Abstract
The HIV-1 envelope protein (Env) has evolved to subvert the host immune system, hindering viral control by the host. The tryptophan metabolic enzyme kynureninase (KYNU) is mimicked by a portion of the HIV Env gp41 membrane proximal region (MPER) and is cross-reactive with the HIV broadly neutralizing antibody (bnAb) 2F5. Molecular mimicry of host proteins by pathogens can lead to autoimmune disease. Here, we demonstrate that neither the 2F5 bnAb nor HIV MPER-KYNU cross-reactive antibodies elicited by immunization with an MPER peptide-liposome vaccine in 2F5 bnAb VHDJH and VLJL knock-in mice and rhesus macaques modified KYNU activity or disrupted tissue tryptophan metabolism. Thus, molecular mimicry by HIV-1 Env that promotes the evasion of host anti-HIV-1 antibody responses can be directed towards non-functional host protein epitopes that do not impair host protein function. The 2F5 HIV Env gp41 region is therefore a key and safe target for HIV-1 vaccine development.
Introduction
HIV-1 infection remains a serious threat to health world-wide, and developing an effective HIV-1 vaccine is a global priority. A major challenge in achieving an effective vaccine is the inability of vaccines to induce broadly neutralizing antibodies (bnAbs) that recognize conserved epitopes on a majority of circulating HIV-1 strains (1-4). To date, bnAbs have not been elicited by vaccination, yet up to 50% of chronically individuals infected with HIV-1 develop bnAbs (5). Isolation of bnAbs from infected individuals has revealed six conserved sites of vulnerability on the HIV-1 envelope protein (Env): the gp120 CD4 binding site (CD4bs) (6-11); gp120 V1/V2 loop epitopes (12-14); gp120 V3-glycan epitopes (15-17); the gp41 membrane-proximal external region (MPER) (18-21), the gp41 fusion domain (22) and; the gp120-gp41 interface (23, 24). At high concentrations, passive administration of bnAbs that target these conserved epitopes (2F5, 4E10, 2G12, b12, VRC01, 3BNC117 or 10-1074) have prevented simian-human immunodeficiency virus (SHIV) infection in monkeys (25-29).
Molecular mimicry of antigenic determinants between pathogens and host molecules can result in tissue damage, such as the antibodies against a region of the hepatitis B virus that cross-react with myelin basic protein that can lead to tissue damage in the central nervous system (30-32). Similarly, Campylobacter jejuni lipo-oligosaccharides mimic host nervous system gangliosides and cause Guillain-Barre syndrome following infection (33, 34). Studies in mice have demonstrated that these host ganglioside antibodies are under tolerance control (33). Autoantibodies have also been shown to penetrate the blood brain barrier and affect intracellular enzyme function of neuronal cells during autoimmune disease (35). In retinopathy anti-enolase antibodies can alter the function of the enolase enzyme in neuronal cells (36). Furthermore, in multiple sclerosis (MS) anti-hnRNP antibodies were found to traffic into neurons and lead to apoptosis, contributing to MS pathology (37, 38).
HIV-1 bnAbs have unusual features such as extensive somatic hypermutation (SHM) and long third heavy chain complementarity regions (CDRH3s) that are associated with poly- or autoreactive B-cell antigen receptors (BCR)---all traits that can make antibodies subject to immune tolerance control (39-41). We previously demonstrated that many HIV-1 bnAbs are autoreactive and/or polyreactive and the MPER-reactive bnAb, 2F5, binds to the tryptophan metabolism enzyme kynureninase (KYNU) (42, 43). A second MPER bnAb, 4E10, cross-reacts with the RNA splicing factor 3b subunit 3 and also with anionic lipids. Because of the lipid reactivity, the 4E10 bnAb has anticoagulant activity and when administered to humans, it was biologically active and prolonged the partial thromboplastin time (44). Knock-in (KI) mice expressing the 2F5 or 4E10 bnAb VHDHJH and VLJL rearrangements exhibit profound deletion of immature B cells in the bone marrow, demonstrating first-tolerance checkpoint control of these autoreactive, HIV-1 bnAbs (45-50).
KYNU is a phylogenetically conserved enzyme of tryptophan metabolism that contains the 2F5 core epitope (ELDKWA) within the KYNU H4 domain; the 2F5 bnAb binds this site with nM affinity (42). A key goal of HIV vaccine design is to induce gp41 MPER 2F5-like antibodies, yet the effects of MPER antibodies on KYNU function are unknown. Interestingly, disregulated tryptophan metabolism has been implicated in AIDS-related dementia, thus it is of critical importance to determine if MPER antibodies that cross-react with KYNU disrupt tryptophan metabolism (51).
Here, we have determined the effect of 2F5 and 2F5-like anti-HIV gp41 Abs on KYNU enzymatic activity using an MPER peptide-liposome vaccine that activates anergic B cells in 2F5 bnAb double KI (VHDHJH + VLJL) mice. This vaccine also elicited MPER-specific Abs in rhesus macaques (50, 52). Thus, it is essential to determine if antibodies to the KYNU cross-reactive gp41 epitope inhibit KYNU or are deleterious in vivo. We demonstrate that immunization with MPER peptide-liposomes in 2F5 double KI mice and in rhesus macaques elicited KYNU-reactive, 2F5 epitope-targeted Abs, but that these Abs did not inhibit KYNU enzymatic activity in vitro nor perturb tryptophan metabolism or tissue pathology in vivo.
Materials and Methods
In vitro Kynureninase enzyme assay
Recombinant human kynureninase (rndsystems #4877-KH) and 10nM 3-hydroxy-DL-kynurenine (Sigma #H1771) substrate are incubated in assay buffer (50mM tris, 0.05% (w/v) Brij-35, 5μM pyridoxal phosphate, pH 8.0 in a F16 black maxisorp plate (Nunc, #475515). Fluorescence is read at excitation and emission wavelengths of 315nm and 415nm, respectively, in kinetic mode for 5 minutes by a fluorescent plate reader (SpectraMax Genini EM). Calibration standard 3-hydroxyanthranilinic acid (Sigma, #H9391) is used as a calibration control to calculate the conversion factor. Specific activity is calculated (Fig. S2) (42).
Recombinant antibodies are added at 0, 6, 60, 600, or 6000 ng and immune sera added at a 1:10 dilution to determine the inhibition or enhancement of KYNU enzyme activity compared with no inhibitor control.
Recombinant proteins and binding
Polyclonal anti-KYNU antibody was purchased (rndsystems #AF4887) and the V(D)J gene fragments of antibodies 2F5, CH65 were synthesized and cloned (GenScript) into plasmids containing human or rhesus IgG1/IgK/IgL constant regions. Recombinant mAbs were produced in 293 F cells (Life Technologies) by cotransfection with plasmids expressing the Ig heavy and light chain genes and were purified from the culture supernatant by protein A column chromatography (53, 54). Human KYNU and human KYNU with a single aspartic acid to glutamic acid mutation genes were synthesized with a C-terminal 6-histadine tag and cloned into pcDNA3.1 (GenScript). Plasmids were transfected into 293F cells and culture supernatants purified by nickel and size exclusion chromatography (55). Biotin-labeled SP62 (652QQEKNEQELLELDKWASLWN671) peptide and Biotin-labeled MPER656 (656NEQELLELDKWASLWNWFNITNWLWYIK683) peptide were synthesized (CPC scientific).
Recombinant and serum antibody binding was determined using ELISA as described (55, 56).
Mouse immunization
The mature 2F5 (m2F5 DKI) and germline 2F5 (gl2F5 DKI) double knock-in mice were generated on the C57BL/6 background previously described (46, 50). Mature 2F5 DKI mice were immunized six times with intraperitoneal injections (200μl) administered every 14 days in 3 groups of 4 mice each with the MPER peptide-liposome (25μg) formulated with GLA (0.625μg), Alhydrogel (Alum;12.5μg;QD565) or a combination. MPER peptide-liposomes contained a version of the 2F5 epitope-containing MPER peptide 656 (656NEQELLELDKWASLWNWNITNWLWIK683) that was synthesized with the C-terminal hydrophobic membrane anchor tag GTH1 (YKRWIILGLNKIVRMYS) previously described (57). All mice were 8 to 12 weeks old at the start of the immunization study and were housed in the Duke University Vivarium in a pathogen-free environment with 12-hour light/dark cycles at 20-25°C in accordance with all the Duke University Institutional Animal Care and Use Committee (IACUC)-approved animal protocols.
Mouse tissue processing, chemistry and pathology
Serum was collected and analyzed 10 days post immunization and animals were necropsied after completion of the study. Two groups of 6 (12 total) non-immunized aged-matched B6 control mice and unimmunized 2F5 mature (5 mice) and 2F5 germline (6 mice) double knock-in mice were necropsied as unimmunized controls.
Serum and brain tryptophan, kynurenine and kynurenic acid levels were performed on immunized and naïve mice as previously described (58).
Prepared stained tissue sections were delivered to a board certified veterinary pathologist for assessment at Duke University Department of Pathology. The tissues were fixed in 10% neutral buffered formalin, paraffin embedded, cut on microtome at 5 microns, and stained with hematoxylin and eosin.
Rhesus macaque immunization and pathology
23 healthy, adult Chinese-origin rhesus macaques were housed at BIOQUAL Inc (Rockville, MD) in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. The protocol was approved by BIOQUAL's Institutional Animal care and Use Committee under OLAW Assurance Number A-3086-01. BIOQUAL is IAAALAC accredited. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH) and with the recommendations of the Weatherall report; “The use of non-human primates in research”. All procedures were performed under anesthesia using ketamine hydrochloride, and all efforts were made to minimize stress, improve housing conditions, and to provide enrichment opportunities (e.g., social housing when possible, objects to manipulate in cage, varied food supplements, foraging and task-oriented feeding methods, interaction with caregivers and research staff). Animals were euthanized by sodium pentobarbital injection in accordance with the recommendations of the panel on Euthanasia of the American Veterinary Medical Association.
Macaques were immunized (250μl × 2 sites) with MPER peptide-liposomes formulated with GLA (8 animals), GLA + Alum (8 animals) or Alum alone (7 animals) 6 times in 6 week intervals. Blood was collected preimmunization and 2 weeks after each immunization. Animals were necropsied at the completion of the study animals.
Throughout the immunization study all animals were clinically monitored. That included Complete Blood Count (CBC), serum chemistry and hematology that were performed by the attending veterinarian using commercial automated hematology and serum chemistry analyzers (BIOQUAL Inc.). Prepared stained splenic tissue sections were delivered to a board certified veterinary pathologist for assessment at Duke University Department of Pathology. The tissues were fixed in 10% neutral buffered formalin, paraffin embedded, cut on microtome at 5 microns, and stained with hematoxylin and eosin.
Isolation and characterization of MPER-reactive antibodies
MPER-specific memory B cells of a macaque immunized 3× with the MPER peptide-liposome vaccine formulated in GLA + Alum were sorted by flow cytometry as described (21, 50). Briefly, ∼ 1 × 107 PBMCs were decorated with B cell antibody panel: CD14 (BV570), CD3 (PerCPCy5.5), CD20 (FITC), CD27 (APC-Cy7), and IgD (PE) (BD Biosciences) and Alexa Fluor 647 and Brilliant Violet 421–tagged MPER.03 peptides (KKKNEQELLELDKWASLWNWFDITNWLWYIRKKK). HIV gp41-specific memory B cells were gated as CD3−CD14−CD20+CD27+sIgD−MPER.03 (AF647)+ MPER.03 (BV421)+ and sorted into 96-well PCR AQ22 plates containing 20 μl of reverse transcription reaction buffer that included 5 μl of 5× first-strand complementary DNA (cDNA) buffer, 1.25 μl of dithiothreitol, 0.5 μl of RNaseOUT (Life Technologies), 0.0625 μl of Igepal (Sigma-Aldrich), and 13.25 μl of ultrapure distilled water (Life Technologies).
Rhesus macaque VHDJH and VLJL segments were isolated by single-cell reverse transcription PCR (RT-PCR) using the method as described (54). The isolated V(D)J gene fragments were used for the construction of linear expression cassettes for production of recombinant mAbs in 293T cells for small scale ELISA screening (59).
Cloanalyst (Cloanalyst. http://www.bu.edu/computationalimmunology/research/software/) was used to annotate isolated VHDJH and VLJL sequences with immunogenetic information and to test for clonal lineage membership (50, 54).
Select V(D)J gene fragments were synthesized, expressed and purified as rhesus IgG1 recombinant mAbs described above.
Rhesus macaque serum and recombinant mAbs were screened by ELISA for binding to SP62 and select SP62 alanine mutants, recombinant KYNU and KYNU Mut (55).
Polyreactivity analysis of antibodies
The polyreactivity of rhesus mAbs was assessed with the AtheNA Multi-Lyte System (ZEUS Scientific) and HEp-2 cells immunofluorescence assay (Inverness Medical Professional Diagnostics) as described (50, 60).
Statistical analysis
Statistical analysis was performed in SAS version 9.4 (SAS Institute). All statistical tests performed were Wilcoxon-Mann-Whitney tests with Benjamini-Hochberg false discovery rate correction for multiple testing. GraphPad Prism version 6.01 was used for graphical representation.
Results
The 2F5 bnAb does not inhibit KYNU enzymatic activity
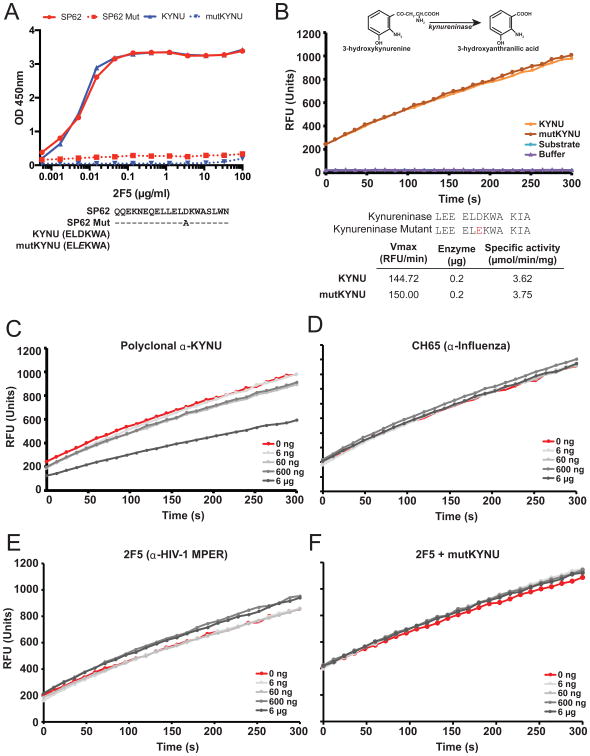
We previously identified that KYNU cross-reacted with the HIV-1 gp41 membrane-proximal targeting bnAb 2F5 (42). 2F5 binds to the HIV-1 gp41 peptide (652QQEKNEQELLELDKWASLWN671), but does not bind this peptide when position 664 is mutated within the 2F5 epitope (652QQEKNEQELLELAKWASLWN671; Fig. 1A). The KYNU H4 domain in most mammals (ELDKWA) exactly replicates this MPER epitope, and this motif is conserved within humans, mice and rhesus monkeys (42). Opossums, however, carry a rare substitution in the KYNU H4 ELDKWA motif where aspartic acid (D) is replaced by glutamic acid (E) (42). The same replacement in human recombinant KYNU (mutKYNU) completely abrogates binding by the 2F5 bNAb (Fig. 1A).
Figure 1. 2F5 does not inhibit KYNU enzymatic activity.
(A) ELISA binding of 2F5 to the HIV-1 gp41 peptide SP62 that includes the 2F5 epitope (red), SP62 Mut (red dashed), KYNU (blue) and mutKYNU (blue dashed). (B) In vitro KYNU enzymatic assay measuring levels of 3-hydroxyanthranilic acid being converted by kynureninase from 3-hydroxykynurenine measured as RFU over time with the addition of KYNU, mutKYNU, substrate alone and buffer alone. Sequences of KYNU and mutKYNU that were tested in the in vitro KYNU enzymatic assay. Vmax and specific activity of wild-type KYNU and mutKYNU in the in vitro KYNU enzymatic assay using 0.2μg of each enzyme (C-F) KYNU enzymatic assay by adding increasing amounts of (C) polyclonal α-KYNU antibody (D) CH65 antibody (E) 2F5 antibody or (F) 2F5 antibody with mutKYNU enzyme.
KYNU catabolizes 3-hydroxy-kynurenine into 3-hydroxyanthranilic acid. We used a standard in vitro enzymatic assay to measure the production of 3-hydroxyanthranilic acid at saturating substrate concentrations (42) (Fig S1A). Addition of recombinant KYNU or mutKYNU resulted in identically increased rates of 3-hydroxyanthranilic acid production, whereas addition of substrate alone or assay buffer did not (Fig. 1B). As a control for Ab-mediated enzyme inhibition, we added increasing concentrations of polyclonal anti-KYNU IgG Ab and demonstrated a concentration-dependent inhibition of 3-hydroxyanthranilic acid production (Fig. 1C). Increasing amounts of the CH65 influenza IgG Ab had no detectable effect on KYNU/mutKYNU enzyme activity (Fig. 1D & S1E).Both KYNU and mutKYNU were identically inhibited by KYNU IgG and neither were affected by the CH65 rIgG (Fig. S1B-E).
In contrast, at no concentration did the addition of 2F5 bNAb inhibit KYNU or mutKYNU enzymatic activity (Fig. 1E & 1F). Likewise, the 2F5 bnAb variable region inserted into a rhesus macaque IgG1 Fc backbone showed no activity on KYNU or mutKYNU enzymatic activity (Fig. S1D).
Vaccine-elicited gp41 bnAbs in mutated 2F5 knock-in mice do not inhibit KYNU
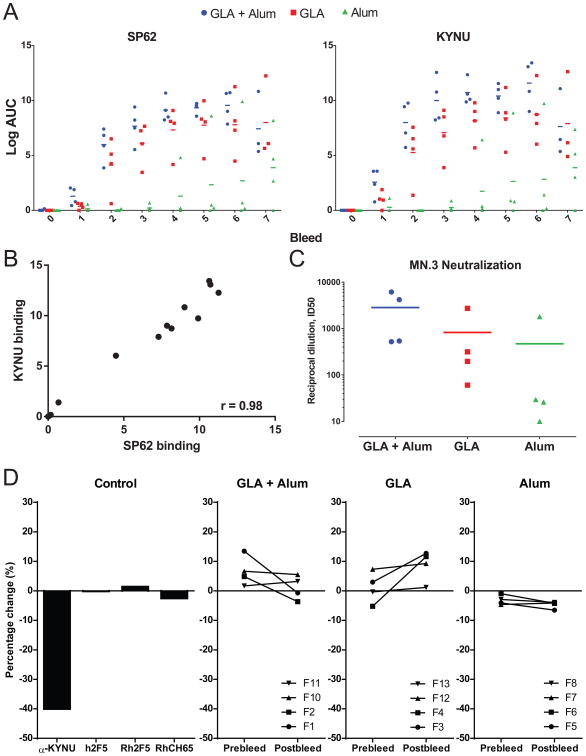
We immunized mice that express only the mutated (m)2F5 bnAb VH +VL termed m2F5 double KI mice with MPER peptide-liposomes formulated with alum, glucopyranosyl lipid adjuvant (GLA;TLR4 agonist), or a combination of alum and GLA to determine which adjuvant elicited the highest titers of 2F5 bnAbs. After two immunizations, GLA and GLA + alum groups had induced antibodies that targeted the HIV gp41 MPER peptide (652QQEKNEQELLELDKWASLWN671) with ELISA Log AUC values of 4 and 6, respectively, while no animal in the alum alone group had titers of MPER-binding mAbs (Fig. 2A). Similarly, after two immunizations, GLA and GLA + alum immunized mice had titers of anti-KYNU antibodies (Fig. 2A). Ab to MPER peptide and KYNU protein was boosted with subsequent immunizations for the three adjuvant groups, but GLA or GLA + alum immunized mice maintained higher antibody titers compared to alum immunized mice (Fig. 2A). Ab to MPER and KYNU were strongly correlated with one another in all adjuvant groups demonstrating the relatedness of the two responses (Fig. 2B). At bleed 6, plasma antibody neutralization was observed against the HIV virus MN in the TZM-bl neutralization assay (Fig. 2C). The magnitude of neutralization titers corresponded with the HIV binding antibody titers with the GLA+Alum and GLA adjuvant groups having the highest neutralization titers, while only 1 animal in the Alum immunized group had ID50 titers greater than 50 (Fig. 2C).
Figure 2. Immunization of m2F5 DKI mice with MPER peptide-liposomes.
(A) Plasma antibody binding of SP62 and KYNU at pre-immunization (bleed 0) and after 7 immunizations (bleeds 1-7) with MPER peptide-liposomes formulated with Alum, GLA or GLA + Alum adjuvants measured by ELISA. Binding for each individual animal displayed, bar indicates group mean. (B) Plot of plasma antibody binding titers to SP62 and KYNU for each mouse in all adjuvant groups at bleed 6. Axis are Log AUC in ELISA. Spearman correlation shown (r = 0.98; P < 0.0001). (C) Plasma antibody HIV neutralization of MN virus measured in the TZM-bl neutralization assay at bleed 6 for mice immunized with MPER peptide-liposomes formulated with Alum, GLA or GLA + Alum adjuvants. (D) KYNU enzymatic assay indicating the percent change in KYNU activity for control antibodies (left panel; h = human antibody, Rh = rhesus macaque antibody) and mice from all three adjuvant groups pre and post 5th immunization (right three panels). Binding for each individual mouse displayed.
We then examined the effects of plasma IgG Ab from immunized m2F5 double KI mice on KYNU enzymatic activity. No significant inhibition of KYNU enzymatic activity was observed for any of the three immunization groups, including the GLA and GLA + alum groups with the highest titers of vaccine-elicited KYNU IgG Abs (Fig. 2D). Indeed, there was no significant difference in the inhibitory activity between pre- and post-immunization plasma IgG samples. Thus, vaccine-elicited 2F5 bnAbs from m2F5 double KI mice had no effect on KYNU activity.
Induction of KYNU Ab has no deleterious effects on m2F5 double KI mice
Although we did not detect any inhibition of KYNU activity by the 2F5 bNAb or plasma IgG in immunized m2F5 double KI mice (Fig. 1E & 2D), we recovered tissue samples from naïve B/6 control mice, 2F5 double KI germline mice (gl2F5) that expressed the 2F5 unmutated ancestor (UA), m2F5 double KI mature mice and from immunized m2F5 double KI mature mice, and compared brain and serum levels of kynurenic acid, kynurenine, and tryptophan, the products of tryptophan metabolism upstream of KYNU (Fig. 3A). Ab that inhibited KYNU activity in vivo would result in increased levels of these metabolites. No significant differences in kynurenic acid, kynurenine or tryptophan levels in the brains of naïve or immunized mice was observed (Fig. 3B & S2A). No significant differences were found in serum kynurenic acid levels, but slightly higher levels of kynurenine and tryptophan were present in mice immunized with all forms of MPER peptide-liposome versus control B/6 mice (Fig. 3C & S2A). The elevations of kynurenine and tryptophan in immunized mice were not specifically caused by elicited KYNU Abs, since the alum-formulated liposome-immunized group with the lowest levels of KYNU Ab titers among the three immunization groups exhibited levels of serum kynurenine and tryptophan that were indistinguishable from those animals receiving the more potent GLA and GLA + alum immunogens (Fig. 3C).
Figure 3. Tryptophan metabolites and spleen histology of 2F5 VHDHJH + VLJL knock-in mice.
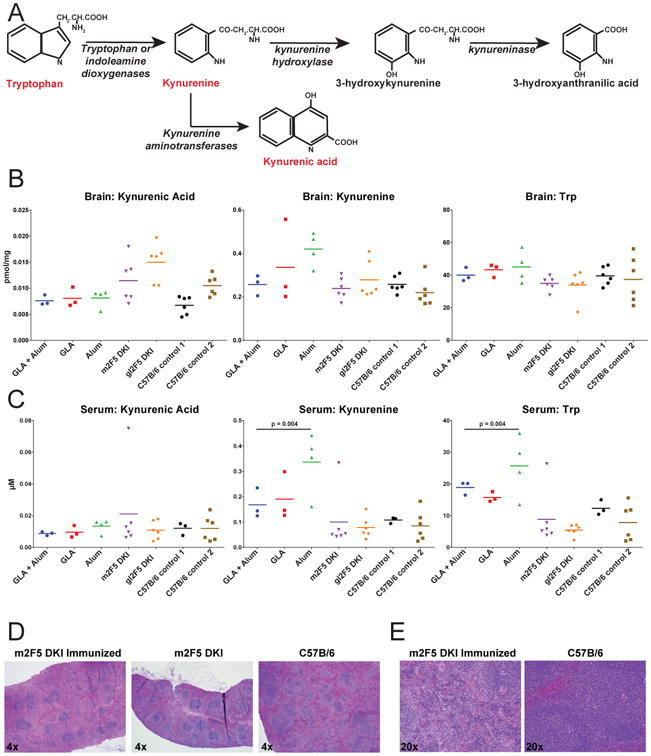
(A) The first 3 steps of tryptophan metabolic pathway with monitored metabolites highlighted in red. (B-C) Tryptophan metabolite levels in (B) brain and (C) serum of MPER peptide-liposome-immunized m2F5 DKI mice formulated with GLA + Alum, GLA or Alum, unimmunized 2F5 DKI mice with the mutated (m2F5 DKI) and germline 2F5 knocked-in (gl2F5 DKI) and two groups of C57B/6 control mice (P-values determined by Wilcoxon-Mann-Whitney). Levels of individual animals graphed. Line graphed at group mean. (D) HE stains at 4× magnification of spleens from m2F5 DKI MPER peptide-liposome-immunized mice, unimmunized m2F5 DKI mice and control C57B/6 mice. (E) HE stains at 20× magnification of spleens from m2F5 DKI immunized and C57B/6 mice.
Histopathology assessment of spleen tissue sections from m2F5 double KI mice were performed. Consistent with the deletional tolerance characteristic of m2F5 double KI mice (46), both naïve and immunized KI mice exhibited comparably low numbers of splenic B lymphocytes when compared to B/6 controls resulting in small spleens and diminished white pulp areas (Fig. 3D). Furthermore, spleens were analyzed for vaccine-related lesions and no pathologic abnormalities to marginal zone, periarteriolar lymphoid sheath areas or red pulp were observed in vaccinated animals with high titers of KYNU cross-reactive antibodies (Fig. 3D & 3E). Immunized germline (gl) VH + VL 2F5 double KI mice without anti-KYNU antibodies also exhibited B cell deletion and tolerance control (50) and our analysis revealed that similar to the m2F5 KI mice, gl2F5 KI mice also had small spleens that were similarly lymphodepleted (Fig. S2B). Thus, the lymphodepletion seen in m2F5 and gl2F5 KI mice was due to deletion in bone marrow of greater than 95% of B cells due to immune tolerance.
We also performed histopathology of the brains from m2F5 double KI, gl2F5 double KI and B/6 control mice, and there was no evidence of neural degeneration, demyelination or inflammation in any of the brain sections studied (Fig. S2C).We concluded that despite the induced 2F5 bnAb levels and high titers of KYNU binding IgG Ab resulting from induced 2F5 bnAb levels elicited by immunizing m2F5 double KI mice with MPER peptide-liposome, did not perturb tryptophan metabolism in the brain or serum and induced no pathological changes in peripheral lymphoid tissues (Fig. 3).
Adjuvanted MPER peptide-liposome vaccine-elicited, non-pathogenic 2F5-epitope targeting, KYNU-reactive antibodies in rhesus macaques
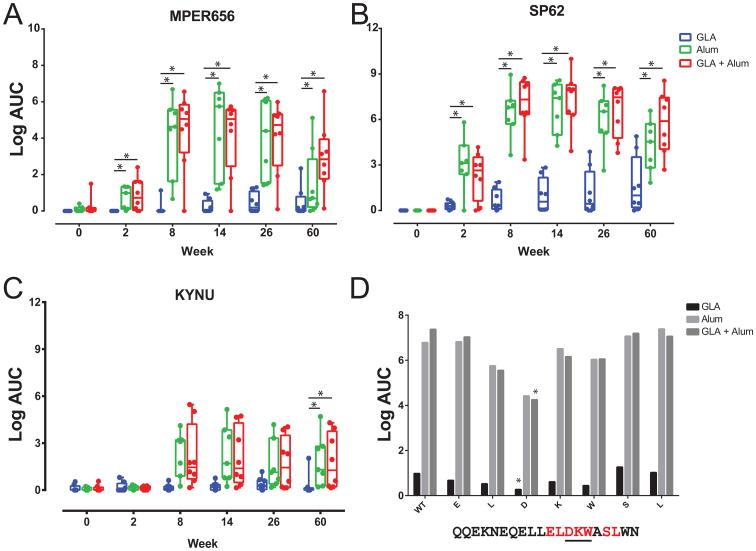
Three groups of rhesus macaques (RMs) were immunized six times with MPER peptide-liposomes formulated with GLA, alum, or GLA + alum. In contrast to immunization of 2F5 knock-in mice, GLA alone was poorly immunogenic in RMs and elicited the lowest MPER IgG plasma Ab (Fig. 4A). In contrast, MPER liposomes formulated in alum or GLA + alum elicited higher titers of MPER plasma Ab (Fig. 4A & 4B). Immunization with alum or GLA + alum MPER peptide-liposomes also elicited higher titers of cross-reactive KYNU antibodies in macaques when compared to the GLA alone group (Fig. 4C). The 2F5 core epitope is the DKW motif within the HIV-gp41 MPER peptide SP62 (652QQEKNEQELLELDKWASLWN671), and plasma antibody binding to SP62 mutated peptides within the D, K or W was reduced for all three groups, and significantly reduced for the SP62 D664A mutant in the GLA and GLA + Alum groups (Fig. 4D; P < 0.05; Wilcoxon-Mann-Whitney Test). Thus, MPER peptide-liposome-induced plasma antibody was targeted near the 2F5 bnAb epitope in all RMs, with alum alone or GLA + alum adjuvants eliciting the highest titers of MPER and KYNU-reactive antibodies.
Figure 4. Adjuvanted MPER-peptide-liposome-immunized rhesus macaques elicits KYNU-reactive antibodies.
(A-C) Plasma antibody levels measured by ELISA targeting (A) MPER656, (B) SP62 and (C) KYNU analyzed after immunization with MPER peptide-liposomes formulated with GLA, Alum and GLA + Alum adjuvants in rhesus macaques (*P < 0.05; Wilcoxon-Mann-Whitney). Animals were immunized at weeks 0, 6, 12, 18, 24, and 58. (D) Average plasma antibody binding at week 60 to wild-type SP62 and alanine mutated versions (red; core 2F5 epitope underlined) for all vaccine groups (*P < 0.05; Wilcoxon-Mann-Whitney).
In all three adjuvant groups, the KYNU RM plasma IgG had no effect on KYNU enzyme activity in vitro (Fig. 5A). We analyzed immunized macaque spleen histopathology and found it was normal in all groups (Fig. 5B). These data confirmed that the splenic lymphopenia of naïve gl2F5 and vaccinated m2F5 double KI mice is the consequence of B cell deletion due to immune tolerance and not a pathologic consequence of anti-KYNU antibody production.
Figure 5. Adjuvanted MPER peptide-liposome-immunized rhesus macaques do not inhibit KYNU or display tissue abnormalities.
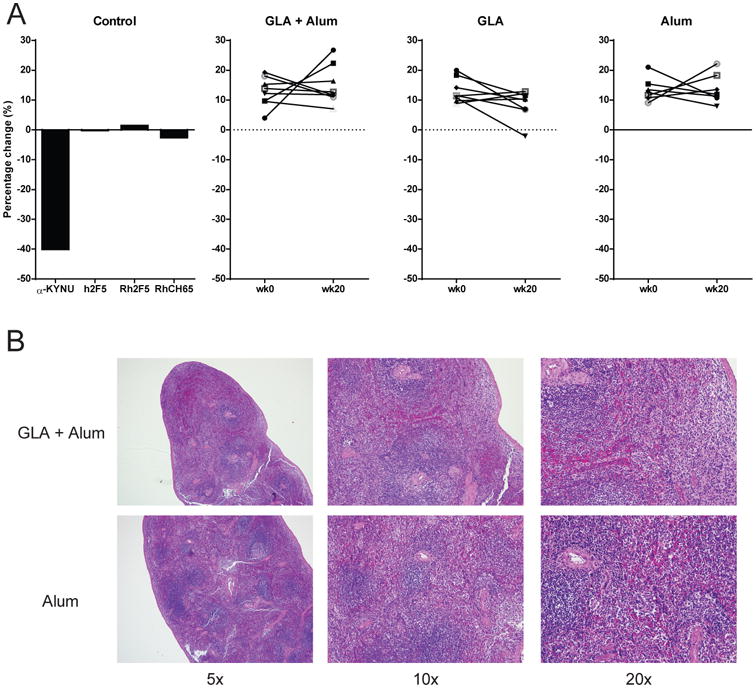
(A) Percent inhibition of KYNU enzymatic activity measured by the in vitro KYNU enzyme assay for positive and negative control antibodies (left panel) and for plasma antibodies from rhesus macaques measured pre and post immunization with MPER peptide-liposomes formulated with GLA + Alum, GLA or Alum (right 3 panels). (B) HE stained spleens from MPER peptide-liposome formulated with GLA + Alum or Alum alone immunized macaques at 5×, 10× and 20× magnification.
RMs blood levels for electrolytes, glucose, renal and liver function and complete blood counts denoted no changes related to MPER peptide-liposome vaccination (Fig. S3A & S3B).
Vaccine-elicited MPER recombinant MAbs in RMs targeted the 2F5 epitope
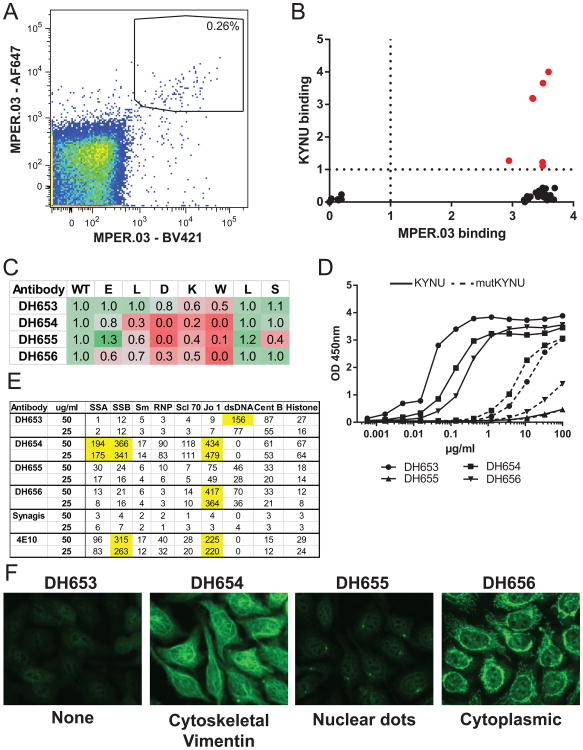
Gp41 Env-reactive Memory B cells (CD3−CD14−CD20+CD27+sIgD−) from PBMCs from a RM immunized with MPER-liposome formulated in GLA and alum that were decorated with MPER peptide tetramers (MPER.03) labeled with two fluorophores were sorted for sequencing of the B cell receptor (BCR VH + VL) genes. MPER-reactive memory B cells represented 0.26% of total memory B cells (Fig. 6A). BCR genes were RT-PCR amplified, sequenced and fused to cassettes containing a CMV promoter, Ig constant region and polyA tail by overlapping PCR. Heavy and light chain amplicon pairs were transiently transfected into 293F cells and supernatants containing recombinant antibody were screened for binding by ELISA. Using this approach, 31 MPER-reactive antibodies were identified that bound clade B MN gp41 recombinant protein, 27 bound recombinant HIV-1 gp140 Env (Con-S or B. JRFL gp140; Fig S4A), and 6 MPER-reactive rAbs that in addition to the MPER, also bound human KYNU (Fig. 6B).
Figure 6. Isolation of MPER-targeting KYNU cross-reactive antibodies from MPER peptide-liposome-immunized rhesus macaques.
(A) Memory B cells decorated with an MPER tetramer (MPER.03) conjugated with two fluorophores (AF647 and BV421) from PBMCs of an MPER peptide-liposome-immunized rhesus macaque 2-weeks after the third immunization. Single-cells were sorted into 96-plates for immunoglobulin gene amplification and sequencing shown within the black sort gate. (B) Individual wells with positive immunoglobulin sequences were amplified and transiently transfected for small-scale production of recombinant antibody and tested for MPER and KYNU binding by ELISA (O.D. at 450nm shown on X and Y axis). (C) Epitope mapping of MPER and KYNU reactive antibodies on the wild-type (WT) HIV-1 SP62 peptide and alanine mutants in the core 2F5 epitope (ELDKW) and 2 amino acids outside the 2F5 core epitope (LS). Values are ratio of binding of alanine mutants compared to WT. Red shading indicates decrease in binding. (D) Isolated antibody binding to KYNU (solid) and mutKYNU (dashed) measured by ELISA. (E) Autoreactivity of MPER antibodies isolated from rhesus macaques in the AtheNA autoantibody assay. Values >150 are positive and highlighted in yellow. Synagis and the MPER bnAb 4E10 were used as negative and positive controls, respectively. (F) Hep-C IFA staining of antibodies targeting the HIV-1 MPER and cross-reactive with KYNU. Antibody concentration was 50 μg/ml and exposed time was 5 seconds. Original magnification 40×, representative section displayed. Cellular staining pattern for each antibody shown below.
VDJ sequence analysis revealed that the isolated MPER antibodies used diverse VH gene segments, and 3 of the 6 KYNU-cross-reactive Abs used a VH gene segment orthologous to the human VH2-5 gene used in the 2F5 bNAb (Table 1; Fig. S4B) (61). The remaining KYNU binders carried rearrangements of VH4 family gene segments (Table 1; Fig. S4B).
Table I. Immunogenetic characteristics of KYNU cross-reactive antibodies. DH653, DH654, DH655 and DH656 studied as purified mAbs. DH656.2 and DH673 identified by transient transfection.
| Antibody | VH (human) | HCDR3 (AA) | Mut. Freq (%) | VL (human) | LCDR3 (AA) | Mut. Freq (%) |
|---|---|---|---|---|---|---|
| DH653.2 | 2-A (2-5) | 12 | 1.1 | κ2-S10 (2-28) | 9 | 8.7 |
| DH653 | 2-A (2-5) | 12 | 3.4 | κ2-S10 (2-28) | 9 | 7.5 |
| DH656 | 2-A (2-5) | 20 | 2.7 | λ3-B (3-19) | 11 | 2.7 |
| DH655 | 4-H (4-39) | 14 | 4.9 | λ1-F (1-36) | 11 | 1.1 |
| DH654 | 4-L (4-4) | 16 | 4.9 | λ3-B (3-19) | 11 | 2.7 |
| DH673 | 4-L (4-4) | 18 | 2.6 | λ3-B (3-19) | 11 | 4.6 |
The 31 VDJ rearrangements recovered from memory B cells that bound the MPER.03 tetramer exhibited varying levels of somatic hypermutation (SHM) and diverged from the germline RM VH gene segments by 0.38% to 11.8% ; rearrangements from KYNU-reactive Abs exhibited a similar range of mutation frequencies, 1.1% to 5.2% (Table 1; Fig. S4C). Most MPER bnAbs have long third heavy chain complementarity regions (CDRH3) that are greater than 15 amino acids, and the RM antibodies had CDRH3 lengths ranging from 12 to as long as 21 amino acids (Table 1; Fig. S4D).
We selected four mAbs, based on gp41 and KYNU cross-reactivity, including 2 that used the RM VH2-5 gene segment orthologue, for further study. All 4 mAbs bound to HIV-1 gp41 peptide (SP62; 652QQEKNEQELLELDKWASLWN671) and 3 of the 4 mAbs had greater than 60% reduction in binding to peptides containing alanine substitutions within the 2F5 bnAb DKW epitope (Fig. 6C). The mAb DH563 showed a more moderate 20-50% reduction on the DKW alanine mutants (Fig. 6C). Similarly, all 4 antibodies bound to KYNU, and had reduced binding to mutKYNU that contains ELEKWA instead wild-type KYNU, ELDKWA (Fig. 6D). None of the mAbs were capable of neutralizing HIV-1 in the TZM-bl pseudovirus inhibition assay (data not shown).
In addition to KYNU-cross-reactivity, 3 of the 4 antibodies also were cross-reactive with other host proteins associated with autoimmune disease; DH653 reacted with dsDNA, DH654 reacted with SSA, SSB and Jo1 proteins and DH656 reacted with Jo1 protein (Fig. 6E). Three of the four KYNU reactive Abs also exhibited antinuclear antibody phenotypes by reacting with nuclear antigens in HEp-2 cells; DH654 decorated cytoskeletal proteins, DH655 bound to nuclear punctae (dots), and DH656 diffusely bound within the cytoplasm (Fig. 6F). Even though these KYNU binding Abs exhibited reactivity for HEp-2 cell components, none of them were capable of inhibiting KYNU enzymatic activity in vitro (Fig. 7). Thus, adjuvanted MPER-liposome immunogens were capable of inducing a polyclonal and polyreactive IgG Ab responses to MPER epitopes in RMs, but whereas these responses included anti-KYNU antibodies, they did not result in tissue pathology nor did they inhibit KYNU enzyme function.
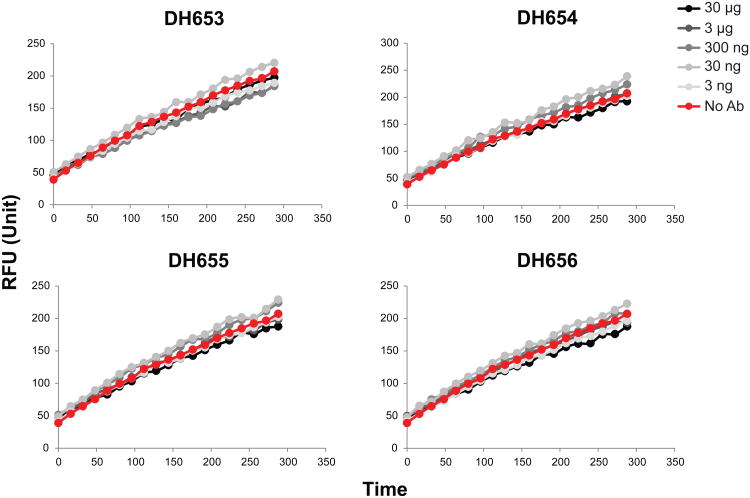
Figure 7. MPER and KYNU cross-reactive antibodies from MPER peptide-liposome immunized rhesus macaques do not inhibit KYNU enzymatic activity.
KYNU enzyme activity over time with no Ab and increasing concentration of mAbs isolated from MPER peptide-liposome immunized rhesus macaques that cross-reacted with KYNU.
Discussion
Mimicry of host proteins by microbial pathogens can subvert the host immune response (30-34, 62, 63). In this study, we have demonstrated that the HIV-1 bnAb 2F5 had no inhibitory effect on KYNU enzyme activity. Moreover, a single-residue change within the 2F5 epitope in KYNU that abolished 2F5 binding also did not effect KYNU enzyme function. The 2F5 epitope (ELDKWA) within KYNU is in the H4 domain that mediates KYNU homodimerization (64), likely explaining the inability of anti-ELDKWA antibodies to be able to inhibit KYNU activity.
B cell development in 2F5 VHDJH and VLJL knock-in mice was blocked by clonal deletion in bone marrow, because of host cross-reactivity with both lipids and KYNU resulting in bnAb immune tolerance control (46, 50). MPER bnAbs 2F5, 4E10 and 10E8 all interact with the virion membrane as a part of their epitopes (47, 65, 66). We have developed a minimal peptide-liposome immunogen that has been designed to present MPER bnAb epitopes in the same manner as on an HIV-1 virion (57). Immunization of 2F5 knock-in mice with the MPER peptide-liposome immunogen rescued anergic B cells that escaped deletion to produce high-levels of bnAbs (46). Immunization of macaques showed that the MPER peptide-liposome can initiate antibodies with 2F5-like characteristics (50).
In this study, bnAb 2F5 knock-in mouse plasma antibodies recognized KYNU, but like the human 2F5 monoclonal antibody, did not inhibit KYNU activity. Moreover, we examined brain and serum levels of tryptophan and its metabolites after MPER peptide-liposome immunization in vivo and did not detect any tryptophan pathway metabolite perturbations.
Immunization of rhesus macaques with MPER peptide-liposomes formulated in Alum or GLA + Alum elicited high-titers of antibodies that co-recognized the 2F5 epitope and KYNU. In macaques, plasma antibody inhibition of KYNU was not observed and no significant pathology was observed in macaque spleen or blood hematology or chemistry tests. Isolation of monoclonal antibodies by antigen-specific single-cell flow cytometry revealed that the 2F5-epitope (ELDKWA) in HIV-1 gp41 and KYNU could be recognized by antibodies that used diverse immunoglobulin gene segments, and these mAbs did not inhibit KYNU activity. We did not detect plasma HIV-1 neutralization in the TZM-bl assay for any of the vaccine-induced MPER-KYNU cross-reactive antibodies, indicating that further immunizations with affinity maturation, selecting antibodies with recognition of lipid, will be required to achieve neutralization activity (50).
In summary, host mimicry is a strategy utilized by HIV-1 to escape antibody responses to functionally conserved bnAb epitopes. Efforts to induce bnAbs by vaccination and to use bnAbs as therapeutic antibodies are critical to control of the AIDS epidemic. Thus, determining the impact of potentially protective antibodies on host protein function is an important safety consideration. Our study demonstrates that the 2F5 bnAb and vaccine-elicited 2F5-epitope targeted antibodies elicited in mice and macaques that cross-reacted with the host enzyme KYNU, did not inhibit enzymatic function in vitro nor cause tissue or enzyme activity abnormalities in vivo. While cross-reactivity with host antigens may impede bnAb development and impact therapeutic antibody pharmacokinetics, the 2F5-KYNU interaction does not impair host enzyme function or mediate adverse events. Thus, the 2F5 HIV-1 Env gp41 region therefore appears to be a safe target on HIV-1 Env for vaccine development.
Supplementary Material
Acknowledgments
We would like to acknowledge Dawn J. Marshall, John F. Whitesides, Joshua Eudailey, Tarra A. Von Holle, Thaddeus C. Gurley, Lawrence C. Armand, Andrew Foulger, Giovanna Hernandez, Jamie Pritchett, Krissey Lloyd and Christina Stolarchuk for expert technical assistance. We also would like to acknowledge Kelly Soderberg and Samantha Bowen for project coordination and management.
This work was supported by the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID; UMI-AI100645) grant from NIH/NIAID/DAIDS, and a Collaboration for AIDS Vaccine Discovery Grant from the Bill and Melinda Gates Foundation.
Abbreviations used in this article
- KYNU
kynureninase
- MPER
membrane-proximal external region
- bnAb
broadly neutralizing antibody
- SHM
somatic hypermutation
- VH
variable heavy chain
- VL
variable light chain
- KI
knock-in
- UA
Unmutated ancestor
Footnotes
GenBank accession numbers : KX914888-KX914899 (http://www.ncbi.nlm.nih.gov/geo/)
Author Contributions: T.B isolated and characterized antibodies, designed assays, analyzed and interpreted data and wrote and edited the manuscript. G.Y., T.M.H. and G.K. designed and analyzed KYNU in vitro enzyme assay, interpreted data and edited the manuscript. O.I. and C.B.N. performed tryptophan metabolite analysis. R.Z. and H.X.L. designed and assisted with rhesus macaque antibody gene sequencing. J.Z., H.B. C.M.B. and L.V. provided 2F5 knock-in mice and carried out immunization and tissue collection. S.S. L.L.S., R.M.S. assisted with rhesus macaque immunizations and sample collection. S.M.A., C.B.F and S.G.R. provided MPER peptide-liposome immunogen. R.P. performed immunoassays. N.V. performed statistical analysis. T.B.K designed software and performed analysis of antibody sequences. M.A.M. produced fluorophor-labeled MPER proteins and oversaw flow cytometry experiments. R.M. and J.I.E. performed histopathology analysis. B.F.H. designed the study, oversaw all experiments, analyzed all data and wrote and edited the manuscript.
References
- 1.Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annual review of immunology. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Taeye SW, Moore JP, Sanders RW. HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends in immunology. 2016;37:221–232. doi: 10.1016/j.it.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF. New approaches to HIV vaccine development. Current opinion in immunology. 2015;35:39–47. doi: 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, Mascola JR, Nabel GJ. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harbor perspectives in medicine. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. Aids. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang KK, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, N. C. S. Program. Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao HX, Kwong PD, Haynes BF. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 8.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, N. C. S. Program. Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, N. C. S. Program. Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, N. C. S. Program. Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. Journal of virology. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, N. C. S. Program. Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol GPI, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang CH, Ramos A, Bian CB, Wickramasinghe L, Kong L, Eren K, Wu CY, Wong CH, I. P. C. Investigators, I. A. H. I. V. R. N. The. Kosakovsky Pond SL, Wilson IA, Burton DR, Poignard P. Early Antibody Lineage Diversification and Independent Limb Maturation Lead to Broad HIV-1 Neutralization Targeting the Env High-Mannose Patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, Sarkar A, Liang CH, Shivatare SS, Wu CY, Schief WR, Wong CH, Wilson IA, Ward AB, Zhu J, Poignard P, Burton DR. A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans. Immunity. 2016;45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. Journal of virology. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, Donathan M, Bilska M, Gray ES, Abdool Karim SS, Kepler TB, Whitesides J, Montefiori D, Moody MA, Liao HX, Haynes BF. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PloS one. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, Cale EM, Chen L, Choi CW, Chuang GY, Doria-Rose NA, Druz A, Georgiev IS, Gorman J, Huang J, Joyce MG, Louder MK, Ma X, McKee K, O'Dell S, Pancera M, Yang Y, Blanchard SC, Mothes W, Burton DR, Koff WC, Connors M, Ward AB, Kwong PD, Mascola JR. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scharf L, Scheid JF, Lee JH, West AP, Jr, Chen C, Gao H, Gnanapragasam PN, Mares R, Seaman MS, Ward AB, Nussenzweig MC, Bjorkman PJ. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell reports. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, Mankoff Z, Schmidt SD, Lifson JD, Mascola JR, Nussenzweig MC, Martin MA. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. Journal of virology. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature medicine. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 28.Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, Marx PA, Burton DR. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 29.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, Forthal DN, Koff WC, Poignard P, Watkins DI, Burton DR. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. Journal of virology. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnett LA, Fujinami RS. Molecular mimicry: a mechanism for autoimmune injury. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1992;6:840–844. doi: 10.1096/fasebj.6.3.1740233. [DOI] [PubMed] [Google Scholar]
- 31.Silvestris F, Williams RC, Jr, Dammacco F. Autoreactivity in HIV-1 infection: the role of molecular mimicry. Clinical immunology and immunopathology. 1995;75:197–205. doi: 10.1006/clin.1995.1072. [DOI] [PubMed] [Google Scholar]
- 32.Fujinami RS, Oldstone MB. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: mechanism for autoimmunity. Science. 1985;230:1043–1045. doi: 10.1126/science.2414848. [DOI] [PubMed] [Google Scholar]
- 33.Bowes T, Wagner ER, Boffey J, Nicholl D, Cochrane L, Benboubetra M, Conner J, Furukawa K, Furukawa K, Willison HJ. Tolerance to self gangliosides is the major factor restricting the antibody response to lipopolysaccharide core oligosaccharides in Campylobacter jejuni strains associated with Guillain-Barre syndrome. Infection and immunity. 2002;70:5008–5018. doi: 10.1128/IAI.70.9.5008-5018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Human antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 35.Diamond B, Honig G, Mader S, Brimberg L, Volpe BT. Brain-reactive antibodies and disease. Annual review of immunology. 2013;31:345–385. doi: 10.1146/annurev-immunol-020711-075041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magrys A, Anekonda T, Ren G, Adamus G. The role of anti-alpha-enolase autoantibodies in pathogenicity of autoimmune-mediated retinopathy. Journal of clinical immunology. 2007;27:181–192. doi: 10.1007/s10875-006-9065-8. [DOI] [PubMed] [Google Scholar]
- 37.Douglas JN, Gardner LA, Salapa HE, Levin MC. Antibodies to the RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein A1 Colocalize to Stress Granules Resulting in Altered RNA and Protein Levels in a Model of Neurodegeneration in Multiple Sclerosis. Journal of clinical & cellular immunology. 2016;7:402. doi: 10.4172/2155-9899.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douglas Joshua N, G LA, L MC. Antibodies to an Intracellular Antigen Penetrate Neuronal Cells and Cause Deleterious Effects. Journal of Clinical and Cellular Immunology. 2013;4 [Google Scholar]
- 39.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature immunology. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haynes BF, Shaw GM, Korber B, Kelsoe G, Sodroski J, Hahn BH, Borrow P, McMichael AJ. HIV-Host Interactions: Implications for Vaccine Design. Cell host & microbe. 2016;19:292–303. doi: 10.1016/j.chom.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, Spicer L, VandeBerg JL, Haynes BF, Kelsoe G. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. The Journal of experimental medicine. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, Kelsoe G. Polyreactivity and autoreactivity among HIV-1 antibodies. Journal of virology. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vcelar B, Stiegler G, Wolf HM, Muntean W, Leschnik B, Mehandru S, Markowitz M, Armbruster C, Kunert R, Eibl MM, Katinger H. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. Aids. 2007;21:2161–2170. doi: 10.1097/QAD.0b013e328285da15. [DOI] [PubMed] [Google Scholar]
- 45.Doyle-Cooper C, Hudson KE, Cooper AB, Ota T, Skog P, Dawson PE, Zwick MB, Schief WR, Burton DR, Nemazee D. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. Journal of immunology. 2013;191:3186–3191. doi: 10.4049/jimmunol.1301285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, Kelsoe G, Haynes BF. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH x VL knockin mice reveals multiple tolerance controls. Journal of immunology. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Zhang J, Hwang KK, Bouton-Verville H, Xia SM, Newman A, Ouyang YB, Haynes BF, Verkoczy L. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. Journal of immunology. 2013;191:1260–1275. doi: 10.4049/jimmunol.1300770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Alam SM, Bouton-Verville H, Chen Y, Newman A, Stewart S, Jaeger FH, Montefiori DC, Dennison SM, Haynes BF, Verkoczy L. Modulation of nonneutralizing HIV-1 gp41 responses by an MHC-restricted TH epitope overlapping those of membrane proximal external region broadly neutralizing antibodies. Journal of immunology. 2014;192:1693–1706. doi: 10.4049/jimmunol.1302511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, Liao HX, Kelsoe G, Haynes BF. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R, Verkoczy L, Wiehe K, Munir Alam S, Nicely NI, Santra S, Bradley T, Pemble CWt, Zhang J, Gao F, Montefiori DC, Bouton-Verville H, Kelsoe G, Larimore K, Greenberg PD, Parks R, Foulger A, Peel JN, Luo K, Lu X, Trama AM, Vandergrift N, Tomaras GD, Kepler TB, Moody MA, Liao HX, Haynes BF. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Science translational medicine. 2016;8:336ra362. doi: 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clinical chemistry. 1998;44:858–862. [PubMed] [Google Scholar]
- 52.Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, Lockwood B, Scearce RM, Montefiori DC, Dennison SM, Xia SM, Hwang KK, Liao HX, Alam SM, Haynes BF. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. Journal of immunology. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, Stolarchuk C, Lloyd KE, Parks R, Eaton A, Foulger A, Nie X, Karim SS, Barnett S, Kelsoe G, Kepler TB, Alam SM, Montefiori DC, Moody MA, Liao HX, Morris L, Santra S, Harrison SC, Haynes BF. Structural Constraints of Vaccine-Induced Tier-2 Autologous HIV Neutralizing Antibodies Targeting the Receptor-Binding Site. Cell reports. 2016;14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiehe K, Easterhoff D, Luo K, Nicely NI, Bradley T, Jaeger FH, Dennison SM, Zhang R, Lloyd KE, Stolarchuk C, Parks R, Sutherland LL, Scearce RM, Morris L, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Sinangil F, Phogat S, Michael NL, Kim JH, Kelsoe G, Montefiori DC, Tomaras GD, Bonsignori M, Santra S, Kepler TB, Alam SM, Moody MA, Liao HX, Haynes BF. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41:909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, Gao F, Markowitz M, Heath SL, Bar KJ, Goepfert PA, Montefiori DC, Shaw GC, Alam SM, Margolis DM, Denny TN, Boyd SD, Marshal E, Egholm M, Simen BB, Hanczaruk B, Fire AZ, Voss G, Kelsoe G, Tomaras GD, Moody MA, Kepler TB, Haynes BF. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. The Journal of experimental medicine. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dennison SM, Stewart SM, Stempel KC, Liao HX, Haynes BF, Alam SM. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. Journal of virology. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coppola A, Wenner BR, Ilkayeva O, Stevens RD, Maggioni M, Slotkin TA, Levin ED, Newgard CB. Branched-chain amino acids alter neurobehavioral function in rats. American journal of physiology Endocrinology and metabolism. 2013;304:E405–413. doi: 10.1152/ajpendo.00373.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang R, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. Journal of virology. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, Nicely N, Tomaras GD, Kelsoe G, Chen B, Kepler TB, Haynes BF. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. Journal of virology. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kain R, Exner M, Brandes R, Ziebermayr R, Cunningham D, Alderson CA, Davidovits A, Raab I, Jahn R, Ashour O, Spitzauer S, Sunder-Plassmann G, Fukuda M, Klemm P, Rees AJ, Kerjaschki D. Molecular mimicry in pauci-immune focal necrotizing glomerulonephritis. Nature medicine. 2008;14:1088–1096. doi: 10.1038/nm.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu RK, Ariga T, Usuki S, Kaida K. Pathological roles of ganglioside mimicry in Guillain-Barre syndrome and related neuropathies. Advances in experimental medicine and biology. 2011;705:349–365. doi: 10.1007/978-1-4419-7877-6_17. [DOI] [PubMed] [Google Scholar]
- 64.Lima S, Khristoforov R, Momany C, Phillips RS. Crystal structure of Homo sapiens kynureninase. Biochemistry. 2007;46:2735–2744. doi: 10.1021/bi0616697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. Journal of immunology. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Frey G, Peng H, Rits-Volloch S, Garrity J, Seaman MS, Chen B. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. Journal of virology. 2014;88:1249–1258. doi: 10.1128/JVI.02664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.