Abstract
Multiple myeloma (MM) is incurable once osteolytic lesions have seeded at skeletal sites, but factors mediating this deadly pathogenic advance remain poorly understood. Here we report evidence of a major role for the cell adhesion molecule CD166, which we discovered to be highly expressed in MM cell lines and primary bone marrow (BM) cells from patients. CD166+ MM cells homed more efficiently than CD166− cells to the BM of engrafted immunodeficient NSG mice. CD166 silencing in MM cells enabled longer survival, a smaller tumor burden and less osteolytic lesions, as compared to mice bearing control cells. CD166 deficiency in MM cell lines or CD138+ BM cells from MM patients compromised their ability to induce bone resorption in an ex vivo organ culture system. Further, CD166 deficiency in MM cells also reduced formation of osteolytic disease in vivo after intra-tibial engraftment. Mechanistic investigation revealed that CD166 expression in MM cells inhibited osteoblastogenesis of BM-derived osteoblast progenitors by suppressing RUNX2 gene expression. Conversely, CD166 expression in MM cells promoted osteoclastogenesis by activating TRAF6-dependent signaling pathways in osteoclast progenitors. Overall, our results define CD166 as a pivotal director in MM cell homing to the BM and MM progression, rationalizing its further study as a candidate therapeutic target for MM treatment.
Keywords: Multiple myeloma, CD166, disease progression, osteolytic lesions, osteoclastogenesis
Introduction
Multiple myeloma (MM) is a malignancy characterized by uncontrolled neoplastic plasma cells growing in the bone marrow (BM) and causing osteolytic bone diseases (1). The BM microenvironment is crucial for MM survival, proliferation, migration and resistance to drugs (2,3). Up to 90% of MM patients develop bone disease, which not only affects patients’ quality of life, but also their longevity. MM bone disease is characterized by multiple osteolytic lesions throughout the skeleton, suggesting that trafficking of MM cells to secondary sites is important for disease progression. The mechanisms of trafficking and homing of MM cells into the BM microenvironment have been previously investigated (4–6). However, the exact mechanisms have not been well understood.
CD166 or activated leukocyte cell adhesion molecule (ALCAM) is a member of the immunoglobulin superfamily capable of mediating homophilic (CD166-CD166) and heterophilic (CD166-CD6) interactions (7,8). Expression of CD166 is conserved across species (9) with 93% homology between murine and human (10), suggesting that CD166 from both species can interact with each other and modulate mouse or human cell activities.
CD166 is involved in various physiologic and pathologic processes including cell adhesion, cell migration, hematopoiesis and tumor progression (11,12). Expression of CD166 is positively correlated with the disease progression in breast cancer and melanoma (13–15). However, the role of CD166 in MM has not been investigated. We previously demonstrated that CD166 plays an important role in sustaining the ability of osteoblasts (OB) to support the maintenance and function of HSC (16). We also recently reported that CD166 is an important molecule on normal murine and human HSC and is critical for HSC homing to the BM and engraftment (17). Interestingly, our studies demonstrated that CD166 is a functional marker on normal HSC and OB since CD166− HSC engrafted poorly in normal hosts and the microenvironment of CD166− KO mice did not support the long-term engraftment of normal HSC. Taken together, these data prompted us to investigate whether CD166 is involved in the trafficking of MM cells or in modulating MM disease progression and osteolytic diseases.
Methods
Cells, cell culture, and mice
The H929 and RPMI 8226 human MM cell lines were purchased from ATCC and was authenticated by ATCC using the COI assay and STR analysis in June 2008 and April 2010, respectively. Authenticated OPM2, MM1.S and JJN3 cell lines were provided by Dr. G. David Roodman (18) in 2014(IUSM, Indianapolis, IN). BM aspirates of myeloma patients were kindly provided by Dr. Rebecca Silbermann (IUSM, Indianapolis, IN). All studies were approved by the Institutional Review Board of IUSM.
Adult NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (6–8-week-old), C57BL/6 mice and CD166−/− mice (6–8-week-old or 10-day old pups) were used. Mice were bred and housed in the animal facility at IU. For MM injection studies, NSG mice received 275 cGy ionizing radiation from a cesium source followed by cell injection 2h later. Procedures were approved by the Institutional Animal Care and Use Committee of IUSM and followed National Institutes of Health guidelines.
For Ex Vivo Organ Culture Assay (EVOCA), calvariae from 10-day old neonatal C57BL/6 mice and global CD166−/− mice were dissected as described (19). Half calvarial pieces were cocultured with myeloma cells in a-MEM/RPMI1640 50/50 medium supplemented with 1% P/S for 10 days and the medium was changed every 72h thereafter. When calvariae were cocultured with patient MM cells, a-MEM/RPMI1640 50/50 medium with 1% P/S and 5% BSA was used. For histology, calvariae were removed from culture and dipped in PBS then fixed with 10% neutral buffered formalin, decalcified with 10% w/v EDTA, embedded in paraffin, sectioned, and stained H&E or tartrate-resistant acid phosphatase (TRAP). Sections were viewed on Leica DMLB microscope equipped with Q-imaging micropublisher camera. Osteolytic resorption surface area was measured by Bioquant image analysis software (Bioquant, Nashville, TN)
MM primary BM CD138+ cell selection, flow cytometry and sorting
MM primary BM CD138+ cells (>97% hematopoietic cells, supplemental Figure 1) were selected by immunomagnetic separation using anti-CD138 MicroBeads (Miltenyi Biotec). Briefly, MM patients’ BM cells were cultured in petri dishes overnight to remove adherent stromal cells. Non-adherent BM mononuclear cells were then harvested, counted and incubated with anti-CD138 Microbeads at 10ul microbeads/1×107 cells followed by magnetic separation.
BM cell suspensions or MM cell lines were labeled with monoclonal CD166-PE antibody (Biolegend). Cells were analyzed on an LSRII (BD Biosciences) flow cytometer, and events (0.1–2×106) were analyzed with FlowJo (Ver.9.7.6, FlowJo). Using cells stained with CD166-PE, CD166+ and CD166− cells were sorted on a BD FACSAria cell sorter (BDbiosciences) from either MM cell lines or from patient CD138+ MM.
In Vitro osteoclast assays
Primary mouse bone marrow macrophages (BMM) were used for in vitro osteoclast studies as described (20). BMM were first cultured in 48-well plates in α-MEM, 10% FBS, 1% P/S and 10ng/ml mM-CSF (Peprotech) for 3 days after which 50ng/ml mRANKL (Biolegend) were added for 7 days. Mock transfected or CD166KD H929 cells were added to the culture on day3. On day7, H929 cells were washed off with PBS. The adherent cells were fixed and stained with TRAP (Sigma-Aldrich). TRAP-positive cells (≥3 nuclei) were counted as osteoclasts.
Stimulation of BMM with H929 cells and Western blotting
BMM were plated in 6-well plates in α-MEM with 10ng/ml mM-CSF for 3 days. BMM were serum starved for 2h before stimulation with mock or CD166KD H929 cells for 30 min. H929 cells were washed off with cold PBS on ice. BMM were lysed with RIPA buffer (Santa Cruz Biotechnology). Total protein was extracted and extracts were subjected to SDS-PAGE and Western blotting. Primary Abs used included TRAF6 (Biolegend), phospho-Akt, Akt, (Cell Signaling Technology), phospho-p38, p38, IkBα, NFATc1 (Santa Cruz), actin (Sigma-Aldrich). HRP-conjugated secondary Abs (Cell Signaling Technology) were probed and developed with ECL solution (Millipore).
Real-time PCR
Quantitative PCR (qPCR) was performed on an ABI7900 using a SYBR Green PCR Core Kit (Applied Biosystems). Primers for quantitative PCR are listed in Supplemental Table 1. Relative expression was calculated using the comparative 2−ΔΔCt method, with GAPDH as the internal control.
Transfection and infection studies
Lentiviral vectors (hCD166shRNA on pLKO.1-puro-CMV-tGFP vector from Sigma Aldrich, Supplemental table 1) were transfected into the packaging cell line 293T using lipofectamine 2000 (invitrogen). Viral supernatant was collected 48h after transfection. H929 cells were incubated with viral supernatant for 12h then cultured in fresh RPMI1640 for 72h. GFP-positive cells were sorted on a BD FACSAria cell sorter.
Homing assay and mice xenograft human MM model
Homing of inoculated MM cells to the BM of irradiated recipients was performed as previously described (21). Briefly, 2×107 H929-GFP cells were washed with PBS and IV injected into sub-lethally irradiated NSG mice. Recipients were killed 14h post-inoculation, and BM cells were recovered for analysis.
To generate a murine xenograft model for human MM, 200ul containing 1×105 H929-GFP cells were IV injected into sub-lethally irradiated NSG mice. To evaluate disease progression, blood was drawn from individual mice every two weeks and serum human IgA (huIgA)-kappa levels were assayed using ELISA. Mouse survival was monitored for 220 days. In another experiment (14 mice/group), 6 mice were euthanized at 8 weeks post-transplantation, and the calvariae were dissected and processed for histology analysis. Tibiae were dissected and imaged with x-ray and micro-CT. BM was flushed and analyzed by flow cytometry. The remaining mice were monitored for survival.
For intratibial tumor inoculation, 10ul containing 1×105 H929-GFP cells (mock or CD166KD) were injected into sub-lethally irradiated NSG mice under anesthesia. X-ray of the mice tibiae was performed at 4 and 8 weeks later under anesthesia. Mice were euthanized at 9 weeks. Bilateral tibiae were dissected and imaged with micro-CT. BM in the tibiae was then flushed and analyzed by flow cytometry.
Radiography and micro-computed tomography (micro-CT)
Osteolytic lesions were analyzed by radiography using a Kubtec digital X-ray imager with energy range: 10–90kV and tube current up to 1.0mA (Kubtec, Milford, CT). Mice were x-rayed in a prone at 2.7x magnification. Osteolytic lesion area was measured by Bioquant image analysis software (Bioquant, Nashville, TN). Bone osteolytic lesions were quantified using a VIVACT-40 micro-CT system (Scanco Medical, Brüttisellen, Switzerland) at voxel size of 15μm; scanner settings: 55kVp, 145μA and 350ms integration time, by an observer blinded to the groups. Total bone volume was evaluated from 2mm below the most distal layer of the proximal growth plate and spanned 5mm distally along the diaphysis. A threshold of 230 was used to manually delineate bone from surrounding soft tissue.
Statistical analysis
Experiment were repeated at least 3 times. Quantitative data are presented as mean ± SEM unless otherwise stated. Statistical difference of Kaplan-Meier survival curves was determined by long-rank test. For comparisons between two groups, a two-sided student t-test were used. When more than two groups were compared simultaneously, analysis of variance (ANOVA) followed by Bonferroni’s post ttests were used Results were considered significantly different for P < .05.
Results
Expression of CD166 on MM cells and the role of CD166 in cell migration
We previously showed that CD166 is a critical functional marker on murine and human HSC and on elements of the hematopoietic niche (17). We therefore evaluated CD166 expression on MM cells. Five cell lines tested by flow cytometry had varying levels of CD166 expression on their cell surface (Fig 1A). Moreover, CD166 was also expressed on CD138+ cells obtained from 6 MM patients (Figure 1A). H929 and OPM2, expressed median levels of CD166 mRNA according to the CCLE data-base (Supplemental Figure 1B). Therefore these 2 cell lines were chosen for these studies since they did not represent an extreme bias in terms of CD166 expression. Supplemental Figure 1B also shows mRNA expression in MM cell lines relative to that in breast cancer cell lines. More than 50% of CD138+ BM cells from all patients expressed CD166. We also examined the expression of CD6, the only other known CD166 ligand, and found that cells from both MM cell lines and from patient BM did not express CD6 (supplemental Figure 2A).
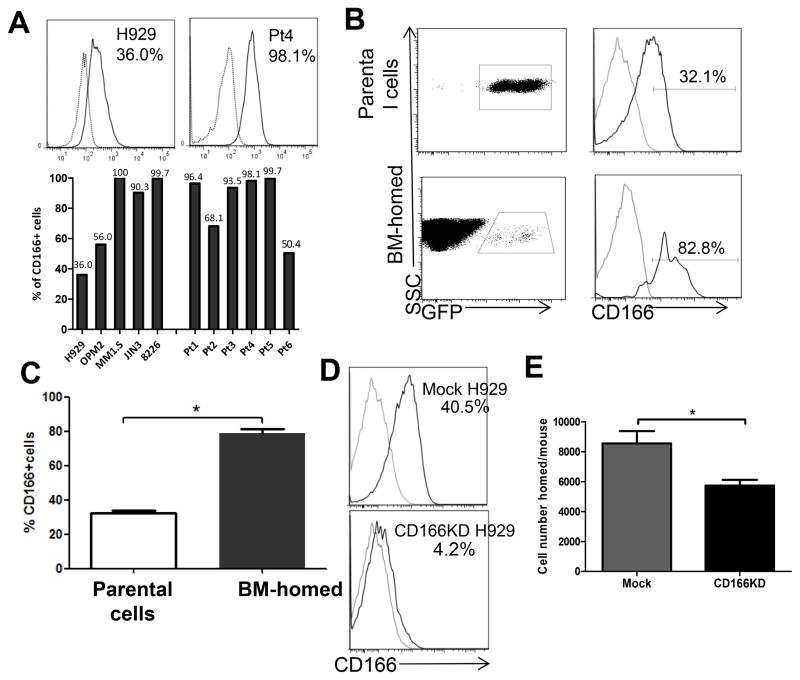
Figure 1. CD166 is expressed in both MM cell lines and primary MM CD138+ cells and is critical for homing of MM cells to the BM of NSG mice.
(A) Representative flow cytometric analysis of CD166 expression level on H929 cells and MM patients’ CD138+ cells (upper panel). The levels of CD166 expression on MM cell lines and 6 MM patients’ CD138+ cells (lower panel). (B–C) A total of 2×107 GFP-labeled H929 cells were intravenously injected into sub-lethally irradiated NSG mice and GFP+ cells were recovered from mice BM 14h later. The percentage of CD166+ cells within parental H929 cells and from BM-homed cells were compared flow cytometrically. Data are represented as mean ± SEM from 3 pooled experiments (N = 3 mice/group/experiment, each assayed individually). (D) Flow cytometric assessment of the level of CD166 knockdown with lentiviral shRNA for huCD166. (E) 2×107 GFP-labeled mock or CD166KD H929 cells were intravenously injected into sub-lethally irradiated NSG mice. GFP cells were recovered from mouse BM 14h later and the number of H929 cells homed to the BM from two femurs and two tibiae was calculated. Data are represented as mean ± SEM from 3 pooled experiments (N = 3 mice/group/experiment, each assayed individually). *p<0.05. For histograms in A and D: grey line = isotype control, black line = sample.
We next examined the role of CD166 in the migration of MM cells to the BM using previously described protocols (21). H929-GFP cells were IV injected into NSG mice and 14h post-injection, mice were euthanized and the percentage of MM cells (GFP+) in the BM was determined flow cytometrically (Figure 1B). While the percentage of CD166+ H929 cells was originally 29.9%±1.4% among parental cells, that among BM-homed cells increased to 80.0%±2.5% (Figure 1C, p<0.05). To examine whether the increased CD166+ MM population in the BM is due to up-regulation of CD166 in the BM microenvironment, we sorted CD166+ and CD166− cells from H929 and repeated the homing studies described above. Analyses of BM-homed cells showed that while both populations homed to the BM, CD166− cells remained negative in the BM (supplemental Figure 2B), indicating that the increased number of CD166+ cells in the BM is not due to CD166 upregulation. Furthermore, no significant difference was observed in the expression of four other markers (VLA-4, CXCR4, VLA-5 and CD44) involved in the trafficking of hematopoietic cells between CD166+ and CD166− fractions of H929 (supplemental Figure 3A). Collectively, these results suggest that CD166 directs the homing of MM cells to the marrow microenvironment.
To investigate the impact of CD166 on MM homing, we used GFP-tagged lentiviral shRNA for huCD166 to reduce CD166 protein level in H929 cells up to 90% (Figure 1D). huCD166 shRNA-expressing H929 cells displayed significantly reduced (45%) homing efficiency compared with the mock shRNA-expressing cells (Figure 1E). The use of huCD166 shRNA did not alter the growth kinetics (supplemental Figure 3B) of H929 cells or alter the surface expression of adhesion molecules on these cells (data not shown).
Knockdown (KD) of CD166 on myeloma cells delays disease progression in vivo
We used a xenograft model to determine the role of CD166 in the initiation of MM and in disease progression. NSG mice were intravenously injected with mock or CD166KD H929 cells, and disease progression was monitored over time. Disease progression is described in these studies as appearance and kinetics of increase of MM-derived Ig in serum, survival, and the relative degree of severity of osteolytic lesions in inoculated mice. Serum levels of human IgA (huIgA) kappa in mice receiving mock H929 were detected earlier and increased faster than in mice receiving CD166KD H929 cells (Figure 2A). Survival was monitored for 220 days post-inoculation and was plotted in a Kaplan-Meier curve. NSG mice bearing CD166KD H929 cells showed a significant delay in disease progression and prolonged survival (p= 0.0005, Figure 2B).
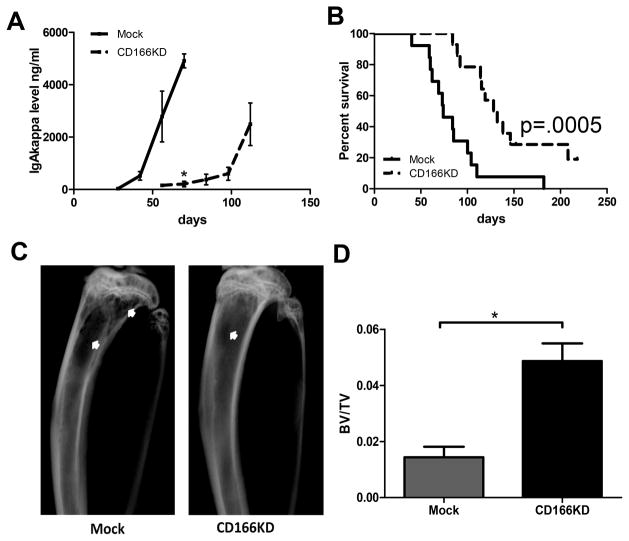
Figure 2. CD166 is critical for MM disease progression.
(A&B) 1×105 Mock or CD166KD H929 cells were intravenously injected into NSG mice. (A) Human IgA-kappa levels in the serum of individual mice were measured by ELISA every two weeks. (B) Survival was monitored for a period of 220 days and Kaplan-Meier survival curves were plotted. Data are pooled from 2 independent experiments (N = 6–8 mice/group/experiment were followed until day 220 post-inoculation). (C) Eight weeks after inoculation, some of the mice were euthanized. Tibiae were imaged with radiography and representative images are shown. Bone lesions area were indicated with white arrows. (D) Eight weeks after inoculation, some of the mice were euthanized. Tibiae were analyzed with micro-CT and trabecular bone volume (BV/TV fraction) were analyzed by an analyzer blinded to the experimental groups. Data are representative of 2 separate experiments (mean±SEM, N = 6 mice/group/experiment, each assayed individually). *p<0.05.
In a separate cohort of mice treated similarly, we euthanized some mice at 8 weeks. Flow cytometric analysis detected GFP-labeled H929 cells in the BM of mice transplanted with mock or CD166KD H929 cells (Supplemental Figure 3C). Radiography demonstrated significantly larger osteolytic lesion areas on the tibiae in mice inoculated with mock cells than with CD166KD cells (16.3±5.5mm2 vs 5±4.5mm2, p<0.05; Figure 2C). Furthermore, microCT analysis of the tibiae confirmed that these mice had significantly lower trabecular bone volume (BV/TV fraction) compared to those transplanted with CD166KD H929 cells (Figure 2D). Consistent with these results, calvariae from mice receiving CD166KD cells had significantly lower osteoclast numbers per bone surface (1.6±0.2) compared to those from mice inoculated with mock H929 (4.4±1.0, p<0.05) as indicated by TRAP staining (Supplemental Figure 3D). On the other hand, downregulation of osteocalcin mRNA, an OB differentiation marker, in calvariae was decreased in mice that received CD166KD H929 cells (supplemental Figure 3E).
CD166 is critical for the pathobiology of lytic bone disease in MM
To further investigate the impact of CD166 on MM-associated osteolytic lesions, we used an ex vivo organ culture system (22) designed to allow for the formation of bone resorption ex vivo. Flow cytometrically sorted CD166+ or CD166− H929 cells were cocultured with calvariae from 10 day-old WT or global CD166 knockout (KO) mice for 10 days after which the calvariae were processed for histological analysis. Mean resorption surface to total bone surface ratio was quantified using Bioquant image analysis software which demonstrated that absence of CD166 on MM cells (CD166− H929 cells) or bone (CD166−/− calvariae), or both, significantly decreased the resorption surfaces on calvariae (Figure 3A–B, different magnification images in supplemental Figure 4). Similar results (supplemental Figure 5A–B) were observed when another MM cell line, OPM2 was used in similar studies.
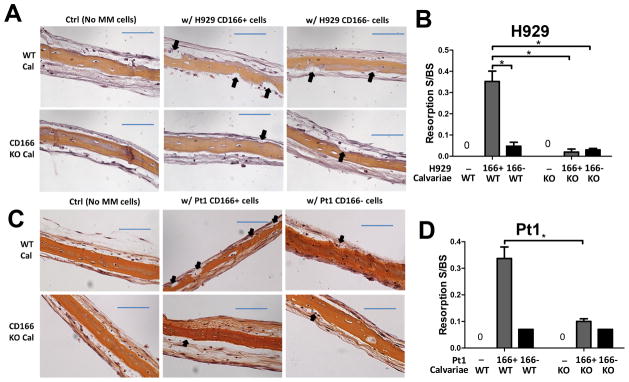
Figure 3. Absence of CD166 on myeloma cells reduces bone resorption on calvariae ex vivo.
(A–D) Ex vivo organ culture assay (EVOCA) was used to examine the effect of CD166 expression on myeloma bone resorption lesions. After culture of MM cells on calvariae as described in Methods, calvariae were fixed, decalcified, sectioned and processed for H&E staining. Bone resorption (black arrow) on calvariae was analyzed from three non-overlapping fields per bone under 20 x magnification. The quantitative representation of EVOCA assay was performed by measuring and calculating resorption surface to bone surface (BS) ratio with Bioquant software 2014. (A) H&E staining of Calvariae from WT or CD166KO pups cultured with flow sorted 2×104 CD166+ or CD166− H929 cells for 10 days (×20, scale bar=100 μm). (B) Quantitative representation of EVOCA results with H929; Data represent 3 separate experiments done in triplicates for each group and are expressed as mean± SEM, *p<0.05. (C) H&E staining of calvariae from WT or CD166KO pups cultured with 2×104 flow cytometrically sorted CD166+ or CD166− primary MM CD138+ cells for 10 days (×20, scale bar=100 μm). (D) Quantitative representation of EVOCA results shown in (C).
Next, we determined whether the absence of CD166 on MM patient cells decreased bone resorption surface. Primary CD138+CD166+ or CD138+CD166− MM cells from patients were separated by flow cytometric cell sorting (expression of CD166 on patients’ CD138+ cells is shown in supplemental Figure 5C) and cocultured with calvariae from 10 day-old WT or CD166 KO pups for 10 days. The lowest resorption surface to bone surface ratio was observed when CD138+CD166− MM cells were cocultured with CD166KO calvariae (Pt1 Figure 3C–D). We found similar results with MM cells from two other patients (supplemental Figure 5D–E).
To confirm these ex vivo observations in vivo, NSG mice were intra-tibially inoculated with mock transduced or CD166KD H929 cells. Radiographic analysis revealed that mice receiving mock H929 cells developed osteolytic lesions 4 weeks after tumor inoculation. The bone deteriorated further through the course of 8 weeks. On the contrary, although tibiae from CD166KD H929 inoculated mice showed osteolytic lesions at 4 weeks, these lesions worsened to a lesser extent than that observed in the mock group over the next 8 week period (Figure 4A). In agreement, micro-CT images at 9 weeks demonstrated that mock H929 cells induced much more sever lesions on the tibiae compared to CD166KD cells (Figure 4B).
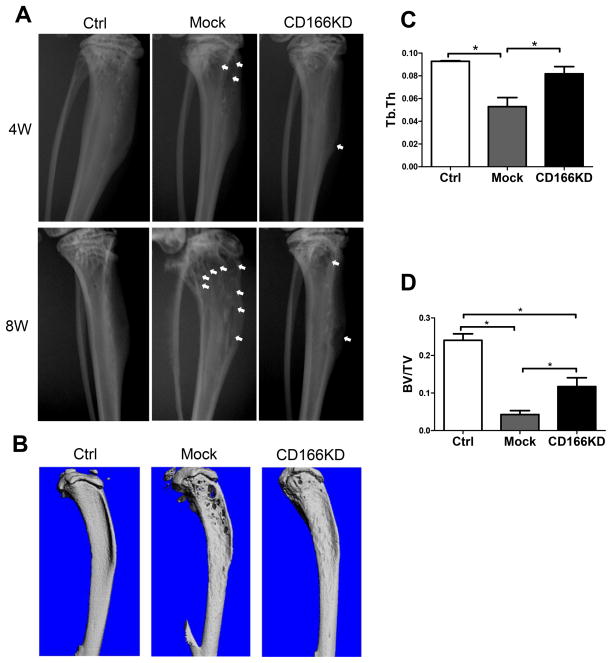
Figure 4. Absence of CD166 on MM cell leads to less bone osteolytic lesions.
(A) Representative radiographic images of tibiae from mice inoculated with mock or CD166KD H929 cells at 4 weeks and 8 weeks after inoculation. Bone lesion areas are indicated with white arrow. (B) Nine weeks after inoculation, mice were euthanized and tibiae were collected and scanned using micro-CT for 3D reconstruction. Bones from 3 representative mice are shown. (C) Trabecular thickness Tb.Th, and (D) trabecular bone volume (BV/TV fraction) were determined by micro-CT readings. n=5–6/group, mean± SEM, *p<0.05.
Both trabecular thickness (Figure 4C) and trabecular bone volume (Figure 4D) at 9 weeks post-inoculation were significantly decreased in the tibiae from mice receiving mock H929 cells. Flow cytometric analysis of the BM from the inoculated tibiae did not reveal differences in the percentage or cell number between mice inoculated with mock or CD166KD H929 cells (supplemental Figure 6A–C). However, 2 out of 5 contralateral tibiae in the mock group had detectable H929 cells (>0.1% by flow cytometric analysis). In contrast, none of the 6 contralateral tibiae of mice inoculated with CD166KD H929 cells had detectable GFP+ cells (supplemental Figure 6D). These results are consistent with our homing assay data that CD166 is critical for MM cells trafficking in the BM.
CD166 expression on MM cells alters bone remodeling
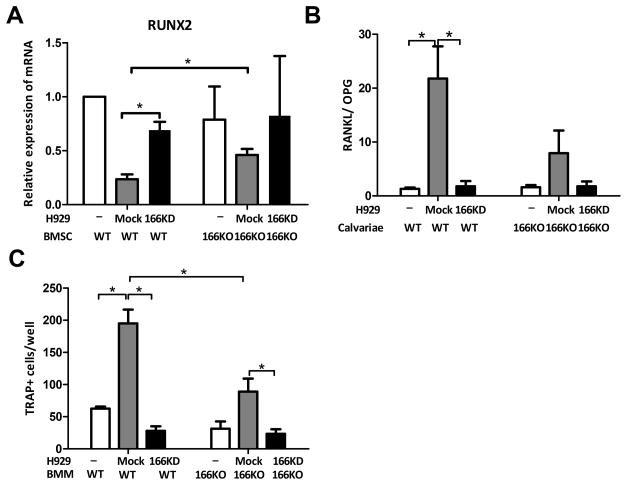
To examine how CD166 induces bone osteolytic lesions in MM, we explored the mechanisms involved in MM-induced osteoblast suppression. BMSC were isolated from WT or CD166KO mice as described before (23) and were cocultured with CD166KD or mock H929 cells for 48h in osteoblast differentiating medium. H929 cells were washed off by cold PBS, and mRNA was isolated from BMSC. qPCR results revealed that gene expression of RUNX2, a critical transcription factor in osteoblastogenesis, was significantly suppressed when CD166 was present in the culture. (Figure 5A).
Figure 5. Expression of CD166 on myeloma cells alters bone remodeling balance.
(A) BMSC were cocultured with H929 cells (as indicated) for 48h. H929 cells were removed and RNA was isolated from BMSC. Relative expression of RUNX2 RNA in BMSC was detected by quantitative PCR relative to GAPDH and the “WT BMSC alone” sample. (B) Calvariae were cocultured with H929 cells (as indicated) for 7 days. H929 cells were removed. Relative RANKL and OPG RNA expression in calvariae cells were examined by quantitative PCR and RANKL/OPG ratio was calculated. (C) Non-adherent BM derived macrophage from WT or CD166KO mice were cultured for 3 days to enrich for adherent BM macrophages (BMM). BMM were then cultured in α-MEM in the presence of recombinant mouse M-CSF (10 ng/ml) and RANKL (50ng/ml) for 7 days and mock H929 or CD166KD H929, followed by TRAP staining. TRAP-positive cells with 3 or more nuclei were scored under an inverted microscope. Data represent 3 separate experiments done in triplicates for each group and are expressed as mean± SEM, *p<0.05.
We then examined the mechanisms involved in MM-induced osteoclast activation. We measured by qPCR analysis gene expression levels of RANKL and OPG in calvariae cocultured with CD166KD H929 or mock H929 cells for 7 days. At day 7, H929 cells were washed off by cold PBS and calvariae were subjected to RNA isolation (24). Gene analysis with mouse-specific primers showed that RANKL/OPG ratio, an important indicator for osteoclastogenic activity (25), was significantly decreased when CD166 was absent from either MM cells (CD166− H929 cells) or from bone cells (CD166−/− calvariae) (Figure 5B). Gene expression of MCSF which is critical for osteoclastogenesis, was not significantly impacted (supplemental Figure 7A). Furthermore, we used a previously described assay (26) to measure osteoclastogenesis of BMM from WT or CD166KO mice cultured with mock or CD166 KD H929 cells. The degree to which osteoclasts were generated in culture was visibly reduced when either expression of CD166 on H929 cells was blocked or was genetically abrogated on BMM (Supplemental Figure 7B). Importantly, quantification of osteoclast formation (TRAP+ cells; ≥3 nuclei) demonstrated that the number of TRAP+ cells was significantly compromised in cultures where CD166 was absent from H929 cells or when BMM were collected from CD166KO mice (Figure 5C).
TRAF6 upregulation is a possible target in the CD166-mediated enhancement of osteoclastogenesis during MM
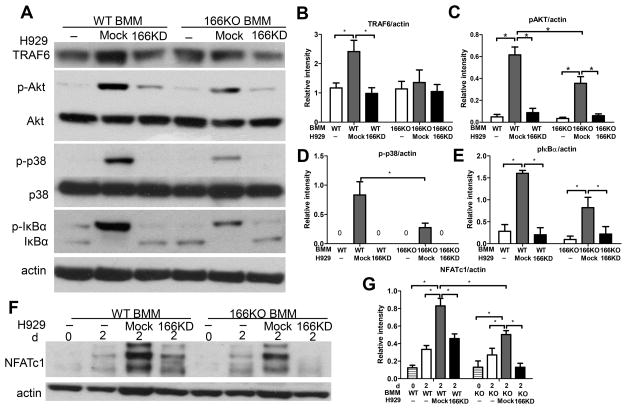
To further delineate the mechanisms involved in CD166-induced osteoclastogenesis in MM, we starved BMM for 2h (27) then stimulated these cells with mock H929 or CD166KD H929 cells for 30 mins. MM cells were then washed off and BMM were subjected to Western blot analysis for tumor necrosis factor (TNF) receptor associated-factor (TRAF) 6, which plays a critical role in the signal transduction pathway of NF-kappaB in osteoclastogenesis (28). While BMM from WT mice upregulated TRAF6 expression upon stimulation with control H929 cells, WT BMM stimulated by CD166KD H929 or BMM from CD166−/− mice failed to upregulate TRAF6 expression (Figure 6A–B). In MM, TRAF6 mediates osteolytic bone lesions through the regulation of several downstream signaling pathways, including Akt, NFkB and p38MAPK (27,29). We next examined the activation of these downstream targets of TRAF6 in BMM treated as described above. Western blot analysis demonstrated that the absence of CD166 on H929 or BMM decreased phospho AKT, phospho p38, and phospho IkBa levels (Figure 6A, C–E), indicating that loss of CD166 in MM cells decreased their ability to activate AKT, p38/MAPK and NFkB in BMM and likely responsible for suppressing osteoclastogenesis.
Figure 6. Absence of CD166 expression on MM cells downregulates key signaling pathways in osteoclastogenesis.
BMM were derived from bone marrow cells by culturing in the presence of M-SCF (10 ng/ml) for 3 days. (A) BMM were serum starved for 2h before exposure to mock H929 or CD166KD H929 for 30 min. Protein was extracted from BMM by RIPA lysis buffer for Western Blot analyses after H929 cells were washed off with cold PBS. Whole cell extracts were subjected to Western blot analysis with specific Abs as indicated. (F) BMM were cultured with mock H929 or CD166KD H929 in the presence of M-CSF (10 ng/ml) and RANKL (50ng/ml) for the 2 days. Protein was extracted from BMM by RIPA lysis buffer for Western Blot analyses after H929 cells were washed off with cold PBS. Whole cell extracts were subjected to Western blot analysis with specific Abs as indicated. (B–E, G) Quantitative densitometry of the expression of the indicated proteins was normalized to actin protein expression. Data were collected from three separate observations and are expressed as mean± SEM. *p<0.05.
While Akt, NFkB and p38MAPK are downstream of TRAF6 in osteoclastogenesis pathways, these regulators function through a common transcription factor, the nuclear factor of activated T cells (NFAT)c1 which is a key modulator in osteoclastogenesis (30). So we further examined NFATc1 protein levels during osteoclastogenesis. NFATc1 expression was strongly attenuated by the absence of CD166 expression on either H929 cells or BMM (Figure 6F–G).
Discussion
Interactions between MM cells and cells of the BM microenvironment are vital for MM cell growth and bone destruction (2). MM is characterized by multiple bone lesions throughout the skeleton (31), indicating that trafficking of MM cells is important for MM disease progression. In this study, we show that CD166 is expressed on both MM cell lines and CD138+ primary BM cells from 6 out of 6 patients analyzed. Studies unequivocally demonstrate that CD166 is expressed on cells in the BM microenvironment (32,33). CD166 is reported to contribute to bone metastasis in prostate cancer (34). Results from our homing assays (Figure 1D–G), demonstrate for the first time that CD166 is involved in MM cell trafficking and suggest that CD166 may be critical for MM pathogenesis. Interestingly, none of the classical adhesion molecules associated with homing of hematopoietic cells to the BM (35) could be implicated in the preferential homing of CD166+ MM cells.
To examine the possibility of involvement of CD166 in MM pathogenesis, we used shRNA constructs to generate CD166 KD H929 cells and established MM via IV inoculation in NSG mice using mock or KD H929 cells. Our data demonstrated a statistically significant difference in the survival of mice harboring mock H929 compared to those inoculated with KD H929 cells. Of interest is that osteolytic lesion areas in tibiae from mice with KD H929 cells were significantly smaller than those in mice inoculated with mock H929 cells. Consistently, TRAP staining of calvariae from mice in mock groups showed significantly higher osteoclast number/bone surface, suggesting the direct involvement of CD166 in MM pathogenesis and the associated bone osteolytic disease. It would be interesting to examine if inducible downregulation of CD166 after establishment of the disease in mice can lead to tumor regression, decreased osteolytic lesions, and enhanced survival.
To address the problem that IV inoculation may result in more mock cells homing to the BM thus causing more bone osteolytic lesions, we first applied a novel (22) ex vivo organ culture assay (EVOCA) in which MM cells grow in direct contact with calvarial cells. We found that the absence of CD166 on any cell type present in the EVOCA culture diminished the severity of bone resorption of the calvarial surfaces. CD166 can mediate homophilic (CD166-CD166) interactions and heterophilic interactions (CD166-CD6) (36). Since we have shown that MM cells do not express CD6, data from the EVOCA assay suggest that homophilic interactions between MM cells and osteolineage cells expressing CD166 are critical for the progression of osteolytic disease. Furthermore, using an intra-tibial inoculation model (Figure 4), we confirmed that the expression of CD166 on MM cells directly impacts osteolytic lesions in vivo.
Knocking down CD166 in H929 cells does not alter the proliferation kinetics of these cells in vitro or in vivo (supplemental Figure 3B, 6A–B). These observations demonstrate that CD166 may not be required for the survival, proliferation, or apoptosis of these cells, but that it is critical for the interaction of MM cells with other cells in the BM microenvironment that also express CD166. CD166KD did not alter the expression of adhesion markers (data not shown) suggesting that altered osteoclast/osteoblast function is not mediated by the lack of adherence between CD166KD MM cells and osteolineage cells through these markers.
Bone remodeling is balanced between bone formation by osteoblasts and bone resorption by osteoclasts. However, this balance is broken during bone destruction caused by the presence of cancer cells (37,38). Therefore, MM-associated change in bone remodeling may result from CD166-mediated suppression of osteoblastogenesis or enhancement of osteoclastogenesis. Using in vitro assays, we demonstrated that expression of CD166 on MM cells repressed RUNX2 expression, the key transcription factor in osteoblast differentiation from BM-derived mesenchymal stem cells (39). In contrast, loss of CD166 on MM cells reduced the osteoclastogenic potential of BMM when these two cell types were co-cultured together, suggesting that osteoclastogenesis was enhanced by CD166+ MM cells. In determining the molecular mechanisms underlying the CD166+ MM cell regulation of osteoclast formation, we observed that CD166 expression stimulated overexpression of RANKL, while it failed to affect expression of OPG, an inhibitor of osteoclastogenesis. Together, these observations implicate CD166 in the regulation of osteoclastogenesis through upregulation of RANKL and disturbing the critical RANKL/OPG ratio.
Additional investigation revealed a possible novel positive regulatory pathway involving CD166-TRAF6 mediated osteoclast formation. TRAF6 is regulated by the RANKL-RANK signaling pathway in osteoclastogenesis (28,40). Our study indicates that CD166 may regulate TRAF6 directly, suggesting that inhibition of CD166 may represent an effective way to control bone osteolytic disease in MM. Also CD166 expression on MM cells regulates NFATc1 expression as well as many other intermediates involved in osteoclastogenesis (Figure 6).
In conclusion, our data implicate CD166 in the progression of MM in vivo and suggest that this molecule is intimately involved in the pathogenesis of MM. In a very recent paper, Paiva et al (41) reported that CD166 is downregulated in MM minimal residual disease tumor cells suggesting that persistence of MM is mediated by therapy-induced clonal selection of cells with low CD166 expression. While these data appear to be inconsistent with our findings, it is important to point out that the studies of Paiva et al relied on gene expression profiling (GEP) of minimal residual disease cells rather than assessment of protein surface expression. As shown in Suppl Figure 1B, GEP data do not necessarily translate to expression levels suggesting that our data and those from Paiva et al may not be contradictory. CD166 is a functional molecule that impacts HSC function and the competence of the normal hematopoietic niche (17). Therefore, the question arises whether CD166 is a marker that identifies a group of MM initiating cells that are responsible for sustaining this disease. Given the involvement of CD166 in the pathophysiology of MM, CD166 could be a potential target for inhibiting the MM-induced osteolytic disease as well as tumor progression. In fact anti-CD166 antibody has already been used in the breast cancer setting to reduce cancer cell invasion and tumor growth, two criteria investigated in our work in homing and disease progression studies (42).
Supplementary Material
Acknowledgments
This work was supported in part by the Indiana University Simon Cancer Center (P30 CA082709; E.F. Srour, core leader) and the Indiana Center for Excellence in Molecular Hematology (National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK090948; E.F. Srour, Co-PI).
The authors thank Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility for their outstanding technical help and support. We thank Dr. G. David Roodman (Indiana University School of Medicine, Indianapolis, IN) for critical suggestions and review of the manuscript. We acknowledge the In Vivo Therapeutics Core of the Indiana University Simon Cancer Center (partially funded by National Cancer Institute grant P30 CA082709 and National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK090948).
Footnotes
The authors declare no conflict of interest.
References
- 1.Slovak ML. Multiple myeloma: current perspectives. Clinics in laboratory medicine. 2011;31(4):699–724. x. doi: 10.1016/j.cll.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Mitsiades CS, McMillin DW, Klippel S, Hideshima T, Chauhan D, Richardson PG, et al. The role of the bone marrow microenvironment in the pathophysiology of myeloma and its significance in the development of more effective therapies. Hematology/oncology clinics of North America. 2007;21(6):1007–34. vii–viii. doi: 10.1016/j.hoc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2009;23(1):10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Camp B, Van Riet I. Homing mechanisms in the biology of multiple myeloma. Verhandelingen - Koninklijke Academie voor Geneeskunde van Belgie. 1998;60(3):163–94. [PubMed] [Google Scholar]
- 5.Alsayed Y, Ngo H, Runnels J, Leleu X, Singha UK, Pitsillides CM, et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109(7):2708–17. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79(5):885–92. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 7.van Kempen LC, Nelissen JM, Degen WG, Torensma R, Weidle UH, Bloemers HP, et al. Molecular basis for the homophilic activated leukocyte cell adhesion molecule (ALCAM)-ALCAM interaction. The Journal of biological chemistry. 2001;276(28):25783–90. doi: 10.1074/jbc.M011272200. [DOI] [PubMed] [Google Scholar]
- 8.Bowen MA, Aruffo AA, Bajorath J. Cell surface receptors and their ligands: in vitro analysis of CD6-CD166 interactions. Proteins. 2000;40(3):420–8. [PubMed] [Google Scholar]
- 9.Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139(6):2072–82. e5. doi: 10.1053/j.gastro.2010.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen MA, Bajorath J, D’Egidio M, Whitney GS, Palmer D, Kobarg J, et al. Characterization of mouse ALCAM (CD166): the CD6-binding domain is conserved in different homologs and mediates cross-species binding. European journal of immunology. 1997;27(6):1469–78. doi: 10.1002/eji.1830270625. [DOI] [PubMed] [Google Scholar]
- 11.Cayrol R, Wosik K, Berard JL, Dodelet-Devillers A, Ifergan I, Kebir H, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nature immunology. 2008;9(2):137–45. doi: 10.1038/ni1551. [DOI] [PubMed] [Google Scholar]
- 12.Masedunskas A, King JA, Tan F, Cochran R, Stevens T, Sviridov D, et al. Activated leukocyte cell adhesion molecule is a component of the endothelial junction involved in transendothelial monocyte migration. FEBS letters. 2006;580(11):2637–45. doi: 10.1016/j.febslet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Medical science monitor: international medical journal of experimental and clinical research. 2006;12(8):BR263–73. [PubMed] [Google Scholar]
- 14.Swart GW, Lunter PC, Kilsdonk JW, Kempen LC. Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer metastasis reviews. 2005;24(2):223–36. doi: 10.1007/s10555-005-1573-0. [DOI] [PubMed] [Google Scholar]
- 15.van Kempen LC, van den Oord JJ, van Muijen GN, Weidle UH, Bloemers HP, Swart GW. Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. The American journal of pathology. 2000;156(3):769–74. doi: 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitteti BR, Cheng YH, Kacena MA, Srour EF. Hierarchical organization of osteoblasts reveals the significant role of CD166 in hematopoietic stem cell maintenance and function. Bone. 2013;54(1):58–67. doi: 10.1016/j.bone.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitteti BR, Kobayashi M, Cheng Y, Zhang H, Poteat BA, Broxmeyer HE, et al. CD166 regulates human and murine hematopoietic stem cells and the hematopoietic niche. Blood. 2014;124(4):519–29. doi: 10.1182/blood-2014-03-565721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teramachi J, Silbermann R, Yang P, Zhao W, Mohammad KS, Guo J, et al. Blocking the ZZ domain of sequestosome1/p62 suppresses myeloma growth and osteoclast formation in vitro and induces dramatic bone formation in myeloma-bearing bones in vivo. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2016;30(2):390–8. doi: 10.1038/leu.2015.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammad KS, Chirgwin JM, Guise TA. Assessing new bone formation in neonatal calvarial organ cultures. Methods Mol Biol. 2008;455:37–50. doi: 10.1007/978-1-59745-104-8_3. [DOI] [PubMed] [Google Scholar]
- 20.Wei S, Wang MW, Teitelbaum SL, Ross FP. Interleukin-4 reversibly inhibits osteoclastogenesis via inhibition of NF-kappa B and mitogen-activated protein kinase signaling. The Journal of biological chemistry. 2002;277(8):6622–30. doi: 10.1074/jbc.M104957200. [DOI] [PubMed] [Google Scholar]
- 21.Yusuf RZ, Scadden DT. Homing of hematopoietic cells to the bone marrow. Journal of visualized experiments: JoVE. 2009;(25) doi: 10.3791/1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtin P, Youm H, Salih E. Three-dimensional cancer-bone metastasis model using ex-vivo co-cultures of live calvarial bones and cancer cells. Biomaterials. 2012;33(4):1065–78. doi: 10.1016/j.biomaterials.2011.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nature protocols. 2009;4(1):102–6. doi: 10.1038/nprot.2008.221. [DOI] [PubMed] [Google Scholar]
- 24.Rollig C, Knop S, Bornhauser M. Multiple myeloma. Lancet. 2015;385(9983):2197–208. doi: 10.1016/S0140-6736(14)60493-1. [DOI] [PubMed] [Google Scholar]
- 25.Ross FP. M-CSF, c-Fms, and signaling in osteoclasts and their precursors. Annals of the New York Academy of Sciences. 2006;1068:110–6. doi: 10.1196/annals.1346.014. [DOI] [PubMed] [Google Scholar]
- 26.Kim K, Kim JH, Lee J, Jin HM, Kook H, Kim KK, et al. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood. 2007;109(8):3253–9. doi: 10.1182/blood-2006-09-048249. [DOI] [PubMed] [Google Scholar]
- 27.Moon JB, Kim JH, Kim K, Youn BU, Ko A, Lee SY, et al. Akt induces osteoclast differentiation through regulating the GSK3beta/NFATc1 signaling cascade. J Immunol. 2012;188(1):163–9. doi: 10.4049/jimmunol.1101254. [DOI] [PubMed] [Google Scholar]
- 28.Lamothe B, Webster WK, Gopinathan A, Besse A, Campos AD, Darnay BG. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochemical and biophysical research communications. 2007;359(4):1044–9. doi: 10.1016/j.bbrc.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Tamashiro S, Baritaki S, Penichet M, Yu Y, Chen H, et al. TRAF6 activation in multiple myeloma: a potential therapeutic target. Clinical lymphoma, myeloma & leukemia. 2012;12(3):155–63. doi: 10.1016/j.clml.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayanagi H. The role of NFAT in osteoclast formation. Annals of the New York Academy of Sciences. 2007;1116:227–37. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 31.Roodman GD. Pathogenesis of myeloma bone disease. Blood cells, molecules & diseases. 2004;32(2):290–2. doi: 10.1016/j.bcmd.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Chitteti BR, Bethel M, Kacena MA, Srour EF. CD166 and regulation of hematopoiesis. Current opinion in hematology. 2013;20(4):273–80. doi: 10.1097/MOH.0b013e32836060a9. [DOI] [PubMed] [Google Scholar]
- 33.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer genomics & proteomics. 2010;7(5):231–43. [PubMed] [Google Scholar]
- 34.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. The Journal of experimental medicine. 2007;204(3):645–55. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahin AO, Buitenhuis M. Molecular mechanisms underlying adhesion and migration of hematopoietic stem cells. Cell adhesion & migration. 2012;6(1):39–48. doi: 10.4161/cam.18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. European journal of cell biology. 2002;81(6):313–21. doi: 10.1078/0171-9335-00256. [DOI] [PubMed] [Google Scholar]
- 37.Roodman GD. Mechanisms of bone metastasis. The New England journal of medicine. 2004;350(16):1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 38.Reagan MR, Liaw L, Rosen CJ, Ghobrial IM. Dynamic interplay between bone and multiple myeloma: emerging roles of the osteoblast. Bone. 2015;75:161–9. doi: 10.1016/j.bone.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marie PJ. Transcription factors controlling osteoblastogenesis. Archives of biochemistry and biophysics. 2008;473(2):98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 40.Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408(6812):600–5. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 41.Paiva B, Corchete LA, Vidriales MB, Puig N, Maiso P, Rodriguez I, et al. Phenotypic and genomic analysis of multiple myeloma minimal residual disease tumor cells: a new model to understand chemoresistance. Blood. 2016;127(15):1896–906. doi: 10.1182/blood-2015-08-665679. [DOI] [PubMed] [Google Scholar]
- 42.Wiiger MT, Gehrken HB, Fodstad O, Maelandsmo GM, Andersson Y. A novel human recombinant single-chain antibody targeting CD166/ALCAM inhibits cancer cell invasion in vitro and in vivo tumour growth. Cancer immunology, immunotherapy: CII. 2010;59(11):1665–74. doi: 10.1007/s00262-010-0892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.