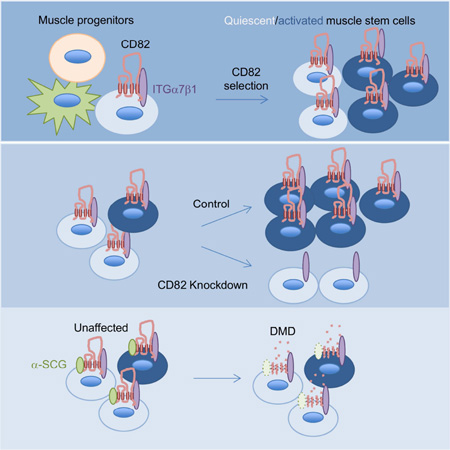
Summary
Cell surface markers for prospective isolation of stem cells from human skeletal muscle have been difficult to identify. Such markers would be powerful tools for studying satellite cell function during homeostasis and in pathogenesis of diseases such as muscular dystrophies. In this study, we show that the tetraspanin KAI/CD82 is an excellent marker for prospectively isolating stem cells from human fetal and adult skeletal muscle. Human CD82+ muscle cells robustly engraft into a mouse model of muscular dystrophy. shRNA knockdown of CD82 in myogenic cells reduces myoblast proliferation, suggesting it is functionally involved in muscle homeostasis. CD82 physically interacts with alpha7beta1 integrin (α7β1-ITG) and with α-sarcoglycan, a member of the Dystrophin-Associated Glycoprotein Complex (DAPC), both of which have been linked to muscular dystrophies. Consistently, CD82 expression is decreased in Duchenne muscular dystrophy patients, together suggesting that CD82 function may be important for muscle stem cell function in muscular disorders.
Graphical Abstract
Introduction
The muscular dystrophies are progressive disorders affecting children and adults (Chelly and Desguerre, 2013; Mercuri and Muntoni, 2013). One example of this class of diseases is Duchenne muscular dystrophy (DMD), which is caused by mutations in dystrophin (Monaco et al., 1986), a large cytoplasmic protein located at the sub-sarcolemma of myofibers (Hoffman et al., 1987). Dystrophin functions in muscle by interacting with a group of proteins known collectively as the Dystrophin-Associated Glycoprotein Complex (DAPC) (Yoshida and Ozawa, 1990; Ibraghimov-Beskrovnaya et al., 1992). In the absence of dystrophin, the cellular levels of many DAPC proteins are severely reduced (Ervasti et al., 1990), thus when dystrophin is mutated in DMD the function of other proteins is compromised.
A second protein complex located at the sarcolemma of myofibers is the α7β1 integrin. This protein complex is thought to provide membrane stabilization by linking the cytoskeleton to the extracellular matrix (Burkin and Kaufman, 1999). Mutations α7 integrin (α7-ITG) cause muscle disease in humans (Mayer et al., 1997; Hayashi et al., 1998). Overexpression of α7-ITG in dystrophic mdx mice, a mouse model for DMD (Bulfield et al., 1984), significantly ameliorates the dystrophic pathology via increased stability of the link between α7-ITG and laminin (Burkin et al., 2005). The tetraspanin sarcospan, an associated member of the DAPC, interacts with the α7β1 integrin (Marshall et al., 2015), however whether other proteins are also associated with this complex or link the DAPC and α7β1 integrin protein complexes is not entirely known. In the present study, we demonstrate that the tetraspanin KAI/CD82 is an excellent prospective marker for purification of stem cells from human fetal and adult skeletal muscles. CD82+ human muscle cells successfully engraft in vivo in an immune-deficient mouse model of muscular dystrophy. CD82 interacts with α7β1-ITG in human myogenic cells and it is linked to the DAPC complex via interaction with α-sarcoglycan. Expression of CD82 is decreased in muscle tissue and myoblasts from DMD patients, suggesting that CD82 function may be linked to muscular dystrophies.
Results
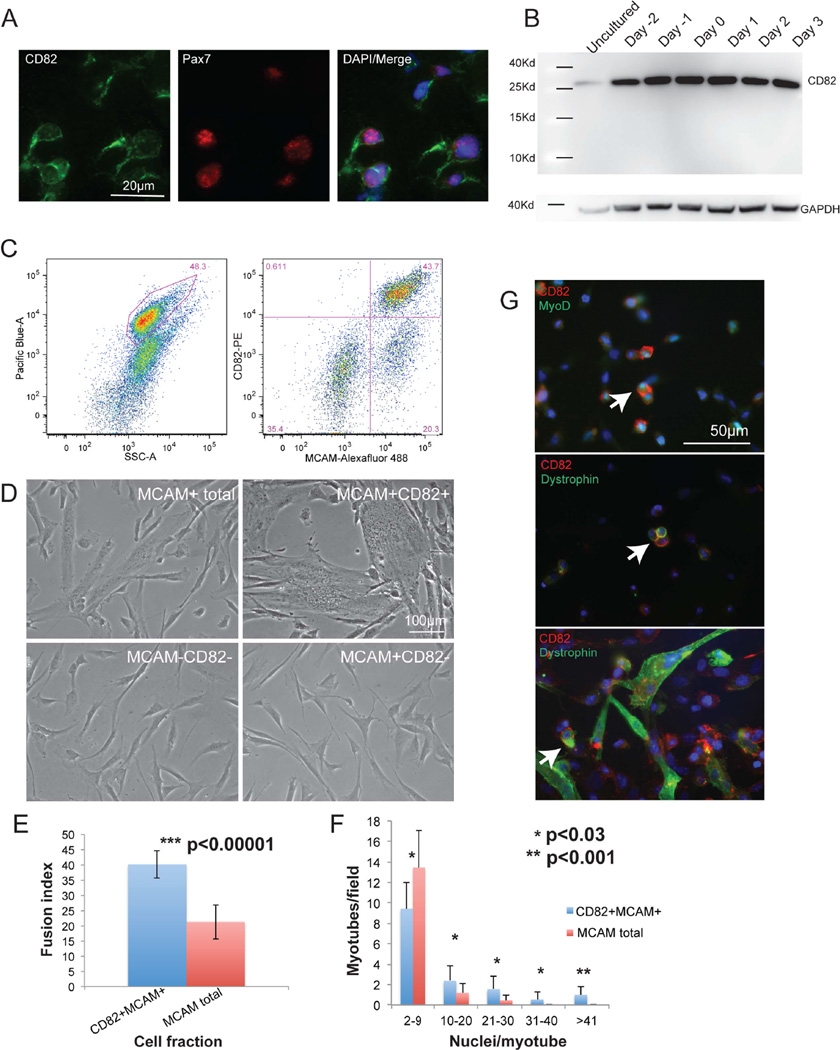
To uncover regulators of human myogenesis, we sought to identify markers that label myogenic cells in developing human muscle. Melanoma Cell-Adhesion Molecule (MCAM) enriches for myogenic cells in human fetal muscle (Lapan and Gussoni, 2012), however MCAM is expressed in both myogenic Pax7+ satellite cells, mature myofibers and a subfraction (~25%) of Pax7− cells (Figure S1A, B). To further refine the myogenic from non-myogenic cells within the MCAM-positive fraction, comparison of the transcriptome of MCAM+ versus MCAM− cells identified CD82 as one candidate preferentially expressed in MCAM+ cells (Table S1). CD82 showed partial myofiber staining and it outlined cells that co-stained with Pax7 in vivo (Figure 1A). By western blot, CD82 protein was detected as a band of ~30Kd in uncultured, proliferating and differentiating human fetal myogenic cells (Figure 1B). FACS analysis of freshly dissociated human fetal muscle cells using CD82 and MCAM confirmed that CD82 marked a subpopulation of MCAM+ cells (Figure 1C). Sorted cell populations were induced to differentiate in vitro and myotube forming activity was restricted to the CD82+ subpopulation of MCAM+ cells, while neither double negative, nor MCAM+CD82− retained myotube-forming potential (Figure 1D). To confirm enrichment in myogenic activity when CD82 and MCAM were used in conjunction, the myotube formation ability of MCAM+CD82+ cells was compared to MCAM+ (total) cells. The fusion index (Figure 1E) and myotube size (Figure 1F) were significantly higher for MCAM+CD82+ compared to MCAM+ cells. By immunofluorescence, co-staining of CD82, dystrophin and MyoD revealed CD82 expression at the membrane of mononuclear MyoD+ and dystrophin+ cells (Figure 1G). Dystrophin is most known for its function in myofibers (Hoffman et al., 1987), although a role in muscle stem cell division has recently been reported (Dumont et al., 2015).
Figure 1. CD82 is a prospective marker for muscle satellite cells.
A) Cross sections of human fetal skeletal muscle immunostained for CD82 (green) and Pax7 (red). Co-staining confirmed that satellite cells express CD82. Nuclei are stained in blue with DAPI. Scale bars=50µm. B) Western blot of differentiating human fetal cells showing expression of CD82 (~30Kd) at all timepoints analyzed. C) FACS purification of myogenic cells from human fetal tissue using CD82 and MCAM. Live cells were gated based on Calcein Blue signal (left) and 4 cell fractions were isolated: MCAM+CD82+, MCAM+CD82−, MCAM−CD82− and MCAM+ total (MCAM+CD82+ and MCAM+CD82− combined). D) Cells fractions were plated at the same density and induced to differentiate for 48 hours (scale bar: 100µm). MCAM−CD82− and MCAM+CD82− never fused, suggesting that selection for both MCAM and CD82 enriches for myogenic activity. E) The fusion index of MCAM+CD82+ at 48hrs following differentiation was significantly higher than in MCAM total (p<0.00001, n=5 samples). F) Myotube size is significantly increased in MCAM+CD82+ cells compared to MCAM total. For each sample, 10 independent microscopic images with 250–540 nuclei/image were analyzed. Small myotubes (2–9 nuclei in size) are significantly increased in MCAM total cells, while large myotubes containing >41 nuclei were only seen in the MCAM+CD82+ cell fraction. Data are represented as mean ± SEM and p values were calculated via t-test. G) Co-expression of CD82, MyoD and dystrophin in cultured primary human fetal muscle cells (P1). CD82 localizes to the cell membrane of mononuclear myogenic cells expressing MyoD or dystrophin (arrows) (Dumont et al., 2015) and it is maintained in differentiating cultures. Scale bar: 50 microns.
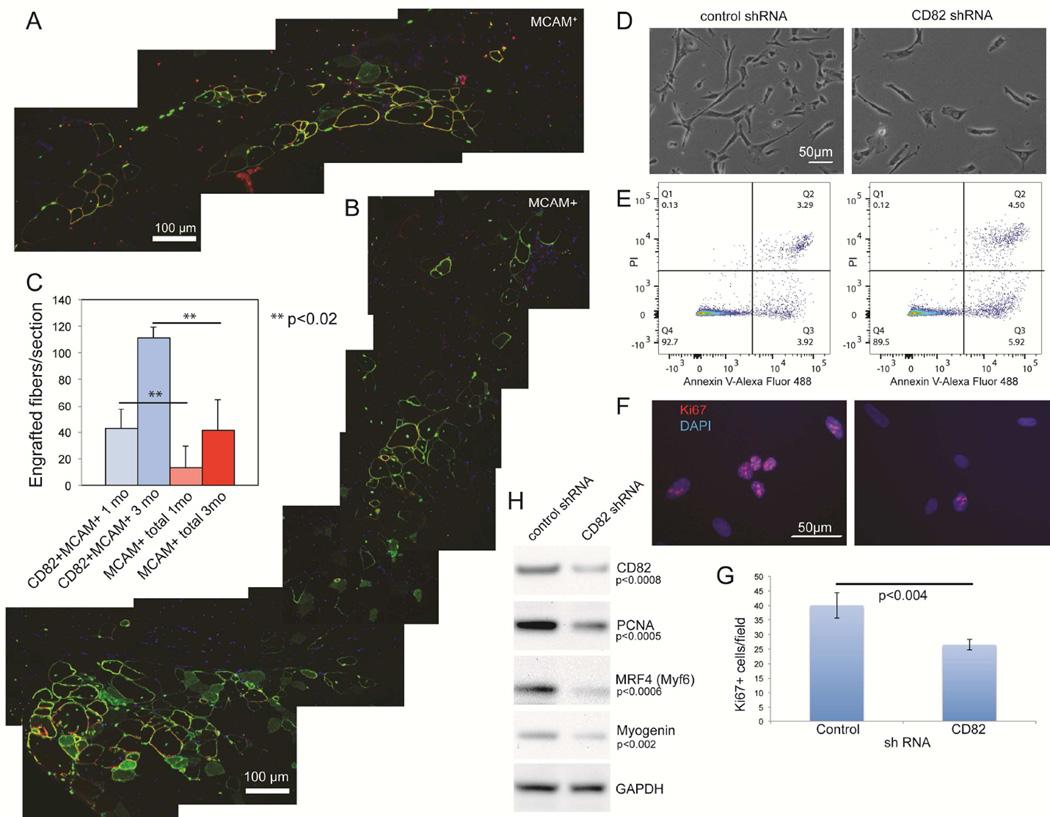
To determine if increased cell fusion in vitro translates into efficient myogenic cell engraftment in vivo, immune-deficient mice with muscular dystrophy (NODRag1nullmdx5cv) (Rozkalne et al., 2014) were injected intramuscularly with human fetal CD82+MCAM+ and MCAM+ total cells. Tissues were harvested 1 and 3 months after transplantation and cell engraftment was quantified based on co-expression of human-specific spectrin, human lamin A/C and dystrophin (Figure 2A, B) (Rozkalne et al., 2014). At both timepoints, significantly more myofibers of human origin were found in recipient muscles injected with MCAM+CD82+ cells compared to MCAM+ cells (Figure 2C, p<0.02). To investigate if CD82 would be a useful marker for myogenic cell selection in human adult muscle, mononuclear cells were stained for CD56 (NCAM) and CD82 (Figure S2A). FACS analyses showed co-expression of CD82 and CD56 and only the CD82+ cells in the CD56+ fraction exhibited myogenic activity in vitro (Figure S2B) and in vivo (Figure S2D). Immunofluorescence co-staining of CD82 and Pax7 on muscle tissue sections detected CD82 expression on the membrane of Pax7+ satellite cells (Figure S2C). To study the function of CD82 in myogenic cells, CD82-sh or control-sh RNA silencing was performed on primary human fetal MCAM+CD82+ myogenic cells using lentiviral vectors (Figure 2D). CD82-sh cultures showed a significantly decreased number of total cells, which was not due to apoptosis (Figure 2E) but to impaired cell proliferation, as confirmed by Ki-67 immunostaining (Figure 2F–G) and decreased levels of PCNA expression (Figure 2H). Additionally, myogenic differentiation appeared decreased in CD82-silenced cells, as indicated by decreased MRF4 (Myf6) and myogenin expression.
Figure 2. CD82+MCAM+ human muscle cells engraft in immune-deficient mice with muscular dystrophy.
IM injections were performed in the tibialis anterior (TA) muscle of recipient NODRag1nullmdx5cv mice. 100,000 cells positive for MCAM alone (MCAM+) were injected in one TA, while the contralateral TA received 100,000 double-positive cells (MCAM+CD82+). A) Merged images showing the entire injection site 3 months after transplantation of human fetal MCAM+ cells. B) Merged images showing the entire injection site using human fetal MCAM+CD82+ cells 3 months following transplantation. Red: human spectrin; green: human lamin A/C (nuclei contour) and dystrophin (myofiber contour); blue: nuclei (DAPI) C) Engraftment quantification was performed based on expression of both human specific spectrin and dystrophin (Rozkalne et al., 2014). At 4 weeks after transplantation we observed significantly more myofibers of human origin in muscles injected with MCAM+CD82+cells compared to MCAM+ cells (**p<0.02). The same was true for muscles harvested 3 months after transplantation (n=7 per each group, **p<0.02). D) CD82-shRNA and control shRNA on primary human muscle cells using commercial lentiviruses (Santa Cruz Biotechnology). The total number of cells was significantly decreased in CD82-sh compared to control-sh cultures (t-test p<0.03). Apoptosis assays utilized Annexin V detection (D), since caspase 3 activity can regulate other functions, including self-renewal of satellite cells (Dick et al., 2015). No significant differences in apoptosis were seen between control- and CD82-sh cultures. Ki67 immunostaining (F) was used to determine the number of proliferating cells in the cultures, which was significantly decreased in CD82shRNA cultures via t-test (G). H) Western blot analyses in control- and CD82-shRNA cultures confirmed decreased expression of CD82, PCNA, MRF4 and myogenin. GAPDH was used as a loading control.
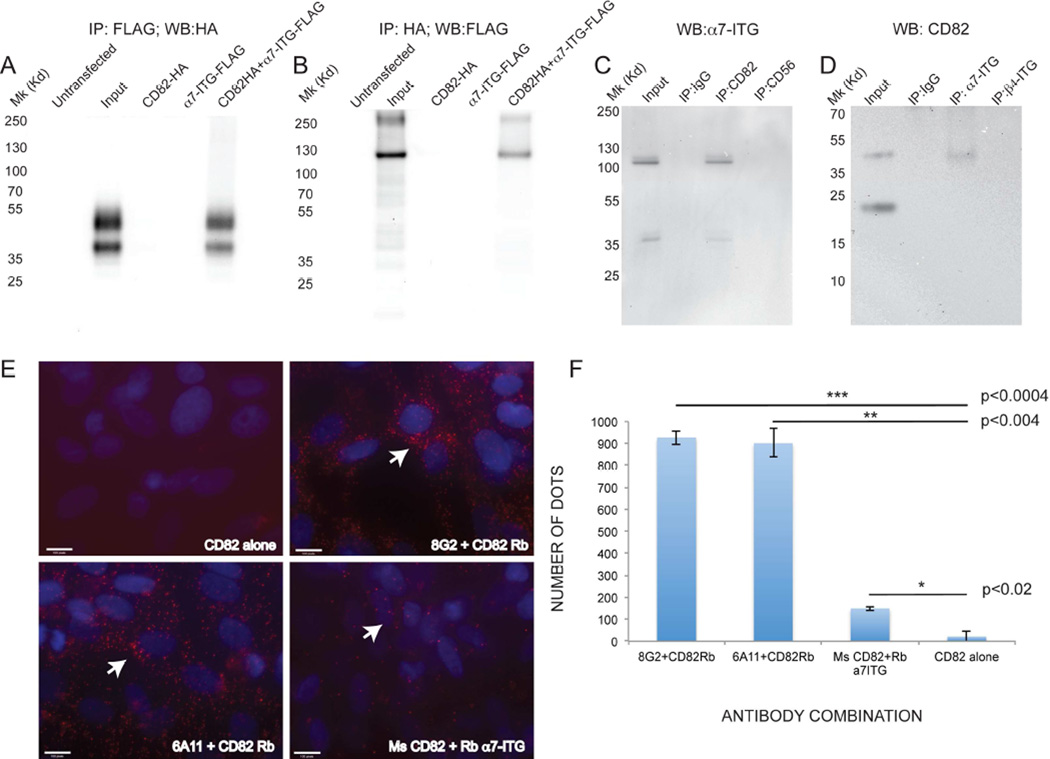
Tetraspanins are known to form complexes with other proteins at the cell membrane (Hemler, 2005). CD82 is known to interact with integrins, including α3, α4 and α6 integrin in a cell-specific manner (Mannion et al., 1996; Okochi et al., 1997). We hypothesized that CD82 may interact with α7 integrin (α7-ITG), given in known expression in fetal and adult muscle satellite cells (Gnocchi et al., 2009). Constructs expressing HA-tagged CD82 and FLAG-tagged α7-ITG were transfected into HEK293 cells individually or together (Figure 3A, B). Following immunoprecipitation (IP) using anti-FLAG and western blot using anti-HA, only cells transfected with both constructs exhibited two protein bands, consistent with the expected sizes for non-glycosylated and glycosylated CD82 (Cannon and Cresswell, 2001), suggesting interaction (Figure 3A). Reciprocal IPs using anti-HA for pulldown followed by western blot using anti-FLAG revealed a band at the expected molecular weight (MW) for α7-ITG (Figure 3B). To confirm these findings, endogenous IPs using primary human fetal myoblasts were performed (Figure 3C–D). Control protein lysates (input) detected bands of ~120Kd and 37Kd, consistent with the expected MW of isoforms α7-ITGB and α7-ITGA, respectively (Velling et al., 1996). CD82 successfully co-immunoprecipitated with α7-ITGA and α7-ITGB, while negative control pulldowns using IgG or anti-CD56 did not co-immunoprecipitate e isoform (Figure 3C). Reverse co-IP’s using anti-α7-ITG resulted in CD82 protein pulldown, while immunoprecipitation using IgG or an antibody to another integrin (anti-β4 integrin) did not (Figure 3D). To confirm the vicinity of CD82 and α7-ITG, in situ proximity ligation assays (PLA) were performed on human fetal MCAM+ myogenic cells (Figure 3E) (Fredriksson et al., 2002; Soderberg et al., 2008). A negative control using anti-CD82 alone showed minimal background signal, while antibodies to CD82 and α7-ITG from different species and in different combinations yielded numerous positive red signals, confirming proximity (Figure 3E–F).
Figure 3. CD82 and α7-integrin co-immunoprecipitate.
A, B) protein lysates of HEK293 cells overexpressing CD82-HA-tagged, α7-ITG-FLAG-tagged alone or combined. A) immunoprecipitation with anti-FLAG pulls down CD82-HA only when both proteins are overexpressed. B) Reciprocal co-immunoprecipitation pulls down α7-integrin-FLAG (expected MW ~100–130Kd) and an additional band at ~250 Kd. C) Co-immunoprecipitation of endogenous α7-integrin using anti-CD82 on human primary fetal muscle cell protein lysates. Input lane is a positive control (non-immunoprecipitated lysate). Lysates immunoprecipitated with IgG and anti-CD56 are negative controls for binding specificity, only anti-CD82 immunoprecipitated α7-integrin (MW 100–130Kd). D) Reverse co-IP showing immunoprecipitation of CD82 with anti α7-integrin. IgG and β4-integrin are negative control pulldowns, while the input lane is a positive control (non-immunoprecipitated lysate). E) Proximity ligation assays on purified cultured MCAM+ human fetal myogenic cells. Positive red signals are seen only when both CD82 and α7-ITG antibodies are added (arrows). Nuclei are stained in blue with DAPI. Scale bars: 100 pixels. F) Quantifications of number of dots (points of contact indicating proximity) in PLA assays were compared using a t-test.
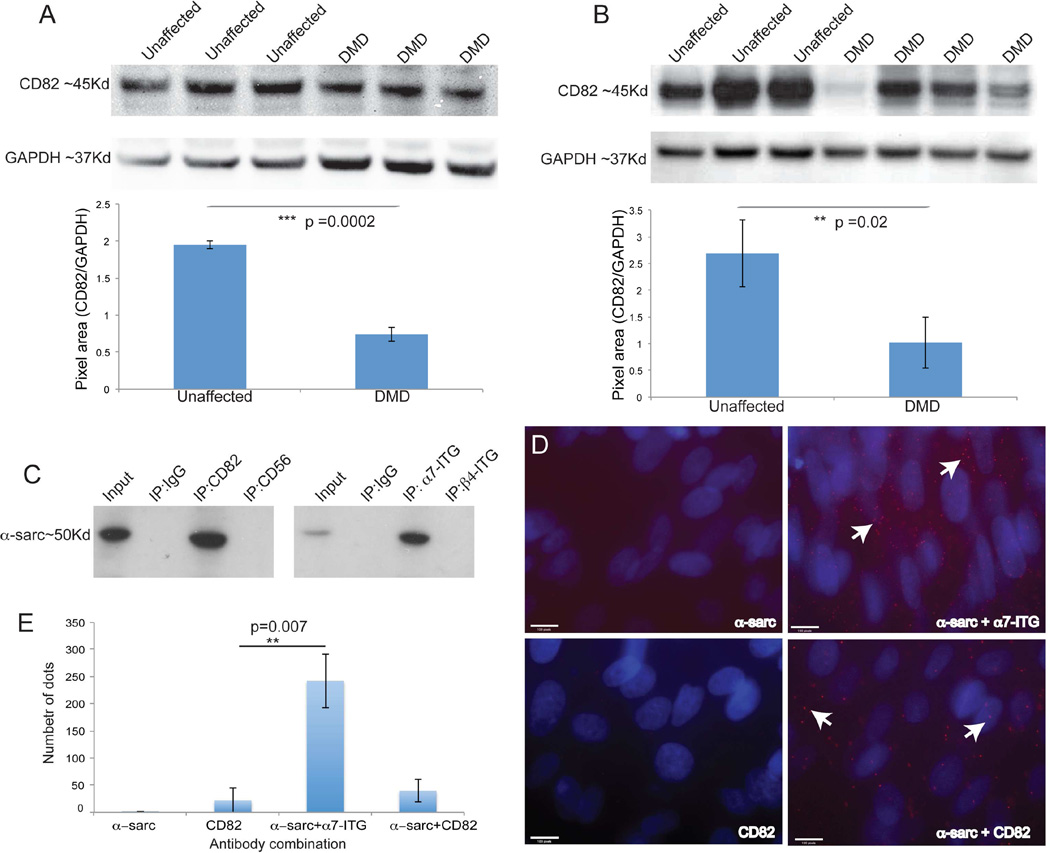
We then sought to examine the relationship of CD82 expression and diseased muscle. Localization of CD82 in human muscle tissue sections from unaffected individuals revealed CD82 immunostaining localized to the membrane of Pax7+ satellite cells (Figure S3A). In addition to Pax7+ satellite cells, affected muscles including DMD muscle showed expression of CD82 in regenerating fibers, which appeared lobulated as if the myofiber cytoplasm was subdivided (arrows). Co-localization of Pax7+ and CD82+ cells was quantified in all conditions (Figure S3B). To determine if the overall expression of CD82 was affected in DMD muscle protein lysates of tissue (Figure 4A) or purified myogenic cells (Figure 4B) were analyzed by western blot. Bands of the expected size for CD82 were seen in unaffected and DMD samples, with variable but significantly decreased expression in DMD. Decreased CD82 expression in dystrophin deficiency raised the possibility that CD82 is linked to disease pathology. We hypothesized that if CD82 is associated with dystrophin or a member of the DAPC, its expression might be decreased in diseased muscle. A candidate DAPC-interacting protein could be α-sarcoglycan, since its expression is reduced in DMD muscle cells compared to normal ones (Cassano et al., 2011) as a result of instability of the entire DAPC complex when dystrophin is mutated (Ervasti et al., 1990). CD82 and α7-ITG protein pulldowns were analyzed by western blot using anti-α-sarcoglycan (Figure 4C) and confirmed that α-sarcoglycan co-immunoprecipitated with CD82 and α7-ITG. Additionally, PLA assays using purified human fetal MCAM+ myogenic cells supported the finding that CD82, α-sarcoglycan and α7-ITG are likely part of a protein complex (Figure 4D–E).
Figure 4. CD82 expression in DMD muscle and co-immunoprecipitation with α–sarcoglycan.
A) Western blot of skeletal muscle tissue lysates of unaffected individuals and DMD patients. CD82 (~37Kd) is present in all samples, although it is significantly decreased in DMD patients. GAPDH (~37Kd) is used as a loading control. B) Western blot of myoblast protein lysates from unaffected controls and DMD patients. CD82 expression is variable, but overall significantly reduced in DMD cells. C) Co-immunoprecipitation of α–sarcoglycan with CD82 and α7-integrin suggest the proteins are in a complex in human myogenic cells. D) PLA assay and positive red signals (seen only when antibodies to two proteins were added) confirm physical proximity of α–sarcoglycan, α7-integrin and CD82. Nuclei are stained in blue with DAPI. E) Quantification of the number of dots seen by PLA assay using different antibody combinations.
To determine if overexpression of CD82 in control or DMD myogenic cells leads to changes in proliferation or differentiation, a doxycycline-inducible lentivirus expressing V5-tagged CD82 was used to infect normal and DMD primary cells (Figure S4). Overexpression was confirmed by FACS (Figure S4A) and by western blot in myoblasts and myotubes (Figure S4B, D). EdU incorporation assayed by FACS revealed no significant changes between control (no Dox) and CD82 overexpressing (+Dox) cultures. Interestingly, DMD myoblasts intrinsically incorporated more EdU than normal cells, suggesting they are hyperactivated. CD82 overexpression in normal myoblasts (Figure S4C) and myotubes (Figure S4E) increased MRF4 expression, in agreement with the silencing data that had shown decreased MRF4 expression following CD82 silencing. In addition, α-sarcoglycan expression in myotubes increased significantly following CD82 overexpression, while α7-integrin expression did not (Figure S4E).
Discussion
The tetraspanins are an important family of proteins known to recruit other proteins at the cell membrane including integrins and cell adhesion molecules, thus initiating important cell decisions (Hemler, 2005). In skeletal muscle, tetraspanins have been associated with muscle differentiation and/or muscle function. The tetraspanins CD9, CD81 and CD53 have each been implicated in muscle cell fusion (Tachibana and Hemler, 1999; Liu et al., 2012). Transgenic overexpression of sarcospan, a tetraspanin associated with the DAPC, leads to dystrophic sarcolemma stabilization through upregulation of utrophin and activation of AKT signaling (Marshall et al., 2012; Marshall et al., 2013). CD82 has been mostly studied in cancer, as its expression is associated with inhibition of metastasis formation (Dong et al., 1995). Recently, CD82 has been reported to maintain the dormancy of hematopoietic stem cells (HSC), suggesting a role for this protein in normal cells, specifically stem cells (Hur et al., 2016). Our studies demonstrate that CD82 is expressed by human muscle stem cells (Pax7+ progenitors), but its expression is also maintained in activated and differentiating myogenic cells. Unlike the hematopoietic system, CD82 expression in muscle is not restricted to the stem cells, suggesting tissue-specific function of this tetraspanin, possibly mediated by unique interacting partners. Downregulation of CD82 expression in purified myogenic cells leads to decreased proliferation and differentiation, while its overexpression increases myogenic cell differentiation. CD82 is likely in a protein complex with α7 integrin, a known muscle satellite cell marker (Blanco-Bose et al., 2001; Pasut et al., 2012) and with α-sarcoglycan, a member of the DAPC complex (Ervasti et al., 1990). Our studies show that CD82 expression is overall reduced in DMD myogenic cells and muscle tissue lysates, while a lobulated pattern of expression is seen in regenerating myofibers, suggesting presence of residual protein following incorporation of a CD82-expressing cell in the myofiber. In patients with mutations in α-sarcoglycan, expression of α7 Integrin is not reduced, but its localization appears in a more internalized position, possibly the costameres (Anastasi et al., 2004). In agreement with these findings, α-sarcoglycan expression appears to be unaffected in α7 integrin null mice (Guo et al., 2006). These findings suggest that the expression of α7 integrin and α-sarcoglycan might not be functionally linked, while our data raises the possibility that CD82 is the protein that supports the stability of the complex.
Upregulation of α7 integrin expression in skeletal muscle is known to increase stability of the sarcolemma and ameliorate the function of dystrophic muscle (Burkin et al., 2001; Burkin et al., 2005). Interestingly, the number of satellite cells in Tg MCK-α7-ITG:mdx muscles is increased (Burkin et al., 2005), leaving open the possibility that CD82 might be involved in some of these observed changes. In addition, drug compounds which upregulate α7 integrin expression might also target CD82 function. Future studies in these models will be able to further elucidate the functional ties among CD82, α7 integrin and α-sarcoglycan.
Experimental Procedures
Please refer to detailed experimental procedures in the Supplemental Information.
Human samples
Human de-identified, discarded fetal tissue and human adult tissue were collected under protocols approved by the Committee of Clinical Investigation at Boston Children’s Hospital (IRB-P00020286 and 03-12-205R), respectively.
Immunofluorescence staining
Frozen muscle tissue sections were fixed in cold acetone at −20°C for 5 min. Antigen retrieval was performed at 95°C for 5 min, slides were then blocked for 45 min and incubated overnight with primary antibodies at 4°C. Slides were washed in PBS-Tween 20 (PSBT), incubated with secondary antibody at RT for 1hr, washed again in PBST and mounted for analysis.
Primary muscle cell purification by FACS
Dissociated mononuclear fetal and adult muscle cells were filtered through a 40µm filter and primary antibodies were diluted in HBSS supplemented with 0.5% BSA (see Supplemental Table). To gate for live cells, calcein blue was incubated along with primary antibodies for 30 min on ice. Samples were washed once and filtered again prior to cell sorting.
In vitro fusion assays
Sorted cells were plated for 18h and incubated in differentiation medium changed daily. Images were taken on a Nikon Eclipse TS100 microscope fitted with a Spot RT3 camera starting at D0 and every 24 hours. The fusion index (FI) was calculated as the percentage of nuclei fused within myotubes/total nuclei.
Cell transplantation in vivo and engraftment analyses
100,000 MCAM+CD82+ or MCAM+ cells were injected IM in opposite TA muscles of immune-deficient dystrophic NODRag1nullmdx5cv recipients. Tissues were harvested, snap-frozen at 4 and 12 weeks post transplantation (Meng et al., 2014) and the entire muscles were sectioned and analyzed as described (Rozkalne et al., 2014). All fibers positive for spectrin and dystrophin were counted as engrafted with human cells. Engrafted myofibers were compared between MCAM+ and MCAM+CD82+ injected muscles using a paired t-test.
RNA silencing
Human CD82 and control sh-RNA lentiviral particles were purchased from Santa Cruz Biotechnology. Purified CD82+ CD56+ human cells were infected with lentiviral particles and infected cells were selected with puromycin according to the manufacturer’s instructions. Apoptosis assays were performed using a commercial kit (Thermo-Fisher).
Co-Immunoprecipitations
400 µg protein cell lysates were pre-cleared of IgG fractions, then incubated with 5µg of immunoprecipitating antisera. The immunoprecipitated lysates were incubated with 30µl of anti-IgG magnetic beads followed by magnetic separation. Bound fractions were electrophoresed on 4–12% NuPage gradient gels. Western blots were incubated with primary antisera overnight at 4°C, washed, incubated with secondary antisera for 1 hour at room temperature, washed and incubated with ECL Reagent.
PLA assays
PLA assays were performed using the Duolink® In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich) using the manufacturer’s instructions. Briefly, 15,000–20,000 MCAM+ human fetal myogenic cells were plated in 8 or 16-well slide chambers. Cells were fixed with 4%PFA, permeabilized in 0.5% Triton X-100 in PBS and washed 3×5 min in 0.05% Tween20 in TBS. Cells were blocked in Duolink II blocking solution for 30 min at 37°C; primary antibodies were added for 1hr at 37°C, diluted 1:50 in Duolink II diluent. Slides were washed 2×5 min at RT, then probes (mouse and rabbit) were applied incubated at 37°C for 1hr. Slides were washed 2×5 min, ligase was added incubated at 37°C for 30 min. Slides were washed 2×2 min, incubated in Duolink polymerase in the dark at 37°C for 100 min. Slides were washed 2×10 min at RT protected from light, mounted, coverslipped and imaged.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Jamie Marshall and Natassia Vieira for helpful discussions. Pax7, myogenin and embryonic myosin monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD and maintained at The University of Iowa. This work was supported by a grant (199642) from the Muscular Dystrophy Association (USA) and from NIH (R01AR06317) to E.G. Additional funding was generously provided by the Bernard F. and Alva B. Gimbel Foundation to L.M.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.S.A, A.R. and E.G. planned the experiments and wrote the manuscript. M.S.A., A.R. and A.C. performed most of the experiments; S.J., H.M., M.W.L., J.M.S., F.R. performed some experiments and provided technical expertise; E.E. and L.M.K. planned some of the experiments and contributed research material.
References
- Anastasi G, Cutroneo G, Rizzo G, Arco A, Santoro G, Bramanti P, Vitetta AG, Pisani A, Trimarchi F, Favaloro A. Sarcoglycan and integrin localization in normal human skeletal muscle: a confocal laser scanning microscope study. Eur J Histochem. 2004;48:245–252. [PubMed] [Google Scholar]
- Blanco-Bose WE, Yao CC, Kramer RH, Blau HM. Purification of mouse primary myoblasts based on alpha 7 integrin expression. Exp Cell Res. 2001;265:212–220. doi: 10.1006/excr.2001.5191. [DOI] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PAL, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci (USA) 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Kaufman SJ. The alpha7beta1 integrin in muscle development and disease. Cell Tissue Res. 1999;296:183–190. doi: 10.1007/s004410051279. [DOI] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Milner DJ, Chaney EJ, Mulligan JA, Kaufman SJ. Transgenic expression of {alpha}7{beta}1 integrin maintains muscle integrity, increases regenerative capacity, promotes hypertrophy, and reduces cardiomyopathy in dystrophic mice. Am J Pathol. 2005;166:253–263. doi: 10.1016/s0002-9440(10)62249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Cresswell P. Quality control of transmembrane domain assembly in the tetraspanin CD82. EMBO J. 2001;20:2443–2453. doi: 10.1093/emboj/20.10.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano M, Dellavalle A, Tedesco FS, Quattrocelli M, Crippa S, Ronzoni F, Salvade A, Berardi E, Torrente Y, Cossu G, et al. Alpha sarcoglycan is required for FGF-dependent myogenic progenitor cell proliferation in vitro and in vivo. Development. 2011;138:4523–4533. doi: 10.1242/dev.070706. [DOI] [PubMed] [Google Scholar]
- Chelly J, Desguerre I. Progressive muscular dystrophies. Handb Clin Neurol. 2013;113:1343–1366. doi: 10.1016/B978-0-444-59565-2.00006-X. [DOI] [PubMed] [Google Scholar]
- Dick SA, Chang NC, Dumont NA, Bell RA, Putinski C, Kawabe Y, Litchfield DW, Rudnicki MA, Megeney LA. Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells. Proc Natl Acad Sci U S A. 2015;112:E5246–E5252. doi: 10.1073/pnas.1512869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barrett JC. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, Rudnicki MA. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Gnocchi VF, White RB, Ono Y, Ellis JA, Zammit PS. Further characterisation of the molecular signature of quiescent and activated mouse muscle satellite cells. PLoS One. 2009;4:e5205. doi: 10.1371/journal.pone.0005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Willem M, Werner A, Raivich G, Emerson M, Neyses L, Mayer U. Absence of alpha 7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Hum Mol Genet. 2006;15:989–998. doi: 10.1093/hmg/ddl018. [DOI] [PubMed] [Google Scholar]
- Hayashi YK, Chou FL, Engvall E, Ogawa M, Matsuda C, Hirabayashi S, Yokochi K, Ziober BL, Kramer RH, Kaufman SJ, et al. Mutations in the integrin alpha7 gene cause congenital myopathy. Nat Genet. 1998;19:94–97. doi: 10.1038/ng0598-94. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Hur J, Choi JI, Lee H, Nham P, Kim TW, Chae CW, Yun JY, Kang JA, Kang J, Lee SE, et al. CD82/KAI1 Maintains the Dormancy of Long-Term Hematopoietic Stem Cells through Interaction with DARC-Expressing Macrophages. Cell Stem Cell. 2016;18:508–521. doi: 10.1016/j.stem.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- Lapan AD, Gussoni E. Isolation and characterization of human fetal myoblasts. Methods Mol Biol. 2012;798:3–19. doi: 10.1007/978-1-61779-343-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QC, Zha XH, Faralli H, Yin H, Louis-Jeune C, Perdiguero E, Pranckeviciene E, Munoz-Canoves P, Rudnicki MA, Brand M, et al. Comparative expression profiling identifies differential roles for Myogenin and p38alpha MAPK signaling in myogenesis. J Mol Cell Biol. 2012;4:386–397. doi: 10.1093/jmcb/mjs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion BA, Berditchevski F, Kraeft SK, Chen LB, Hemler ME. Transmembrane-4 superfamily proteins CD81 (TAPA-1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29) J Immunol. 1996;157:2039–2047. [PubMed] [Google Scholar]
- Marshall JL, Kwok Y, McMorran BJ, Baum LG, Crosbie-Watson RH. The potential of sarcospan in adhesion complex replacement therapeutics for the treatment of muscular dystrophy. FEBS J. 2013;280:4210–4229. doi: 10.1111/febs.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JL, Chou E, Oh J, Kwok A, Burkin DJ, Crosbie-Watson RH. Dystrophin and utrophin expression require sarcospan: loss of alpha7 integrin exacerbates a newly discovered muscle phenotype in sarcospan-null mice. Hum Mol Genet. 2012;21:4378–4393. doi: 10.1093/hmg/dds271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JL, Oh J, Chou E, Lee JA, Holmberg J, Burkin DJ, Crosbie-Watson RH. Sarcospan integration into laminin-binding adhesion complexes that ameliorate muscular dystrophy requires utrophin and alpha7 integrin. Hum Mol Genet. 2015;24:2011–2022. doi: 10.1093/hmg/ddu615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Absence of integrin alpha 7 causes a novel form of muscular dystrophy. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- Meng H, Janssen PM, Grange RW, Yang L, Beggs AH, Swanson LC, Cossette SA, Frase A, Childers MK, Granzier H, et al. Tissue triage and freezing for models of skeletal muscle disease. J Vis Exp. 2014 doi: 10.3791/51586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Muntoni F. Muscular dystrophies. Lancet. 2013;381:845–860. doi: 10.1016/S0140-6736(12)61897-2. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Okochi H, Kato M, Nashiro K, Yoshie O, Miyazono K, Furue M. Expression of tetra-spans transmembrane family (CD9, CD37, CD53, CD63, CD81 and CD82) in normal and neoplastic human keratinocytes: an association of CD9 with alpha 3 beta 1 integrin. Br J Dermatol. 1997;137:856–863. [PubMed] [Google Scholar]
- Pasut A, Oleynik P, Rudnicki MA. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol Biol. 2012;798:53–64. doi: 10.1007/978-1-61779-343-1_3. [DOI] [PubMed] [Google Scholar]
- Rozkalne A, Adkin C, Meng J, Lapan A, Morgan JE, Gussoni E. Mouse regenerating myofibers detected as false-positive donor myofibers with anti-human spectrin. Hum Gene Ther. 2014;25:73–81. doi: 10.1089/hum.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Tachibana I, Hemler ME. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol. 1999;146:893–904. doi: 10.1083/jcb.146.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velling T, Collo G, Sorokin L, Durbeej M, Zhang H, Gullberg D. Distinct alpha 7A beta 1 and alpha 7B beta 1 integrin expression patterns during mouse development: alpha 7A is restricted to skeletal muscle but alpha 7B is expressed in striated muscle, vasculature, and nervous system. Dev Dyn. 1996;207:355–371. doi: 10.1002/(SICI)1097-0177(199612)207:4<355::AID-AJA1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.