Abstract
Iron is an essential nutrient for life. During infection, a fierce battle of iron acquisition occurs between the host and bacterial pathogens. Bacteria acquire iron by secreting siderophores, small ferric iron binding molecules. In response, host immune cells secrete lipocalin 2 (also known as siderocalin), a siderophore-binding protein, to prevent bacterial reuptake of iron-loaded siderophores. To counter this threat, some bacteria can produce lipocalin 2-resistant siderophores. This review discusses the recently described molecular mechanisms of siderophore iron trafficking between host and bacteria, highlighting the therapeutic potential of exploiting pathogen siderophore machinery for the treatment of antibiotic-resistant bacterial infections. As the latter reflect a persistent problem in hospital settings, siderophore-targeting or siderophore-based compounds represent a promising avenue to combat such infections.
Keywords: siderophore, iron, Lcn2, siderocalin, mitophagy, hypoxia
Siderophores: Important Virulence Factors and Promising Molecular Targets
Nosocomial infections (see Glossary), also known as hospital-acquired infections (HAI), are a rising health concern worldwide. In the United States alone, there were at least 721,800 HAIs estimated in 2011 [1] caused by various pathogens. Gram positive Staphylococcus aureus (S. aureus) is of particular concern, estimated to be responsible for about 10% of all HAIs. The development of hyper-virulent and antibiotic-resistant strains makes treating these infections more difficult. For instance, carbapenemase-producing pathogens, such as Klebsiella pneumoniae and Escherichia coli, are resistant to the carbapenem-class of antibiotics widely used to treat multidrug-resistant bacteria. Many HAIs can infect a variety of organs, including the gastrointestinal system and urinary tract, and may also infect surgical sites. These pathogens are becoming more widespread and constitute an immediate public health problem [2].
Virulence factors may aid pathogens in colonizing the host as well as enhance disease. These factors consist of a wide variety of substances including bacterial toxins, adherence factors, protective capsules, and, relevant to this review, siderophores. Iron is a necessary element in virtually all living organisms and is utilized to catalyze a wide variety of indispensable enzymatic reactions [3]. Early microorganisms were able to utilize soluble ferrous iron (Fe2+), abundant due to an oxygen-poor atmosphere; however, as oxygen-rich conditions arose, ferrous iron was oxidized to insoluble ferric iron (Fe3+), removing an easily bioavailable source of iron. Responding to this challenge, microorganisms evolved, utilizing siderophores which are small ferric iron (Fe3+) chelating molecules [4]. In a pathogenic context, microbes secrete siderophores to acquire and solubilize ferric iron from the host. In fact, comparisons of different Acinetobacter genomes have shown that the presence of genes involving siderophore biosynthesis is predictive of high or low virulence [5]. Consistently, in bacteria including Acinetobacter baumannii, siderophores were shown as necessary components for the development of surface attachment and extracellular polysaccharide synthesis (termed biofilm formation) [6–8] and the establishment of mutually-beneficial, iron-sufficient microbial communities [9]. Given that siderophores are involved in biofilm formation which promotes antibiotic resistance [10, 11], targeting siderophores by blocking siderophore synthesis or function provides a promising alternative antimicrobial approach.
Iron is also essential to host cells, and is tightly regulated under various physiological conditions [12]. Hosts utilize various major iron transport systems, such as iron-loaded transferrin, lactoferrin, and heme [3]. However, the affinities of bacterial siderophores to iron are generally much higher than those of host proteins/molecules [4], allowing pathogens to outcompete the host in iron acquisition. In response to the siderophore threat, mammalian immune cells (e.g. macrophages, neutrophils) can secrete a siderophore binding protein, lipocalin 2 (Lcn2) (a 24 kDa glycoprotein also known as siderocalin, 24p3, and NGAL (neutrophil gelatinase associated lipocalin)), to intercept bacterial uptake of iron-loaded siderophores (ferric-siderophores) [13]. This battle over limited iron is an important host-pathogen interaction for survival during infections (Box 1).
Box 1. Siderophore-Lipocalin 2 Interface during Infection.
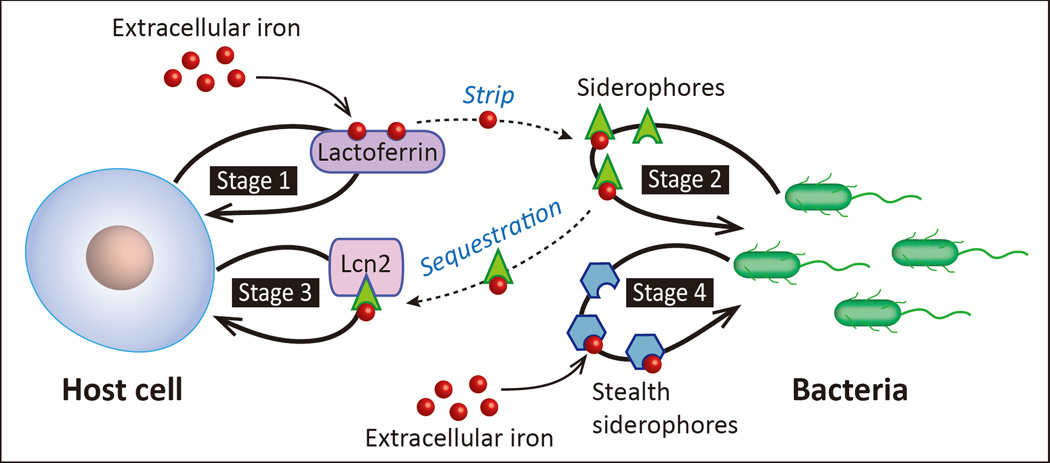
One of the major challenges for pathogenic bacteria is acquiring sufficient iron during infection and post-infection inside the host. Competition between the host and pathogen for this coveted transition metal occurs at the host-pathogen interface, a conceptual framework describing the exchange of signals and battle for common resources between host and pathogen. The fight for iron represents a robust “tug-of-war” occurs, which begins with the host secreting iron-binding proteins such as lactoferrin to sequester any readily available iron (Fig. I, stage 1).
When deprived of an easy iron source, pathogens upregulate siderophore biosynthesis and iron trafficking pathways (Fig. I, stage 2). Siderophores have much higher affinity for iron [4] and can strip iron from lactoferrin and other host iron-binding proteins, restoring access to iron for pathogens. In response, host cells secrete lipocalin 2 (important in innate immunity). In neutrophils, Lcn2 is stored within granules and rapidly secreted as part of a “first-response” to infection. Infection also drives massive increases in de novo Lcn2 production through Toll-like Receptor (TLR) and cytokine signaling [65]. Lcn2 complexes with ferric-siderophores and prevents reuptake by pathogens, once again denying bacterial iron acquisition (Fig. I, stage 3). The Lcn2 threat is so severe that some pathogens have evolved chemically-modified, Lcn2-resistant siderophores, expressing them in response to Lcn2 secretion [66]. The siderophore enterobactin produced in E. coli is readily bound by Lcn2, while glycosylated enterobactin (salmochelin) is not (Table 1, Fig. 1B). These “stealth siderophores” (i.e. chemically modified and unable to bind Lcn2) are important tools for preventing host’s Lcn2 function (Box Fig. I, stage 4).
Beyond simply denying iron to pathogens, Lcn2 can maintain host immune protein function. Recent data show that enterobactin can inactivate host myeloperoxidase (e.g. in neutrophils), a critical, iron-containing enzyme that generates bactericidal hypochlorous acid from H2O2 and Cl− [67]. Enterobactin attacks the heme prosthetic group and inactivates myeloperoxidase; Lcn2 can preserve myeloperoxidase function by binding to enterobactin [67].
Another bacterial strategy is to produce siderophores that natively cannot be recognized by Lcn2, as in the case of K. pneumonia, which can cause major respiratory infections, and produces yersiniabactin (Table 1, Fig. 1B) [68]. it is becoming clear that the host environment plays an important role in Lcn2-mediated immunity. Alterations in urine pH and composition can have significant impacts on the ability of Lcn2 to combat E. coli in a urinary-tract infection context. Alkalinization of urine (elevated pH above 6.45) increases the ability of Lcn2 to restrict iron and, subsequently, inhibit the growth of E. coli [69]. Given that inducing urinary alkalinization in a clinical setting is trivial, modulating urinary chemistry may be an attractive option in combating uropathogenic E. coli infections and requires further investigation.
Siderophores are becoming better appreciated for their role in virulence beyond simple iron chelation, acting as signals leading to a robust host defense, by inducing for instance, mitophagy, hypoxic responses, and cytokine production. This review discusses recent findings in siderophore biology, with highlights on the therapeutic potential of siderophore pharmacological targeting in microbial infections (e.g. S. aureus and A. baumannii) through the use of siderophore-antibiotic drug conjugates, gallium containing compounds, and siderophore biosynthesis inhibitors. We begin with a brief overview of nosocomial siderophore biology and import/export, followed by recent findings in host-pathogen interactions. Finally, we summarize recent advances in siderophore biology in an infection context, examining inhibitors of siderophore biosynthesis as novel antimicrobial therapeutics. Indeed, over the past few years, studies have revealed several components in siderophore production and trafficking, exposing potential vulnerabilities that can be exploited clinically. Targeting such vulnerabilities could potentially allow clinicians to circumvent the problem of traditional antibiotic-resistance in their patients, ideally reducing the burden of HAIs and improving health outcomes.
Nosocomial Bacterial Siderophore Import and Export Mechanisms
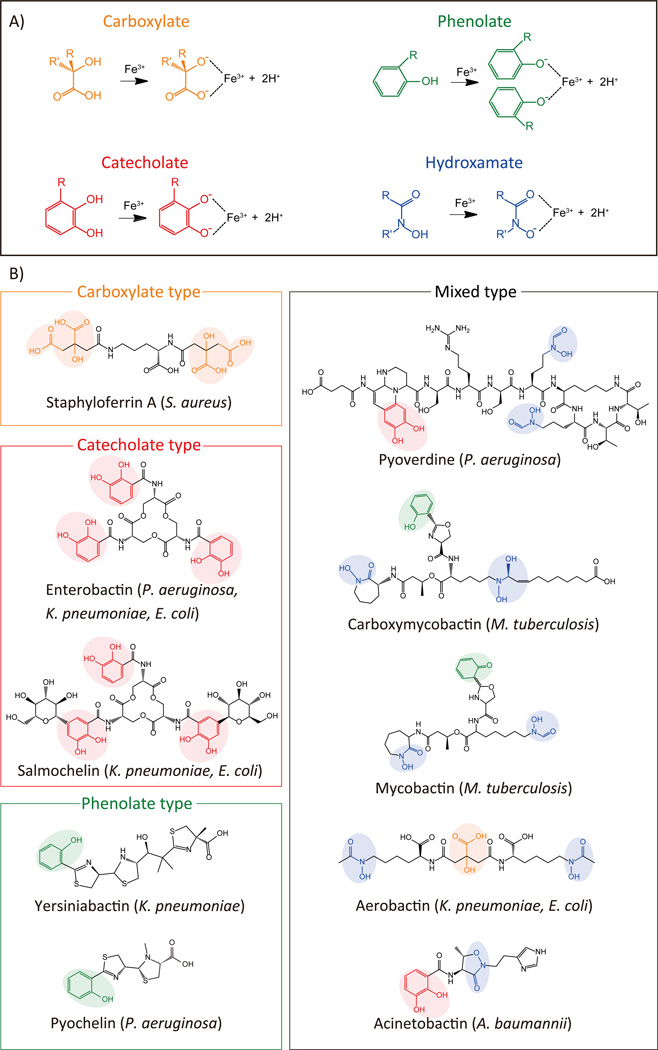
Major pathogens that cause nosocomial infections and acquire multidrug resistance are summarized in Table 1. Gram-negative bacteria (e.g. Pseudomonas aeruginosa, Klebsiella pneumonia, A. baumannii; E. coli), Gram-positive bacteria (e.g. Staphylococcus aureus), and acid-fast bacilli (e.g. Mycobacterium tuberculosis) [14] all utilize siderophores as important augmenters of infection and commit an abundance of resources to their production. Utilizing metabolites such as proteogenic and nonproteogenic amino acids, chorismate, and citrate, bacteria create a diverse repertoire of molecules through a ribosomal independent process [15]. Siderophores may be classified according to their iron-binding moieties: catecholate, hydroxamate, phenolate, carboxylate, and mixed-type (containing more than one of the aforementioned moieties) (Fig. 1A). For example, staphyloferrin A, produced by S. aureus, is a carboxylate-type with four carboxylate (R-COO−) moieties for iron binding (Fig. 1B). More complicated structures also exist: pyoverdine, a mixed-type siderophore produced by P. aeruginosa, is composed of eight amino acids, including: D-Ser, L-Arg, L-hf-Orn (N5-formyl-N5-hydroxyornithine), L-Lys, and L-Thr. Pyoverdine binds iron via two hydroxamate (O=CH-NROH) and one catecholate moiety (Fig. 1B). Following iron capture, ferric-siderophores are imported into bacteria through siderophore-specific receptors (Table 1, Box 2) for utilization of iron in growth and colonization during infection.
Table 1.
Nosocomial Pathogens and their Siderophores.
| Nosocomial Pathogens |
Types of Disease |
Antibiotic Resistance |
Siderophores | Siderophore Receptor (OMR) |
|
|---|---|---|---|---|---|
| Gram-negative |
Pseudomonas aeruginosa |
|
Carbapenem | Pyoverdine | FpvA |
| Pyochelin | FptA | ||||
|
Klebsiella pneumoniae |
|
Carbapenem | Aerobactin | IutA | |
| Yersiniabactin | FyuA | ||||
| Enterobactin | FepA | ||||
| Salmochelin | IroN | ||||
|
Acinetobacter baumannii |
|
Carbapenem | Acinetobactin | BauA | |
| Fimsbactin | n.d. | ||||
| Baumannoferrins | n.d. | ||||
|
Escherichia coli |
|
Fluoroquinolone Carbapenem Cephalosporin |
Enterobactin | FepA | |
| Salmochelin | IroN | ||||
| Aerobactin | IutA | ||||
| Yersiniabactin | FyuA | ||||
| Gram-positive |
Staphylococcus aureus |
|
Methicillin Vancomycin |
Staphyloferrin A | HtsA |
| Staphyloferrin B | SirA | ||||
| Staphylopine | CntA | ||||
| Mycobacteria |
Mycobacterium tuberculosis |
|
Isoniazid Rifampicin Ethambutol Pyrazinamide Streptomycin Ofloxacin Kanamycin |
Mycobactin | n.d. |
| Carboxy- mycobactin |
n.d. |
n.d: not determined
Figure 1. Siderophore Structure.
A. Four moieties confer iron-binding capacity to siderophores: carboxylate (orange), phenolate (green), catecholate (red), and hydroxamate (blue). B. Siderophore structures from selected nosocomial pathogens are shown (see Table 1): Mixed-type siderophores contain more than one type of iron-binding moiety.
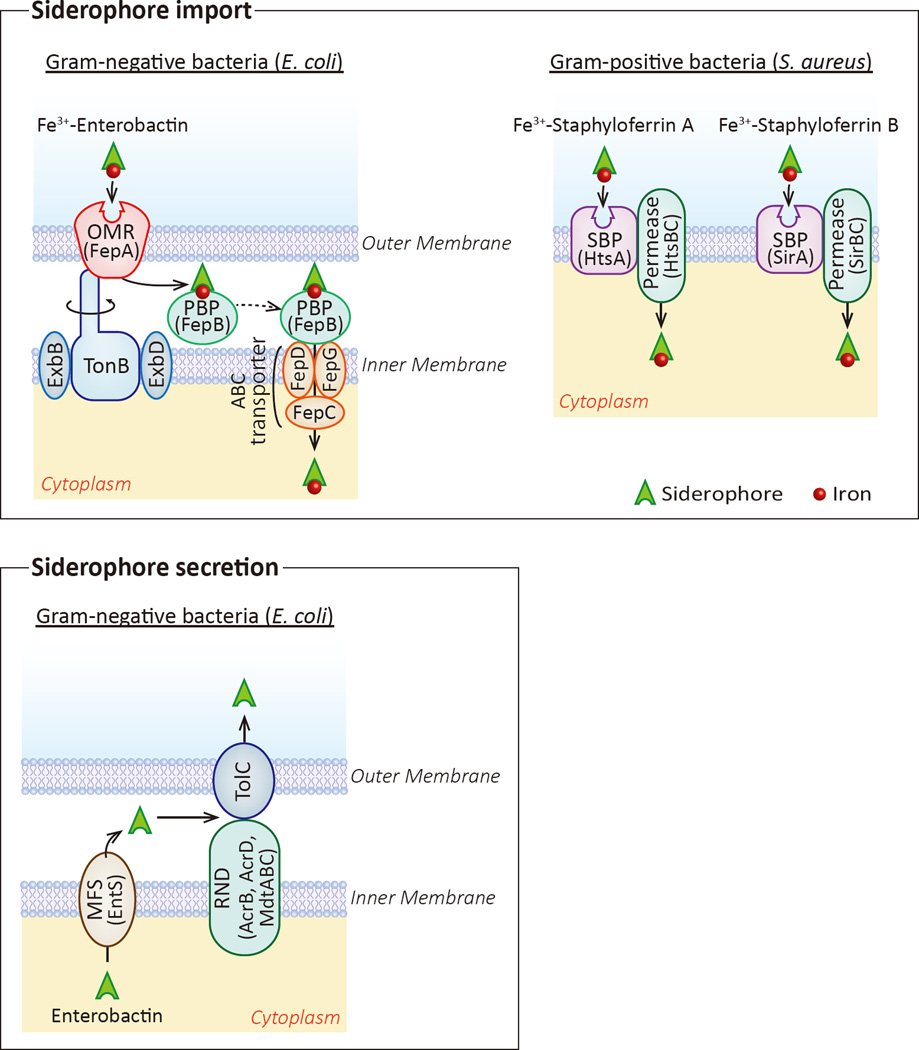
Box 2. Siderophore Import and Secretion Mechanisms.
In gram-negative bacteria, each ferric-siderophore complex is recognized by specific outer membrane receptor(s) (OMR) (Fig. II, top left, specific OMRs in Table 1). In E. coli, enterobactin is recognized by the OMR FepA. While OMRs are incredibly diverse, with different bacterial species and different siderophore classes having specific receptors (Table 1), in general, OMRs interact with the inner-membrane protein TonB to facilitate uptake of the ferric-siderophore complex. The current model (termed “Rotational Surveillance and Energy Transfer”, or ROSET) suggests that TonB, driven by the inner membrane proteins ExbB and ExbD as well as the electrochemical proton motive force generated in the periplasm during normal cellular respiration, physically rotates within the inner membrane, causing a conformational shift in the OMR. The conformational shift of TonB in the OMR promotes internalization of the ferric-siderphore complex [70]. Once enterobactin is transported across the outer membrane, the periplasmic binding protein (PBP) FepB shuttles it to the inner membrane, where a complex consisting of FepC, FepD, and FepG transport enterobactin into the cytoplasm (Fig. II, top left). Other siderophores may have slightly different uptake mechanisms.
Following uptake by the OMR, the ferric-pyoverdine complex in P. aeruginosa is split into its constituent components within the periplasm by the action of the PBPs FpvC and FpvF. The iron is pumped into the cytoplasm by the ABC transporter complex FpvDE and iron-free pyoverdine is secreted back into the environment to gather more iron (reviewed in [16]).
Gram-positive bacteria, such as S. aureus, have only a single membrane and therefore possess a simpler siderophore uptake system. In general, gram-positive bacteria express siderophore-binding protein (SBP), associated with permease. SPB binding to an extracellular ferric-siderophore causes a conformational change in a SBP-permease complex, allowing transport of the ferric-siderophore across the membrane into the cytoplasm. In S. aureus, carboxylate-type siderophores staphyloferrin A and staphyloferrin B are recognized by membrane-bound lipoproteins HtsA and SirA proteins, respectively. Upon staphyloferrin binding, these proteins undergo a conformational change that activates the permeases HtsBC or SirBC, allowing staphyloferrin to cross the membrane (Fig. I top right).
Siderophore secretion is a necessary step in bacterial iron acquisition. Perhaps the best characterized siderophore secretion pathway is enterobactin in E. coli, which is a multi-step process. First, enterobactin is transported from the cytoplasm to the periplasm via EntS (MFS-class efflux pump). Next, the concerted action of one inner-membrane RND-class efflux pump (AcrB, AcrD, or MdtABC) and the outer-membrane protein channel TolC transport enterobactin across the outer membrane [71] (Fig. I bottom).
Siderophore trafficking is different in Gram-positive bacteria, which have a single membrane, and Gram-negative bacteria, which have inner and outer membranes separated by a periplasmic space. Gram-negative bacteria, with dual membranes, require a multistep process for siderophore uptake. First, ferric-siderophores are recognized by a specific outer membrane receptor (OMR). After transport across the outer membrane, the ferric-siderophore is trafficked to the inner membrane by a periplasmic binding protein (PBP) where the ferric-siderophore is directly pumped into the cytoplasm or the iron is removed and the iron-free siderophore recycled [16] (Box 2). In contrast, Gram-positive pathogens only have a single membrane and, therefore, possess a comparatively simple uptake mechanism involving a siderophore-binding protein (SBP) and an associated permease located on the cell membrane (Box 2).
In order to maintain a constant supply of iron for infection, siderophores need to be exported from the bacterium following biosynthesis to find and chelate environmental iron. Siderophore secretion requires the activity of at least one of the following efflux pump families: major facilitator superfamily (MFS), ATP binding cassette (ABC), and the Gram-negative-specific Resistance-Nodulation-Cell Division (RND). In E. coli, for example, enterobactin secretion requires MFS- and RND-family proteins (Box 2).
Host and Pathogens Alter Many Iron-regulated Pathways to Fight for Survival
Siderophore mediated iron uptake becomes imperative for many pathogens during infection, as ablation of this system significantly reduces a pathogen’s ability to colonize a host [4]. Recent findings have shown that siderophores have several pivotal effects on the host that go beyond the frontlines of the host-pathogen interface discussed in Box 1. Bacterial siderophores can elicit strong cellular effects on the host by disrupting organelle iron homeostasis, activating transcriptional pathways, and affecting cellular behavior. Recently, it was shown that iron chelation could trigger cells to undergo mitophagy, the controlled autophagy of mitochondria [17]. Mitophagy appears to be a positive adaptation during iron deficiency, contributing to overall mitochondrial integrity and increased lifespan in pathogenic yeast Candida glabrata [18] and the nematode Caenorhabditis elegans [19]. Pyoverdine, the P. aeruginosa siderophore (Fig. 1, Table 1), is toxic to mitochondria in C. elegans, resulting in mitochondrial fragmentation and loss of mitochondrial membrane potential, leading to cell and (if severe enough) organismal death; notably, C. elegans mortality is attenuated by pre-incubating pyoverdine with iron [20]. This suggests that pyoverdine-induced mortality is dependent on the ability of pyoverdine to sequester host iron. After pyoverdine exposure, C. elegans undergoes mitophagy to remove the damaged mitochondria [21]. Inhibition of mitophagy makes C. elegans more susceptible to P. aeruginosa infection and subsequent death, indicating that mitophagy is an important cellular defense process to ameliorate/prevent cell injury and death after infection [21]. Interestingly, it appears that mitophagy after iron depletion can occur in either a PINK1/Parkin-dependent [21] or -independent [17] mechanism. PINK1 and Parkin encoded by the PARK6 and PARK2 genes, respectively, are the best characterized mediators of mitophagy, and mutations in these genes have been implicated in the development of Parkinson’s disease [22]. To date, however, no studies have been performed to investigate the effect of siderophores on mitophagy in mammalian systems.
In addition to causing mitophagy, bacterial siderophores can generate a hypoxic response in host cells, even in normoxic conditions, by stabilizing the host transcription factor hypoxia-inducible factor 1α (HIF-1α) [20, 23, 24]. Under normal cellular conditions in mammals, HIF-1α is hydroxylated by oxygen- and iron-dependent HIF prolyl-hydroxylases, allowing for HIF-1α ubiquitylation by the E3 ubiquitin ligase von Hippel-Lindau (pVHL) and subsequent downstream proteasomal degradation [25]. In low oxygen and/or low iron conditions, HIF prolyl hydroxylases lack these critical co-factors and cannot function, leading to stabilization of HIF-1α and upregulation of HIF-1α target genes. HIF-1α upregulates a myriad of genes including those encoding immune effector molecules such as granule proteases, antimicrobial peptides, and inflammatory cytokines [26, 27]. This is a beneficial response as it allows the host cells to better combat infection. In cultured human lung epithelial cells, a combination of enterobactin and lipocalin 2 were shown to stabilize HIF-1α and lead to the upregulation of critical genes for host immunity, including interleukin-6 (IL-6) [24]. In addition, activation of HIF-1α signaling has been shown to increase bacterial phagocytosis by macrophages in a p38 MAPK-dependent manner, assisting in pathogen clearance during infection [28]. Another HIF-1α target, inducible nitric oxide synthase (iNOS), is an important infection mediator that produces reactive nitrogen species (RNS) , and has been shown to be a critical factor in decreasing intracellular bacterial load during the early stages of mycobacterial infection in a zebrafish model [29]. Consistent with these findings, intestine-specific knockout of Hif-1α in mice results in worse animal survival following infection with Yersinia enterocolitica when compared to wild-type mice [23]. In addition, Hif-1-deficient C. elegans worms have been found to be more susceptible to P. aeruginosa pathogenesis when compared to wild-type worms [20]. Lack of intestinal HIF-1α likely prevents upregulation of important immune genes in response to siderophores [24, 25]. In summary, siderophores induce important host defense pathways, such as mitophagy and HIF-1α stabilization, both of which are critical for host survival.
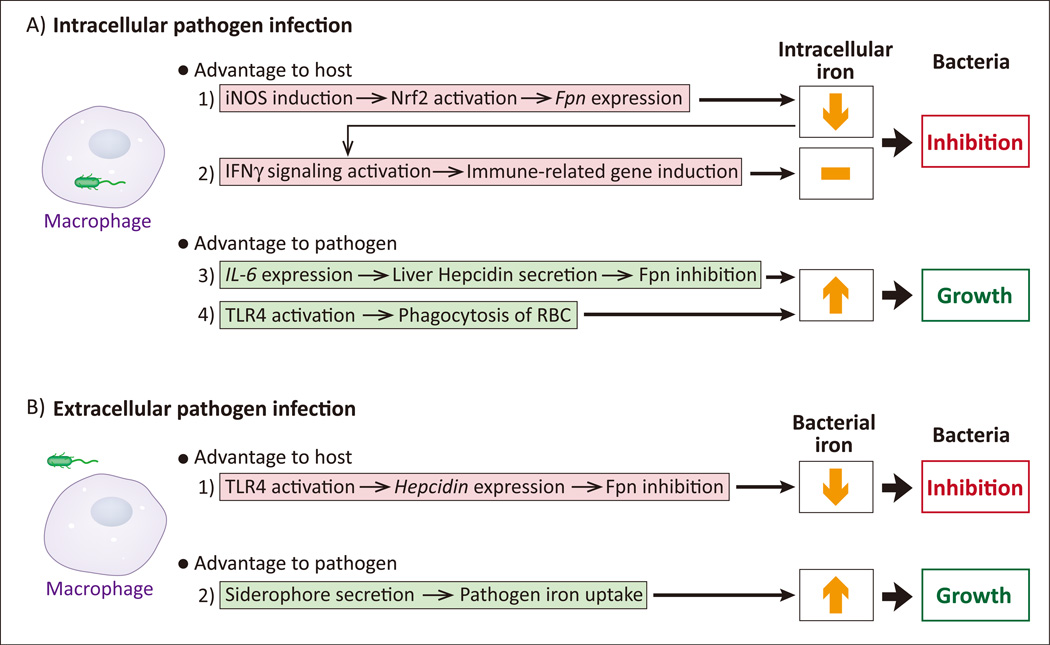
Intracellular pathogens, such as Mycobacterium and Salmonella species, can infect and reside inside of macrophages. As discussed above and in Box 1, a “tug-of-war” develops between host and bacteria over the iron contents of the host cell, with the pathogen attempting to acquire as much iron as possible and the macrophage attempting to deny pathogen access to iron. To that end, infected macrophages upregulate the iron export protein ferroportin (Fpn) through iNOS-mediated NFE2-related factor 2 (Nrf2) signaling [30] (Key Figure, Fig. 2A, #1). Reduction of intracellular iron has two antibacterial effects: the removal of an essential nutrient (for pathogens) and enhanced interferon γ (IFNγ) pathway signaling [31, 32] (Key Figure, Fig. 2A, #2). IFNγ is an inflammatory cytokine that upregulates a suite of immune-related genes and activates macrophages for antibacterial and antiviral activity [33]. In response, pathogens can inhibit Fpn-mediated iron export by inducing hepcidin expression. Hepcidin, a mammalian peptide hormone mainly produced in the liver and in macrophages, binds to Fpn and induces Fpn degradation, decreasing iron export activity and conserving intracellular iron stores on target cells [34]. Salmonella can induce hepatic hepcidin production via inflammatory IL-6 signaling and downstream estrogen-related receptor γ (ERRγ) activation [35] (Key Figure, Fig. 2A, #3); this is beneficial for Salmonella, an intracellular pathogen, as it maintains the intracellular iron pool. Furthermore, Salmonella can stimulate macrophages to specifically phagocytose erythrocytes in an effort to acquire more iron [36] through Toll-like receptor 4 (TLR4) signaling [37] (Key Figure, Fig. 2A, #4). This is a particularly interesting response that is well-conserved throughout vertebrates in response to severe infection, and is speculated to have evolved as a countermeasure to prevent potentially lethal levels of inflammation [36, 38]. In essence, the host will cede iron to the pathogen in order to prevent a worse outcome. Other pathogens, such as the extracellular P. aeruginosa, also upregulate hepcidin expression in macrophages through activation of TLR4 signaling [39] (Key Figure, Fig. 2B, #1), but this is a host defense response, as it removes an easily available source of iron for bacterial siderophores (Key Figure, Fig. 2B, #2).
Key Figure, Figure 2. Host and Pathogens Alter Many Iron-regulated Pathways to Fight for Survival.
A. 1) In response to intracellular pathogens, host macrophages upregulate inducible nitric oxide synthase (iNOS) to stimulate ferroportin (Fpn) expression. Fpn is an iron exporter, denying the pathogen access to the intracellular iron pool. 2) The lowered intracellular iron concentration also activates interferon-γ (IFN-γ), a cytokine that stimulates expression of a suite of antipathogenic, immune-related genes. 3) Intracellular pathogens induce IL-6 in host macrophages, leading to Fpn inhibition and resulting in increased intracellular iron. 4) Intracellular pathogens activate Toll-like Receptor 4 (TLR4) to induce the host macrophage to phagocytose erythrocytes (red blood cells; RBCs) in an effort to increase the intracellular iron pool. B. 1) Extracellular pathogens activate TLR4 on the host macrophage, resulting in downregulation of Fpn and lessened extracellular iron access to the pathogen. 2) Extracellular pathogens secrete siderophores to acquire as much iron from the environment as possible.
In summary, bacterial siderophores have wide-ranging biological effects on both pathogen and host, simultaneously helping pathogens acquire iron and damage host cells (e.g. mitochondrial damage) while causing host compensatory reactions (e.g. mitophagy and upregulation of immune genes). However, there are several key questions about the physiological effects of siderophores that remain unanswered (see Outstanding Questions). Further research will give a clearer portrait of the mechanisms by which siderophores alter host processes to benefit the pathogen and how the host responds in kind.
Outstanding Questions.
What are the identities of the major candidate proteins in nosocomial pathogen siderophore biology (mycobactin OMR, siderophore biosynthetic enzymes, etc.)? Are they promising therapeutic targets?
How else do pathogens modulate host pathways/processes to gain access to iron?
What other host defense pathways are activated by siderophores?
How effective are siderophore conjugates (gallium, antibiotics) in treating nosocomial infections in a clinical setting? How can their efficacy be improved?
Do (currently controversial) mammalian siderophores really exist? If so, which compounds are they? What are their functions in host iron homeostasis? How do they differ from other siderophores in different organisms?
Using Siderophores to Combat Bacterial Infection
The emergence of multidrug-resistant bacteria is a major concern that has stimulated development of new antibiotics and approaches [40, 41]. Nosocomial pathogens appear to be particularly resilient, having spread across the world at an alarming rate [1] and causing many types of disease (see Table 1). For example, P. aeruginosa is intrinsically resistant to many antibiotics and easily develops acquired resistance during chronic lung infection in cystic fibrosis patients [42]. Novel treatment strategies have been investigated to combat these pathogens, including the use of non-ferrous metals to interrupt important metabolic functions. Siderophores have been envisioned as “Trojan horses” with a capacity to deliver therapeutic agents into drug-resistant bacteria, targeting siderophore biosynthesis pathways to deny pathogens access to iron.
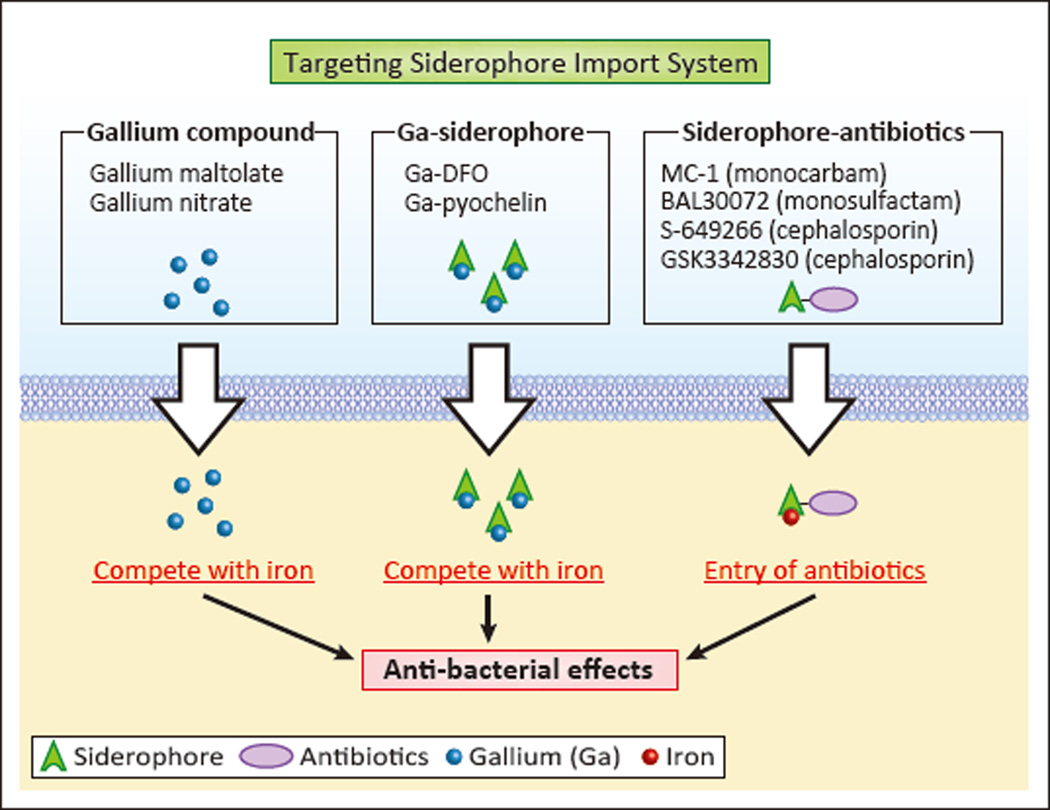
Gallium (Ga3+) is used in combating bacterial infections because it can compete with iron (Fe3+) to bind siderophores, while being unable to participate in biologically necessary redox chemistry. In fact, gallium arrests metabolic processes that depend on iron-mediated redox reactions. Gallium compounds such as gallium maltolate have been evaluated as a potential therapeutic agents for methicillin-resistant S. aureus [43] which can cause pneumonia and other infections (summarized in Table 1). In addition, a commonly-used drug for hypercalcemia, Ganite ® (gallium nitrate), results in antimicrobial activity against A. baumannii (Table 1) in vivo and in vitro [44]. In these approaches, gallium compounds presumably outcompete iron to bind siderophores, and are subsequently taken up by bacteria (Figure 3, left).
Figure 3. Novel Antimicrobials utilizing the Siderophore Import System.
Both gallium (Ga) salts (Gallium maltolate and Gallium nitrate) and Ga-conjugated siderophores (Ga-DFO and Ga-pyochelin) are used to deliver Ga to bacterial cells through siderophore import machinery. Ga interrupts important metabolic processes by competing with iron in critical redox reactions. Antibiotics can also be conjugated to siderophores (MC-1, BAL30072, S-649266, and GSK3342830), allowing enhanced uptake utilizing siderophore import machinery to boost anti-bacterial effects.
Gallium-loaded siderophores present another approach for gallium-based therapeutics. Desferrioxamine (DFO, Desferal®), a siderophore produced by the Streptomyces pilosus, has been used for the treatment of iron overload in diseases such as hemochromatosis and β-thalassemia [45]. Gallium-DFO (Ga-DFO) was demonstrated to be therapeutic for P. aeruginosa infection, such as in septicemia and infected pressure ulcers (bed sores) (Table 1), due to its bactericidal activity and prevention of biofilm formation [46]. Topical administration of Ga-DFO, in combination with gentamicin, also decreased the severity of P. aeruginosa infection in a rabbit cornea infection model [46]. In another study, pre-loading pyochelin -- a siderophore produced by P. aeruginosa (Table 1)-- with gallium, consistently led to a stronger inhibitory effect on P. aeruginosa growth than gallium nitrate in in vitro bacterial cultures [47], suggesting that treatment with gallium-siderophore complexes could be a more effective therapeutic option than using simple gallium salts (Figure 3, middle).
In a similar vein, conjugation of siderophores to antibiotics is an exciting treatment strategy. Siderophore moieties used for conjugation include catecholates and hydroxamates (Fig. 1) [48, 49]. Recently, several siderophores conjugated with β-lactam antibiotics have been evaluated in preclinical and clinical phases, including MC-1 (monocarbam), BAL30072 (monosulfactam) [50], S-649266 (cephalosporin) [51, 52] and GSK3342830 (cephalosporin), which target multidrug-resistant Gram-negative bacteria including A. baumannii and P. aeruginosa (Figure 3, right). P. aeruginosa is a particularly challenging pathogen to treat as it is intrinsically resistant to many antibiotics and easily acquires resistance (Table 1) [42, 53]. Porin channels in the outer membrane of Gram-negative bacteria including P. aeruginosa are essential for the entry of antibiotics such as β-lactams [54]. However, the porin channels of P. aeruginosa are narrower than other pathogens, resulting in minimal antibiotic uptake and increased drug resistance [55]. Consequently, antibiotic-conjugated siderophores are used as a ‘Trojan horse’ [48, 49] exploiting the siderophore-uptake system instead of porin proteins to efficiently deliver antibiotics to P. aeruginosa [48].
The siderophore biosynthesis pathway is another attractive target for therapeutics. Multidrug resistance in M. tuberculosis, the well-known bacterium that causes tuberculosis (Table 1), is dramatically increasing [56]. The biosynthesis pathways of siderophores produced by M. tuberculosis, mycobactin and the carboxylated form carboxymycobactin (Table 1), are well-understood and have been pharmacologically targeted [57]. The second step of mycobactin biosynthesis is mediated by 2,3-dihydroxybenzoate-AMP ligase (MbtA), an adenylate-forming enzyme [58]. The MbtA inhibitor salicyl-AMS [5'-O-(N-salicylsulfamoyl) adenosine] has been shown to suppress the growth of M. tuberculosis in the lungs of Mycobacterium-infected mice [58]. Inhibitors of other steps in mycobactin biosynthesis, such as benzimidazole-2-thione 4 (inhibitor of salicylate synthase MbtI) have been identified by high-throughput screening of more than 100,000 commercial compounds [59], but any utility in a clinical setting remains to be investigated.
Mechanistic insight into other aspects of siderophore biology gives future direction to therapeutic studies. For example, inhibiting siderophore export by deleting MmpS4/S5 (siderophore export proteins) in M. tuberculosis strains has been found to cause cytotoxicity upon exposure to mycobactin and carboxymycobactin [60]. Intracellular siderophore accumulation due to a defect in siderophore recycling seems to be the cause of toxicity, suggesting that MmpS4/S5 might be considered putative therapeutic targets against M. tuberculosis.
Inhibitors of siderophore biosynthesis have been tested in other pathogens as well. In S. aureus, although both staphyloferrin A and staphyloferrin B are produced (Table 1), staphyloferrin B appears to be more critical for survival in iron-limiting conditions [61]. A recent study revealed that SbnG is the citrate synthase responsible for supplying citrate for staphyloferrin B biosynthesis [62]. SbnG homologs are conserved across several pathogens, including P. aeruginosa. The structure of SbnG is more similar to bacterial class II aldolases than to the citrate synthase in the tricarboxylic acid (TCA) cycle [63], giving rise to the possibility of an inhibitor that can block SbnG citrate synthesis and subsequent staphyloferrin B production while not affecting the host TCA cycle. In addition, two molecules, baulamycin A and baulamycin B extracted from Streptomyces tempisquensis, have been identified as strong inhibitors of siderophore biosynthesis pathways in several bacteria, including S. aureus and E. coli [64]. Collectively, more understanding of siderophore biosynthesis might expand the lineup of potential targets for novel antimicrobial drugs.
Concluding Remarks
Future Directions in Siderophore Biology
The field of siderophore biology is a growing and exciting field of research, and new siderophores are being discovered regularly. Siderophores, constitute a key component of pathogenicity in nosocomial infections, but require careful study. While much work remains to characterize the molecular mechanisms of bacterial siderophore metabolism (see Outstanding Questions and Box 3), the future of siderophore research lies in extending basic findings into medical applications. A greater mechanistic knowledge of the back-and-forth struggle for iron between pathogen and host will provide clinical targets for physicians to treat nosocomial infections, such as inhibiting siderophore metabolism (biosynthesis, secretion, import) or utilizing siderophore conjugates to specifically deliver antibiotics (or other therapeutic agents) to the pathogen.
Box 3. The Clinician’s Corner.
Nosocomial infections, such as pneumonia, urinary tract infections, and surgical site infections, are on the rise and often result in high morbidity and mortality. Thus, novel approaches to controlling these infections are warranted.
Many nosocomial infections are caused by antibiotic-resistant pathogens.
Siderophores – small, pathogen-derived molecules utilized in iron acquisition – are important virulence factors for many pathogens.
Several potential siderophore-related therapeutics are being investigated, including “Trojan horse” compounds, gallium-containing compounds, and siderophore biosynthesis inhibitors.
Moreover, there is increasing evidence for the position of siderophores at the crux of many microbial cellular processes, rendering siderophores even more appealing antimicrobial targets. Rational drug design based on recent findings in siderophore biosynthesis and trafficking has resulted in the clever design of drugs that take advantage of siderophore bacterial dependence during infection. Novel drugs based on this approach will likely continue to be discovered, broadening the clinician’s arsenal in the fight against antibiotic resistant HAIs.
Box 1 Figure I. Iron: A Tug-Of-War.
Host cells secrete iron-binding proteins, such as lactoferrin (stage 1), to prevent pathogens from acquiring iron. Pathogens respond by “stealing” iron from host proteins using high affinity siderophores (stage 2). Host cells secrete the siderophore-binding protein lipocalin 2 (Lcn2) to neutralize the siderophore and prevent pathogen reuptake (stage 3). Pathogens can also produce “stealth siderophores” that cannot be sequestered by Lcn2 (stage 4).
Box 2 Figure II. Siderophore Import and Secretion Mechanisms.
Top, left. Siderophore import in Gram-negative bacteria is a multistep process, involving recognition of the ferric-siderophore by a specific outer membrane receptor (OMR), followed by TonB-dependent uptake into the periplasm. The ferric-siderophore is then trafficked by a periplasmic binding protein (PBP) to the inner membrane, where an ATP-binding cassette (ABC) transporter pumps it into the cytoplasm. Top, right. Siderophore import in Gram-positive bacteria involves recognition by a siderophore binding protein (SBP) located on the cell membrane. An associated permease is responsible for ferric-siderophore transport across the membrane. Bottom. Enterobactin secretion in Gram-negative bacteria involves transport from the cytoplasm to the periplasmic space through Major Facilitator Subtype (MFS) proteins. Transport across the outer membrane involves a TolC complex and an associated Resistance-Nodulated-Cell Division (RND) efflux pump.
Trends.
A “tug-of-war” for survival over available iron supplies can develop between a host and invading pathogens; bacteria attempt to acquire iron through ferric-siderophores, iron-bound transferrin, lactoferrin, and heme while the host can block pathogens to access ferric-siderophores, for instance, by forming a siderophore complex with lipocalin 2.
Ferric-siderophore import into bacteria is mediated through specific receptors localized in the outer membrane.
Secretion of siderophores from pathogens is mediated through efflux pumps including ABC transporters.
Iron chelation by siderophores causes robust responses in host cells through gene expression changes involved in apoptosis, mitophagy, hypoxia, and production of inflammatory cytokines.
Siderophore-conjugated compounds that limit the availability of iron to multi-drug resistant bacteria have recently emerged as new therapeutic approaches to contain nosocomial infections.
Inhibitors of bacterial siderophore biosynthesis are also promising new antimicrobial agents.
Acknowledgments
Briana Wilson was supported by NIH research supplements to promote diversity in health-related research R01GM095550S. Alexander R. Bogdan was supported by NIH Training Grant T32ES007046 from the National Institute of Environmental Health Sciences. This work was in part supported by NIH Research Grants R01GM088392 and R01GM095550 from the National Institute of General Medical Sciences to Y. Tsuji.
Glossary
- Chorismate
a metabolic precursor for many bacterial compounds, including some siderophores (e.g. enterobactin) and aromatic amino acids.
- Hemochromatosis
Disease characterized by iron overload in organs such as heart and liver, which cause arrhythmias and cirrhosis.
- Hypoxia-inducible factor 1α
a transcription factor that upregulates genes important for immune and hypoxic responses. HIF-1α is stabilized and activated by hypoxia or iron-deficiency.
- β-lactam antibiotics
Antibiotics characterized by beta-lactam ring structure including cephalosporins and carbapenems.
- Mitophagy
the regulated and selective process where cells degrade mitochondria via enclosure by double membraned vesicles (autophagosomes) and their subsequent fusion with lysosomes.
- MmpS4/S5
Mycobacterial inner membrane proteins exporting siderophores across the inner membrane to the periplasmic space.
- Nosocomial infections
infections that result from pathogen exposure within the healthcare setting caused by bacteria, viruses, and fungi that are not present at the time of admission. Generally apparent clinically after 48 hours. Also known as hospital-acquired or healthcare-associated infections
- Nrf2 (NFE2-related factor 2)
a transcription factor that upregulates genes important for immune responses and oxidative stress defense. Nrf2 is stabilized and activated by oxidative stress.
- PINK1 and Parkin
two proteins that are the best-characterized mediators of mitophagy. Mutations in these genes are associated with Parkinson’s disease.
- Siderophores
small, ferric iron (Fe3+) chelating molecules secreted by microorganisms in response to iron limitation
- β-thalassemia
Blood disease characterized by loss or reduction of beta-globin, a component of hemoglobin in the blood.
- Toll-like receptor 4 (TLR4)
a cell-surface receptor in humans that recognizes many bacterial products, including the bacterial cell wall component lipopolysaccharide. TLR4 activation initiates many host cell signaling pathways important for immune responses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temkin E, et al. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann N Y Acad Sci. 2014;1323:22–42. doi: 10.1111/nyas.12537. [DOI] [PubMed] [Google Scholar]
- 3.Soares MP, Weiss G. The Iron age of host-microbe interactions. EMBO Rep. 2015;16:1482–1500. doi: 10.15252/embr.201540558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden VI, Bachman MA. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7:986–995. doi: 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 5.Peleg AY, et al. The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS One. 2012;7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison F, Buckling A. Siderophore production and biofilm formation as linked social traits. ISME J. 2009;3:632–634. doi: 10.1038/ismej.2009.9. [DOI] [PubMed] [Google Scholar]
- 7.Gaddy JA, et al. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marti S, et al. Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS One. 2011;6:e26030. doi: 10.1371/journal.pone.0026030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JJ. Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 2015;31:475–482. doi: 10.1016/j.tig.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Su HC, et al. The development of ciprofloxacin resistance in Pseudomonas aeruginosa involves multiple response stages and multiple proteins. Antimicrob Agents Chemother. 2010;54:4626–4635. doi: 10.1128/AAC.00762-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng BS, et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogdan AR, et al. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem Sci. 2016;41:274–286. doi: 10.1016/j.tibs.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty S, et al. The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochim Biophys Acta. 2012;1826:129–169. doi: 10.1016/j.bbcan.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sydnor ER, Perl TM. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 16.Schalk IJ, Guillon L. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids. 2013;44:1267–1277. doi: 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 17.Allen GF, et al. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagi M, et al. Iron-depletion promotes mitophagy to maintain mitochondrial integrity in pathogenic yeast Candida glabrata. Autophagy. 2016;12:1259–1271. doi: 10.1080/15548627.2016.1183080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiavi A, et al. Iron-Starvation-Induced Mitophagy Mediates Lifespan Extension upon Mitochondrial Stress in C. elegans. Curr Biol. 2015;25:1810–1822. doi: 10.1016/j.cub.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 20.Kirienko NV, et al. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe. 2013;13:406–416. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirienko NV, et al. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112:1821–1826. doi: 10.1073/pnas.1424954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann H, et al. Hypoxia-independent activation of HIF-1 by enterobacteriaceae and their siderophores. Gastroenterology. 2008;134:756–767. doi: 10.1053/j.gastro.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Holden VI, et al. Bacterial siderophores that evade or overwhelm lipocalin 2 induce hypoxia inducible factor 1alpha and proinflammatory cytokine secretion in cultured respiratory epithelial cells. Infect Immun. 2014;82:3826–3836. doi: 10.1128/IAI.01849-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci. 2012;37:364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palazon A, et al. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyssonnaux C, et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand RJ, et al. Hypoxia causes an increase in phagocytosis by macrophages in a HIF-1alpha-dependent manner. J Leukoc Biol. 2007;82:1257–1265. doi: 10.1189/jlb.0307195. [DOI] [PubMed] [Google Scholar]
- 29.Elks PM, et al. Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog. 2013;9:e1003789. doi: 10.1371/journal.ppat.1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nairz M, et al. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med. 2013;210:855–873. doi: 10.1084/jem.20121946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wessling-Resnick M. Iron homeostasis and the inflammatory response. Annu Rev Nutr. 2010;30:105–122. doi: 10.1146/annurev.nutr.012809.104804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oexle H, et al. Pathways for the regulation of interferon-gamma-inducible genes by iron in human monocytic cells. J Leukoc Biol. 2003;74:287–294. doi: 10.1189/jlb.0802420. [DOI] [PubMed] [Google Scholar]
- 33.Chandrasekar BS, et al. Interferon-gamma and nitric oxide synthase 2 mediate the aggregation of resident adherent peritoneal exudate cells: implications for the host response to pathogens. PLoS One. 2015;10:e0128301. doi: 10.1371/journal.pone.0128301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michels K, et al. Hepcidin and Host Defense against Infectious Diseases. PLoS Pathog. 2015;11:e1004998. doi: 10.1371/journal.ppat.1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DK, et al. Inverse agonist of estrogen-related receptor gamma controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat Med. 2014;20:419–424. doi: 10.1038/nm.3483. [DOI] [PubMed] [Google Scholar]
- 36.Pilonieta MC, et al. Salmonella enterica infection stimulates macrophages to hemophagocytose. MBio. 2014;5:e02211. doi: 10.1128/mBio.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald EM, et al. Bacterial Stimulation of Toll-Like Receptor 4 Drives Macrophages To Hemophagocytose. Infect Immun. 2016;84:47–55. doi: 10.1128/IAI.01149-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy MW, et al. Hemophagocytic macrophages in murine typhoid fever have an anti-inflammatory phenotype. Infect Immun. 2012;80:3642–3649. doi: 10.1128/IAI.00656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peyssonnaux C, et al. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood. 2006;107:3727–3732. doi: 10.1182/blood-2005-06-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deak D, et al. Progress in the Fight Against Multidrug-Resistant Bacteria? A Review of U.S. Food and Drug Administration-Approved Antibiotics, 2010–2015. Ann Intern Med. 2016 doi: 10.7326/M16-0291. [DOI] [PubMed] [Google Scholar]
- 42.Winstanley C, et al. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baldoni D, et al. In vitro activity of gallium maltolate against Staphylococci in logarithmic, stationary, and biofilm growth phases: comparison of conventional and calorimetric susceptibility testing methods. Antimicrob Agents Chemother. 2010;54:157–163. doi: 10.1128/AAC.00700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antunes LC, et al. In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:5961–5970. doi: 10.1128/AAC.01519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lui GY, et al. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget. 2015;6:18748–18779. doi: 10.18632/oncotarget.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banin E, et al. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc Natl Acad Sci U S A. 2008;105:16761–16766. doi: 10.1073/pnas.0808608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frangipani E, et al. Pyochelin potentiates the inhibitory activity of gallium on Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58:5572–5575. doi: 10.1128/AAC.03154-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorska A, et al. Siderophore-drug complexes: potential medicinal applications of the 'Trojan horse' strategy. Trends Pharmacol Sci. 2014;35:442–449. doi: 10.1016/j.tips.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Mollmann U, et al. Siderophores as drug delivery agents: application of the"Trojan Horse" strategy. Biometals. 2009;22:615–624. doi: 10.1007/s10534-009-9219-2. [DOI] [PubMed] [Google Scholar]
- 50.Coates AR, et al. Novel classes of antibiotics or more of the same? Br J Pharmacol. 2011;163:184–194. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito A, et al. In vitro antimicrobial activity of S-649266, a catechol-substituted siderophore cephalosporin, when tested against non-fermenting Gram-negative bacteria. J Antimicrob Chemother. 2016;71:670–677. doi: 10.1093/jac/dkv402. [DOI] [PubMed] [Google Scholar]
- 52.Ito-Horiyama T, et al. Stability of Novel Siderophore Cephalosporin S-649266 against Clinically Relevant Carbapenemases. Antimicrob Agents Chemother. 2016;60:4384–4386. doi: 10.1128/AAC.03098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int. 2016;2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pages JM, et al. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 55.Sugawara E, et al. Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. FEBS J. 2012;279:910–918. doi: 10.1111/j.1742-4658.2012.08481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen L. Antibiotic resistance mechanisms in M. tuberculosis: an update. Arch Toxicol. 2016;90:1585–1604. doi: 10.1007/s00204-016-1727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sritharan M. Iron homeostasis in Mycobacterium tuberculosis: mechanistic insights into siderophore-mediated iron uptake. J Bacteriol. 2016 doi: 10.1128/JB.00359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lun S, et al. Pharmacokinetic and in vivo efficacy studies of the mycobactin biosynthesis inhibitor salicyl-AMS in mice. Antimicrob Agents Chemother. 2013;57:5138–5140. doi: 10.1128/AAC.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasan M, et al. Inhibitors of the salicylate synthase (MbtI) from Mycobacterium tuberculosis discovered by high-throughput screening. ChemMedChem. 2010;5:2079–2087. doi: 10.1002/cmdc.201000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones CM, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc Natl Acad Sci U S A. 2014;111:1945–1950. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheung J, et al. Discovery of an iron-regulated citrate synthase in Staphylococcus aureus. Chem Biol. 2012;19:1568–1578. doi: 10.1016/j.chembiol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Sheldon JR, et al. TCA cycle activity in Staphylococcus aureus is essential for iron-regulated synthesis of staphyloferrin A, but not staphyloferrin B: the benefit of a second citrate synthase. Mol Microbiol. 2014;92:824–839. doi: 10.1111/mmi.12593. [DOI] [PubMed] [Google Scholar]
- 63.Kobylarz MJ, et al. SbnG, a citrate synthase in Staphylococcus aureus: a new fold on an old enzyme. J Biol Chem. 2014;289:33797–33807. doi: 10.1074/jbc.M114.603175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi A, et al. Baulamycins A and B, broad-spectrum antibiotics identified as inhibitors of siderophore biosynthesis in Staphylococcus aureus and Bacillus anthracis. J Am Chem Soc. 2014;136:1579–1586. doi: 10.1021/ja4115924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li C, Chan YR. Lipocalin 2 regulation and its complex role in inflammation and cancer. Cytokine. 2011;56:435–441. doi: 10.1016/j.cyto.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 66.Steigedal M, et al. Lipocalin 2 imparts selective pressure on bacterial growth in the bladder and is elevated in women with urinary tract infection. J Immunol. 2014;193:6081–6089. doi: 10.4049/jimmunol.1401528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh V, et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat Commun. 2015;6:7113. doi: 10.1038/ncomms8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bachman MA, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shields-Cutler RR, et al. Human Urinary Composition Controls Antibacterial Activity of Siderocalin. J Biol Chem. 2015;290:15949–15960. doi: 10.1074/jbc.M115.645812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klebba PE. ROSET Model of TonB Action in Gram-Negative Bacterial Iron Acquisition. J Bacteriol. 2016;198:1013–1021. doi: 10.1128/JB.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horiyama T, Nishino K. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One. 2014;9:e108642. doi: 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]