Abstract
Background
Congenital toxoplasmosis is an important cause of spontaneous abortion worldwide. However, there is limited information on detection and genotypic characterization of Toxoplasma gondii (T. gondii) in women with recurrent spontaneous abortion (RSA). The aim of this study is the molecular detection and genotypic characterization of T. gondii in formalin-fixed, paraffin-embedded fetoplacental tissues (FFPTs) of women with RSA that have referred to the Avicenna Research Institute in Tehran, Iran.
Materials and Methods
This experimental research was undertaken on 210 FFPTs of women with RSA. The information of the patients was collected from the archives of Avicenna Research Institute in Tehran, Iran. After DNA extraction, the presence of T. gondii was examined by nested polymerase chain reaction targeting the GRA6 gene. Genotyping was performed on positive samples using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) that targeted the GRA6 and SAG3 genes. Sequencing was conducted on two GRA6 positive samples.
Results
T. gondii DNA was detected in 3.8% (8/210) of the samples. Genotyping showed that all positive samples belonged to type III of the T. gondii genotype. Sequencing two genomic DNAs of the GRA6 gene revealed 99% similarity with each other and 99-100% similarity with T. gondii sequences deposited in GenBank. There were six patients with histories of more than three abortions; one patient had a healthy girl and another patient had two previous abortions. Abortions occurred in the first trimester of pregnancy in seven patients and in the second trimester of pregnancy in one patient.
Conclusion
The results of this study have indicated that genotype III is the predominant type of T. gondii in women with RSA in Tehran, Iran. Also, our findings suggest that toxoplasmosis may play a role in the pathogenesis of RSA. However, further studies are needed to elucidate a clear relationship between T. gondii infection and RSA.
Keywords: Toxoplasma gondii, Abortion, Molecular Detection, Genotype, Iran
Introduction
Toxoplasmosis is one of the most common parasitic diseases where approximately one-third of the world’s population is affected (1, 2). Approximately 25 to 30% of the world’s population is infected by Toxoplasma gondii (T. gondii). Nevertheless, the most common form of infection is asymptomatic (2-4). Human infections generally occur by the consumption of undercooked meatthat contains tissue cysts or by water and food contaminated with oocysts present in cat feces. Congenital infection is one of the most important sequels of toxoplasmosis in pregnant women (1). Congenital transmission of T. gondii predominantly occurs at the first time during pregnancy (3, 5). The approximate incidence rate of congenital toxoplasmosis is 1.5 cases per 1000 live births with a global incidence rate of 190,100 cases annually (6). Frequency of transplacental transmission and severity of congenital toxoplasmosis correlates with the gestational age of infected mothers. The highest rates of transplacental transmission occur in the third trimester of pregnancy; which usually results in asymptomatic infections at birth. However, they may develop clinical signs (such as chorioretinitis, slower mental and neurological development) at a later age (1). On the other hand, the severity of congenital toxoplasmosis is highest in the first and second trimesters of pregnancy which usually results in abortion or stillbirth (1, 5, 7, 8).
Recurrent spontaneous abortion (RSA) is the loss of three or more consecutive pregnancies before 20 weeks of pregnancy (9) and affects approximately 1 to 2% of couples trying to conceive (10). Several factors-genetic background, anatomical abnormalities, endocrine disruption, autoimmune disorder, and infectious diseases have been attributed to play roles in the etiology of RSA (9-11). Infectious agents account for 0.5 to 5% of RSA (10). The most common infectious causes of RSA are Chlamydia trachomatis, Ureaplasmaurealyticum, Mycoplasma hominis, cytomegalovirus (CMV), and human papillomaviruses (HPV) (9, 11). Several studies have reported significantly higher seroprevalence of ToRCH infections in women with spontaneous abortion or negative obstetric history, including preterm deliveries, intrauterine deaths or growth retardation (12-16).
Although several studies have reported an association between T. gondii infection and spontaneous abortion (6), the role of toxoplasmosis in the etiology of RSA is less clear. Hence, we have investigated the rate of T. gondii infection in formalin-fixed, paraffin-embedded fetoplacental tissues (FFPTs) of women with RSA that referred to the Avicenna Research Institute in Tehran, Iran.
Materials and Methods
This experimental study was performed on archived FFPTs of women with RSA that referred to the Avicenna Research Institute in Tehran, Iran during 2013-2015. This study was approved by the Ethical Committees of Tarbiat Modares University and Avicenna Research Institute. Avicenna Research Institute was consent about the research on the archived FFPTs of women.
Patients and samples
We collected 210 FFPTs of aborted fetuses or placentas of women with recurrent abortion from the archives of the Avicenna Research Institute in Tehran, a referral center for infertile couples in Iran. Information of clinical symptoms, pathological findings, and genetic background were obtained from patients’ medical records.
DNA extraction
For each FFPT, five 10 μm thick sections were cut and transferred to 1.5 ml microtubes. In order to avoid cross-contamination, we used a new, sterile disposable microtome blade for each block. Sections were deparaffinized by the addition of 1 ml xylene (Merck, Germany) for 15 minutes at 50°C. Subsequently, the tubes were centrifuged at 13000 g for 5 minutes and the supernatant was discarded. This step was repeated twice. The samples were rehydrated in a descending ethanol series (100, 90, 80, 70%) and subsequently washed with distilled water. For DNA extraction, 800 μL of lysis buffer (50 mM tris-HCl, pH=8.0, 25 mM EDTA, and 400 mM NaCl), 100 μL 10% sodium dodecyl sulfate (SDS, Merck, Germany), and 10 μL proteinase K (20 μg/μL, Thermo Fisher Scientific, Wilmington, DE, USA) were added to each tube (17). The suspension was incubated at 55ºC for 72 hours. After overnight, an additional of 10 μL proteinase K (20 μg/μL) was added to each tube (18). In order to precipitate undissolved proteins and debris, we added 300 μL of 6 M NaCl to each tube for 15 minutes at 4°C. After centrifugation (13000 g for 15 minutes), the supernatant was transferred to 1.5 ml microtubes (17). Then, 800 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added to each microtube. The microtubes were centrifuged (13000 g for 15 minutes) and we transferred the supernatants to new microtubes. Subsequently, 1 ml of chloroform was added to each microtube. The microtubes were centrifuged (13000 g for 15 minutes) and the supernatant was transferred to sterile microtubes. DNA was precipitated by the addition of one-tenth the volume of a sodium acetate solution (3 M, pH=5.2) and twice the volume of cold 100% ethanol, kept at -20°C overnight, and subsequently centrifuged at 13000 g for 20 minutes. Finally, the pellet was washed with 70% ethanol, centrifuged (13000 g for 15 minutes), resuspended in 50 μL of distilled water, and stored at -20°C until use.In order to ensure that the DNA was extracted, we used two T. gondii positive tissue samples (GenBank accession numbers. KR809554 and KR809555) which had been detected in our previous study (19). These positive samples were fixed in formalin and embedded in paraffin after which the following procedure for DNA extraction was performed. We also used the Rh strain of T. gondii as a positive control.
Detection of T. gondii infection by nested polymerase chain reaction
PCR was conducted using a pair of T. gondii-specific primers:
GAR6-F1: 5'-ATTTGTGTTTCCGAGCAGGT-3' and R1: 5'-GCACCTTCGCTTGTGGTT-3'.
Nested-PCR was performed with primers:
GAR6-F2: 5'-TTTCCGAGCAGGTGACCT-3' and R2: 5'-TCGCCGAAGAGTTGACATAG-3' (20).
Amplifications were conducted ina final volume of a 20 μL reaction mixturethat contained 10 μL of 2x Taq DNA polymerase Master Mix with 2 mM MgCl2 (Cat. no. A170301, Ampliqon, Denmark), 10 pmol of each primer, 5 μL of distilled water, and 3 μL of template DNA. For nested-PCR, one μL of the first round PCR product was used as the template. For each reaction, two positive controls (DNA extracted from T. gondii paraffin-embedded tissuesand the RH strain of T. gondii) and a negative control (double distilled water) were included. Amplification was performed with initial denaturation for 5 minutes at 95°C, followed by 35 cycles at 95°C for 30 seconds (denaturation), annealing at 59°C in the first round, and 57°C in nested PCR for 30 seconds, extension at 72°C for 30 seconds, and final extension at 72°C for 10 minutes. A total of 5 μl of nested-PCR products along with a 100-bp DNA ladder were electrophoresed in 1.5% safe stain (Sinaclon, Iran) agarose gels and visualized under ultra-violet trans-illumination.
Genotyping of positive samples by restriction fragment length polymorphism
Positive samples were genotyped using GRA6 and SAG3 markers (21). First, we digested the nested-PCR products of GRA6 positive samples using Tru1I (MseI) restriction enzyme (Cat. No. ER0982, Thermo Fisher Scientific, USA) as previously described (20). Digestion was conducted in a final volume of 16 μL reaction mixtures that contained 5 μL of the nested-PCR products, 1μL of Tru1I enzyme, 1 μL of 10X Buffer R, and 9 μL of nuclease-free water. Then, the reaction mixtures were incubated at 65°C for 1 hour according to the manufacturer’s instructions. A total of 10 μl of restriction fragments were electrophoresed by Tris-acetate-EDTA (TAE) buffer through 3% (w/v) agarose gel stained with safe stain and visualized under UV transillumination. We conducted genotyping of the positive samples by the SAG3 marker (21, 22). Nested-PCR was carried out for positive samples using the SAG3 marker as previously described (22). Next, the products were digested using BcnI (NciI) restriction enzyme (Cat. No. ER0061, Thermo Fisher Scientific, USA) at 37°C for 6 hours according to the manufacturer’s protocols. The restriction fragments were electrophoresed and visualized under UV transillumination. The type of T. gondii was determined according to the restriction patterns after digestion with restriction enzymes (21). In order to determine better illustrationpatterns of the genotypes, the GRA6 and SAG3 sequences of three types of T. gondii (RH type I, ME49 type II, and NED type III) were obtained fromGenBank and digested by their restriction enzymes using NEBcutter V2.0 (http://nc2.neb.com/NEBcutter2/).
Nucleotide sequence analysis of the GRA6 gene
We extracted two positive nested-PCR products of the GRA6 gene from the gel (Vivantis Gel Purification kit, Selangor DarulEhsan, Malaysia) according to the manufacturer’s instructions. The products were sequenced in the forward and reverse directions by the Sequetech Corporation (Mountain View, CA, USA), edited with BioEdit software, (23) and compared with GRA6 partial sequences of T. gondii available in GenBank.
Results
Detection of T. gondii in women with recurrent spontaneous abortion
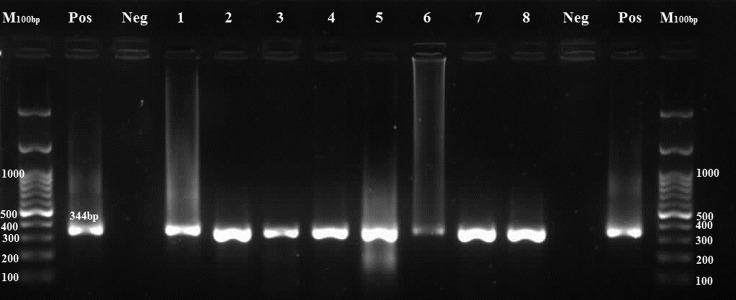
T. gondii DNA was detected in 8 out of 210 samples (3.8%) by the GRA6 marker (Fig .1).
Fig.1.
PCR products of the GRA6 positive samples. Toxoplasma gondii (T. gondii) positive samples give a 344-bp band.
M; 100 bp DNA marker, Pos; Positive control, Neg; Negative control, and Lanes 1-8; Positive samples.
As shown in Table 1, patients had a mean age of 33.5 years (range: 28-39 years). There were seven patients with a history of previous abortion (patients 1-3, 5-8). From these, six occurred in the first trimester and one occurred in the second trimester (patient 1). One patient had a healthy girl (patient 4) with no history of previous abortion. The abortion of this patient (patient 4) was occured in the first trimester of pregnancy. Patient 1had clinical symptoms of fever and severe necrotizing chorioamnionitis before the abortion. Patient 3 reported clinical symptoms such as rapid heart beat, maternal anemia, and edema of the legs and ankles before the abortion. The edema resolved after the abortion. Patient 2 had a history of hypothyroidism. Nonspecific symptoms were reported from other patients before the abortions (Table 1).
Table 1.
Information of the Toxoplasma gondii (T. gondii) infected women with recurrent spontaneous abortion (RSA)
| PatientNo. | Age (Y) CityProvince | Number of gestations (G),Number of abortions (AB) | Week of abortion | Chromosomal aneuploidies§ | Pathological findings in fetoplacental tissues | Symptoms |
|---|---|---|---|---|---|---|
| 1 | 36 Abhar Zanjan | G6, AB6All pregnancies aborted at second trimester | LMP†: 16w(Second trimester) | Not detected | Inflammatory cell infiltration with patchy amnionic necrotizing foci in the membrane | Fever, severe abdominal and back pain,premature rupture of membranes (PPROM) |
| 2 | 31 Tuyserkan Hamedan | G4, AB4All pregnancies aborted at first trimester | LMP: 11w+2dUltrasound: 8w(First trimester) | Not detected | No remarkable pathological findings | Hypothyroidism |
| 3 | 39 Eslamshahr Tehran | G8, AB8All pregnancies aborted at first trimester | LMP: 7w(First trimester) | Not detected | No remarkable pathological findings | No specific symptoms |
| 4 | 38 Tehran Tehran | G2, AB1She has one healthy girl | LMP: 11w(First trimester) | Not detected | No remarkable pathological findings | Rapid heartbeat, anemia andedema of the legs and ankles |
| 5 | 36 Karaj Alborz | G3, AB3All pregnancies aborted at first trimester. | Ultrasound: 6w+4d(First trimester) | MLPA findings compatible with an extra copy of chromosome 15 (trisomy 15) | The membrane showed calcification without inflammation | No specific symptoms |
| 6 | 28 Kashan Isfahan | G5, AB5 | LMP: 13wUltrasound: 11w(First trimester) | Not detected | No remarkable pathological findings | No specific symptoms |
| 7 | 31 Tehran Tehran | G3, AB3All pregnancies aborted at first trimester | LMP: 11wUltrasound: 8w+3d(First trimester) | Not detected | No remarkable pathological findings | No specific symptoms |
| 8 | 29 Tehran Tehran | G2, AB2All pregnancies aborted at first trimester | LMP: 11wUltrasound: 8w(First trimester) | Not detected | No remarkable pathological findings | No specific symptoms |
†; LMP: Last menstrual period and §; Chromosomal aneuploidies were detected using multiplex ligation-dependent probe amplification (MLPA).
Genotyping of positive samples
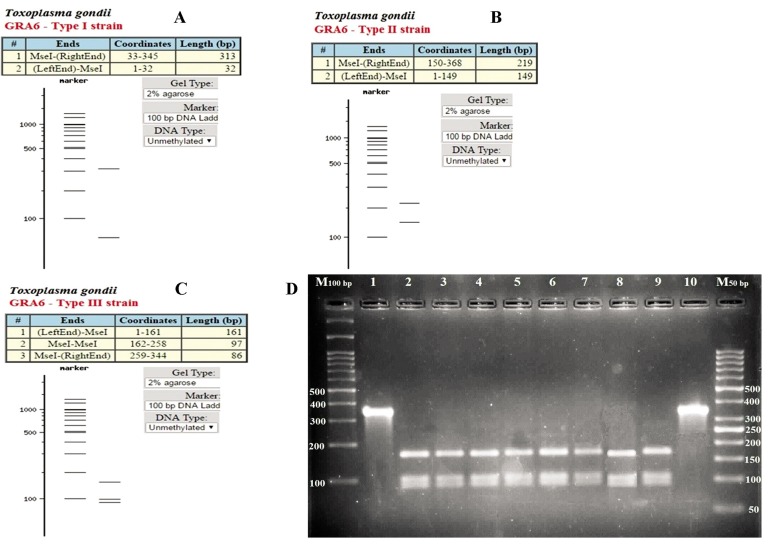
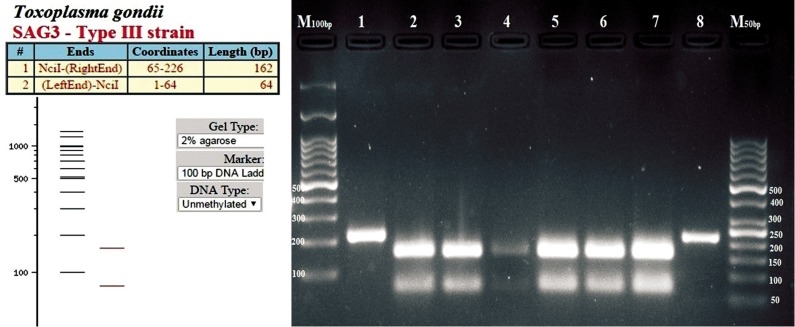
GRA6 completely characterized eight samples as T. gondii genotype III (Fig .2). Genotyping of positive samples were conducted by using the SAG3 marker. The results showed amplification of SAG3 in six out of eight GRA6 positive samples. Digestion of SAG3 PCR products by BcnI enzyme determined that all six products belonged to T. gondii genotype III (Fig .3).
Fig.2.
Genotyping of positive samples with the GRA6 marker. The products were digested with Tru1I enzyme. A, B, C. Patterns of three types of Toxoplasma gondii (T. gondii) genotype, and D. Agarose gel electrophoresis of PCR products digested with Tru1I enzyme.
M; 100 and 50 bp DNA marker, Lanes 1 and 10; Undigested positive samples, and Lanes 2-9; T. gondii genotype type III.
Fig.3.
Genotyping of positive samples with the SAG3 marker. The products were digested with BcnI enzyme. A. Patterns of Toxoplasma gondii (T. gondii) type III genotype and B. Agarose gel electrophoresis of PCR products digested with BcnI enzyme.
M; 100 and 50 bp DNA marker, Lanes 1 and 8; Uundigested positive samples, and Lanes 2-7; T. gondii genotype type III.
Sequencing of the GRA6 gene
We submitted two GRA6 nucleotide sequences with a length of 344 bp to the GenBank database (KT735111, KT735112). The alignment of our sequences revealed the highest similarity (100%) with T. gondii isolated from cat (KP792620, KP792621), sheep (KT735113-19), rat (KP792610, KP792613), and bird (KR809554-8, KP792606-9, KP792600-2) hosts in Iran (Table 2).
Table 2.
Multiple sequence alignment of the GRA6 gene of Toxoplasma gondii (T. gondii) from our samples and other hosts in Iran. Our samples are shown in red as accession numbers KT735111 and KT735112
| KT809309 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KP792604 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTTGTGGTGC |
| KP792605 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTTGTGGTGC |
| KR809555 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTTC |
| KT735112 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAGAACAGCTTCGTGGTGC |
| KP792614 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KR809556 | TTTCCGAGCAGGTGACCTGGGTCGCCTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KR809558 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGTAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735111 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735113 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735114 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735115 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735116 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735117 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735119 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KR809557 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KR809554 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KP792621 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KP792620 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KP792609 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KP792610 | TTTCCGAGCAGGTGACCTGGGTCGCTTTTTTGAAACAGCAGGAAAACAGCTTCGTGGTGC |
| KT735118 | T͙T͙T͙C͙C͙G͙A͙G͙C͙A͙G͙G͙T͙G͙A͙C͙C͙T͙G͙G͙G͙T͙C͙G͙C͙ṬT͙T͙T͙T͙T͙G͙A͙A͙A͙C͙A͙G͙C͙A͙G͙G͙A͙ẠA͙A͙C͙A͙G͙C͙T͙T͙C̣G͙T͙G͙G͙T͙G̣C͙ |
| KT809309 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792604 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792605 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KR809555 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735112 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792614 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KR809556 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KR809558 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735111 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735113 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735114 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735115 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735116 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735117 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735119 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KR809557 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KR809554 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792621 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792620 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792609 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KP792610 | CACGTAGCGTGCTTGTTGGCGACTACCTTTTTTTCTTGGGAGTGTCGGCGAAATGGCACA |
| KT735118 | C͙A͙C͙G͙T͙A͙G͙C͙G͙T͙G͙C͙T͙T͙G͙T͙T͙G͙G͙C͙G͙A͙C͙T͙A͙C͙C͙T͙T͙T͙T͙T͙T͙T͙C͙T͙T͙G͙G͙G͙A͙G͙T͙G͙T͙C͙G͙G͙C͙G͙A͙A͙A͙T͙G͙G͙C͙A͙C͙A͙ |
| KT809309 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTCCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792604 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792605 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809555 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735112 | CGGTGGCACCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792614 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809556 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735112 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792614 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809556 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809558 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735111 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735113 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735114 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735115 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735116 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735117 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735119 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809557 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KR809554 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792621 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792620 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792609 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KP792610 | CGGTGGCATCCATCTGAGGCAGAAGCGTAACTTCTGTCCTTTAACTGTCTCCACAGTTGC |
| KT735118 | C͙G͙G͙T͙G͙G͙C͙A͙ṬC͙C͙A͙T͙C͙T͙G͙A͙G͙G͙C͙A͙G͙A͙A͙G͙C͙G͙T͙A͙A͙C͙T͙ṬC͙T͙G͙T͙C͙C͙T͙T͙T͙A͙A͙C͙T͙G͙T͙C͙T͙C͙C͙A͙C͙A͙G͙T͙C̣G͙C͙ |
| KT809309 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCCGTCGC |
| KP792604 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792605 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KR809555 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735112 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792614 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KR809556 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KR809558 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735111 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735113 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735114 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735115 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735116 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735117 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735119 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KR809557 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KR809554 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792621 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792620 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792609 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KP792610 | TGTGGTCTTTGTAGTTTTCATGGGTGTACTCGTCAATTCGTTGGGTGGAGTCGCTGTCGC |
| KT735118 | T͙G͙T͙G͙G͙T͙C͙T͙T͙T͙G͙T͙A͙G͙T͙T͙T͙T͙C͙A͙T͙G͙G͙G͙T͙G͙T͙A͙C͙T͙C͙G͙T͙C͙A͙A͙T͙T͙C͙G͙T͙T͙G͙G͙G͙T͙G͙G͙A͙G͙T͙C͙G͙C͙ṬG͙T͙C͙G͙C͙ |
| KT809309 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGGGGACAGCA |
| KP792604 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792605 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KR809555 | AGCAGACAGCGGTGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735112 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792614 | AGCAGACAGCGATGGTGTTAAGCAGGCCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KR809556 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KR809558 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735111 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735113 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735114 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735115 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735116 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735117 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735119 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KR809557 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KR809554 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792621 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792620 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792609 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KP792610 | AGCAGACAGCGATGGTGTTAAGCAGACCCCTTCGGAAACCGGTTCGAGCGGTGGACAGCA |
| KT735118 | A͙G͙C͙A͙G͙A͙C͙A͙G͙C͙G͙ẠT͙G͙G͙T͙G͙T͙T͙A͙A͙G͙C͙A͙G͙ẠC͙C͙C͙C͙T͙T͙C͙G͙G͙A͙A͙A͙C͙C͙G͙G͙T͙T͙C͙G͙A͙G͙C͙G͙G͙ṬG͙G͙A͙C͙A͙G͙C͙A͙ |
| KT809309 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792604 | AGAAGCAGTGGGGACCCCTGAAGACTATGTCAACTCTTCGGCGA |
| KP792605 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KR809555 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735112 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792614 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KR809556 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KR809558 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735111 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735113 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735114 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735115 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735116 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735117 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735119 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KR809557 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KR809554 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792621 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792620 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792609 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KP792610 | AGAAGCAGTGGGGACCACTGAAGACTATGTCAACTCTTCGGCGA |
| KT735118 | A͙G͙A͙A͙G͙C͙A͙G͙T͙G͙G͙G͙G͙A͙C͙C͙ẠC͙T͙G͙A͙A͙G͙A͙C͙T͙A͙T͙G͙T͙C͙A͙A͙C͙T͙C͙T͙T͙C͙G͙G͙C͙G͙A͙ |
*; Exact match between all sequences and .; Mismatch with at least one sequence.
Discussion
In the current study, we detected T. gondii DNA in 3.8% of women with RSA. To our knowledge, this study was the first report of molecular diagnosis of T. gondii in women with RSA. In previous studies in Iran, Ghasemi et al. (8, 24) detected T. gondii DNA in 7.3% (6/82) of women with spontaneous abortion and in 3.6% (1/28) of women with stillbirth in Tehran. Asgari et al. (25) detected T. gondii DNA in 14.4% (78/542) of paraffin-embedded tissue samples from women with spontaneous abortion in Shiraz, Southern Iran. Hoveyda et al. (26) detected T. gondii DNA in 15.48% (10/65) of paraffin-embedded tissue samples from Iranian women with abortions by PCR. Genotyping of positive samples by PCR-restriction fragment length polymorphism (RFLP) has indicated that all eight positive samples belonged to genotype III of T. gondii. is classified into three main genotypes (type I, II, and III) with some differences in virulence and epidemiological patterns (27, 28). Genotype III is the most prevalenttype of T. gondii worldwide (27, 29). However, type I has the highest virulence of among T. gondii genotypes (28). In Iran, genotype III is the most prevalent type of T. gondii (30), however genotype II (30, 31) and in some studies genotype I has been reported in different hosts (32, 33).
Association of T. gondii seropositivity with infertility or bad obstetric outcomes has been reported in different studies. In this regard, El-Tantawy et al. (34) observed significantly higher seroprevalcenc of T. gondii in infertile women. In that study, 61.85% (193/312) of infertile and 44% (44/100) of fertile women had T. gondii IgG seropositivity in Egypt. Malik et al. (35) demonstrated a significantly higher seroprevalence of T. gondii in 417 women with unfavorable obstetric history such as intrauterine deaths, intrauterine growth retardation, and preterm deliveries in India. According to the results, T. gondii IgM antibody was detected in 28% (120/417) of women with negative obstetric history, which 57% (68/120) had a history of previous abortion. Interestingly, T. gondii IgM antibody was found in 76.5% of women with two or more abortions and 23.5% of women with a single abortion. Toxoplasmosis was diagnosed in 6 out of 9 (66.7%) patients with secondary infertility and 3 (33.3%) with primary infertility (35). Aral et al. (36) did not find a significant association between T. gondii seropositivity and infertility in women in Turkey.
In recent years, several studies were conducted about the influences of latent (asymptomatic) toxoplasmosis on mothers and their offspring (3, 4, 37). In this regard, Kaňková and Flegr (38) reported that pregnant women with latent toxoplasmosis (IgG seropositive women) had developmentally younger fetuses (based on ultrasound scan) comparedto T. gondii negative women at week 16 of pregnancy. Kaňková et al. (39) also demonstrated that infants of mothers with latent toxoplasmosis had significantly slower postnatal motor development than mothers without latent toxoplasmosis during the first year of life. Another study by the same group revealed that T. gondii-infected pregnant women had used significantly more assisted reproductive technology to conceive compared to T. gondii-negative women. T. gondii-infected women had a longer time to conceive and more fertility problems than T. gondii-negative women (40).
This study was the first molecular detection of T. gondii in fetoplacental tissues of women with RSA, however it had some limitations. We did not access the previous abortion samples of the patients. In addition, we were unable to follow the patients and their future pregnancies. Hence, our study only suggested that toxoplasmosis might play a role in the pathogenesis of RSA. Additional investigations with larger groups of patients should be conducted in order to elucidate a clear relationship between T. gondii infection and RSA.
Conclusion
The results of this study have indicated that genotype III is the predominant type of T. gondii in women with RSA in Tehran, Iran. Our results also indicated that T. gondii infection might play a role in the pathogenesis of RSA. However, more research should be conducted in this regard to elucidate a clear relationship between T. gondii infection and RSA.
Acknowledgments
This study is a part of a Ph.D. thesis of the first author that financially supported by Tarbiat Modares University. The authors would like to sincerely thank Miss Azadeh Soltani (Avicenna Research Institute, Iran) for her help in preparation of tissue sections and technical assistance. The authors also appreciate all members of the Pathology and Molecular Genetics laboratories at Avicenna Research Institute for their sincere assistance throughout this study. The authors report no conflict of interest.
References
- 1.Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363(9425):1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- 2.Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25(2):264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdoli A, Dalimi A, Arbabi M, Ghaffarifar F. Neuropsychiatric manifestations of latent toxoplasmosis on mothers and their offspring. J Matern Fetal Neonatal Med. 2014;27(13):1368–1374. doi: 10.3109/14767058.2013.858685. [DOI] [PubMed] [Google Scholar]
- 4.Dalimi A, Abdoli A. Latent toxoplasmosis and human. Iran J Parasitol. 2012;7(1):1–17. [PMC free article] [PubMed] [Google Scholar]
- 5.Paquet C, Yudin M. Toxoplasmosis in pregnancy: prevention, screening, and treatment. J Obstet Gynaecol Can. 2013;35(1):78–79. doi: 10.1016/s1701-2163(15)31053-7. [DOI] [PubMed] [Google Scholar]
- 6.Torgerson PR, Mastroiacovo P. The global burden of congenital toxoplasmosis: a systematic review. Bull World Health Organ. 2013;91(7):501–508. doi: 10.2471/BLT.12.111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XL, Wei HX, Zhang H, Peng HJ, Lindsay DS. A meta analysis on risks of adverse pregnancy outcomes in Toxoplasma gondii infection. PLoS One. 2014;9(5):e97775–e97775. doi: 10.1371/journal.pone.0097775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghasemi FS, Rasti S, Piroozmand A, Bandehpour M, Kazemi B, Mousavi SG, et al. Toxoplasmosis-associated abortion and stillbirth in Tehran, Iran. J Matern Fetal Neonatal Med. 2016;29(2):248–251. doi: 10.3109/14767058.2014.996127. [DOI] [PubMed] [Google Scholar]
- 9.Pandey MK, Rani R, Agrawal S. An update in recurrent spontaneous abortion. Arch Gynecol Obstet. 2005;272(2):95–108. doi: 10.1007/s00404-004-0706-y. [DOI] [PubMed] [Google Scholar]
- 10.Ford HB, Schust DJ. Recurrent pregnancy loss: etiology, diagnosis, and therapy. Rev Obstet Gynecol. 2009;2(2):76–83. [PMC free article] [PubMed] [Google Scholar]
- 11.Nigro G, Mazzocco M, Mattia E, Di Renzo GC, Carta G, Anceschi MM. Role of the infections in recurrent spontaneous abortion. J Matern Fetal Neonatal Med. 2011;24(8):983–989. doi: 10.3109/14767058.2010.547963. [DOI] [PubMed] [Google Scholar]
- 12.Aali bibi S, Fasihi Harandi M, Nazari E, Salari Z. Comparison of Toxoplasma gondii IgG and IgM seropositivity between women with spontaneous abortions and ongoing pregnancies. Iranian Journal of Obstetrics, Gyneocology and Infertility. 2011;14(1):1–6. [Google Scholar]
- 13.Alsamarai AM, Hassan HMM, Alsalihi FG, Alobaidi AH, Aljumaili ZKM. Toxoplasma gondii, Rubella and Cytomegalovirus co-infections as risk factors for abnormal pregnancy outcomes. Middle East Journal of Family Medicine. 2014;12(3):16–23. [Google Scholar]
- 14.Aljumaili ZKM, Alsamarai AM, Najem WS. Seroprevalence of Herpes Simplex Virus Type 2 (HSV 2) in women with bad obstetric history. American Journal of Dermatology and Venereology. 2013;2(3):31–38. [Google Scholar]
- 15.Ramana B, Reddy BK, Murty D, Vasudevanaidu K. Seroprevalance of rubella in women with bad obstetric history. J Family Med Prim Care. 2013;2(1):44–46. doi: 10.4103/2249-4863.109943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turbadkar D, Mathur M, Rele M. Seroprevalence of torch infection in bad obstetric history. Indian J Med Microbiol. 2003;21(2):108–110. [PubMed] [Google Scholar]
- 17.Biase FH, Franco MM, Goulart LR, Antunes RC. Protocol for extraction of genomic DNA from swine solid tissues. Genet Mol Biol. 2002;25(3):313–315. [Google Scholar]
- 18.Bonin S, Groenen PJ, Halbwedl I, Popper HH. DNA extraction from formalin-fixed paraffin-embedded (FFPE) tissues. In: Stanta G, editor. Guidelines for molecular analysis in archive tissues. New York: Springer; 2011. pp. 33–36. [Google Scholar]
- 19.Abdoli A, Dalimi A, Soltanghoraee H, Ghaffarifar F. Molecular detection of Toxoplasma gondii in house sparrow (Passer domesticus) by LAMP and PCR methods in Tehran, Iran.J Parasitic Dis. J Parasitic Dis; 2015. (A head of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan A, Su C, German M, Storch G, Clifford D, Sibley LD. Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains. J Clin Microbiol. 2005;43(12):5881–5887. doi: 10.1128/JCM.43.12.5881-5887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137(01):1–11. doi: 10.1017/S0031182009991065. [DOI] [PubMed] [Google Scholar]
- 22.Grigg ME, Ganatra J, Boothroyd JC, Margolis TP. Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis. 2001;184(5):633–639. doi: 10.1086/322800. [DOI] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 24.Ghasemi FS, Rasti S, Bandehpour M, Kazemi B, Piroozmand A, Mousavi GA. Molecular diagnosis of Toxoplasma gondii in aborted women. Jundishapur J Microbiol. 2015;8(1):e15925–e15925. doi: 10.5812/jjm.15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asgari Q, Fekri M, Monabati A, Kalantary M, Mohammadpour I, Motazedian MH, et al. Molecular genotyping of Toxoplasma gondii in human spontaneous aborted fetuses in Shiraz, Southern Iran. Iran J Public Health. 2013;42(6):620–625. [PMC free article] [PubMed] [Google Scholar]
- 26.Hoveyda L, Shanehsazzadeh M, Behbahani M. Toxoplasmosis in Iranian abortion cases with appendectomy. J Applied Biol Sci. 2012;6(2):31–36. [Google Scholar]
- 27.Sibley LD, Khan A, Ajioka JW, Rosenthal BM. Genetic diversity of Toxoplasma gondii in animals and humans. Philos Trans R Soc Lond B Biol Sci. 2009;364(1530):2749–2761. doi: 10.1098/rstb.2009.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao J, Yolken RH. Strain hypothesis of Toxoplasma gondii infection on the outcome of human diseases. Acta Physiologica. 2015;213(4):828–845. doi: 10.1111/apha.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shwab EK, Zhu XQ, Majumdar D, Pena HF, Gennari SM, Dubey JP, et al. Geographical patterns of Toxoplasma gondii genetic diversity revealed by multilocus PCR-RFLP genotyping. Parasitology. 2014;141(4):453–461. doi: 10.1017/S0031182013001844. [DOI] [PubMed] [Google Scholar]
- 30.Zia-Ali N, Fazaeli A, Khoramizadeh M, Ajzenberg D, Dardé M, Keshavarz-Valian H. Isolation and molecular characterization of Toxoplasma gondii strains from different hosts in Iran. Parasitol Res. 2007;101(1):111–115. doi: 10.1007/s00436-007-0461-7. [DOI] [PubMed] [Google Scholar]
- 31.Saki J, Khademvatan S, Yousefi E, Tavalla M, Abdizadeh R. Detection and genotyping of Toxoplasma gondii isolated from soil in Ahvaz, southwest of Iran. J Parasit Dis. 2016 doi: 10.1007/s12639-016-0778-1. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavalla M, Oormazdi H, Akhlaghi L, Shojaee S, Razmjou E, Hadighi R, et al. Genotyping of Toxoplasma gondii isolates from soil samples in Tehran, Iran. Iran J Parasitol. 2013;8(2):227–233. [PMC free article] [PubMed] [Google Scholar]
- 33.Danehchin L, Razmi G, Naghibi A. Isolation and genotyping of toxoplasma gondii strains in ovine aborted fetuses in Khorasan Razavi Province, Iran. Korean J Parasitol. 2016;54(1):15–20. doi: 10.3347/kjp.2016.54.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Tantawy N, Taman A, Shalaby H. Toxoplasmosis and female infertility: is there a co-relation? American Journal of Epidemiology and Infectious Disease. 2014;2(1):29–32. [Google Scholar]
- 35.Malik A, Rizvi M, Khan F, Khan N, Rabbani T, Khan HM. Toxoplasma gondii in women with bad obstetric history and infertility: a five-year study. Asian Pac J Trop Dis. 2014;4(Suppl 1):S236–S239. [Google Scholar]
- 36.Aral AG, Elhan HA, Akarsu C. Retrospective evaluation of Toxoplasma gondii seropositivity in fertile and infertile women. Mikrobiyol Bul. 2011;45(1):174–180. [PubMed] [Google Scholar]
- 37.Flegr J, Prandota J, Sovičková M, Israili ZH. Toxoplasmosis-a global threat.Correlation of latent toxoplasmosis with specific disease burden in a set of 88 countries. PloS One. 2014;9(3):e90203–e90203. doi: 10.1371/journal.pone.0090203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaňková Š, Flegr J. Longer pregnancy and slower fetal development in women with latent" asymptomatic" toxoplasmosis. BMC Infect Dis. 2007;7:114–114. doi: 10.1186/1471-2334-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaňková S, Sulc J, Křivohlavá R, Kuběna A, Flegr J. Slower postnatal motor development in infants of mothers with latent toxoplasmosis during the first 18 months of life. Early Hum Dev. 2012;88(11):879–884. doi: 10.1016/j.earlhumdev.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Kaňková S, Flegr J, Calda P. The influence of latent toxoplasmosis on women’s reproductive function: four crosssectional studies.Folia Parasitol (Praha) Folia Parasitol (Praha); 2015. pp. 041–041. [DOI] [PubMed] [Google Scholar]