Abstract
The oncogenic transcription factor Gli1 is a critical effector in the Hedgehog (Hh) pathway which is necessary for the development and progression of pancreatic ductal adenocarcinoma (PDAC). While TGF-β and K-Ras are known regulators of Gli1 gene transcription in this setting, it is not understood how Gli1 functional activity is regulated. Here we report the identification of Gli1 as a substrate for the protein arginine N-methyltransferase PRMT1 in PDAC. We found that PRMT1 methylates Gli1 at R597, promoting its transcriptional activity by enhancing the binding of Gli1 to its target gene promoters. Interruption of Gli1 methylation attenuates oncogenic functions of Gli1 and sensitizes PDAC cells to gemcitabine treatment. In human PDAC specimens, the levels of both total Gli1 and methylated Gli1 were correlated positively with PRMT1 protein levels. Notably, PRMT1 regulated Gli1 independently of the canonical Hh pathway as well as the TGF-β/Kras-mediated non-canonical Hh pathway, thereby signifying a novel regulatory mechanism for Gli1 transcriptional activity. Taken together, our results identifed a new posttranslational modification of Gli1 that underlies its pivotal oncogenic functions in PDAC.
Introduction
The Hedgehog (Hh) signaling pathway plays critical roles in normal tissue patterning and differentiation during mammalian embryogenesis (1). Although the Hh pathway is inactivated in adults, it is reactivated during tumorigenesis in various organs, including pancreas (2). Canonically, inhibitory engagement of the Hh membrane receptor, Patched 1 (PTCH1), by Hh ligands releases Smoothened (SMO) from PTCH. Activated SMO then removes Suppressor of Fused (SuFu) from Gli transcriptional factors and allows Gli proteins to translocate into the nucleus to drive transcription of the target genes of Hh pathway. Three Gli proteins (Gli1, Gli2, and Gli3) have been identified, with Gli1 possessing the strongest transcriptional activity (3).
Besides SuFu, other Gli1-binding proteins have been reported to regulate Gli1 activities. For example, we previously demonstrated that p70S6K phosphorylates Gli1 and releases Gli1 from SuFu, leading to Gli1 activation (4). DYRK1 (5) and aPKC-ι/λ (6) also enhance Gli1 activities via Gli1 phosphorylation. In contrast, protein kinase A-mediated Gli1 phosphorylation negatively regulates Gli1 functions (7). In addition, the Numb/Itch complex and the p300/CBP-associated factor interact with Gli1 and mediate Gli1 ubiquitination and degradation (8,9). Snf5 also interacts with and suppresses Gli1 (10). Together, these findings demonstrate that Gli1 activity can be regulated through various non-canonical Hh pathways and suggest that identification of novel Gli1-associated proteins may shed new light on the regulation of Gli1.
The canonical Hh pathway (cHh) is known to play significant roles in pancreatic ductal adenocarcinoma (PDAC), one of the most intractable cancers, with the poorest survival rate of all cancers (11). Using a paracrine mechanism (12), PDAC cells produce Hh ligands to activate the cHh pathway in tumor-associated stromal cells but not in PDAC cells. However, Gli1 is also functionally required for PDAC cells to survive and proliferate (13,14), and high Gli1 protein levels are related to poor survival in patients with PDAC (15). Although TGF-β and Kras are known regulators of Gli1 gene transcription in PDAC (14), how the Gli1 activity is regulated in PDAC remains to be clarified. Thus, improving our understanding on the mechanism of Gli1 regulation in PDAC may lead to the development of a novel targeted therapy for PDAC.
Materials and Methods
Human tissues
Human PDAC tissues from patients treated at The University of Texas MD Anderson Cancer Center were obtained retrospectively for immunohistochemical microarray analysis. The tissues were collected in accordance with the protocols approved by the Institutional Review Board at MD Anderson Cancer Center, and written informed consent had been obtained from all patients at the time of enrollment. PDAC xenografts were obtained as previously described (16).
Antibodies and reagents
The antibodies used in this study were Gli1 (#3538, Cell Signaling Technology, for western blotting and chromatin immunoprecipitation (ChIP) assay, and#sc-20687, Santa Cruz Biotechnology, for immunohistochemistry), actin (#A2066, Sigma-Aldrich), Flag (#F3165, Sigma-Aldrich), Flag M2 magnetic beads (#M8823, Sigma-Aldrich), PRMT1 (#2449, Cell Signaling Technology), and tubulin (#T5168, Sigma-Aldrich). GDC-0449, NVP-LDE225, and RAD-001 were purchased from Selleck Chemicals LLC, GANT58 and GANT61 from Tocris Bioscience, and AMI-1 from Sigma-Aldrich. The SYBR Green real-time PCR kit was obtained from Bio-Rad and TGF-β from Peprotech (#100-21). The antibody against R597-methylated Gli1 was developed using the following synthetic peptide with asymmetric dimethylation at R597 as antigen: RARYASA-[R597(aMe2)]-GGGTS (C-terminal amidation and N-terminal KLH conjugation). Other peptides used in the study included the following:
Cold Gli1 peptide without R597 methylation [Gli1-R597]: RARYASA-[R597]-GGGTS;
Hot Gli1 peptide with R597 monomethylation [Gli1-R597(Me)]: RARYASA-[R597(Me)] -GGGTS;
Hot Gli1 peptide with R597 asymmetric dimethylation [Gli1-R597(aMe2)]: RARYASA-[R597(aMe2)]-GGGTS;
Hot Gli1 peptide with R597 symmetric dimethylation [Gli1-R597(sMe2)]: RARYASA-[R597(sMe2)]-GGGTS.
Plasmids
Flag-tagged Gli1 was generated from pCMV10-3xFlag-Gli1 (4) and inserted into pCDH-CMV-MCS-EF1-Neo (System Biosciences). The R597 mutant was generated by using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies) with pCDH-CMV-MCS-EF1-Neo-Gli1 as a template. The pCDH-RFP-Luciferase plasmid was constructed by inserting the luciferase gene into the pCDH-RFP vector (System Biosciences). The hemagglutinin (HA) -tagged PRMT1 plasmid was generated by inserting PRMT1 complementary DNA into pCMV5. PRMT1 knockdown was carried out by shRNA with the sequence of CCGGCAGTACAAAGACTACAA (#1), GTGTTCCAGTATCTCTGATTA (#2), GCAAGTGAAGCGGAATGACTA (#3) in the vector pLKO.1. #1 shRNA-resistant PRMT1 was produced through mutation of the target sequence of #1 shRNA in PRMT1-expressing plasmid (mutating CCGGCAGTACAAAGACTACAA to TAGACAATATAAGGATTATAA) without changing amino acid. Lentiviral shRNA system in pGIPZ vector targeting Kras was purchased from Thermo Scientific. pGEX-6P-1 (GE Healthcare) was used to purify proteins for the in vitro methylation assay.
Cell culture
The human PDAC cell lines AsPC-1, MIA PaCa-2, and CFPAC-1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained at 37 °C in a 5% CO2 incubator in Dulbecco modified Eagle medium/F12 or RPMI 1640 plus 10% fetal bovine serum. Human PDAC-associated stromal cell line HPSC was previously described (17). All cell lines were characterized as mycoplasma negative and validated by STR DNA fingerprinting using the AmpFLSTR Identifiler kit (ThermoFisher) according to manufacturer’s instructions semiannually. The STR profiles were compared with known ATCC fingerprints (www.ATCC.org) and with the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/) (Nucleic Acids Research 37:D925-D932, PMCID: PMC2686526). The STR profiles matched known DNA fingerprints or were unique.
Transfection and lentiviral infection
The plasmids were transfected using Lipofectamine 2000 according to the manufacturer’s instructions. The cells were harvested for mRNA extraction or protein extraction after 48 h of transfection. For lentiviral infection, vector plasmids and packaging plasmids were co-transfected into 293T cells using Lipofectamine 2000, and the lentiviruses were concentrated as described previously (18). The generated viruses were used for cell infection in the presence of polybrene (EMD Millipore). After infection, stable cells were isolated by selection for resistance to puromycin or neomycin or by fluorescence-activated cell sorting. Specifically, to generate stable cells with luciferase, MIA PaCa-2 cells were infected with lentivirus containing pCDH-RFP-Luciferase followed by RFP sorting. MIA PaCa-2 luciferase-expressing stable (MIA PaCa-2-Luc) cells were then used to establish wild-type Gli1 or R597-mutantGli1 stable cells by infection with lentivirus containing pCDH-CMV-MCS-EF1-Neo-Gli1WT or pCDH-CMV-MCS-EF1-Neo-R597K under G418 selection. To generate Gli1 knockout cells, AsPC-1 cells were co-transfected with Gli1 CRISPR/Cas9 KO plasmid (Santa Cruz Biotechnology, sc-400266) and Gli1 HDR plasmid (Santa Cruz Biotechnology, sc-400266-HDR). After 48 hours, cells were selected with puromycin for 1 week and then analyzed for knockout efficiency.
TCGA gene expression data sets and data processing
TCGA mRNA expression (RNASeq V2 RSEM) data for lung adenocarcinoma, prostate adenocarcinoma, breast invasive carcinoma, colorectal adenocarcinoma, PDAC, liver hepatocellular carcinoma, and ovarian serous cystadenocarcinoma were downloaded from the cBioPortal Web site (19,20). Box plots showing 5th/95th percentiles and medians were generated with SigmaPlot software.
Animal studies
All animal experiments were approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. 6-week nude mice were housed under standard conditions. For orthotopic tumorigenicity assay, MIA PaCa-2 cells with luciferase only, luciferase plus wild-type Gli1, or luciferase plus R597K-mutant Gli1 were suspended in 50% Matrigel (#354230, BD Biosciences) in phosphate-buffered saline at a concentration of 5 × 106 cells/ml. The viability of the cells was > 98%. The number of mice per group was 5. General anesthesia was administered using isoflurane (#029405, Henry Schein Animal Health). A left lateral laparotomy was performed, and the spleen and distal pancreas were mobilized. Approximately 50 µl of the cells (0.25 × 106) were injected into the pancreas. The abdominal incision was closed using a surgical suture, and analgesia was administered for immediate pain relief. Palpable tumors were detected after 14 days and imaged twice a week using the Xenogen IVIS in vivo imaging system (Caliper Life Sciences) as described elsewhere (21). For subcutaneous tumorigenicity assay, 1 × 105 cells were subcutaneously injected in right flank. The resulting tumors were measured with calipers weekly, and tumor volume was determined using the formula (Length) × (Width)2, where l is the longest diameter and w is the shortest diameter. The tumors were measured with calipers every week. Data were presented as tumor volume (mean ± SD). Statistical analysis was done using the Student's t-test by the program SPSS for Windows.
Statistical analyses
Statistical analyses were performed with the Student’s t-test, Spearman rank correlation test, or Fisher exact test. A P value of < 0.05 was considered statistically significant. All data analyses were performed using the analytic software SPSS (IBM) for Windows.
Results and Discussion
PRMT1 interacts with Gli1 and promotes Gli1 transcription
We first analyzed The Cancer Genome Atlas (TCGA) database and found that among of the seven deadliest cancers in 2014 in the United States (22), PDAC had the highest mean Gli1 mRNA expression (Figure 1A), which supports the reports that Gli1 plays critical roles in PDAC. Then, we profiled Gli1-associated proteins to identify potential regulators of Gli1 in PDAC cells. For this purpose, Flag-tagged Gli1 was stably transfected into MIA PaCa-2PDAC cells. Gli1-associated proteins were isolated using anti-Flag antibody and analyzed by mass spectrometry (Supplemental Figure 1A). Comparison of Flag-Gli1 and Flag-vector results profiled a total of 471 potential Gli1-binding proteins, including several well-known Gli1 interaction partners, e.g. SuFu and protein kinase A (3) (Table S1). Because Gli1 regulation in the non-canonical Hh pathway often requires enzyme-induced posttranslational modifications such as phosphorylation, ubiquitination, and acetylation (4–9), we thus specially focused on the Gli1-binding proteins with enzymatic activities (Table S2). Besides kinases, ubiquitin-conjugating enzymes, and acetyltransferases, which are responsible for modifications known to occur in Gli1 (4–8,23,24), we also noticed a methyltransferase, PRMT1, which catalyzes protein methylation (25) but has not yet been reported to be associated with Gli1. PRMT1 is a type I protein arginine methyltransferase that catalyzes asymmetric dimethylation on arginine residues. In mammalian cells, about 85% of all occurrences of asymmetric dimethylation are produced by PRMT1 (25). The arginine methylation mediated by PRMT1 positively or negatively affects protein functions depending on the biological contexts (26,27), and our recent work further revealed an important function of PRMT1 on EGFR regulation in colon cancer (28).
Figure 1. Correlation between the Expression Levels of Gli1 and PRMT1 in PDAC Cells.
(A) GLI1 mRNA levels according to TCGA data for patients with the seven deadliest cancers in the United States in 2014. The data are medians with the 5th and 95th percentiles and standard deviations (error bars).
(B) Western blot analysis of immunoprecipitation (IP) of endogenous Gli1 and PRMT1 in AsPC-1 cells. IgG, immunoglobulin G.
(C) AsPC-1 cells expressing scrambled shRNA (sh-Ctrl), PRMT1-targeting shRNA (sh-PRMT1 #1, 2, or 3), or PRMT1-targeting shRNA (sh-PRMT1 #1) with reconstituted shRNA-resistant PRMT1.
(D) Western blot analysis of Gli1 and PRMT1 in three PDAC cell lines infected with control (Ct) or PRMT1 (Pr) shRNA. The results were quantified using ImageJ software, and normalized to the values for tubulin. The experiments were performed at least two times to assure reproducibility of the results.
(E) mRNA expression of endogenous Gli1 and PRMT1 measured by quantitative real-time PCR in the indicated cell lines transfected with control shRNA or sh-PRMT1. The data are means with standard deviations (n = 3). *P< 0.05, **P< 0.01 (paired two-tailed Student’s t-test).
(F) Representative immunohistochemistry staining of Gli1 and PRMT1 in human PDAC tissues. All immunostained slides were scanned on the ACIS III automated cellular image system for quantification by digital image analysis. The percentage of positive cells (X) and signal intensity (Y) are shown. The number from X × Y represents an arbitrary quantitative score. Tumor (T) area was labeled with dash line. The positive staining in stroma is labeled with arrowhead.
(G) Analysis of correlation between Gli1 and PRMT1 levels on the basis of immunohistochemistry results for 122 human PDAC tissue samples. Protein expression was calculated from both the percentage of stained cells and the immunostaining intensity. Protein expression levels above and below the mean for all samples were categorized as high and low, respectively. There are 4 categories based on the Gli1 and PRMT1 scores on all of the immunostained slides: 1) Gli1 and PRMT1 high; 2) Gli1 and PRMT1 low; 3) Gli1 high and PRMT1 low; 4) Gli1 low and PRMT1 high. Fisher’s exact test was used to evaluate the correlation between Gli1 and PRMT1 in the 122 human tissue slides (P < 0.05).
Then, we confirmed the interaction between Gli1 and PRMT1 in AsPC-1 and MIA PaCa-2 cells (Figure 1B), but neither Gli2 nor Gli3 interacted with PRMT1 (Supplemental Figure 1B). To address the potential relationship between PRMT1 and Gli1 and the role of Gli1 in PDAC, we first knocked down PRMT1 in AsPC-1 cells by using three different small hairpin RNAs (shRNAs) and observed decreases in Gli1 protein levels. The decreases were reversed by shRNA-resistant ectopic expression of PRMT1 (Figure 1C). Knockdown of PRMT1 in MIA PaCa-2 and CFPAC-1 also decreased Gli1 protein and mRNA levels (Figures 1D and 1E). In contrast, ectopic expression of PRMT1 enhanced Gli1 expression (protein and mRNA)in MIA PaCa-2 cells (Supplemental Figure 1C). Interestingly, Gli1 protein stability did not change following such enhancement in AsPC-1 cells (Supplemental Figure 1D). These results suggested that PRMT1 upregulates Gli1 protein levels through enhanced Gli1 transcription. Microarray analysis of patient-derived PDAC tissues (n = 122) by immunohistochemistry staining also supports a positive correlation between Gli1 and PRMT1 protein expression levels (P = 0.008) (Figures 1F and 1G).
PRMT1 methylates Gli1 at R597
To determine whether Gli1 is a substrate for PRMT1, we performed an in vitro methylation assay and detected a major methylation signal in a Gli1 fragment (Gli1 F2) containing amino acids 354 to 753 (Supplemental Figure 2A). We analyzed four segments of this fragment to identify potentially methylated arginine residues. Of the four subfragments, only F2-3, containing amino acids 543 to 620, produced methylation signals (Supplemental Figure 2B). When we mutated all six arginines in Gli1 F2-3 to lysine (K), only mutation of Arg597 (R597) eliminates the methylation signal (Supplemental Figure 2C). We obtained a similar result with this mutation in the full-length Gli1, in which the methylation signal was completely eliminated (Figure 2A). These results indicated that Gli1 is a substrate of PRMT1 in vitro and that R597 is the primary site methylated by PRMT1.
Figure 2. Methylation of Gli1 by PRMT1.
(A) In vitro methylation assay with PRMT1 and wild-type (WT) or R597K-mutant Gli1. Left panel, Coomassie Blue staining. Right panel, fluorography.
(B) Western blot analysis of immunoprecipitation (IP) with antibody specific to meGli1R597 (meGli1), antibody to total Gli1, and other antibodies as indicated in MIA PaCa-2 cells transfected with a plasmid carrying Flag (FL)-tagged WT Gli1 or R597K-mutant Gli1 (RK).
(C) Western blot analysis of meGli1, Gli1, and PRMT1 in MIA PaCa-2 cells transfected with an empty vector or a hemagglutinin (HA)-tagged PRMT1 plasmid.
(D) Western blot and correlation analysis of meGli1 and PRMT1 in human PDAC xenografts maintained in mice. Each set of samples was subjected to two independent Western blotting (upper panels), and the bands were quantified using ImageJ software. Mean expression levels were used to determine Pearson coefficients for correlation between PRMT1 and meGli1, between PRMT1 and Gli1, and between Gli1 and meGli1 (lower panels).
We then purified Gli1 from AsPC-1 cells for mass spectrometry analysis, and validated dimethylated R597 also occurred in vivo (Supplemental Figure 2D). Sequence alignment of Gli1 from different mammals indicated that R597 within the RG rich motif, which is a typical feature of PRMT1 substrates (29), is highly conserved from mice to humans (Supplemental Figure 2E), suggesting a potentially important role of R597. To further characterize this methylated residue in Gli1 and to study the correlation between R597-dimethylated Gli1 (meGli1R597) and PRMT1, we developed a meGli1R597-specific antibody, which recognizes peptides with asymmetrically dimethylated R597 in Gli1 but not peptides with non-modified, monomethylated, or symmetrically dimethylated R597 (Figure Supplemental 2F). This antibody recognized only wild-type Gli1 but not the Gli1R597K mutant (Figure 2B) and therefore is suitable for detection of meGli1R597.
In MIA PaCa-2 cells, ectopic expression of PRMT1 led to enhanced Gli1 methylation (Figure 2C). Since the antibody was unable to detect meGli1R597 by immunohistochemistry (data not shown), we instead performed Western blot analysis of protein lysates from 30 human PDAC xenografts maintained in mice (30). Our results indicated a positive correlation between the levels of methylated Gli1 and PRMT1 (Figure 2D), further validating our in vitro finding that Gli1 is a substrate of PRMT1. However, PRMT1 and total Gli1 or Gli1 and meGli1 only showed weak positive correlation, which might be attributed to multiple regulators of Gli1 besides PRMT1 (Figure 2D).
Methylation of R597is required for regulation of Gli1 by PRMT1
Because PRMT1 increases Gli1 mRNA transcription and Gli1 is a target gene of itself (31), we asked whether Gli1R597 is required for PRMT1-enhanced Gli1 transcriptional activity. As expected, the transcripts of several Gli1 target genes, including endogenous GLI1, PTCH1, IGFBP6, CCND1, BCL2, and SNAIL1 (32), were lower in MIA PaCa-2 cells stably expressing R597K-mutant Gli1 (MIA/Gli1RK cells) than in MIA PaCa-2 cells expressing wild-type Gli1 (MIA/Gli1WT cells) (Figure 3A and Supplemental Figure 3A). Consistently, results from a transient reporter assay in 293T cells using Gli binding sequence (GliBS) reporter plasmid (31) confirmed that wild-type Gli1 harbored stronger transcriptional activity compared with R597K-mutant Gli1 (Supplemental Figure 3B). To further investigate the significance of R597 in Gli1 functions, we knocked out Gli1 in AsPC-1 cells, which have a high basal level of Gli1 protein, by CRISPR/Cas9, followed by reconstitution with wild-type Gli1 or R597K-mutant Gli1 (Figure 3B). We found that the loss of Gli1 decreased the expression of Gli1 target genes. Interestingly, restoring wild-type Gli1 expression also rescued the expression of all five target genes, but restoring the R597K mutant Gli1 only rescued PTCH1 and CCND1 (Figure 3B). Hence, the R597 site may be required for stronger transcriptional activity of Gli1, and without methylation, Gli1 transcriptional activity on some of Gli1 target genes may be suppressed. In addition, ectopic expression of PRMT1 in MIA/Gli1WT cells, but not in MIA/Gli1RK cells, enhanced Gli1 target gene expression (Figure 3C). Likewise, depletion of PRMT1 repressed the expression of Gli1 target genes in MIA/Gli1WT cells but not in MIA/Gli1RKcells (Supplemental Figure 3C), which implied that PRMT1 affects Gli1 transcriptional activities through the site of R597.
Figure 3. R597 Methylation Positively RegulatesGli1 Transcriptional Activity.
(A) Left panel, Western blot analysis of meGli1 and total Gli1 in MIA PaCa-2 luciferase cells stably transfected with an empty vector (Vec), wild-type Gli1 (Gli1WT), or R597K-mutant Gli1 (Gli1RK). Right panel, mRNA expression levels, measured by quantitative real-time PCR, of Gli1 target genes in Vec-, Gli1WT (WT)-, and Gli1RK (RK)-transfected MIA PaCa-2 cells. Error bars represent SD (n = 3). *P < 0.05, **P < 0.01 (paired two-tailed Student’s t-test).
(B) Left panel, Western blotting of Gli1 protein levels in AsPC-1 parental cells (PA), AsPC-1 cells with Gli1 knockout (Gli1−/−), and Gli1−/− AsPC-1 cells reconstituted with Gli1 (WT) or Gli1 RK mutant (RK). The intensity of the bands was quantified and normalized to that of tubulin. Right panel, mRNA expression of Gli1 target genes by qRT-PCR in PA, Gli1−/−, WT, and RK cells. The expression levels of target genes were normalized to that of ACTIN. Statistical significance was determined by paired, two-tailed Student’s t-test. Error bars represent SD (n = 3). *P < 0.05, **P < 0.01.
(C) mRNA levels of IGFBP6 and BCL2 in MIA PaCa-2 cells transfected with wild-type Gli1 and an empty vector (WT/Vec), wild-type Gli1 and PRMT1 (WT/Prm), R597K-mutant Gli1 and an empty vector (RK/Vec), or R597K-mutant Gli1 and PRMT1 (RK/Prm). The value of WT/Prm was normalized to that of WT/Vec. The value of RK/Prm was normalized to that of RK/Vec. Error bars represent SD (n = 3). *P < 0.05 (paired two-tailed Student’s t-test).
(D) Left panel, ChIP assay using Gli1 antibody for immunoprecipitation (IP) and promoter-specific primers for quantitative qRT-PCR to confirm Gli1 binding regions in promoters. IgG, immunoglobulin G; SP, primers specific to Gli1 binding regions; NSP, primers not specific to Gli1 binding regions. Middle panel, protein expression of hemagglutinin-tagged wild-type Gli1 (HA-WT) and Flag-tagged R597K-mutant Gli1 (Flag-RK) in 293T cells. Right panel, quantitative results of ChIP assay. WT, wild-type Gli1; RK, R597K-mutant Gli1. Error bars represent SD.
(E) Quantitative results of ChIP assay for Gli1-bound promoters of IGFBP6 and BCL2 from qRT-PCR analyses in MIA PaCa-2 cells carrying wild-type (WT) or R597K-mutant (RK) Gli1 and infected with virus carrying control shRNA (shC) or shRNA targeting PRMT1 (shP). Error bars represent SD (n = 3). *P < 0.05, **P < 0.01 (paired two-tailed Student’s t-test).
(F) Analysis of IGFBP6 and BCL2 promoters bound by Gli1 by ChIP-qPCR in AsPC-1 cells expression scrambled (sh-Ctrl) shRNA, PRMT1-targeting (sh-PRMT1 #1 and #2) shRNA, or PRMT1-targeting shRNA with reconstituted wild type PRMT1 (sh#1-Rsc). Statistical significance was determined by paired, two-tailed Student’s t-test. Error bars represent SD from triplicate experiments. *P < 0.05, **P < 0.01.
Because Gli1 is a transcriptional factor, we asked whether the methylation status of Gli1R597 affects the occupancy of Gli1 on the promoter of its target genes. To this end, we examined the occupancy of Gli1 variants in natural promoters of CCND1, IGFBP6, and BCL2 (33,34) by ChIP assay. The results indicated that wild-type Gli1 occupied the promoter of BCL2 and IGFBP6 more than did the R597K-mutant Gli1, whereas both wild-type and mutant Gli1 similarly occupied the promoter of CCND1 (Supplemental Figure 3D). Similar results were observed from PCR analysis in which wild-type Gli1 promoted the transcription of BCL2 and IGFBP6 more than did the R597K-mutant Gli1 whereas both wild-type or R597K-mutant Gli1 promoted similar transcription of CCND1. These results suggested that Gli1 R597 methylation is required for Gli1 occupancy on promoters of some target genes but not for the others. Guendel et al. previously reported that methylation of BRCA1 by PRMT1 alters the occupancy of BRCA1 on its target gene promoters (35). Collectively, our results indicated that methylation of Gli1 at R597 rendered stronger transcriptional activity on some of Gli1 target genes, including SNAIL1, BCL2, and IGFBP6.
Next, we specifically focused on IGFBP6 and BCL2 promoters, which were affected by Gli1 methylation status. Wild-type Gli1 and R597K-mutant Gli1 were transfected separately or together into MIA PaCa-2cells with adjustment of plasmid amounts to ensure similar levels of total Gli1 in different samples. The results indicated that the occupancy of Gli1 on the promoters is positively correlated with the expression of wild-type Gli1 but not R597K-mutant Gli1 (Figure 3D). Depletion of PRMT1 by shRNA decreased the accumulation of Gli1 to the promoters in MIA/Gli1WT but not MIA/Gli1RK cells (Figure 3E). In AsPC-1 cells, knockdown of PRMT1 reduced the binding of Gli1 to the promoters of IGFBP6 and BCL2, and rescue of PRMT1 restored the accumulation of Gli1 at the promoters (Figure 3F). Similar results were observed when we treated cells with the pan-PRMT inhibitor AMI-1 (arginine methyltransferase inhibitor 1) (36), which attenuated Gli1 methylation (Supplemental Figures 3E). Together, the results suggested that methylation of R597 in Gli1 by PRMT1 promotes Gli1 accumulation at the promoters of Gli1 target genes and enhances their transcriptions.
Gli1 oncogenic functions are inhibited by loss of R597 methylation
Since Gli1expression is associated with malignant transformation, we assessed the effects of methylation on Gli1 oncogenic functions. The results from bromodeoxyuridine (BrdU) incorporation assay showed that MIA/Gli1RK cells grew slower than MIA/Gli1WT cells (Supplemental Figure 4A). Cell viability (Supplemental Figure 4B), anchorage-independent growth (Figure S4C), and migration and invasion abilities (Supplemental Figures 4D and 4E) were lower in MIA/Gli1RK cells than in MIA/Gli1WT cells. In AsPC-1 cells, knockout of Gli1 inhibited cell proliferation, as measured by BrdU incorporation and cell counting. The inhibition was completely reversed by reconstitution with wild-type Gli1 but not R597K-mutant Gli1 (Supplemental Figure 4F and 4G). These results suggested that R597 methylation is necessary for full oncogenic effects of Gli1.
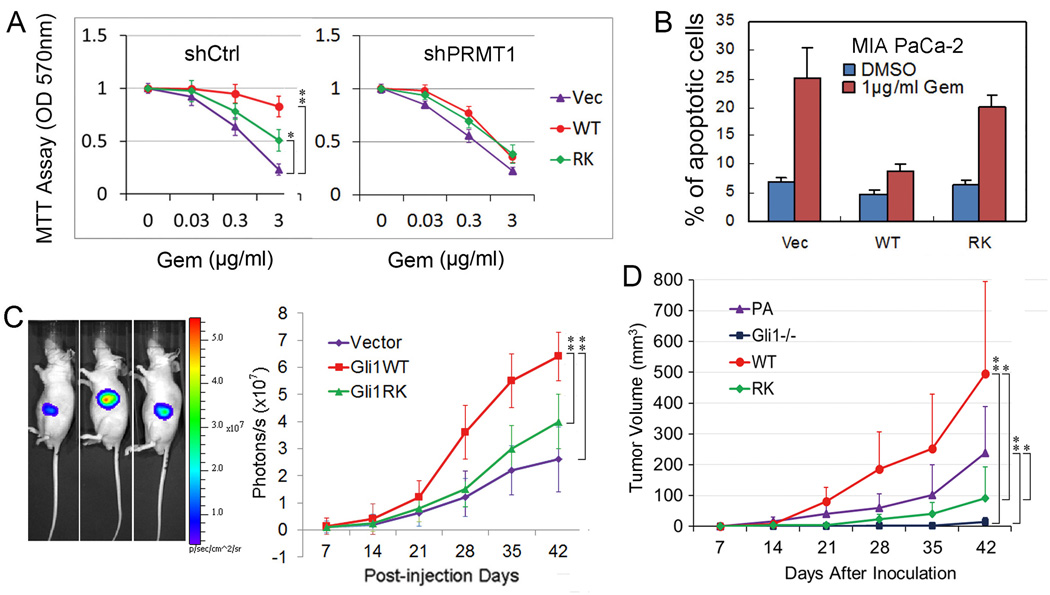
Given that Gli1 has been implicated in resistance to gemcitabine, a chemotherapy agent widely used to treat PDAC (37–39), we investigated whether PRMT1-methylated R597 contributes to gemcitabine resistance. Indeed, even though MIA/Gli1WT cells were more resistant to gemcitabine than MIA/Gli1RK cells, the differences were marginal after PRMT1 knockdown (Figure 4A). Similar results were obtained with CFPAC-1 cells carrying wild-type or R597-mutant Gli1 (Supplemental Figure 4H). In addition, the basal level of apoptosis and the percentage of cells showing gemcitabine-induced apoptosis were higher in MIA/Gli1RK cells than in MIA/Gli1WT cells (Figure 4B).
Figure 4. R597 methylation positively regulates Gli1 oncogenic functions.
(A) Responses of MIA PaCa-2 stable clones to gemcitabine with or without PRMT1 depletion. shCtrl: control shRNA; shPRMT1: PRMT1 shRNA; Vec: MIA PaCa-2 stable clone with empty vector; WT: stable clone with wild-type Gli1; RK: stable clone with Gli1R597 mutant. Error bars represent SD (n = 4).
(B) Propidium iodide staining by fluorescence-activated cell sorting to determine the percentage of apoptosis in different stale clones with or without gemcitabine. Error bars represent SD (n = 3).
(C) The indicated MIA PaCa-2 stable clones were inoculated into the pancreas of 6-week nude mice. Tumor volume was measured at the indicated time points using the formula (Length) × (Width)2. Vector: MIA PaCa-2-Luc stable cells; Gli1WT: MIA PaCa-2-Luc Gli1WT stable cells; Gli1RK: MIA PaCa-2-Luc Gli1R597K stable cells. Error bars represent SD (n = 5).
(D) AsPC-1 parental cells (PA), AsPC-1 cells with Gli1 knockout (Gli1−/−), or Gli1−/− AsPC-1 cells reconstituted with Gli1 (WT) or Gli1 RK mutant were subcutaneously injected into the right flank of nude mice (n = 7). Tumor volume was measured once a week. Statistical significance was determined by paired, two-tailed Student’s t-test. Error bars represent SD. *P < 0.05, **P < 0.01.
Results from an orthotopic model of human PDAC in nude mice indicated that inhibition of R597 methylation in Gli1 attenuated tumor growth (Figure 4C). Likewise, AsPC-1 cells with Gli1 knockdown formed very few subcutaneous tumors, and cells reconstituted with wild-type Gli1 formed much larger tumors than cells reconstituted with R597K-mutant Gli1 (Figure 4D and Supplemental Figure 4I). Collectively, these results indicated that inhibition of R597 methylation in Gli1 reduces the oncogenic activities of Gli1 in vitro and in vivo and that such inhibition may provide a novel strategy for reducing gemcitabine resistance in PDAC.
Regulation of Gli1 by PRMT1 is a novel pathway in PDAC
Since Gli1 can be regulated via a SMO-dependent (cHh) or SMO-independent (non-canonical Hh) pathway (4,14), we investigated whether the regulation of Gli1 by PRMT1 in PDAC relies on SMO. We treated MIA PaCa-2, MIA/Gli1WT, and MIA/Gli1RK cells with the SMO inhibitor GDC-0449 (marketed as Vismodegib by Roche) or NVP-LDE225 (marketed as Erismodegib by Novartis) and found that none of the cells were sensitive to the inhibitors (Supplemental Figure 5A). These results are consistent with previous studies showing that PDAC cells lack the cHh pathway (23). Gli1 is known to be transcriptionally regulated by TGF-β and Kras in PDAC (14). Thus, we also examined the possibility that TGF-β and/or Kras regulate PRMT1-mediated methylation of Gli1. To avoid interference of endogenous Gli1 transcription, we used the previously mentioned Gli1−/− with reconstitution of exogenous wild-type Gli1, which does not carry endogenous Gli1 promoters. In this cell line, knockdown of Kras had no effects on PRMT1-mediated Gli1 methylation (Supplemental Figure 5B), and TGF-β treatment, which induces Gli2 upregulation as expected (14), did not affect Gli1R597 methylation either (Supplemental Figure 5C). Therefore, regulation of Gli1 by PRMT1 is a novel pathway in PDAC. Because tumor-associated stromal cells play important roles in the tumor microenvironment of PDAC (40,41), we further investigated if the regulation of Gli1 by PRMT1 exists in PDAC-associated stromal cells. We detected both Gli1 and meGli1 in a human PDAC-associated stromal cell line, HPSC (17) and treatment of HPSC cells with PRMT inhibitor (AMI) substantially reduced meGli1 (Supplemental Figure 5D). Therefore, in tumor-associated stromal cells, methylation of Gli1 may also be dependent on PRMT1. Finally, we treated PDAC cells with GANT58 and GANT61, which interfere with the binding of Gli1 to the promoters of Gli1 target genes (42). Remarkably, GANT58 and GANT61 inhibited the viability of both MIA/Gli1WT and MIA/Gli1RK cells (Figure 5A). These results suggested that direct targeting of Gli1 may be an alternative to targeting the upstream cHh pathway for treatment of PDAC.
Figure 5. PRMT1Regulates Gli1 via a Novel Non-Canonical Hh Pathway.
(A) MTT assay of the indicated MIA PaCa-2 stable cells treated with GANT58 or GANT61. The data are means (relative to the value for day 1) with standard deviations (n = 3). *P < 0.05, **P < 0.01 (paired two-tailed Student’s t-test).
(B) A schematic diagram illustrating the regulation of Gli1 via SMO-dependent (cHh) or SMO-independent (non-canonical Hh) pathways in PDAC.
Gli1 function is regulated by post-translational modifications, e.g., phosphorylation and acetylation (4,5,7,24,43). In this study, we have identified Gli1 as a new substrate for PRMT1 and characterized a novel post-translational modification of Gli1 in PDAC cells that was independent of cHh or TGF-β/Kras (Figure 5B). Methylation of Gli1 by PRMT1 promotes Gli1 transcriptional activity by enhancing Gli1’s accumulation on its target gene promoters, thereby activating the expression of those genes. In addition, even though R597K-mutant Gli1 retained its transcriptional activity without R597 methylation, the activity was much weaker than that of wild-type Gli1 for some of target genes. These findings implied that although R597 methylation is not absolutely required for Gli1 function, it does increase Gli1 transcriptional activities, which is sufficient to enhance the oncogenic functions of Gli1 by increasing transcription of its target genes, such as SNAIL1, BCL2, and IGFBP6. It would be of interest to comprehensively profile all the genes that are sensitive to Gli1 R597 methylation. In vivo, Gli1 functions are constitutively inhibited by SuFu (via inhibitory interaction with Gli1) or by a repressor form of Gli3 (via competitive binding to the same promoters as those bound by Gli1). However, increased Gli1 proteins and/or increased Gli1 binding to its promoters can overcome inhibition by SuFu or Gli3 repressor through either cHh pathway activation or the newly identified methylation of R597 in Gli1. Thus, methylation of R597 in Gli1 is potentially an important alternative mechanism of promoting Gli1 oncogenic functions in vivo. We also observed that the regulation of Gli1 by PRMT1 was not confined in the tumor cells, and tumor-associated stromal cell may also have this pathway, which implied that PRMT1-mediated Gli1 methylation has broader involvement in various cell types.
The Hh signaling pathway plays a pivotal role in PDAC development and progression via a paracrine loop (12). However, a cHh pathway inhibitor based on this paracrine model did not produce successful results against PDAC in a clinical trial (40,41,44). In the current study, we demonstrated that elimination of Gli1 methylation by PRMT1 substantially attenuated Gli1-related oncogenic functions and sensitized PDAC cells to gemcitabine, a front-line drug for PDAC. In addition, and our result that GANT61 attenuated proliferation of MIA/Gli1WT and MIA/Gli1RK PDAC cells complements previous reports that GANT61 suppresses PDAC by inhibiting growth of cancer stem cells and promoting autophagy of tumor cells (45,46). Collectively, the findings in this study indicated that the targeting both PRMT1 and Gli1 is a viable new direction for treatment of PDAC.
Supplementary Material
Acknowledgments
We thank Arthur Gelmis at the Department of Scientific Publications at MD Anderson for editing the manuscript. This work was supported in part by the following: National Institutes of Health (CA109311, CA099031, and CCSG CA016672); The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (to M.-C.H.); Ministry of Science and Technology, International Research-intensive Centers of Excellence in Taiwan (I-RiCE; MOST 105-2911-I-002-302); Ministry of Health and Welfare, and China Medical University Hospital Cancer Research Center of Excellence (MOHW105-TDU-B-212-134003); Center for Biological Pathways; and MD Anderson Cancer Center Sister Institute Network Fund.
Footnotes
Conflicts of Interest
The authors have declared that no conflict of interest exists.
References
- 1.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends in molecular medicine. 2010;16(8):337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng JM, Curran T. The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11(7):493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Science signaling. 2012;5(246):re6. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer cell. 2012;21(3):374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. The Journal of biological chemistry. 2002;277(38):35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 6.Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013;494(7438):484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng T, Chi S, Zhang X, Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. The Journal of biological chemistry. 2006;281(1):9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- 8.Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nature cell biology. 2006;8(12):1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 9.Mazza D, Infante P, Colicchia V, Greco A, Alfonsi R, Siler M, et al. PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell death and differentiation. 2013;20(12):1688–1697. doi: 10.1038/cdd.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nature medicine. 2010;16(12):1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 12.Dimou A, Syrigos K, Saif MW. Rationale for inhibition of the hedgehog pathway paracrine loop in pancreatic adenocarcinoma. JOP : Journal of the pancreas. 2011;12(1):1–5. [PubMed] [Google Scholar]
- 13.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, et al. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(17):E1038–E1047. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes & development. 2009;23(1):24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marechal R, Jean-Baptiste B, Annabelle C, Pieter D, Jean Robert D, Magali SM, et al. Sonic Hedgehog and Gli1 expression predict outcome in resected pancreatic adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-14-0667. [DOI] [PubMed] [Google Scholar]
- 16.Kang Y, Zhang R, Suzuki R, Li SQ, Roife D, Truty MJ, et al. Two-dimensional culture of human pancreatic adenocarcinoma cells results in an irreversible transition from epithelial to mesenchymal phenotype. Laboratory investigation; a journal of technical methods and pathology. 2015;95(2):207–222. doi: 10.1038/labinvest.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer research. 2008;68(3):918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nature protocols. 2009;4(4):495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang JY, Hsu JL, Meric-Bernstam F, Chang CJ, Wang Q, Bao Y, et al. BikDD eliminates breast cancer initiating cells and synergizes with lapatinib for breast cancer treatment. Cancer cell. 2011;20(3):341–356. doi: 10.1016/j.ccr.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.American Cancer Society. Cancer Facts & Figures 2014. 2014 [Google Scholar]
- 23.Lauth M, Bergstrom A, Shimokawa T, Tostar U, Jin Q, Fendrich V, et al. DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat Struct Mol Biol. 2010;17(6):718–725. doi: 10.1038/nsmb.1833. [DOI] [PubMed] [Google Scholar]
- 24.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nature cell biology. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacological research : the official journal of the Italian Pharmacological Society. 2009;60(6):466–474. doi: 10.1016/j.phrs.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Molecular cell. 2013;50(5):613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Molecular cell. 2005;18(3):263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. The Journal of clinical investigation. 2015 doi: 10.1172/JCI82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gayatri S, Bedford MT. Readers of histone methylarginine marks. Biochimica et biophysica acta. 2014;1839(8):702–710. doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nature protocols. 2009;4(11):1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124(7):1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 32.Zhu H, Lo HW. The Human Glioma-Associated Oncogene Homolog 1 (GLI1) Family of Transcription Factors in Gene Regulation and Diseases. Current genomics. 2010;11(4):238–245. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu XF, Guo CY, Liu J, Yang WJ, Xia YJ, Xu L, et al. Gli1 maintains cell survival by up-regulating IGFBP6 and Bcl-2 through promoter regions in parallel manner in pancreatic cancer cells. Journal of carcinogenesis. 2009;8:13. doi: 10.4103/1477-3163.55429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Molecular and cellular biology. 2006;26(16):6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guendel I, Carpio L, Pedati C, Schwartz A, Teal C, Kashanchi F, et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PloS one. 2010;5(6):e11379. doi: 10.1371/journal.pone.0011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases. The Journal of biological chemistry. 2004;279(23):23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 37.Kim MP, Gallick GE. Gemcitabine resistance in pancreatic cancer: picking the key players. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(5):1284–1285. doi: 10.1158/1078-0432.CCR-07-2247. [DOI] [PubMed] [Google Scholar]
- 38.Xin Y, Shen XD, Cheng L, Hong DF, Chen B. Perifosine inhibits S6K1-Gli1 signaling and enhances gemcitabine-induced anti-pancreatic cancer efficiency. Cancer chemotherapy and pharmacology. 2014;73(4):711–719. doi: 10.1007/s00280-014-2397-9. [DOI] [PubMed] [Google Scholar]
- 39.Peng Z, Ji Z, Mei F, Lu M, Ou Y, Cheng X. Lithium inhibits tumorigenic potential of PDA cells through targeting hedgehog-GLI signaling pathway. PloS one. 2013;8(4):e61457. doi: 10.1371/journal.pone.0061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lunardi S, Muschel RJ, Brunner TB. The stromal compartments in pancreatic cancer: are there any therapeutic targets? Cancer letters. 2014;343(2):147–155. doi: 10.1016/j.canlet.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 42.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(20):8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coni S, Di Magno L, Canettieri G. Determination of Acetylation of the Gli Transcription Factors. Methods in molecular biology. 2015;1322:147–156. doi: 10.1007/978-1-4939-2772-2_13. [DOI] [PubMed] [Google Scholar]
- 44.Gore J, Korc M. Pancreatic cancer stroma: friend or foe? Cancer cell. 2014;25(6):711–712. doi: 10.1016/j.ccr.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, An Y, Wang X, Zha W, Li X. Inhibition of the Hedgehog pathway induces autophagy in pancreatic ductal adenocarcinoma cells. Oncol Rep. 2014;31(2):707–712. doi: 10.3892/or.2013.2881. [DOI] [PubMed] [Google Scholar]
- 46.Fu J, Rodova M, Roy SK, Sharma J, Singh KP, Srivastava RK, et al. GANT-61 inhibits pancreatic cancer stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice xenograft. Cancer letters. 2013;330(1):22–32. doi: 10.1016/j.canlet.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.