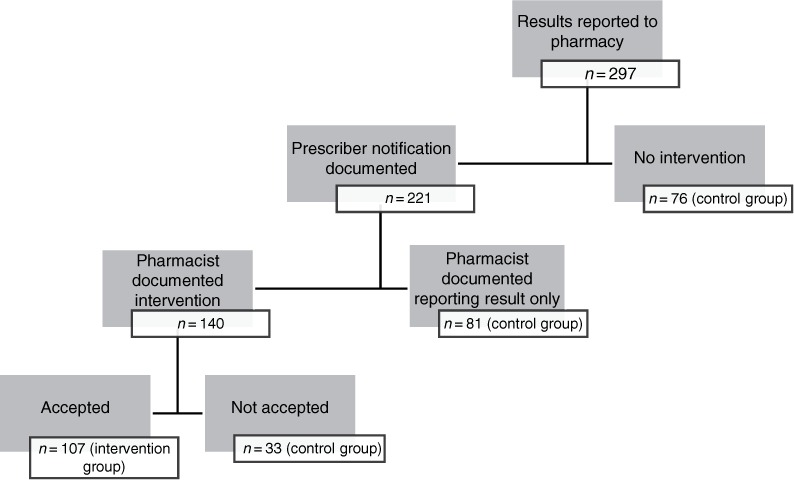
Flowchart of included results.
Data collection included age, gender, LOS, date/time of blood sample collections, date/time/results of rapid array, date/time/results of traditional culture, date/time of antibiotic orders, date/time of antibiotic discontinuation, physician notification, infectious disease (ID) consultation, and pharmacist intervention. The purpose of this study was to demonstrate the impact of using rapid diagnostic technology to identify gram-positive bacteria in combination with a pharmacist-directed ASP in a tertiary-care community hospital.
Stewardship Protocol
All 35 dispensing and clinical pharmacists received a 1-hour training from the ID pharmacist on the automated microarray-based nucleic acid test, interpreting results, and making recommendations to the prescriber. A table was developed to outline the interpretation of array results and provide appropriate antibiotic choices and to prompt communication to the prescriber. An example of the prompted communication was “The results indicate that the organism in the patient's blood culture is likely methicillin sensitive staph aureus as staphylococcus aureus was detected but mecA was not detected. Based on these results, I would recommend changing vancomycin to cefazolin.”
A protocol was concurrently developed and approved by the Pharmacy & Therapeutics and Medical Executive Committees, addressing when the test would be performed, who was responsible for communicating results, and the automatic initiation of antibiotic therapy when appropriate. The pharmacist could automatically initiate one dose of vancomycin 25 to 30 mg per kilogram actual body weight if the rapid array results indicated methicillin-resistant Staphylococcus aureus (MRSA) and the patient was not already on an antibiotic with MRSA coverage.
The protocol stated that the microarray test would be automatically initiated if gram-positive growth was detected between 0600 and 2400 (18-hour operational time) in blood cultures collected from all adult inpatients. The microbiology department provided 24-hour service for reporting results, but initial set-up of the microarray test only occurred between 0600 and 2400. If growth was detected during the remaining 6 hours, the initial set-up of the microarray test occurred at 0600. When the microarray test was completed, the microbiology associate called the charge pharmacist and reported the identification of the organism(s) and resistance genes detected. The time to identify bacteria was calculated as time between blood culture collection and array result.
The pharmacist communicated results to the prescriber along with recommendations for antibiotic changes and/or ID consult if S. aureus were detected. If no antibiotic changes were needed, that was communicated to the prescriber and documented as the recommendation from the pharmacist. Recommendations for ID consults were considered a pharmacist intervention. Pharmacists provided 24-hour coverage for notification of results and antibiotic recommendations. Based on the pharmacist's discretion, results obtained after usual physician office hours could be reported to prescribers the following morning. However, if immediate action was needed based on the results, the pharmacist was instructed to contact the prescriber immediately. Microarray results were also available immediately in the patient's electronic medical record. If no pharmacist communication to the prescriber was documented, the results were included in the control group.
The protocol included an algorithm that dictated specific actions to take if certain results from the microarray were found. For example, the protocol gave staff pharmacists the authority to independently initiate a one-time dose of vancomycin if S. aureus plus mecA was detected, the patient had no contraindications, and the patient was not currently on vancomycin or another antibacterial appropriate for MRSA bacteremia.
The microbiology lab performed a traditional culture on all blood samples to identify organisms and determine susceptibilities. This was performed using VITEK 2 automated system (bioMérieux, Marcy-l'Étoile, France). Time to traditional culture was determined by calculating the time between when the blood culture was obtained and the time when culture and sensitivity report was finalized.
Outcomes
The primary outcomes were mean time to optimal antibiotic therapy, mean time antibiotics were avoided before traditional culture results, and percent of patients with time to optimal antibiotic therapy reached in less than or equal to 2 hours. Time to optimal antibiotic therapy was defined as time from array results to time when targeted therapy was ordered for the patient if antibiotics were changed. Mean time antibiotics were avoided before traditional culture results is defined as time therapy was discontinued following array results to time culture results were finalized. Array time was used as starting time versus culture collection time, because the focus of this study was on pharmacist intervention. Optimal antibiotic therapy was defined as antibiotic targeted to the offending pathogen and included de-escalation or discontinuation of unnecessary antibiotics targeting other organisms. Antibiotic discontinuation for contaminated blood cultures was considered optimal antibiotic therapy. A contaminated blood culture was suspected if only 1 of 2 or more blood culture sets were positive for commonly identified contaminants such as coagulase-negative staphylococci.
Statistical Analysis
For inferential statistics for categorical data, either Pearson's chi-square with Yates correction or Fisher's exact test (for small samples) was applied. For continuous data, Student's t test with 95% confidence interval was applied.
RESULTS
The retrospective analysis identified 297 patients with positive blood cultures during the study period. The accepted, documented recommendation group had 107 patients whereas the control group had 190 patients (Figure 1). There was no statistical difference for age, sex, and LOS between both groups (Table 1). Mean time to identify bacteria was 26.8 hours with nucleic acid array versus 75.3 hours with traditional culture, which is a statistically significant difference of 48.5 hours (p < .0001) (Figure 2). Sixty-seven (23%) of the tests were completed during the hours from 1900 to 0600.
Table 1.
Study population demographics
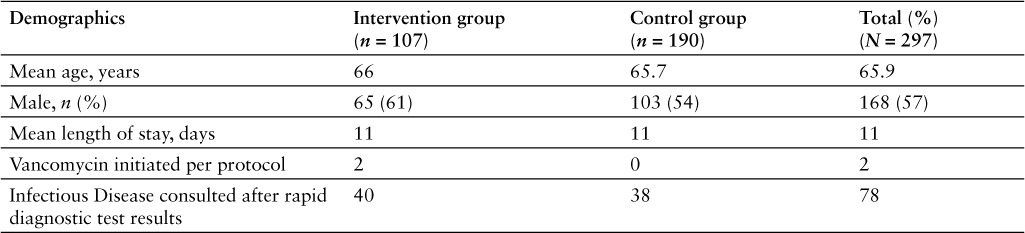
Figure 2.
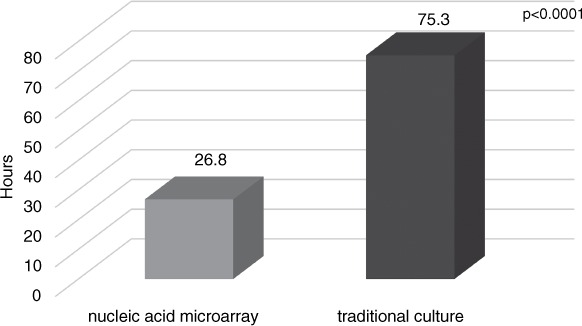
Mean time to identify bacteria by nucleic acid microarray versus traditional culture.
Antibiotics were discontinued following array results (prior to traditional culture results) in 39% (117/297) of patients. Mean number of hours overall that antibiotics were avoided (prior to traditional culture results) was 38.8 hours. Antibiotics were avoided for 39.5 hours before traditional culture with pharmacist intervention versus 37.2 hours in the group without intervention (p > .05) (Figure 3).
Figure 3.
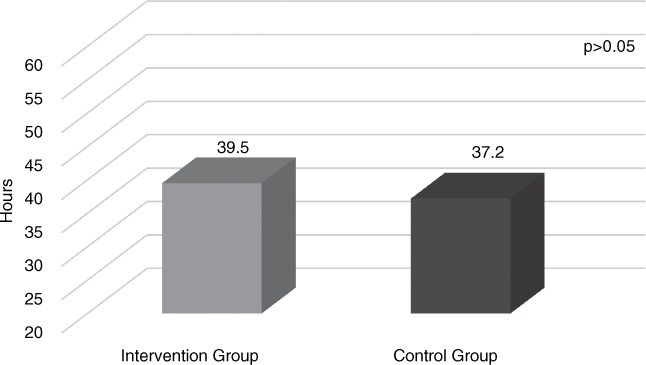
Mean time antibiotics were avoided before final culture.
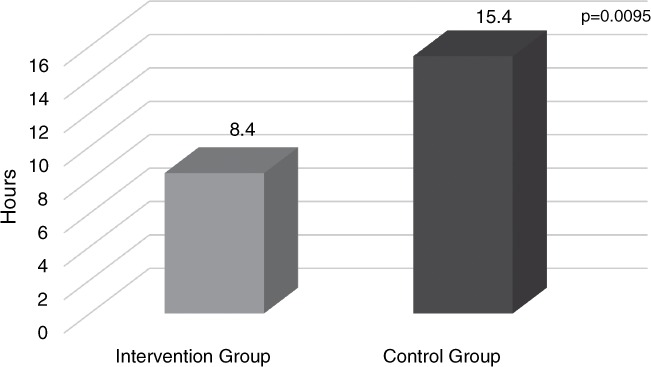
Antibiotics changed following array results (prior to traditional culture results) in 40.4%. If antibiotics changed, the mean time overall to optimal antibiotic following array result was 11.5 hours. The rapid identification of gram-positive bacteria combined with accepted pharmacist intervention improved time to optimal antibiotic therapy (8.4 vs 15.4 hours, p = .0095) (Figure 4).
Figure 4.
Mean time to optimal antibiotic following nucleic acid microarray results.
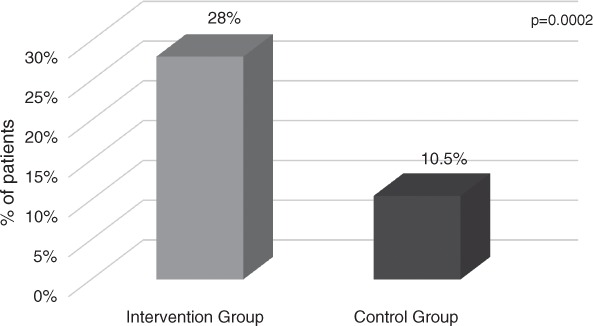
In 50 patients (16.8%), the time to optimal antibiotic order occurred at less than or equal to 2 hours after array result was available. Antibiotic change occurred in less than or equal to 2 hours in more patients in the pharmacist intervention group (28% vs 10.5%, p = .0002) (Figure 5).
Figure 5.
Antibiotic change occurred in 2 hours or less following nucleic acid microarray results.
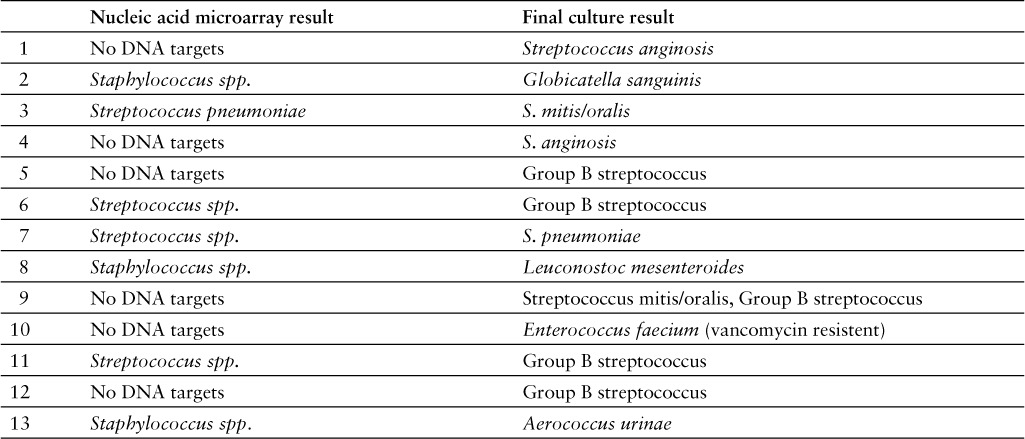
Table 2 shows targets identified by nucleic acid microarray. When comparing rapid identification test with traditional cultures there were 24 (8%) results that did not match the traditional culture results, which resulted in sensitivity and specificity of 94% (95% CI, 91%–97%) and 69% (95% CI, 49%–84%), respectively (n = 298). This specificity result was significantly different (p < .05) than company-supported literature that reported the sensitivity and specificity for the 12 genus or species targets ranged between 92.6% and 100% and 95.4% and 100%, respectively.8 After further review, 54% (13/24) of the discordant results were considered clinically relevant; they would have led to an alternative recommendation based on the table the pharmacists followed for recommendation guidance (Table 3). Discordant results that were considered clinically significant are the following: In 1 sample, no DNA targets were detected by rapid microarray but traditional culture grew vancomycin-resistant E. faecium; in 3 samples, Streptococcus spp. only was detected by rapid microarray but the traditional cultures grew Group B streptococcus, S. pneumoniae, or S. anginosus; in 5 samples, no DNA targets were detected but the traditional cultures grew Group B streptococcus, S. pneumonia, or S. anginosus, which all should have originally been detected by microarray.
Table 2.
Targets identified by nucleic acid microarray
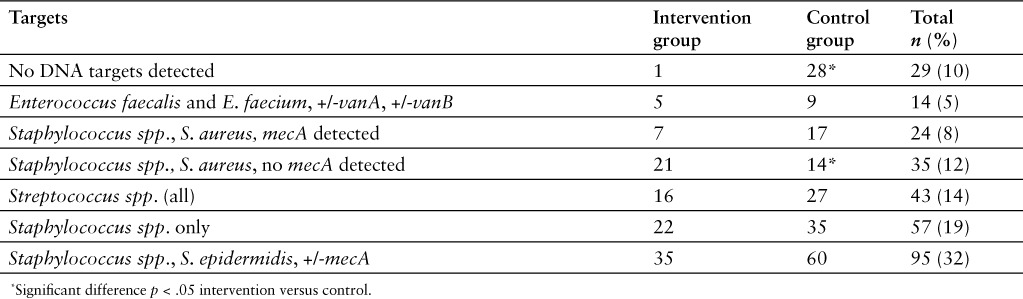
Table 3.
Discordant results
DISCUSSION
Our results are consistent with recent studies that have shown that rapid identification combined with ASPs improves patient outcomes.4–7,9 The outcomes we found are likely due to a combination of the rapid nucleic acid array and pharmacist intervention. The results of our study showed that a pharmacist-directed protocol within the hospital decreased the time to optimal antibiotics.
Pharmacist documentation of prescriber notification should have occurred in all patients but was documented in only 74% of patients. Pharmacist intervention accompanied the results in only 140 of the 221 cases. This may have been due to data collection for this study being started very early in process implementation. Some pharmacists may not have been comfortable with reporting antimicrobial recommendations to prescribers or may not have been documenting accurately since it was early in implementation. If no intervention was documented, it is unknown if and what the pharmacist communicated to the prescribers, and this may have impacted the final results. Education for the pharmacists about the rapid diagnostic technology, making antimicrobial recommendations based on the results, and proper documentation is an important part of implementation. The results of our study suggest that the education prior to implementation could have been improved. Competency assessment prior to implementation is also an important consideration.
Inexperience of laboratory staff may have influenced results. This study was conducted early in the implementation of nucleic acid microarray testing. Technical performance of the microbiologists could have influenced the number of discordant results. Further study should be considered to determine whether the number of discordant results is related to the length of experience with this technology.
As discussed, 23% of the results were completed between 1900 and 0600. In addition to the 24-hour pharmacist coverage, running the nucleic acid array from 0600 to 2400 expanded our patient population, our ability to improve patient care, and outcomes. These results show the need for at least an 18-hour and optimally a 24-hour run nucleic acid array in institutions. Because pharmacists were allowed the discretion to call on the results immediately or during usual physician office hours, time to optimal therapy may be more conservative than if the prescribers were notified immediately for all results. It is not known what percentage of the interventions were called on immediately, although this is an important consideration for future research.
Inadequate prescriber education may have also influenced the results. ID physicians were educated about the technology prior to initiation, but most prescriber education was given by the pharmacist at the time of result notification. As the study began only 3 months after nucleic acid array testing was initiated, many prescribers were unfamiliar with the technology at the time of result notification. This may have affected the acceptance rate of the pharmacist's intervention.
Our study found that 8% of the patients studied had array results that did not match the traditional culture. After review, the recommended antibiotic therapy would have been changed in 54% of these cases. Rapid identification techniques should be evaluated for sensitivity and specificity in clinical practice to see if there may be a difference between the manufacturers' reported results. More research is needed comparing rapid nucleic acid array results to traditional cultures in clinical settings, as discordant results could potentially have a considerable impact on patient outcomes.
There are several limitations to our study. The retrospective design of our study suggests a link between pharmacist-directed stewardship coupled with rapid identification technology and decreased time to optimal antibiotic therapy, but a randomized, prospective design is best suited to confirm causality. Although our results found a significant association between pharmacist interventions and primary outcomes, the lack of an ideal comparator group, such as a group that only received traditional blood cultures, may have led to underestimated results. However, considering the high mortality rate associated with gram-positive bacteremia and potential ethical conflict of withholding rapid identification, we decided this design was the best to minimize potential patient harm.
The array program described in this study could be implemented at other institutions. Rural hospitals and hospitals with limited resources (lack of ID physicians or pharmacists) could have positive outcomes from implementation of a similar program. Additionally, initiation of antibiotics by trained pharmacists based on rapid diagnostic test results could lead to earlier targeted therapy for patients with gram-positive bacteremia.
CONCLUSION
Nucleic acid array reduced the time to grampositive bacteria identification in blood culture by 48.5 hours over traditional culture methods. Rapid identification of gram-positive bacteria in blood cultures combined with pharmacist intervention significantly improved time to optimal antibiotic therapy over rapid identification alone. A pharmacist-directed protocol combined with rapid identification technology enhanced antimicrobial stewardship for patients with gram-positive bacteremia.
ACKNOWLEDGMENTS
The authors declare no conflicts of interest. The authors thank Sara Jones, PharmD, Garrett New, PharmD, Trenton Shoda, PharmD, Nathan Cole, PharmD, Nathan Mullins, MT, Erin Gentry, PharmD, Molly Grasberger, PharmD, and Matthew Becker, PharmD, for their help in the planning, execution, and writing of this study.
REFERENCES
- 1. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004; 39: 309– 317. [DOI] [PubMed] [Google Scholar]
- 2. Kollef MH. Inadequate antimicrobial treatment: An important determinant of outcome for hospitalized patients. Clin Infect Dis. 2003; 31( suppl 4): S131– 138. [DOI] [PubMed] [Google Scholar]
- 3. Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. The clinical significance of positive blood cultures: A comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. Ii. Clinical observations, with special reference to factors influencing prognosis. Rev Infect Dis. 1983; 5: 54– 70. [DOI] [PubMed] [Google Scholar]
- 4. Box MJ, Sullivan EL, Ortwine KN. et al. Outcomes of rapid identification for gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy. 2015; 35( 3): 269– 276. [DOI] [PubMed] [Google Scholar]
- 5. Huang AM, Newton D, Kunapuli A. et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013; 57( 9): 1237– 1245. [DOI] [PubMed] [Google Scholar]
- 6. Perez KK, Olsen RJ, Musick WL. et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med. 2013; 137: 1247– 1254. [DOI] [PubMed] [Google Scholar]
- 7. Messina AP, van den Bergh D, Goff DA. Antimicrobial stewardship with pharmacist intervention improves timeliness of antimicrobials across thirty-three hospitals in South Africa. Infect Dis Ther. 2015; 4: 5– 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchan BW, Ginocchio CC, Manii R. et al. Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: Evaluation of an automated microarray-based nucleic acid test. PLoS Med. 2013; 10( 7): e1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vardakas KZ, Anifantaki FI, Trigkidis KK, Falagas ME. Rapid molecular diagnostic tests in patients with bacteremia: Evaluation of their impact on decision making and clinical outcomes. Eur J Clin Microbiol Infect Dis. 2015; 34( 11): 2149– 2160. [DOI] [PubMed] [Google Scholar]


