Abstract
Only a few Impatiens spp. from South India (one of the five centers of diversity for Impatiens species) were included in the published datum of molecular phylogeny of the family Balsaminaceae. The present investigation is a novel attempt to reveal the phylogenetic association of Impatiens species of South India, by placing them in the global phylogeny of Impatiens based on a combined analysis of two chloroplast genes. Thirty species of genus Impatiens were collected from different locations of South India. Total genomic DNA was extracted from fresh plant leaf, and polymerase chain reaction was carried out using atpB–rbcL and trnL-F intergenic spacer-specific forward and reverse primers. Thirteen sequences of Impatiens species from three centers of diversity were obtained from GenBank for reconstructing the evolutionary relationships within the genus Impatiens. Bayesian inference analysis was carried out in MrBayes v.3.2.2. This analysis supported Southeast Asia as the ancestral place of origin of extant Impatiens species. Molecular phylogeny of South Indian Impatiens spp. based on combined chloroplast sequences showed the same association as that of morphological taxonomy. Sections Scapigerae, Tomentosae, Sub-Umbellatae, and Racemosae showed Southeast Asian relationship, while sections Annuae and Microsepalae showed African affinity.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-016-0574-8) contains supplementary material, which is available to authorized users.
Keywords: Impatiens species, Molecular phylogeny, atpB–rbcL intergenic spacer, trnL-F spacer, Maximum parsimony, Bayesian inference
Introduction
Two genera, namely, Impatiens and Hydrocera, are the sole members of the family Balsaminaceae. The genus Hydrocera is monotypic. Impatiens is a large genus containing more than 1000 species with a distribution pattern in the mountain areas of old world tropics and subtropics (Janssens et al. 2006). The five biodiversity hotspots for this highly endemic genus have been identified as Southeast Asia, Southern India and Sri Lanka, tropical Africa, Madagascar, and the Eastern Himalayas (Yuan et al. 2004; Janssens et al. 2006). Several novel species, belonging to this explosively speciating plant, are recognized in these regions every year (Kuang et al. 2014; Gogoi and Borah 2014; Luo et al. 2015). The genus Impatiens contains more than 210 species in India with amazing localization in two biodiversity hotspots, namely, Himalayas in the north of India and the Western Ghats in the south of India. Over half of these occur in the Western Ghats of India and at least 103 species of Impatiens are endemic to the Western Ghats alone (Bhaskar 2012).
Molecular phylogeny of balsams based on ITS sequences (Yuan et al. 2004) proposed that extant Impatiens species are of Southeast Asian origin, from where it dispersed to other parts of the globe in several dispersal events. Contrarily, atpB–rbcL intergenic spacer sequences based on phylogenetics of Janssens et al. (2006) suggested that Impatiens originated in South China from which it colonized the nearby regions and afterwards dispersed to north America, India, Africa, the Southeast Asian peninsula, and the Himalayan area. All these published data of molecular phylogeny and biogeography of Balsaminaceae inferred from ITS sequences (Yuan et al. 2004) and chloroplast atpB–rbcL spacer sequences (Janssens et al. 2006) contained only a few samples of Impatiens species from South India, creating a gap in the existing phylogeny of balsams. Hence, this work is a novel attempt on the molecular phylogeny of Impatiens species with representatives from six sections of balsams from South India.
Materials and methods
Representative samples from the different sections of Impatiens species were collected from Southern Western Ghats of India. The plants were authenticated, and voucher specimens were deposited in the Herbarium of St. Thomas College (Palai, Kerala, India). The details of the sample collection were summarized in Table 1.
Table 1.
Species used in this study with location, voucher no., and GenBank accession no. of atpB–rbcL and trnL-F sequences
| SI. no. | Species name with section | Locationa | Voucher no. of sample deposited | GenBank accession number | |
|---|---|---|---|---|---|
| atpB–rbcL | trnL-F | ||||
| Section: Scapigerae | |||||
| 1 | I. levingei | Eravikulam National Park | S.P.P.4854 | KU316381 | KU341090 |
| 2 | I. modesta | Eravikulam National Park | S.P.P.4857 | KU530217 | KU341091 |
| 3 | I. pandata | Eravikulam National Park | S.P.P.4856 | KU316383 | KU513967 |
| 4 | I. scapiflora | Vagamon | S.P.P.4502 | KF447374 | KJ746922 |
| Section: Annuae | |||||
| 5 | I. aadishankarii | Wayanad | S.P.P. 4546 | KU316371 | KU341086 |
| 6 | I. chinensis | Munnar | S.P.P.4545 | KU316374 | KU341088 |
| 7 | I. dalzellii | Eravikulam National Park | S.P.P.4852 | KU316375 | KU341089 |
| 8 | I. gardneriana | Wayanad | S.P.P.4520 | KF562062 | KJ746912 |
| 9 | I. herbicola | Neryamangalam | S.P.P.4505 | KF562065 | KJ746914 |
| 10 | I. ligulata | Wayanad | S.P.P.4530 | KF562063 | KJ746916 |
| 11 | I. minor | Neryamangalam | S.P.P.4504 | KF447375 | KJ703108 |
| 12 | I. oppositifolia | Eravikulam National Park | S.P.P.4855 | KU316382 | KU341092 |
| 13 | I. raziana | Eravikulam National Park | S.P.P.4851 | KU316379 | KU341093 |
| 14 | I. tomentosa | Agasthyamala Biosphere Reserve | S.P.P.4861 | KU316386 | KU341094 |
| Section: Microcepalae | |||||
| 15 | I. bababudenensis | Anamudi Hills | S.P.P.4548 | KU316373 | KU341087 |
| 16 | I. balsamina | Munnar | S.P.P.4517 | KF582043 | KJ746906 |
| 17 | I. dasysperma | Neryamangalam | S.P.P.4506 | KM360163 | KJ746909 |
| 18 | I. latifolia | Eravikulam National Park | S.P.P.4549 | KU316378 | KU508414 |
| 19 | I. mysorensis | Wayanad | S.P.P.4534 | KF582048 | KU508416 |
| 20 | I. pulcherrima | Eravikulam National Park | S.P.P.4853 | KU316384 | KU508417 |
| 21 | I. scabriuscula | Wayanad | S.P.P.4531 | KF562058 | KJ746921 |
| 22 | I. walleriana | Munnar | S.P.P.4518 | KF58205 | 0KJ746925 |
| Section: Tomentosae | |||||
| 23 | I. johnii | Wayanad | S.P.P.4543 | KU316377 | KJ746915 |
| 24 | I. munronii | Wayanad | S.P.P.4532 | KF582047 | KU508415 |
| 25 | I. neo-munronii | Wayanad | S.P.P.4523 | KF562061 | KJ746919 |
| Section: Sub-Umbellatae | |||||
| 26 | I. cordata | Munnar | S.P.P.4515 | KF582044 | KU508411 |
| 27 | I. disotis | Wayanad | S.P.P.4528 | KF582042 | KU508412 |
| 28 | I. uncinata | Wayanad | S.P.P.4529 | KF562057 | KJ746923 |
| Section: Racemosae | |||||
| 29 | I. maculata | Devikulam | S.P.P.4507 | KF562056 | KJ746918 |
| 30 | I. wightiana | Wayanad | S.P.P.4522 | KF582052 | KJ746926 |
aAll locations in Kerala, India
Total genomic DNA was extracted using Gen Elute Plant Genomic DNA Miniprep Kit (Sigma Aldrich, St. Louis, USA). For PCR amplification, OrionX h-Taq PCR Smart Mix (Origin, India) was used. The primers used for the amplification of the chloroplast atpB–rbcL intergenic spacer gene were IMP-atpB—5′-ACATCTAGTACCGGACCAATGA-3′ and IMP-rbcL—5′-AACACCAGCTTTGAATCCAA-3′ (10 pM each) (Janssens et al. 2006), and trnL-F region were trnL-F—c:5′-CGAAATCGGTAGACGCTACG-3′ and trnL-F—f:5′-ATTTGAACTGGTGACACGAG-3′ (10 pM each) (Taberlet et al. 1991).
The temperature profile of amplification of atpB–rbcL intergenic spacer region was as per Janssens et al. (2006), and that of trnL-F region was as per Taberlet et al. (1991). Amplification reactions were carried out in an Agilent Sure Cycler 8800 (Agilent Technologies, USA) (ESM Figs. 1S, 2S). Amplicons (atpB–rbcL amplicon of size 900 bp and trnL-F amplicon of size 600–650 bp) were sequenced in AB1 cycle sequencer (Scigenome Labs Pvt. Ltd., Cochin, Kerala, India).
All sequences generated in this study were subjected to a BLAST search (NCBI) against the GenBank nucleotide database and submitted to GenBank (Table 1). I. omeiana was selected as outgroup for phylogenetic analyses of Impatiens (Janssens et al. 2009). Sequences of Impatiens species from three diversity hotspots were collected from GenBank accessions (Table 2). The sequences were multiple aligned and edited using the CLUSTALW (Thomson et al. 1994) program incorporated in BioEdit 7.0.5.2 (Hall 1999).
Table 2.
Details of sequences of atpB–rbcL and trnL-F of Impatiens spp. obtained from GenBank
| SI. no. | Place of origin and species name | Genbank accession number | |
|---|---|---|---|
| atpB–rbcL | trnL-F | ||
| East and Southeast Asia | |||
| 1 | I. aquatilis | DQ147811 | KP776115 |
| 2 | I. davidi | DQ147835 | KP776129 |
| 3 | I. faberi | DQ147841 | KP776132 |
| 4 | I. gongshanensis | KP776024 | KP776135 |
| 5 | I. napoensis | DQ147861 | KP776146 |
| 6 | I. omeiana | KC905619 | KP776152 |
| 7 | I. platychlaena | DQ147867 | KP776154 |
| 8 | I. soulieana | DQ147880 | KP776164 |
| 9 | I. uliginosa | DQ147887 | KP776173 |
| Africa | |||
| 10 | I. hians | DQ147849 | EF649977 |
| 11 | I. keilii | FJ826654 | KP776138 |
| 12 | I. mannii | FJ826660 | EF649980 |
| Himalaya | |||
| 13 | I. scabrida | DQ147877 | KP776162 |
The Akaike information criterion (AIC) implemented in the program jModelTest version 2.1.5 (Darriba et al. 2012) was used to choose substitution models that best fit the data set. Bayesian inference analysis was carried out in MrBayes v.3.2.2 (Ronquist et al. 2012) in two independent runs, each with one heated chain and one cold chain and for one lakh generations. Convergence occurred when standard deviation (SD) of split frequencies fell below 0.05; the first 25% of MCMC generations were discarded as burn-in and a consensus phylogram was created. Posterior probability values were used to estimate branch support. Trees were visualized by Fig Tree, Tree Figure drawing tool version 1.4.2 (Rambaut 2014).
Results and discussion
Phylogenetic analysis of this study included two chloroplast regions (atpB–rbcL, trnL-F) from 30 sequences of South Indian Impatiens species. In addition, 13 sequences of each of these regions were obtained from NCBI database. To assess the level of congruence between these data sets, each data set was analyzed independently to see if they produced a similar topology. The separate analyses produced topologies similar to each other. In comparison with separate analyses, the combined phylogeny had a well-resolved topology.
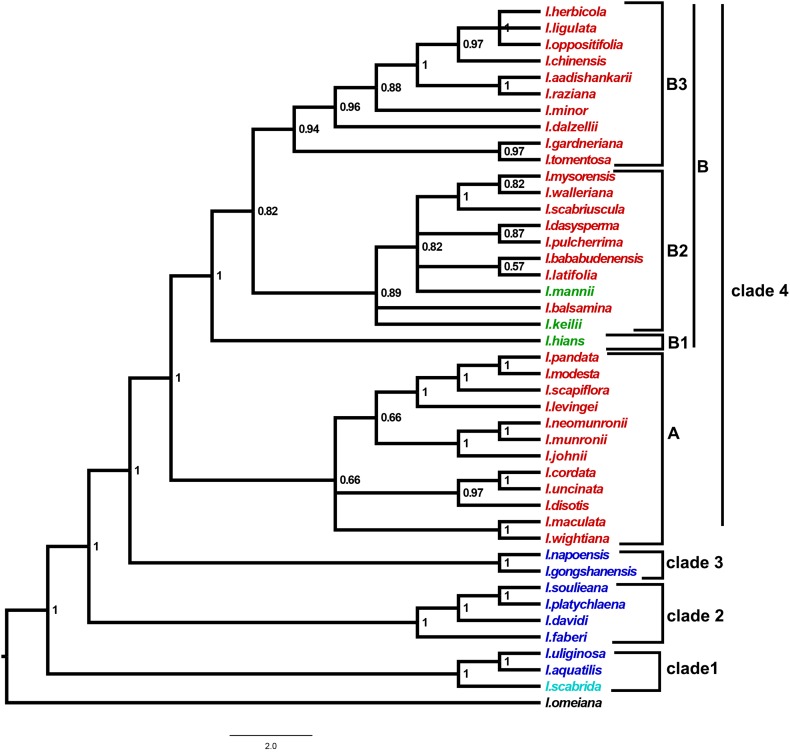
The combined atpB–rbcL and trnL-F data matrix contained 1664 characters. A general time reversible model of evolution with invariant sites and a gamma distribution (GTR + I + G) was selected using jModelTest version 2.1.5. This model was used for the Bayesian inference (BI) analysis. The resulted tree by BI analysis had a well-resolved topology (Fig. 1). The resolved lineages of Impatiens species were grouped into four clades with strong Bayesian posterior probability (BPP) values. Two Southeast Asian species and the Himalayan species formed clade 1. Clade 2 included four Southeast Asian species. Two Southeast Asian species formed clade 3. Clade 4 was divided into two subclades, i.e., A and B (BPP of 1.00). Subclade A contained species of sections Racemosae, Sub-Umbellatae, Tomentosae, and Scapigerae (BPP of 1.00). Subclade B is divided into three subclades, i.e., B1, B2, and B3. African species (I. hians) formed Subclade B1. Subclade B2 included African species (I. keilii and I. mannii) and South Indian species of section Microsepalae with BPP of 0.89. Species of section Annuae produced Subclade B3 with BPP of 0.94.
Fig. 1.
Bayesian consensus cladogram based on combined sequences of chloroplast atpB–rbcL intergenic spacer and trnL-F genes. The numbers by the nodes indicate Bayesian posterior probabilities greater than 0.5
Implications on infrageneric classification and biogeography of Impatiens species of Western Ghats
Impatiens is considered taxonomically as one of the most difficult genera of angiosperms, mainly due to hypervariable structure and fragile nature of its flowers making examinations of dried specimen extremely difficult (Grey-Wilson 1980). The important revision of the African taxa by Grey-Wilson (1980) distinguished six informal infrageneric groups for the African species for practical diagnosis. Based on morphological and molecular data sets, Yu et al. (2015) presented a new classification of Impatiens, with the genus being divided into two subgenera, subgenus Clavicarpa and subgenus Impatiens. The subgenus Impatiens was further subdivided into seven sections.
In the taxonomic treatments of South Indian Impatiens by Bhaskar (2012), balsams of South India were classified under seven sections, i.e., Scapigerae, Epiphyticae, Annuae, Microsepalae, Tomentosae, Sub-Umbellatae, and Racemosae. Based on the present molecular phylogenetic study, species of each section formed monophyletic association with strong BPP support. This study authenticates the morphological classifications of Bhaskar (2012).
Based on several morphological similarities among species endemic to Africa and South India, close affinity between African and South Indian taxa and a possible migration route connecting these two areas were suggested (Grey-Wilson 1980). In this study, species of sections Microsepalae and Annuae showed African affinities with sister–clade relationships. This confirms Grey-Wilson’s (1980) suggestions of affinity between African and South Indian species. Sections Scapigerae, Sub-Umbellatae, Tomentosae, and Racemosae formed a separate clade (Subclade A) with sister–clade relationships with the extant Southeast Asian species.
There are several hypotheses related to the origin of Impatiens (Jones and Smith 1966; Grey-Wilson 1980). Bhaskar (1981) proposed that Western Ghats is the place of origin of the genus Impatiens. His hypothesis was based on the observation that Western Ghats of India contains the phylogenetically old species with primitive radial pollen grains, diploid chromosome number, and shrubby habit.
ITS phylogeny of Yuan et al. (2004) and atpB–rbcL phylogeny of Janssens et al. (2006) revealed that Impatiens spp. colonized African continent from Southwest China in three independent dispersal events. Madagascan species was derived from a single colonization event (Janssens et al. 2009). The present combined chloroplast gene analysis contained only three African and no Madagascan species. In this African species, I. keilii and I. mannii were placed with species of section Microsepalae. Section Annuae formed a sister–clade with this section. Himalayan species (I. scabrida) showed affinity to Southeast Asian species (I. aquatilis and I. uliginosa).
The biogeographical elucidation based on the present study is mainly in accordance with the conclusion of Yuan et al. (2004). The present analysis postulated that South India was colonized by two independent dispersal events, i.e., once by Southeast Asian ancestor as shown by the sister–clade relationships of extant Southeast Asian species and the sections Scapigerae, Tomentosae, Sub-Umbellatae, and Racemosae and a more recent colonization by an ancestor with African affinities (sections Microsepalae and Annuae).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank University Grants commission (UGC), India and Kerala state Council for Science, Technology and Environment (KSCSTE), Kerala, India for instrumentation, and Forest Department of Kerala, India for providing us the permission to collect samples of Impatiens species from Southern Western Ghats. We thank Mahatma Gandhi University, Kerala, India (U.O.No.390.A.II/2009/Academic dated 28.01.2009) for funding the research through PhD research assistance.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Bhaskar V. The genus Impatiens L. in south India: endemism and affinities. Indian Forester. 1981;107:368–376. [Google Scholar]
- Bhaskar V (2012) Taxonomic monograph on Impatiens L. (Balsaminaceae) of Western Ghats south India. The key genus for endemism. Centre for Plant Taxonomic Studies, Bangalore India
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogoi R, Borah S. Impatiens paramjitiana, a new species of Balsaminaceae from Arunachal Pradesh, India. Phytotaxa. 2014;175(3):171–175. doi: 10.11646/phytotaxa.175.3.8. [DOI] [Google Scholar]
- Grey-Wilson C. Impatiens of Africa, Rotterdam Balkema. Kew Bull. 1980;34:221–227. doi: 10.2307/4117018. [DOI] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and program for Windows 95/98/NT. In: Nucleic acids symposium series. 1999;41:95–98. [Google Scholar]
- Janssens S, Geuten K, Yuan YM, Song Y, Kupfer P, Smets E. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Syst Bot. 2006;31(1):171–180. doi: 10.1600/036364406775971796. [DOI] [Google Scholar]
- Janssens SB, Knox EB, Huysmans S, Smets EF, Merckx VS. Rapid radiation of Impatiens (Balsaminaceae) during Pliocene and Pleistocene: result of a global climate change. Mol Phylogenet Evol. 2009;52(3):806–824. doi: 10.1016/j.ympev.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Jones K, Smith J. The cytogeography of Impatiens L. (Balsaminaceae) Kew Bull. 1966;20:63–72. doi: 10.2307/4107885. [DOI] [Google Scholar]
- Kuang RP, Duan LD, Gu JZ, Cai XZ, Cong YY, Liu KM. Impatiens liboensis sp. nov. (Balsaminaceae) from Guizhou, China. Nor J Bot. 2014;32(4):463–467. doi: 10.1111/j.1756-1051.2013.00194.x. [DOI] [Google Scholar]
- Luo Q, Wang TJ, Zhao LH. Impatiens menghuochengensis sp. nov. (Balsaminaceae) from Sichuan, China. Nor J Bot. 2015;32(6):839–843. doi: 10.1111/njb.00503. [DOI] [Google Scholar]
- Rambaut A (2014) FigTree. v. 1.4. 2: Tree drawing tool. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 5 Aug 2014
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17(5):1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thomson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SX, Janssens SB, Zhu XY, Lidén M, Gao TG, Wang W. Phylogeny of Impatiens (Balsaminaceae): integrating molecular and morphological evidence into a new classification. Cladistics. 2015;32(2):179–197. doi: 10.1111/cla.12119. [DOI] [PubMed] [Google Scholar]
- Yuan YM, Song YI, Geuten K, Rahelivololona E, Wohlhauser S, Fischer E, Kupfer P. Phylogeny and biogeography of Balsaminaceae inferred from ITS sequences. Taxon. 2004;53(2):391. doi: 10.2307/4135617. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.