Abstract
Small RNAs have the potential to store a secondary layer of labile biological information in the form of modified nucleotides. Emerging evidence has shown that small RNAs including microRNAs (miRNAs), PIWI-interacting RNAs (piR-NAs) and tRNA-derived small RNAs (tsRNAs) harbor a diversity of RNA modifications. These findings highlight the importance of RNA modifications in the modulation of basic properties such as RNA stability and other complex physiological processes involved in stress responses, metabolism, immunity, and epigenetic inheritance of environmentally acquired traits, among others. High-resolution, high-throughput methods for detecting, mapping and screening these small RNA modifications now provide opportunities to uncover their diagnostic potential as sensitive disease markers.
A Glimpse into RNA Modifications
RNAs are post-transcriptionally altered with a variety of chemical modifications in all domains of life. More than 100 types of RNA modification have been identified to date, in many cases showing context-dependent functions that are only beginning to become clear [1]. Post-transcriptional modifications are known to be especially prevalent in stable, structured RNAs such as tRNA and rRNA where they play important roles in biogenesis, correct structural folding, and function [2]. Emerging evidence indicates that a diverse set of modifications are also found in transient carriers of information, including mRNAs and an expanding catalog of small and large non-coding RNAs. These are exemplified by the recent discovery of reversible 6-methylade-nosine (m6A) modifications in mRNAs [3,4] as well as key enzymes for their dynamic regulation: writer (see Glossary) [5,6], eraser [7,8], and reader [9]. Other studies have documented pseudouridine [10–12], 5-methylcytidine (m5C) [13,14], and, most recently, 1-methyladenosine (m1A) [15,16] in mRNAs. RNA modifications are also prevalent in small RNAs, where they could similarly carry information important for various cellular functions (Figure 1). Here we briefly discuss the roles of representative RNA modifications and RNA editing events in the biogenesis and function of small RNAs. We also highlight the potential clinical relevance of such modified small RNAs and their possible utilization as diagnostic markers of disease. Lastly, we summarize recent technical advances in high-throughput approaches for profiling RNA modifications in small RNAs, with a particular focus on heavily modified tRNAs and tsRNAs. New sequencing-based methods are highly complementary to established mass spectrometry-based analyses, with the advantages of positional resolution and wide availability.
Figure 1. Small RNAs and RNA Modifications.
(A) Schematic depicting a variety of small RNAs. (B) The chemical structures of relatively well-studied modifications, RNA editing events, and noncanonical nucleotides are shown. (C) Known functions and future perspectives for RNA modifications are illustrated. miRNAs, microRNAs; piRNAs, P-element-induced wimpy testis (PIWI)-interacting RNAs; tsRNAs, tRNA-derived small RNAs.
2′-O-Methylation in Small RNAs: Stability and Beyond
miRNAs are key players in RNA silencing pathways inducing mRNA degradation, translational repression, and transcriptional silencing [17]. 2′-O-Methylation, mediated by HUA enhancer 1 (HEN1), was first recognized in plants [18], where it protects miRNAs from small RNA decay pathways that are triggered by 3′-uridylation [19,20], a well-characterized RNA decay pathway [21] (Figure 2A). Although mature miRNAs in animals lack 2′-O-methylation, other mammalian small RNAs such as siRNAs and P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) can be methylated by HEN1 orthologs, which similarly protect these small RNAs from 3′-uridylation [19]. This conserved mechanism provides a rationale in translational research for promoting the stability of small RNA therapeutic agents, with the potential for more efficient RNAi therapy [22]. 2′-O-methylation in plant miRNAs also provides a potential mechanism for prolonging their functional half-life in the human body when ingested, supporting the intriguing – and still controversial – idea that small RNAs derived from plant-based diets can modulate human physiology [23–28].
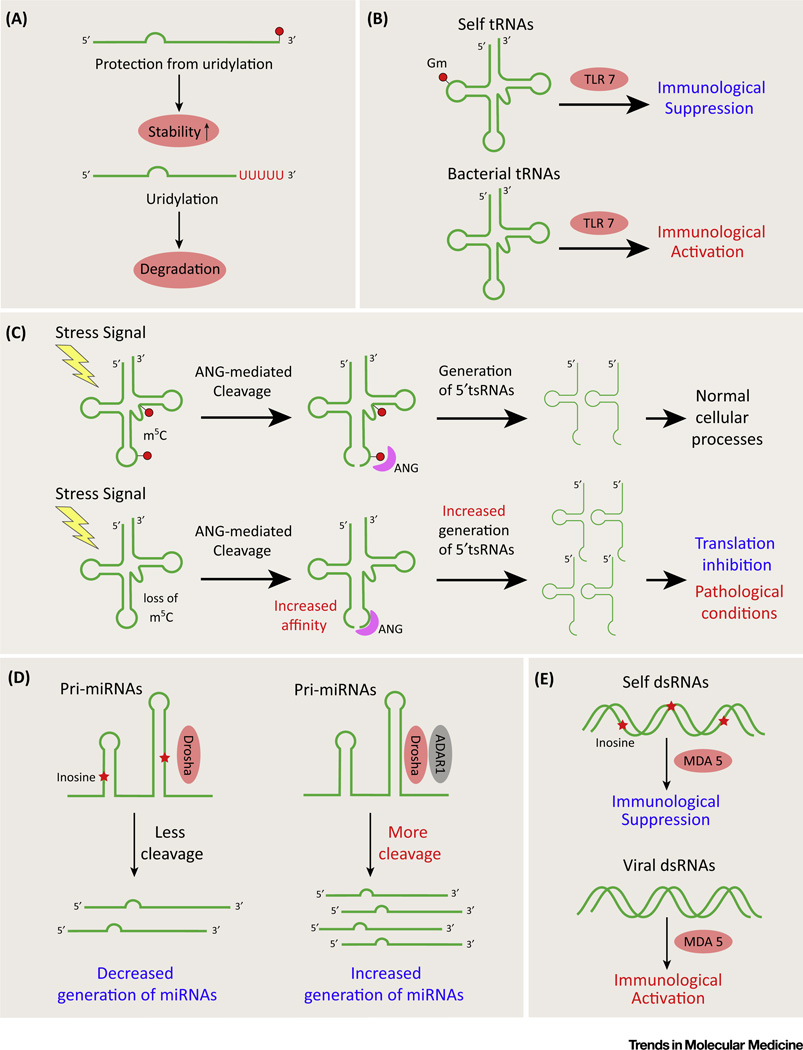
Figure 2. Representative Biological Functions of RNA Modifications and RNA Editing Events in Small RNAs.
(A) 2′-O-Methylation at the 3′ end of small RNAs can protect these from uridylation, preventing degradation by RNA decay pathways. (B) Discrimination of self and non-self RNAs. Bacterial tRNAs lacking 2′-O-methylation at position 18 (Gm) are recognized by Toll-like receptor 7 (TLR-7) on the endosomal surface of relevant immune cells, activating downstream innate immunity. (C) m5C fine-tunes angiogenin (ANG)-mediated tRNA cleavage under conditions of cellular stress (nutritional starvation etc.). tRNAs lacking m5C are preferentially cleaved by ANG, leading to accumulation of tRNA-derived small RNAs (tsRNAs) and associated pathology (as in the case of certain tumors). (D) Adenosine-to-inosine (A-to-I) editing influences miRNA biogenesis. pri-miRNAs harboring inosines [generated by adenosine deaminase acting on RNA 1 (ADAR1)] are resistant to cleavage by double-strand RNA (dsRNA)-specific endoribonuclease (Drosha), the core nuclease in miRNA processing, thus decreasing mature miRNA production. (E) Inosine discriminates endogenous versus viral RNAs. Melanoma differentiation-associated protein 5 (MDA5) is a RIG-I-like receptor dsRNA helicase enzyme that binds viral dsRNAs lacking inosine bases to trigger antiviral responses.
In addition to increasing small RNA stability, specific 2′-O-methylation of guanosine in bacterial tRNA at position 18 (Gm18) is necessary and sufficient to suppress immune activation mediated by Toll-like receptor 7 (TLR-7) during human host–pathogen responses [29,30]. Most bacteria lack the gene responsible for G18 tRNA 2′-O-methylation, TrmH; thus, it may be a positively selected trait in symbiotic and pathogenic species. This example demonstrates an important role for RNA modification in modulating recognition of self- versus non-self-derived RNAs [29,30] (Figure 2B). Overall, the functions of 2′-O-methylation suggest myriad potential translational applications related to the stability and immunogenicity of small RNA-based therapeutics.
m5C: A Stress Sensor and Transgenerational Mark in tRNA and tsRNAs
m5Cs are frequently detected in various non-coding RNAs such as tRNA, rRNA, and, more recently, mRNAs. While the full range of potential functions for m5C in RNA is not completely D understood, bisulfite RNA sequencing, which pinpoints m5C residues [31], has helped reveal the importance of these residues in tRNAs [32–36]. One group has shown that m5C modifications of tRNAs by the methyltransferases DNA methyltransferase 2 (Dnmt2) and NOP2/Sun RNA methyltransferase family member 2 (Nsun2) are required for normal growth and development in mice [33] and flies [36], supporting a close relationship between m5C modification and tRNA stability. Moreover, m5C alters the tRNA-binding activity of the endonuclease angiogenin, as loss of m5C RNA methylation increases angiogenin-mediated cleavage of tRNAs in response to a variety of cellular stresses, generating tsRNAs [32–34]. For instance, accumulation of 5′-tsRNAs reduces protein translation rates [37] and tRNAs lacking Dnmt2-dependent methylation lead to codon mistranslation, which also alters protein expression [35]. These tRNA changes disrupt protein synthesis at a systemic level, negatively affecting body growth as well as neurological and stem cell functions [32–35]. Furthermore, Dnmt2-mediated virus RNA methylation has also been implicated in efficient antiviral defense in Drosophila sp. [38]. Together, these studies highlight an intricate interplay between m5C modification of tRNAs, tsRNA biogenesis, cellular stress responses, and protein synthesis – fundamental processes relevant to diverse disease conditions (Figure 2C).
Recent discoveries have also revealed important and surprising roles for m5C in RNA-mediated transgenerational epigenetic inheritance [39,40]. Studies have demonstrated that the m5C methyltransferase Dnmt2 is required for RNA-mediated epigenetic alterations in fur color and an overgrowth phenotype in mice [39]. Both phenotypes were shown to be induced in mice with wild-type Dnmt2 whereas neither phenotype was transmitted in mutants deficient in Dnmt2 [39]. More recent work has also implicated m5C in intergenerational epigenetic transmission of metabolic alterations arising from paternal dietary exposures. Male mice fed high-fat diets (HFDs) were reported to transmit impaired glucose tolerance and insulin resistance to offspring and some of these effects could also be produced by injecting total RNA or tsRNAs from HFD sperm into normal murine zygotes. Notably, tsRNAs constitute the dominant small RNA species in mature sperm and were shown to exhibit elevated m5C and m2G levels in HFD sperm compared with those from males fed normal diets [40]. These separate lines of evidence involving tRNAs/tsRNAs implicate m5C in transgenerational epigenetic inheritance. Whereas synthetic tsRNAs lacking RNA modifications degrade rapidly when incubated in zygote lysate or serum, endogenous tsRNAs are more stable, suggesting possible roles for modifications in promoting the stability [40,41] and transgenerational effects of tsRNAs after fertilization [40]. These modifications may also contribute to the secondary structures of tsRNAs that lead to altered binding specificities to their targets (DNA, RNAs, or proteins) [40]. Together, these observations suggest that small RNA modifications are important in modulating the efficiency of RNA-mediated transmission from fathers to offspring.
tsRNAs are increasingly recognized as functional signaling and regulatory molecules in a diversity of contexts [32–34,37,40–47]. Mature tRNAs are among the most abundant and most heavily modified RNA species. Aberrations in tRNA modification play roles in cellular glucose metabolism [48] and have been implicated in various pathologies, including cancer, type 2 diabetes, neurological disorders, and mitochondria-linked disorders [49]. A frequently changing population of tsRNAs is also ubiquitous in cells and contains some portion of the RNA modifications made in the parent tRNA molecules. The functions of these tsRNA modifications and the extent to which they are altered after tsRNA biogenesis are largely unexplored but are likely to alter in vivo tsRNA dynamics including stability, secondary structure, and interactions with proteins [50]. Thus, the extensive potential repertoire of tsRNA modifications could be important in a wide diversity of physiological processes and pathological conditions across species. Similarly, rRNAs may also give rise to abundant and highly modified small RNAs (rsRNAs) [51], but these harbor as-yet-unknown functions in normal physiology and disease.
RNA Editing Events in Small RNAs: Biology and Disease Associations
RNA editing is a process that alters the primary nucleotide sequence of RNA by specific enzymes. The chief distinction between RNA editing and RNA modification is that editing is usually irreversible and generally understood to produce altered base-pairing behavior. RNA editing events are widely observed in mRNA, tRNA, rRNA, and miRNA in all kingdoms of life. The most frequent editing events are deaminations that convert adenosine to inosine (A to I) and cytidine to uridine (C to U) [52]. Adenosine deaminase acting on RNA (ADAR) is an enzyme that mediates A-to-I editing by hydrolytic deamination of adenosine in double-strand RNAs (dsRNAs); this can occur within both the coding sequence and the non-coding regions of an mRNA transcript [53].
Deficiency of A-to-I editing activity in certain mRNA transcripts has been implicated in human diseases including amyotrophic lateral sclerosis (ALS) [54] and cancer [55]. miRNAs are also subject to A-to-I editing [53]. Primary miRNA (pri-miRNA) transcripts are typically ~60–70 nt long and can form dsRNA structures that can be edited by ADARs [53]. Deletion of ADAR1 or ADAR2 in mice has been shown to prevent degradation of the miRNA precursor pri-miR-142 by a component of the RNA-induced silencing complex (RISC) resulting in increased levels of the mature miRNA miR-142, thus illustrating a molecular mechanism by which miRNA biogenesis can be controlled [56] (Figure 2D). A-to-I editing also occurs, although less frequently, in mature miRNAs [57]. Edited mature miRNAs (e.g., miR-376) can alter their targeting specificity [58], a change that has been found to promote invasiveness of glioblastoma cells in a mouse model [59]. Moreover, alterations in ADAR editing activity levels have been linked to stem cell maintenance, hereditary autoimmune diseases, and disease progression in cancer, suggesting the potential of using A-to-I editing as a putative biomarker of disease severity for certain conditions [60].
Similar to 2′-O-methylation, the inosines in RNAs produced by A-to-I editing can be used by the innate immune system to discriminate self versus non-self RNAs. When viral dsRNAs lacking inosine enter the cell, RNA sensors such as melanoma differentiation-associated protein 5 (MDA5), a RIG-I-like receptor dsRNA helicase enzyme, can bind dsRNA and activate antiviral responses [61,62] (Figure 2E). By contrast, endogenous dsRNAs edited by ADAR1 contain A-to-I edits that prevent MDA5 binding and antiviral immune responses [61,62] (Fig. 2E).
C-to-U RNA editing is mediated by cytidine deaminases, which was first observed acting on vertebrate apolipoprotein B (apoB) mRNA transcripts [52]. The C-to-U editing sites are usually enriched in the 3′ UTR of mRNAs and influence the targeting specificity of miRNAs [63]. C-to-U editing is also required for the maturation process of mitochondrial tRNAAsp in marsupials [64]. Future high-throughput sequencing studies to systematically map C-to-U RNA editing sites may lead to the discovery of novel biological functions and their roles in disease.
Profiling Small RNA Modifications As Biomarkers of Disease
Several different approaches have been used for high-throughput analysis of RNA modifications; each has different strengths in terms of its ability to monitor specific RNA modifications, characterize global modification profiles in biological samples, or reveal the identity of modified RNAs with sequence-level specificity. Antibody-based detection of modified small RNAs has shown promise as a sensitive diagnostic for identifying early markers of disease. For example, m1A antibodies can detect tRNA conformational changes and circulating tsRNAs that arise from acute cellular stress, potentially providing an earlier indicator of tissue injury than other markers of apoptosis and DNA damage (Figure 3A) [65]. High-throughput liquid chromatography–tandem mass spectrometry (LC-MS/MS) provides a complementary approach that can be used for quantification of multiple RNA modifications within a single RNA sample [40,51,66–68]. LC-MS/MS has uncovered a diversity of previously overlooked RNA modifications among small RNAs (10–30 nt, 30–60 nt) in mouse liver [51], revealing dynamic changes for multiple RNA modifications in mouse models of diabetes (Gm, m5Cm, Cm, Am, Um) [51] (Figure 1B). LC-MS/MS profiling has also identified altered modification profiles, with increased levels of m5C and m2G, in the 30–40-nt fraction of sperm RNAs (predominantly tsRNAs) in mice fed HFDs (described above), suggesting that these chemical modifications may transmit information about paternal diet to offspring [40] (Figure 1C). Similar approaches have been used to detect altered modified nucleosides in the urine of cancer patients, where modification profiles could potentially be utilized as biomarkers for clinical diagnosis in the design of personalized therapy [69]. Thus, the combined use of antibody- and/or LC-MS/MS-based profiling of RNA modifications holds great promise for noninvasive tests (e.g., of blood, sperm, or urine) to help guide the clinical diagnosis and treatment of a variety of disorders.
Figure 3. RNA Modifications As Diagnostic Markers of Pathological Conditions.
(A) m1A-specific antibody detects the m1A modification only in conformationally altered tRNAs and cleaved 3′ end tRNA-derived small RNAs (tsRNAs) that are produced by acute cell damage (as in the kidney). (B) Diabetes can alter the RNA modification profile of tRNAs and small RNAs (10–30 nt and 30–60 nt) in mouse liver. (C) High-fat diet mouse models have demonstrated increases in m5C and m2G modifications in sperm tsRNAs (30–40 nt).
Emerging Methods for Profiling and Mapping RNA Modifications in Small RNAs
While LC-MS/MS is a powerful approach for quantifying the compositional changes of RNA modifications, this method requires digestion of RNAs into short segments or single nucleosides before detection, therefore making it difficult to unambiguously assign modifications to specific transcripts (Figure 4A) [51,66,67]. Thus, complementary approaches to map RNA modifications to specific RNAs are required to gain a more comprehensive view of modified small RNAs.
Figure 4. Emerging Methods for Profiling RNA Modifications in Small RNAs.
(A) Profiling and quantification of RNA modifications using liquid chromatography–tandem mass spectrometry (LC-MS/MS). (B) Identification and mapping of methylated RNAs using Escherichia coli AlkB treatment. AlkB-facilitated RNA methylation sequencing (ARM-seq) measures changes in the abundance (either full read or no read) of tRNA-derived small RNAs (tsRNAs) after AlkB treatment, which removes RNA modifications blocking reverse transcriptase during cDNA library construction. Demethylase-thermostable group II intron reverse transcription tRNA sequencing (DM-tRNA-seq) can identify modifications via comparison of early termination (short) reads in non-treated samples with full reads from samples treated with AlkB; DM-tRNA-seq preferentially sequences full-length tRNAs, which are in much higher abundance than tsRNAs. RT, reverse transcription. TGIRT, thermostable group II intron reverse transcriptase.
Sequencing-based protocols have been developed to identify RNAs modified with m5C, pseudouridine, m6A, and m1A, in some cases also providing precise positional information about the location of these modifications [70]. For example, RNA bisulfite sequencing can detect m5C in both large and small RNAs by converting unmethylated cytosine (C) to uracil (U) with bisulfite, which will be read as thymine (T) during sequencing, while methylated C (m5C) remains unconverted. By comparing the sequencing results of bisulfite-treated samples versus untreated samples, the position of m5C in RNAs can be reproducibly and quantitatively analyzed [31,71]. The mapping of both m6A and m1A modifications in mRNAs is based on immunoprecipitation (IP) of methylated RNAs with antibodies specific to each modification, followed by RNA sequencing of the IP-enriched RNA fragments. When these sequencing reads are mapped to the genome, enrichment peaks correspond to the locations of m6A and m1A in the mRNAs [3,4,16]. These ‘first-generation’ antibody-based approaches can identify modified mRNAs and can localize modified residues to ~100–200-nt regions within transcripts; however, the precise m6A positions are not known to single-nucleotide resolution and this method cannot be applied to mapping modifications on small RNAs. A recent improvement of this technique crosslinks the antibody to the bound RNA fragment with UV light, producing a specific pattern of read truncations and base changes adjacent to the m6A site and thus allowing single-base resolution of m6A sites [72]. This improved method can be used to map human and mouse small nucleolar RNAs (snoRNAs) of approximately 70–90 nt in length [72] and future improvements may lead to precise mapping of even smaller RNAs (<40 nt). Consequently, methodologies based on the combination of chemical treatments with RNA sequencing hold great promise for precisely mapping a variety of RNA modifications in small RNAs.
Notably, other modifications, including m1A, m1G, and m3C, are common in tRNAs (but not miRNAs) and interfere with normal intra- and intermolecular base pairing. Reverse transcription, an essential step in cDNA library preparation for RNA-seq, is halted at a high rate at these so-called ‘hard-stop’ modifications. This significantly impairs efficient detection of small RNAs containing these modifications, suggesting that a large but unknown fraction of biologically important tsRNAs has been missed in all prior studies and large surveys (e.g., ENCODE, TCGA) employing standard small RNA-seq methods to profile small RNA populations.
Recently, two complementary new approaches have been developed to tackle the problem of not overlooking tsRNAs. By using pretreatment of RNAs with the Escherichia coli dealkylating enzyme AlkB to demethylate m1A, m3C, and m1G before sequencing, detection of methyl-modified mature tRNAs [demethylase-thermostable group II intron reverse transcription tRNA sequencing (DM-tRNA-seq)] [73] and tsRNAs [AlkB-facilitated RNA methylation sequencing (ARM-seq)] [74] has been possible (Figure 4B). Comparative methylation analysis using ARM-seq has revealed a wealth of previously undetected methylated tsRNAs and, importantly, demonstrated that these are modified very similarly to the mature tRNAs from which they are derived [74]. Combining these sequencing-based approaches with LC-MS/MS is a promising means of mapping specific types of RNA modifications with both compositional and positional information (Figure 4A,B). A detailed map of the many types of RNA modifications across the major types of small RNAs presents one among many challenges ahead. Charting the dynamics of these modifications in different cell types and physiological processes is another. Together, these new views are critical cornerstones to addressing further questions about the functional roles of modifications of small RNA and their potential use in medicine as both diagnostic markers and therapeutic targets (see Outstanding Questions) (Box 1).
Outstanding Questions.
How evolutionarily conserved are small RNA modifications across different types of small RNAs?
Are there tissue-specific or developmentally regulated small RNA modifications?
Which small RNA modifications are reversible versus irreversible and what are their regulatory machineries?
How do specific modifications alter the structure, stability, and molecular interactions of small RNAs?
Do RNA modifications contribute to the stability and cross-tissue transfer of mobile small RNAs?
Can small RNA modifications be used as general diagnostic markers of disease (metabolic dysfunction, cancer, infections, or others)?
Box 1. The Clinician's Corner.
RNAs extracted from biological samples such as blood and urine have shown great diagnostic value. In addition to the specific profile of small RNAs detected, RNA modifications represent another layer of biological information that can also be harnessed for diagnostic use.
Disease conditions (e.g., those involving acute kidney cell damage) can lead to the release of modified RNA nucleosides (m1A) that are readily detected using antibody-based methods, potentially providing a sensitive diagnostic for early tissue injury and cell damage.
High-throughput detection methods based on mass spectrometry can simultaneously detect multiple types of RNA modifications in one small RNA sample, providing new molecular signatures of disease conditions as in the case of metabolic disorders.
High throughput RNA-seq has been used to show that RNA editing events (A-to-I) in small RNAs correlate with the malignancy grade of some tumors, suggesting the potential use of this approach as a diagnostic screen.
High-resolution profiling of small RNA modifications holds great promise for sensitive and specific detection of aberrant physiological conditions and disease as well as for the development of novel therapeutic approaches designed to manipulate specific RNA modifications.
Concluding Remarks: How Many New Bits of Information?
Compared with the accelerating rate of discovery of many types of small RNAs [75], the study of small RNA modifications in biology and disease remains in its infancy. Emerging evidence indicates that RNA modifications are important molecular markers that can alter the biogenesis, function, and informational capacity of small RNAs. New techniques including high-throughput LC-MS/MS and sequencing-based approaches that specifically target modified RNAs using antibodies, enzymatic treatments, or chemical alterations hold great promise for elucidating the role of tRNAs, tsRNAs, and other modified small RNAs as signaling and regulatory molecules (see Outstanding Questions) (Box 1). These approaches may prove particularly powerful when used in combination, providing new insights into the physiological roles of modified small RNAs as well as new opportunities for diagnosis of small RNA-associated diseases.
Trends.
RNA modifications such as 2′-O-methylation, 5-methylcytidine (m5C), and adenosine-to-inosine (A-to-I) editing modulate the activities of small RNAs in diverse biological processes and play pivotal roles in pathological conditions.
Antibody-based detection modified RNAs and high-throughput liquid chromatography-tandem mass spectrometry (LC-MS/MS) promising diagnostic approaches for distinguishing normal versus diseased conditions.
Novel methods for RNA sequencing facilitate the identification of modified small RNAs that are recalcitrant to sequencing using standard approaches, expanding the diversity of small RNAs that can be detected as potential diagnostic markers of disease.
Acknowledgments
This research was partially supported by the NIH/NHGRI (HG006753-02 to T.M.L, P30GM110767-03 to Q.C.) and start-up support from the University of Nevada, Reno School of Medicine (to Q.C.).
Glossary
- Bisulfite RNA sequencing
method to detect m5C methylation patterns in RNAs by bisulfite treatment of RNAs.
- cDNA library preparation
synthesis of single-strand complementary DNAs from RNA samples by reverse transcription.
- Eraser
a demethylase or associated enzymatic complex that removes RNA modifications.
- P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs)
the largest class of small non-coding RNAs expressed in animal cells; form RNA–protein complexes with PIWI proteins.
- Reader
proteins that bind to specific modified nucleotides to initiate downstream pathways.
- Reverse transcription
synthesis of single-strand complementary DNA using single-strand RNA as the template, mediated by RNA-dependent DNA polymerase (reverse transcriptase).
- RNA silencing pathways
mechanisms to suppress gene expression by RNA-induced degradation of mRNA or inhibition of protein synthesis.
- 3′-Uridylation
the enzymatic addition of uridines to the 3′ end of small RNAs (miRNA, piRNA, etc.); can trigger the small RNA degradation pathway.
- Writer
a methyltransferase or associated enzymatic complex that adds chemical modifications to RNAs.
References
- 1.Frye M, et al. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 5.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia G, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, et al. N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 13.Hussain S, et al. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 16.Dominissini D, et al. The dynamicN1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 18.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G, et al. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl Acad. Sci. U. S. A. 2014;111:6365–6370. doi: 10.1073/pnas.1405083111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M, et al. Emerging roles of RNA modification: m6A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Bouchie A. Companies in footrace to deliver RNAi. Nat. Biotechnol. 2012;30:1154–1157. doi: 10.1038/nbt1212-1154. [DOI] [PubMed] [Google Scholar]
- 23.Chin AR, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, et al. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015;25:517–520. doi: 10.1038/cr.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschi KD, et al. Dietary delivery: a new avenue for micro-RNA therapeutics? Trends Biotechnol. 2015;33:431–432. doi: 10.1016/j.tibtech.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Mlotshwa S, et al. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 2015;25:521–524. doi: 10.1038/cr.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. Honeysuckle-encoded atypical micro-RNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jockel S, et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012;209:235–241. doi: 10.1084/jem.20111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehrig S, et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer M, et al. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanco S, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 34.Blanco S, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuorto F, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov P, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durdevic Z, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013;14:269–275. doi: 10.1038/embor.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiani J, et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 2014;6:172–174. doi: 10.1093/jmcb/mjt052. [DOI] [PubMed] [Google Scholar]
- 42.Goodarzi H, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar P, et al. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng H, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu F, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:816.e16–828.e16. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres AG, et al. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014;20:306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q, et al. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 2016 doi: 10.1038/nrg.2016.106. Published online October 3, 2016 http://dx.doi.org/10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan M, et al. A high-throughput quantitative approach reveals more small RNA modifications in mouse liver and their correlation with diabetes. Anal. Chem. 2013;85:12173–12181. doi: 10.1021/ac4036026. [DOI] [PubMed] [Google Scholar]
- 52.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 53.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 55.Paz N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alon S, et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudhury Y, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zipeto MA, et al. RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 2015;21:549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Z, et al. RNA editing by ADAR1 marks dsRNA as “self”. Cell Res. 2015;25:1283–1284. doi: 10.1038/cr.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu T, et al. Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS One. 2012;7:e33720. doi: 10.1371/journal.pone.0033720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morl M, et al. C to U editing and modifications during the maturation of the mitochondrial tRNAAspin marsupials. Nucleic Acids Res. 1995;23:3380–3384. doi: 10.1093/nar/23.17.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishima E, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014;25:2316–2326. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai WM, et al. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol. 2015;560:29–71. doi: 10.1016/bs.mie.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su D, et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan CT, et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seidel A, et al. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br. J. Cancer. 2006;94:1726–1733. doi: 10.1038/sj.bjc.6603164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen K, et al. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Motorin Y, et al. 5-Methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng G, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cozen AE, et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vickers KC, et al. Mining diverse small RNA species in the deep transcriptome. Trends Biochem. Sci. 2015;40:4–7. doi: 10.1016/j.tibs.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]