Abstract
Many tumors express proteoglycans modified with oncofetal chondroitin sulfate glycosaminoglycan chains (ofCS), which are normally restricted to the placenta. However, the role of ofCS in cancer is largely unknown. The function of ofCS in cancer was analyzed using the recombinant ofCS-binding VAR2CSA protein (rVAR2) derived from the malaria parasite, Plasmodium falciparum. We demonstrate that ofCS plays a key role in tumor cell motility by affecting canonical integrin signaling pathways. Binding of rVAR2 to tumor cells inhibited the interaction of cells with extracellular matrix (ECM) components, which correlated with decreased phosphorylation of Src kinase. Moreover, rVAR2 binding decreased migration, invasion and anchorage-independent growth of tumor cells in vitro. Mass spectrometry of ofCS-modified proteoglycan complexes affinity purified from tumor cell lines on rVAR2 columns, revealed an overrepresentation of proteins involved in cell motility and integrin signaling, such as integrin β1 (ITGB1) and integrin α4 (ITGA4). Saturating concentrations of rVAR2 inhibited downstream integrin signaling, which was mimicked by knockdown of the core CS synthesis enzymes Beta-1,3-Glucuronyltransferase 1 (B3GAT1) and Chondroitin Sulfate N-Acetylgalactosaminyltransferase 1 (CSGALNACT1). The ofCS modification was highly expressed in both human and murine metastatic lesions in situ and pre-incubation or early intravenous treatment of tumor cells with rVAR2 inhibited seeding and spreading of tumor cells in mice. This was associated with a significant increase in survival of the animals. These data functionally link ofCS modifications with cancer cell motility and further highlights ofCS as a novel therapeutic cancer target.
Implications
The cancer specific expression of oncofetal chondroitin sulfate aids in metastatic phenotypes and is a candidate target for therapy.
Keywords: Chondroitin sulfate, VAR2CSA, Cancer, motility, invasion
INTRODUCTION
Glycosaminoglycans (GAGs) are secondary carbohydrate modifications attached to proteoglycans on the cellular plasma membrane or secreted into the extracellular matrix (ECM). During embryogenesis, cell differentiation, and diseases such as cancer, GAGs display radical changes in expression and composition (1-3). Alterations in the GAG component of proteoglycans have been reported in cancer for more than four decades (4-7). As part of the cellular glycocalyx, GAGs are believed to control the information flow from the ECM to signal transduction pathways stemming from the plasma membrane (8). While the function and mechanistic contribution of GAGs in cancer are not fully understood, it is clear that they act as key regulators of the malignant phenotype (9).
Most cancer cells express a distinct chondroitin sulfate (CS) GAG epitope that is normally restricted to trophoblastic cells in the placenta (10). These oncofetal CS (ofCS) chains, previously termed placental type CS, are expressed on CS-modified proteoglycans (CSPGs) of tumor and tumor-infiltrated stromal cells across multiple types of malignancies, indicating a possible broad functional importance of ofCS for the disease pathology (10). CSPGs have been associated with proliferation, migration, invasion, angiogenesis, and metastasis (11-15). In these processes, CSPGs act either alone or in concert with membrane components such as integrins, RTKs or metalloproteases to aid cellular attachment, migration and invasion (16-19). One well-described function of CS on proteoglycans is to capture growth factors and cytokines and present them to adjacent receptors in the membrane. As such, CS can act as a scaffold or reservoir for sustained proliferative and oncogenic signalling (20-24). GAGs are made up of repeated disaccharide units making up long linear polymers. CS consists of alternating glucuronic acid (GlcA) and N-Acetyl-D-galactosamine (GalNac) residues (24). While the base structure is simple, heterogeneity is achieved through secondary modification of the CS carbohydrate backbone, such as alternate sulfation of hydroxyl groups (25). Erythrocytes infected with malaria parasites expressing the VAR2CSA protein adhere to CS only in the placenta, despite the fact that CS is present in most organs of the human host (24, 26-28). This suggests that CS in the placenta is distinct from CS present in other organs. The interaction between malarial VAR2CSA and the placenta is the key molecular event underlying placental malaria (26, 28).
The ofCS motif on cancer and placental cells can be detected using a 72kDa recombinant fragment of the Plasmodium falciparum VAR2CSA protein (rVAR2) (10, 29). VAR2CSA is a large complex multidomain protein (28). The 72kDa functional CS binding domain ID1-ID2a has an unprecedented high specificity and affinity for 4-O-sulfated CS (C4S) (29, 30).
Using rVAR2 as an ofCS binding reagent, we have investigated the role of ofCS in human cancer. We report that ofCS is required for cellular attachment, migration and invasion of tumor cells in vitro and in vivo. Furthermore we identify a number of proteins that are modified or associated with ofCS in human tumor cells including components of the integrin complexes. Our study confirms a pivotal role for ofCS in integrin mediated signalling and supports current efforts using rVAR2 as a broad therapeutic targeting reagent against ofCS in cancer.
METHODS
Reagents and cell culture
Recombinant proteins were expressed in SHuffle T7 Express Competent E. coli (NEB) and purified using HisTrap columns from GE Healthcare followed by size exclusion chromatography, as previosly described (10). Purified monomeric proteins were validated by SDS-PAGE. Purified chondroitin sulfate A (CSA) was obtained from Sigma. Anti-V5-FITC antibodies were obtained from Invitrogen. Most cell lines were obtained from ATCC and grown in their suggested growth media with 1x penicillin and streptomycin cocktail. The Myla2059 Lymphoma cell lines were donated by Niels Ødum at the University of Copenhagen. Mice for animal studies were acquired from Taconic Biosciences.
ECM binding assay
Cells were grown in 10cm dishes to about 70% confluency. The cells were then serum starved in the presence of 450nM rVAR2, rDBL4 (a non-ofCS binding domain of the VAR2CSA protein) or PBS for 18-24hrs. The cells were collected using cellstripper, counted, and adjusted to 0.2×10ˆ6cell/ml in serum free media containing inhibitor as above. 100uL was added to wells in a 96 well plate coated with fibronectin (FN) (10ug/ml, sigma), Collagen-I (23ug/ml, sigma), collagen IV (23ug/ml, sigma), or uncoated plastic. Plastic blocked with BSA was included as a negative control. All samples were run in triplicates. Following a 25min incubation the adherent cells were stained with Methylene Blue in Methanol for 10min. The plates were washed in water and dried. The color was dissolved in 0.2M Sodium Citrate in 50% Ethanol and absorbance was read at 650nM.
Scratch assay
Cells were seeded into 6 well plates and allowed to grow to confluency. The cells were then washed in PBS and serum starved 24hrs in the presence of 450nM rVAR2, rDBL4 or PBS. 400ug/ml CSA (sigma) was used to outcompete rVAR2 effect. A scratch was made in the cell monolayer with a 20uL pipette tip. The cells were washed in PBS and serum free media containing the inhibitors was added. Pictures were taken at 0, 19, 30 and 46hrs at two fixed points per sample.
For the siRNA experiments MG63 cells were transfected with siRNAs (Qiagen) (50nM final) against CSGALNACT1, using RNAiMAX (Invitrogen). Scratch was made 48hrs after transfection.
Boyden Chamber invasion and migration assays
The cells were grown to 70% confluency. They were then serum starved in the presence of 450nM rVAR2 or rDBL4 for 24hrs. The cells were dislodged with cellstripper and counted three times. Then 100.000 cells were added to each insert of a boyden chamber plate (chemicon). Separate kits were used for migration and invasion. Invasion kit included membranes coated in basement membrane extract. Media with or without chemoattractant was added to the lower well. Plates were then incubated 18-36hrs at 37C. The number of migrating cells was determined by a fluorescent probe and comparison to a standard curve.
Identification of ofCS-conjugated CSPGs
Column Based Pull Down
Membrane proteins were extracted with EBC lysis buffer (150mM NaCl, 50mM Tris-HCl, 2.5mM MgCl2, 1mM EDTA, 1% CHAPS and a protease inhibitor cocktail (Roche)). The lysate was loaded onto a Hitrap NHS HP column (GE) containing immobilized rVAR2 or rContrl (rDBL4) control protein. The column was washed in Lysis buffer as well as lysis buffer containing 250mM NaCl. Bound protein was eluded with 0.5M NaCl in lysis buffer and upconcentrated on a vivaspin Column (MWCO 10.000kDa). Protein samples, dissolved in SDS loading buffer, and a high-molecular weight marker (LC5699, Life Technologies) were loaded onto a NuPAGE Tris-acetate gel (Life Technologies). Proteins were subsequently transferred to a nitrocellulose membrane overnight at 4C at 75mA. The membranes were stained with anti-CSPG4 antibody (LHM2, Abcam) or antibodies against the α4, α5 or β1 integrin subunits. The staining was developed in ECL and scanned.
Bead Based pull down
Membrane proteins were extracted in EBC lysis buffer (150mM NaCl, 50mM Tris-HCl, 2.5mM MgCl2, 1mM EDTA, 1% CHAPS and a protease inhibitor cocktail (Roche)). Biotinylated rVAR2 protein was added to the lysate and the mix was incubated overnight at 4C. The rVAR2 and bound protein was pulled down on streptavidin dynabeads (Invitrogen).
Proteomics
The pulled down material was dissolved in non-reducing LDS loading buffer (Invitrogen), reduced in 1mM DTT, and alkylated with 5.5mM Iodoacetamide. The samples were then run 1cm into Bis-Tris gels and stained with commasie blue. The protein was cut out, washed and in-gel digested with trypsin. The resulting peptides were captured and washed using C18 resin. The peptides were sequenced using a Fusion Orbitrap Mass Spectrometer. Sample analysis and hit verification was performed using the MaxQuant software. All samples were verified against control samples being either a control protein coupled columns or empty beads. For the Ingenuity Pathways Analysis proteins that were found to be significantly different between rVAR2 and rControl were analyzed using the Ingenuity Pathways Analysis (IPA) software (Quiagen) against their involvement in cellular function and disease.
Proximity Ligation Analysis
The PLA experiment was done according to the manufactures guidelines (Sigma). In Short, adherent cells were fixed in 4% PFA. The cells were blocked in 1%BSA and 5%FBS in PBS. The cells were then stained with primary antibodies together with rVAR2 or rDBL4 overnight, in these concentrations: rVAR2 (50nM), anti-integrin α4 (MAB16983; 1:100), anti-integrin α5 (H-104, sc-10729; 1:50), anti-NG2 (LHM 2, ab20156; 1:400), anti-panCD44 (2C5, BBA10; 1:400) anti-integrin β1 (EP1041Y, ab52971; 1:200) and anti-integrin β1 antibody (4B7R, sc-9970; 1:50). Cells were washed in Wash Buffer A (DUO82047) between incubations. An anti-V5 (mouse or rabbit) antibody was used for rVAR2 detection. The cells were then stained with Duolink® In Situ PLA® Probe Anti-Mouse MINUS (DUO92004) and Duolink® In Situ PLA® Probe Anti-Rabbit PLUS (DUO92002) diluted in Antibody Diluent (DUO82008). The cells were then treated with the ligation solution, followed by incubation with the amplification solution. Both reagents were provided with the kit Duolink® In Situ Detection Reagents Orange (DUO92007). The cells were washed with Wash Buffer B (DUO82048). Slides were mounted using Duolink® In Situ Mounting Medium with DAPI (DUO82040). Results were analyzed under a Nikon C1 confocal microscope with a 60X oil objective. A total of 75-100 cells were imaged per sample. The images were analyzed using the BlobFinder software (version 3.2.).
Flow cytometry binding analysis
Cells were grown to 70-80% confluency and harvested using Cellstripper®. 200.000 cells were added to each well in a 96 well plate. All incubations were in PBS containing 2%FBS. Cells were incubated with protein (400nM-25nM) 30 min at 4C. Cells were washed 2 times and incubated with secondary antibody (anti-V5-FITC) 30 min at 4C, washed 2 times and analyzed in a FACSCalibur (BD Biosciences) for FL-1 signal intensity. Results were analysis using the FlowJo software.
Signalling stimulation Assays
Cells were seeded into 6-well plates and allowed to adhere overnight. The cells were washed in PBS and serum starved in the presence of 450nM rVAR2 or rDBL4 control for 18-24hrs. The cells were stimulated with 1-3% FBS, 5ug/ml FN (Sigma) or 10-80ug/ml fibronectin CS1 peptide (GeneArt) for the given timepoints. The cells were put on ice, washed 3 times in PBS and lysed in EBC lysis buffer, containing 0.5% NP40 and Phosphatase and protease inhibitor cocktails (Roche). The samples were balanced on protein concentrations (Bradford assay). The samples were run in western and probed for the indicated phosphoproteins. For total protein determination, the membranes were stripped and reprobed with antibodies for the indicated proteins.
For the siRNA experiments cells were transfected with siRNAs (Qiagen) (10nM final) against B3GAT1, CSGALNACT1, using RNAiMAX (Invitrogen) and analyzed for rVAR2 binding by flowcytometry and for mRNA expression by RT-PCR, following 72hr exposure. The evalutation of intracellular signaling in these cells were performed as described above.
The adhesion signaling experiments were performed as follows. The cells were grown to 70% confluency in 10cm dishes. They were serum starved 18-24hrs prior to the assay in the presence of 450nM rVAR2 or rDBL4. The cells were dislodged in cellstripper, counted and seeded into the wells of a 6 well plate 120min. Cell lysates were collected and analysed as described above.
Antibodies used were: α-phospho-Erk1/2 (thr202/tyr204) (cell signaling, 9101), α-Erk 1/2 (Cell signaling, 9102), α-Src (Cell signaling, 2108), α-phospho-Src (Tyr416) (Cell signaling, 2101), α-Akt (Cell signaling, 9272), α-phospho-Akt (Thr308) (Cell signaling, 2965), α-FAK (Cell signaling, 3285), α-phospho-P130Cas (Tyr410) (Cell signaling, 4011), α-p130Cas (Santa Cruz, sc-20029).
Tissue samples and Immunohistochemistry
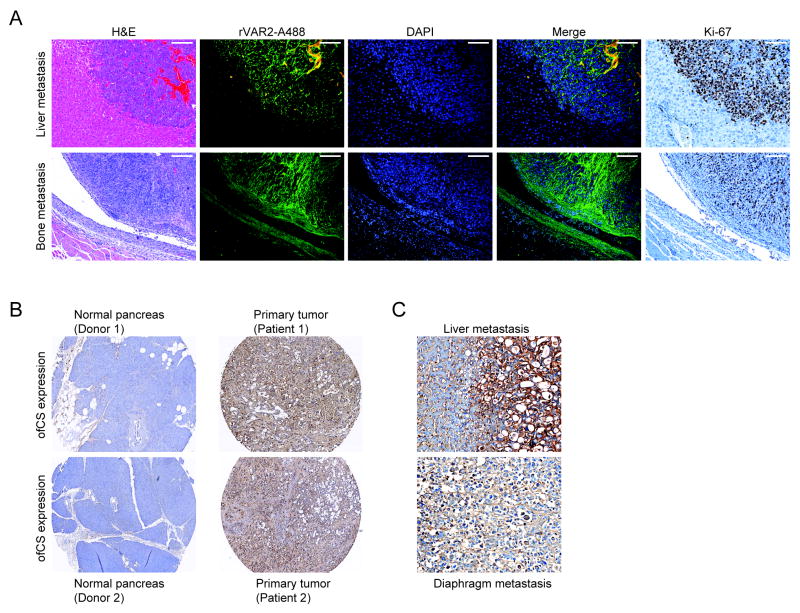
A tissue microarray (TMA) containing 38 patients with primary human pancreatic cancer and corresponding metastatic tissues, as well as control normal pancreas was obtained from the UNMC Rapid Autopsy Pancreas (RAP) program and stained using the Ventana Discovery platform. Sectioned paraffin-embedded TMA was stained with 500 picomolar V5-tagged recombinant VAR2CSA (rVAR2) without antigen retrieval followed by 1:700 (monoclonal anti-V5 step, and an anti-mouse-HRP detection step. Mounted and stained TMA was subsequently scored for membranous staining intensity on a 0-3 scale. Score 2 reflects a staining intensity equal to that of placenta (included as a positive control in each staining run). Expression is considered ‘low’ when ofCS expression is present only in cellular or stromal compartment with intensity score 1 and considered ‘high’ when ofCS expression is present either in cellular or stromal compartment or both with intensity score 2 or 3.
Immunocytochemistry
We obtained FFPE slides of metastatic lesions in murine allografts, created by injecting C57BL/6 mice with 4T1 mammary tumor cells in the left cardiac ventricle, in the animal model published in (10). The slides were deparanifinized and stained with rVAR2-Alexa488. The staining of ofCS was visualized using confocal microscopy.
Animal Studies
The methodologies described were re-viewed and approved by the Institutional Animal Care Committee (IACC) at the University of British Columbia and the animal experiments inspectorate at the University of Copenhagen prior to conducting the study. During the study the care, housing, and use of animals was performed in accordance with the Canadian Council on Animal Care Guidelines and the Danish animal experiments inspectorate guidelines.
For the lewis lung carcinoma seeding model, five to six weeks old C57black/6 female mice were maintained under isoflurane anesthesia and 5 × 105 lewis-luciferase cells suspended in 100 μl of 100 nM of rVAR2 or saline solution were injected into the left ventricle under ultrasound guidance using a 30 gauge needle. The location of the tip of the needle in the left ventricle was confirmed by pulsatile blood flow in the hub of the needle. Animals were monitored until 7 weeks after injection using IVIS imaging system. Metastasis sites were collected at day of sacrifice and fixed in formalin for pathology studies. Mice were sacrificed when they reached the pre-defined humane end point.
For the B16 melanoma model 5 × 105 B16-F10GP cells in 100uL PBS were injected into the right flank of C57BL/6 mice. The animals were randomized into two groups of ten mice. One group was treated by intravenous injection of 100ug rVAR2 at day 0, 6, and 9. The control group was treated with equal volume PBS. Tumor size was monitored by manual measurements using a caliper-measuring tool, taking measurements at the 2 longest perpendicular axes in the x/y plane of each tumor. Tumor volume was calculated according to the standard formula: volume=xy2*0.5236 (31).
Statistical methods
Correlations between clinicopathological parameters and ofCS expression were analyzed by Fisher’s exact test. P<0.05 was considered statistically significant. Statistical analyses were performed with GraphPad Prism (version 6, GraphPad Software, Inc.). Survival of mice was analyzed by Kaplan-Meier survival plot. Statistical significance was determined with Prism GraphPad version 6.0 using the Log-Rank (Mantel-Cox) test. (Chi square 3.84 ; P<0.05)
RESULTS
rVAR2 blocks cellular adhesion to ECM components
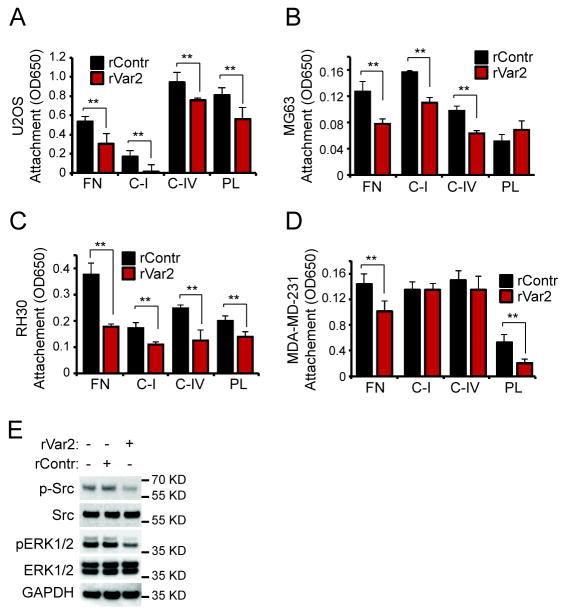
We have shown that placental and tumor cells share the expression of a placental type CS now designated ofCS (10). The expression of this modification in human cancers suggests a pivotal role for ofCS in malignant disease. Based on the preliminary observation that cellular attachment of tumor cells to normal culture flask plastic surfaces was impaired after rVAR2 incubation, we hypothesized that ofCS may play a role in cell motility. To test our hypothesis, we investigated whether rVAR2 targeting of ofCS on U2OS and MG63 osteosarcoma cells would block cellular adhesion to Plastic (PL), Fibronectin (FN) and Collagens I (CI) and IV (CIV). While different binding preferences for individual ECM components could be observed, both cell lines displayed a significant reduction in adhesion to FN, CI, and CIV (Fig 1A and B). The same was true for cell lines representing other cancers, such as RH30 rhabdomyosarcoma cells (Fig 1C) and MDA-MB-231 breast cancer cells (Fig 1D). MDA-MB-231 was not inhibited in binding to collagen, suggesting another mode of binding in this cell type (Fig 1D).
Figure 1. rVAR2 impedes attachment and detachment mechanics in tumor cells.
A, U2OS cells were preincubated with rVAR2 and tested for their capacity to adhere to Fibronectin (FN), Collagen I (C-I), Collagen IV (C-IV), and Plastic (PL). Adherent cells were stained with Methylene Blue and quantified by reading absorbance at 650nm. B, MG63 cells were preincubated with rVAR2 and tested for their capacity to adhere to Fibronectin (FN), Collagen I (C-I), Collagen IV (C-IV), and Plastic (PL). Adherent cells were stained with Methylene Blue and quantified by reading absorbance at 650nm. C, RH30 cells were preincubated with rVAR2 and tested for their capacity to adhere to Fibronectin (FN), Collagen I (C-I), Collagen IV (C-IV), and Plastic (PL). Adherent cells were stained with Methylene Blue and quantified by reading absorbance at 650nm. D, MDA-MB-231 cells were preincubated with rVAR2 and tested for their capacity to adhere to Fibronectin (FN), Collagen I (C-I), Collagen IV (C-IV), and Plastic (PL). Adherent cells were stained with Methylene Blue and quantified by reading absorbance at 650nm. E, U2OS cells were preincubated with rVAR2 or rContrl and allowed to adhere to plastic. The phosphorylation of Src and Erk1/2 kinases in response to the adhesion event was monitored by Western.
The Src and Erk kinases are activated by cellular adhesion (32, 33). Given effects of rVAR2 incubation on functional cell adhesion, we tested whether these pathways were affected in rVAR2 treated U2OS cells. Indeed, we observed a clear inhibition of the phosphorylation of Src, and Erk (Fig 1E), in line with the shown effect on cellular adhesion (Fig 1A-D).
rVAR2 inhibits cellular migration, invasion, and anchorage independent growth in cancer
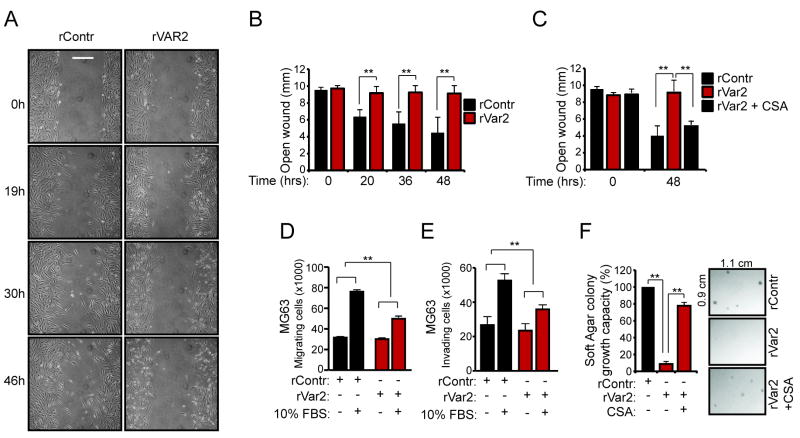
We next wanted to test whether blocking ofCS function impacts cellular migration. We therefore performed scratch assays using the invasive MG63 osteosarcoma cell line. Confluent monolayers of MG63 cells were scratched and incubated with rVAR2 for 24 hrs under serum starvation. We then observed and documented wound closure over time. The cells incubated with rVAR2 failed to close the wound while the control cells effectively filled in the scratch (Fig 2A and B). MG63 cells treated with SiRNA against CSGALNACT1, which is involved key enzyme involved in CS synthesis, showed similar effects (Fig S1). The anti-migratory effect of rVAR2 on the cells could be inhibited by the addition of soluble CSA to outcompete rVAR2 cell binding (Fig 2C). Furthermore, rVAR2 significantly inhibited the migration of MG63 cells across a membrane in a Boyden chamber assay (Fig 2D). Next we tested the ability of rVAR2 to block cancer cell invasion across an ECM modified membrane in the same Boyden chamber assay. This showed that rVAR2 potently inhibited the invasive capacity of cancer cells (Fig 2E). Finally we tested the ability of rVAR2 to inhibit anchorage independent growth of MG63 cells in soft agar colony growth assays (Fig 2F). rVAR2 efficiently reduced the number of colonies formed compared to the control. The effect was shown to be ofCS specific as addition of soluble CSA to outcompete rVAR2 cell binding efficiently rescued the colony formation. Collectively, this shows that blocking the function of ofCS with rVAR2 inhibits the metastatic potential of cancer cells.
Figure 2. CS-dependent motility and achorage-independet growth in tumor cells.
A, MG63 cells were tested for their ability to migrate in the presence of rVAR2 or saline in a scratch-wound heal assay. The figures shows representative pictures of the analysis taken at time 0hr, 19hr, 30hr, and 46hr. B, Column graph showing the statistical analysis of the MG63 scratch-wound heal assay. C, The scratch-wound heal assay was repeated with rVAR2 treatment in the presence of CSA to illustrate ofCS specificity. D, MG63 cells were tested for their ability to migrate across a membrane in a boyden-chamber assay in the presence of rVAR2 or rContr. E, MG63 cells were tested for their ability to invade across an ECM modified membrane in a boyden-chamber assay in the presence of rVAR2 or rContr. F, MG63 cells were tested for their ability to elicit anchorage-independent growth in the presence of rVAR2, rContr, or rVAR2 + CSA in a soft agar colony formation assay.
rVAR2 interacts with ofCS-modified proteoglycans
CS is a posttranslational modification to many CSPGs within the cell membrane. Although these CSPGs differ greatly in their protein cores, they have many overlapping functions in cancer development (11-19). We previously showed that rVAR2, through ofCS chains, interacts with numerous cancer-associated proteoglycans including CSPG4 and CD44, based on overexpression of membrane receptors in HEK cells (10). In continuation of this work, we wanted to explore the significance of ofCS targeting in terms of the number and diversity of CSPG targets in different cancers. To do this we selected five cell lines derived from different cancer types; Melanoma (C32), T-cell Lymphoma (Myla2059), Prostate Cancer (PC3), Osteosarcoma (U2OS), and Rhabdomyosarcoma (RH30). We then performed pull-downs from cell extracts using rVAR2 coupled columns or biotinylated rVAR2 captured on streptavidin coated beads. The elution extracts were then analyzed by mass spectrometry. Using this method we identified 24 proteins previously reported to carry CS (Table 1). Interestingly, all cell lines tested co-expressed different CSPGs and several CSPGs were shared between tumor cell lines of different origin.
Table 1.
List of known CSPGs identified in the rVAR2 pull down screen.
| Protein Name | Gene | Source |
|---|---|---|
| Cell Surface CSPGs | ||
|
| ||
| Chondroitin sulfate proteoglycan 4 | CSPG4 | U2OS, C32, RH30 |
| Amyloid-like protein 2 | APLP2 | U2OS, PC3, Myla2059, C32 |
| Syndecan-1 | SDC1 | U2OS, PC3, C32, RH30 |
| CD44 | CD44 | U2OS, C32, Myla2059, RH30 |
| Integrin beta-1 | ITGB1 | U2OS, C32 |
| Syndecan-4 | SDC4 | U2OS, C32, Myla2059 |
| Glypican-1 | GPC1 | U2OS |
| Sushi repeat-containing protein | SRPX | U2OS, C32 |
| Neuropilin-1 | NRP1 | U2OS, PC3 |
| Sushi repeat-containing protein 2 | SRPX2 | U2OS |
| Syndecan 2 | SDC2 | C32 |
| Neuropilin-2 | NRP2 | C32 |
| Glypican-2 | GPC2 | C32 |
| Glypican-6 | GPC6 | C32 |
| Glypican-4 | GPC4 | C32 |
| Endorepellin | HSPG2 | C32 |
| Delta-sarcoglycan | SGCD | C32 |
| Agrin | AGRN | C32, Myla2059 |
| Syndecan-3 | SDC3 | C32 |
| HLA class II histocompatibility antigen gamma chain | CD74 | C32, Myla2059 |
| Laminin subunit alpha-4 | LAMA4 | C32 |
| Leukocyte surface antigen CD47 | CD47 | Myla2059 |
| Testican-1 | SPOCK1 | Myla2059 |
|
| ||
| Secreted CSPGs | ||
|
| ||
| Serglycin | SRGN | PC3, Myla2059 |
Each CSPG hit is listed at either cell surface or secreted. The hits are given by name and gene. The source of these hits is given in the right column.
Cancer-associated CSPG complexes are involved in cell motility
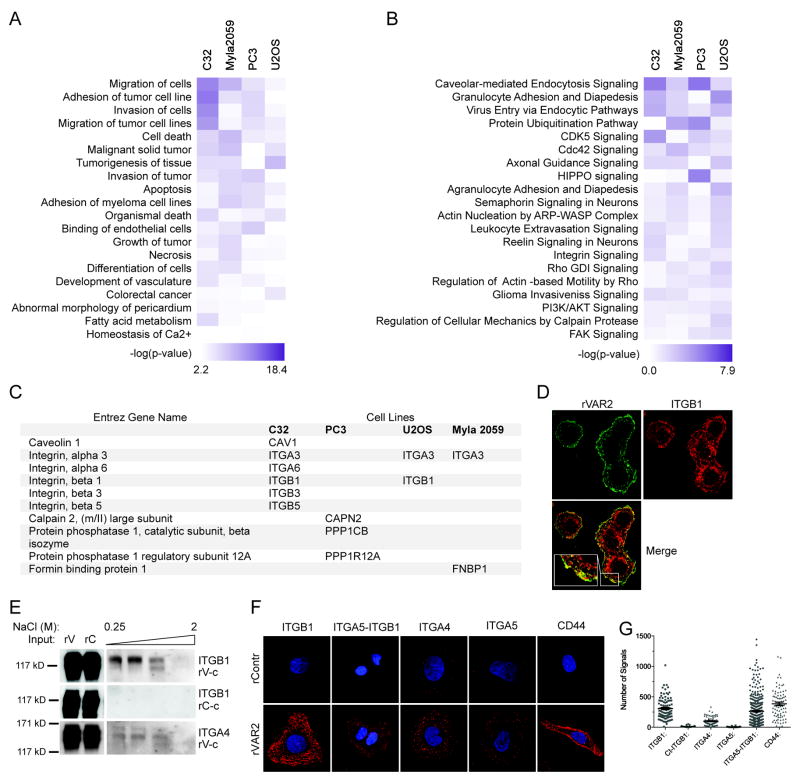
The combined pull down and mass spectrometry analysis identified many ofCS CSPGs across diverse tumor types. Furthermore, we noted that several known CSPG-associated partners were among the identified proteins. To gain insights into the CSPG related proteome and its impact on cellular function, we subjected the full list of pulldown hits to ingenuity pathway analysis (IPA). This revealed an involvement of CSPG complexes in cellular motility and metastasis (Fig 3A and B).
Figure 3. The CS-modified tumor cells proteome in enriched for proteins involved in motility.
A, Network analysis of pulled down proteins using the Ingenuity Pathways Analysis (IPA) software. Proteins that were found to be significantly different between rVAR2 and rControl in the pulldown analysis were analyzed. The top 20 diseases and bio functions involved are shown in a heat Map. We generated the heat map by comparing independent analyses of rVAR2 pull downs in different cell lines. B. IPA heat map analysis of canonical pathways most significantly enriched in proteins pulled-down with rVAR2. C, List of the significant rVAR2 pulled-down proteins involved in the Integrin signaling. D, Co-localization analysis between of-CS (rVAR2 stain (green)) and ITGB1 antibody stain (red). E, Column based pulldown of integrin subunits using rVAR2 (rV-c) from U2OS cells. Figure shows western blot analysis of eluates in increasing NaCl concentration. An rContr coupled column is used as negative control (rC-c). F, PLA analysis of co-localization between ofCS (rVAR2 stain) and integrin subunits. CD44 is used as the positive control. G, Quantification of the PLA analysis.
Several integrin subunits were consistently among the most significant hits in the pulldown analysis (Fig 3C). To verify this we investigated the interaction between rVAR2 staining and integrin β1, integrin α4, and the heterodimeric integrin α5β1 complex by co-localization in immunofluorescence (Fig 3D), co-precipitation using an rVAR2 coupled column (Fig 3E; S2A), and proximity ligation assay (PLA) (34, 35) (Fig 3F and G; S2B and C). This suggested a strong interaction between rVAR2-targeted ofCS and integrin β1, integrin α4, and integrin α5β1.
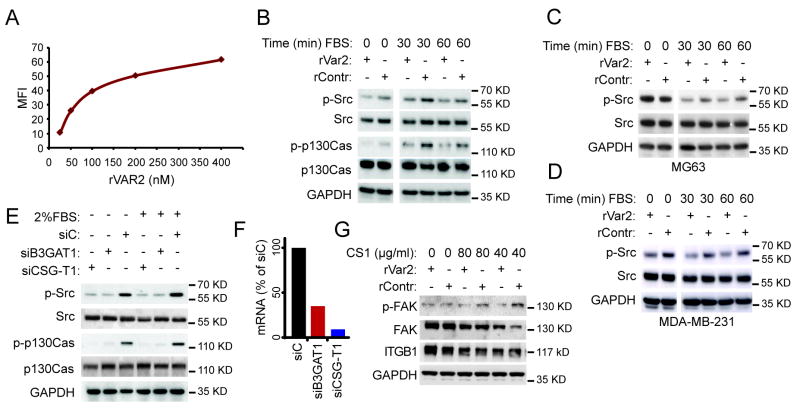
ofCS is involved in integrin related intracellular signalling
Several studies have shown that targeting the CSPG4 core protein on diverse cancer cell types has a direct effect on integrin related intracellular signaling through effectors such as FAK, Src, and Erk1/2 (15, 36, 37). To test whether targeting of the ofCS part of these CSPG/integrin complexes had a similar effect, we incubated the osteosarcoma cell line, U2OS, with rVAR2 and analyzed the intracellular response to FBS stimulation. We used 450nM rVAR2 as this concentration effectively saturated the rVAR2 binding sites on the cells, shown by FACS binding analysis (Fig 4A). A non-CS binding part of VAR2CSA, rDBL4, was used as a recombinant protein negative control (rContr). This showed that phosphorylation of Src, and P130Cas was indeed inhibited in response to FBS stimulation (Fig 4B). There was an effect of rVAR2 on the signaling prior to FBS stimulation as well.
Figure 4. A role for ofCS in signaling downstream integrin beta 1.
A, Titration of rVAR2 staining of U2OS cells in flow cytometry analysis. B, Analysis of downstream integrin signaling in rVAR2 or rContr treated U2OS cells in response to FBS stimulation. Figure shows effect on Src and p130Cas phosphorylation. C, Analysis of downstream integrin signaling in rVAR2 or rContr treated MG63 cells in response to FBS stimulation. Figure shows effect on Src. D, Analysis of downstream integrin signaling in rVAR2 or rContr treated MDA-MB-231 cells in response to FBS stimulation. Figure shows effect on Src. E, Analysis of downstream integrin signaling in U2OS cells treated with siRNA (72hrs) against B3GAT1 and CSGALNACT1 (CSGT-1). A scrambled siRNA is used as control. Figure shows effect on Src and p130Cas phosphorylation. F, RT-PCR analysis of cells treated with siRNA against B3GAT1 and CSGALNACT1 for the analysis shown in E. G, Analysis of FAK phosphorylation in rVAR2 or rContr treated U2OS cells in response to stimulation with the CS1 fibronectin peptide.
To test whether these effects were isolated to U2OS cells, we performed the same analysis on MG63 osteosarcoma (Fig 4C) and MDA-MB-231 breast cancer cells (Fig 4D). The latter has previously been tested in an anti-CSPG4 targeting strategy (15). Again we saw that phosphorylation of Src was inhibited (Fig 4C and D).
We have previously shown that siRNA-mediated knockdown of the enzymes B3GAT1 and CSGALNACT1 which are involved in CS biosynthesis, reduces binding of rVAR2 to the cell surface (10). To test whether the effects seen on intracellular signaling were due to inhibition of the ofCS binding epitope, we tested the effects of B3GAT1 and CSGalNAcT1 knockdown in our FBS stimulation assay (Fig 4E and F). This showed inhibition of phosphorylation of Src and P130Cas with knockdown of both B3GAT1 and CSGALNACT1. This supports our hypothesis that the rVAR2 ofCS binding epitope is involved in integrin mediated signaling in cancer. While stimulation with FBS revealed inhibition of several integrin related effector proteins, such broad stimulation may show effects through other signaling pathways converging on the same effectors. To narrow down the source of stimulation we wanted to investigate the specific cellular activation of integrin complexes, with a ligand such as Fibronectin (FN). Fibronectin is a large complex molecule containing binding sites for numerous cell surface receptors and ECM components. The integrin subunits of various types can interact with several of these sites both with and without CSPGs (38-42). However one region of fibronectin, called the CS1 region, has been shown to support integrin α4β1 binding only when it is in complex with a CSPG (43). We therefore tested whether rVAR2 would inhibit cellular activation in response to stimulation with recombinant CS1 peptide, which has been shown to stimulate the phosphorylation of FAK (44). Accordingly, rVAR2 blocked phosphorylation of FAK (Y397) in response to stimulation with the CS1 peptide on U2OS cells (Fig 4G). This work shows the importance of ofCS in integrin mediated signaling.
ofCS is expressed in metastasis
We have shown the implication of ofCS in integrin mediated cellular function and in cellular adhesion, migration, and invasion, pointing to a potential role for ofCS in metastasis. To investigate the presence of ofCS at metastatic sites, we stained metastatic lesions in murine allografts from C57BL/6 mice injected with 4T1 mammary tumor cells in the left cardiac ventricle (10), using rVAR2-Alexa488. The analysis showed that ofCS was highly and specifically expressed in liver and bone metastasis (Fig 5A). A stain for the Ki-67 proliferation marker showed that the rVAR2 staining metastatic cancer cells were rapidly proliferation (Fig 5A).
Figure 5. Expression of oncofetal CS in metastatic lesions.
A, H&E images (left panel), Immunofluorescent (rVAR2-Alexa 488, DAPI and merged; middle panel) and Ki-67 cell proliferative marker staining (right panel) images of murine liver and bone metastases derived from Balb-C mice inoculated with 4T1 murine mammary cancer cells by intra-cardiac injection. The scale bars represent 100 μm.
B, Immunohistochemical analysis of ofCS, stained with rVAR2-V5 using anti-V5-HRP antibody, in human normal pancreas (left panel), pancreatic cancer tissues (right panel) and, C, liver and diaphragm metastases.
To investigate the presence of ofCS in metastatic sites in human tumors, we stained pancreatic cancer specimens, collected in the UNMC Rapid Autopsy Pancreas (RAP) program, from primary tumor tissue as well as tissue from metastatic sites in different organs. As in the murine model, rVAR2 showed strong staining of the primary tumor as well as all metastatic sites, with little staining of normal pancreatic tissue (Fig 5B and C). Notably, high ofCS expression was unrelated to all annotated clinical parameters (Tabel S1). These data support a role for ofCS in both primary and metastatic tumor compartments.
rVAR2 adhesion to ofCS inhibits metastasis
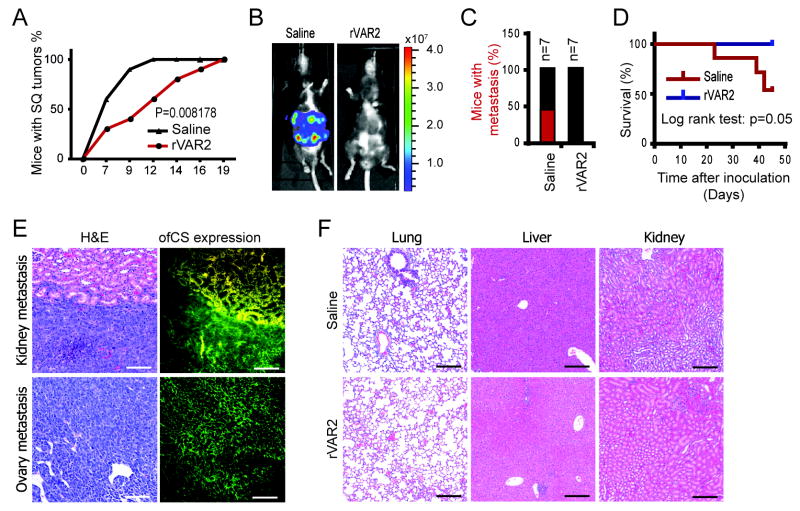
Having shown that ofCS is present in metastatic tumors, we wanted to investigate the effect of rVAR2 treatment on metastasis formation in vivo. First we tested the effect of intravenous administration of rVAR2 on the implementation of B16 melanoma tumors in subcutaneous allotransplanted mice. After B16 cell inoculation the mice were randomized into 2 groups (n=10 each) and treated with rVAR2 at day 0, 6, and 9. The control group was treated with saline. The results showed that 100% of mice in the saline group developed tumors within 12 days while tumor growth in the mice treated with rVAR2 at day 0, 6, and 9 was significantly delayed (Fig 6A; Fig S3, p=0.008178,). These data show that rVAR2 binding to ofCSA on the cell surface of B16 melanoma cells inhibits tumor implantation.
Figure 6. Inhibition of tumor initiation and metastatic spread by rVAR2.
A, Mice were inoculated with subcutanous B16 tumors and randomized into 2 groups (n=10) and treated with rVAR2 at days (0, 6, 9). The control group was treated with saline. Figure shows tumor size progression over time (days). B, two groups of C57black/6 mice were inoculated with murine Lewis lung cancer cells pre-incubated with control (saline) or rVAR2. Representative luciferase images 39 days after inoculation. C, Percentage of C57black/6 mice carrying Lewis cell metastasis (luciferase positive) in saline and rVAR2 group; n=7. D, Kaplan-Meier plot of disease-free survival (p=0.05). E, Metastatic kidney and ovary from control group stained with H&E and immunofluorescence using rVAR2-Alexa488 (right panel). F, Lung, liver and kidney tissue from control group (upper panel) and the rVAR2 group (lower panel) were stained with H& E. The white scale bars represent 100 μm; the black scale bars represent 200 μm.
To analyze the impact of rVAR2-ofCS binding on metastatic spread, luciferase positive Lewis lung carcinoma cells were pre-incubated with 100 nM of rVAR2 or saline and injected into the left cardiac ventricle of mice (n=7 each). In the control arm, 42% (n=3) of the mice formed metastases as visualized by bioluminescence imaging whereas no metastases were found in any of the rVAR2-treated mice (p < 0.05) (Fig 6B and C). Of the mice in the rVAR2 arm, 100% were alive after 45 days. One mouse in the saline group died unexplainedly without signs of metastasis, and is therefore exluded from the survival graph. With this in mind, 50% of the saline control group were dead by day 45 days (n=3) (p < 0.05) (Fig 6D). At the experimental endpoint, autopsy and IHC analysis were performed. The control mice treated with PBS had visible metastases in different organs, including kidney and ovary, which stained strongly for ofCS using rVAR2-alexa 488 (Fig. 6E). No metastases were found in the lung, liver and kidney of the rVAR2-treated mice (Fig. 6F). This shows that interfering with ofCS on cancer cells, significantly (p < 0.05) inhibits the seeding of cancer cells in vivo.
DISCUSSION
We have recently shown that tumor and placental cells carry a common ofCS secondary modification that can be targeted by the malarial VAR2CSA protein (10). The binding of rVAR2 to cancer is nearly universal and highly specific with minimal to absent binding in normal tissue compartments, except for placenta. As such rVAR2 can potentially be utilized in various diagnostic and therapeutic settings (10).
CS in cancer is well described (14, 15, 45-51). The role of CS and CSPGs have been described in many cellular functions including proliferation, migration, invasion, metastasis, angiogenesis, and capture of growth factors, cyto- and chemokines (11-15, 20-24). A well-described function of CS, and of many of the distinct CSPGs, is their interaction with, and potentiation of integrin function (18, 39, 40, 43, 52). In this article we investigated the link between ofCS modification and cellular function in cancer.
There are more than 50 known proteoglycans (PGs) in human tissues. They are found in the cell membranes, in the ECM, excreted into the body fluids, or kept in intracellular granules (24, 53). We recently tested rVAR2 binding in a functional screen where 3500 cell surface receptors were overexpressed in HEK cells. We identified 17 positive binding partners of which some were known PGs (10). Overexpression of receptors in a non-cancer cell line is unlikely to be fully representative of how the receptors are modified during expression in cancer cells. To more directly analyze proteins associated with ofCS we performed co-precipitation from tumor cell extracts using rVAR2. Here we identified 24 known CSPGs as well as known CSPG associated proteins, including integrin subunits (Table 1). Furthermore, the analysis identified several proteins not previously described in relation to CSPGs (data not shown). The co-precipitated proteins included both cell membrane CSPGs as well as secreted Serglycin. Several CSPGs were co-precipitated from each cell type and some were common among tumor cell lines of diverse origin. The broad presence of ofCS on so many CSPGs emphasizes the significance of ofCS substitution in cancer rather than the expression of a specific CSPG. This promotes ofCS-modified CSPG as potential candidate targets in anti-cancer therapy. Our results are refined by the current knowledge of CS-carrying proteoglycans. The part-time glycosylation status of many PGs, meaning that they are not always GAG modified, does however make CSPGs difficult to identify. It is therefore possible that PGs not previously associated with a CS chain are modified with ofCS during transformation to fuel proliferation and tumor cell motility. A thorough and detailed analysis into GAG displacement at the proteome level is needed.
The interaction of CSPGs, including CSPG4, CD44, and the syndecans, with integrin subunits are well described (38-40, 52, 54). Furthermore it has been shown that the α5β1 complex can be modified with a CS chain itself (55, 56). In line with this α4 and β1 integrins were co-precipitated with rVAR2. The association of ofCS with the integrin complexes was further verified using PLA that allows for co-localization analysis at the resolution of a single molecule (35). This method confirmed a strong correlation between integrin α4, β1, and α5β1 and rVAR2 cell binding. These data confirm the findings of others (18, 39, 40, 43), and emphasize the involvement of CSPGs in tumor-associated integrin signalling.
Several articles have shown that targeting the CSPG4 protein core with monoclonal antibodies has an effect on integrin related cellular function (15, 36, 37, 57). Here we show that targeting the ofCS modification present on the proteoglycan component of the integrin complexes has similar effects. These effects include inhibition of intracellular signalling through Src, FAK, P130Cas and Erk in stimulation and adhesion experiments. The mode of action could be two-fold. On one hand, a bulky protein bound to ofCS chains could prevent formation of the CSPG-integrin complexes at the cell surface while, on the other hand shielding of the ofCS epitope could abrogate its cancer promoting effects. We have previously shown that knockdown of key enzymes in the CS biosynthesis pathway reduces rVAR2 binding, suggesting that these enzymes are involved in producing the distinct ofCS epitope (10). In the present study we show that targeting the same enzymes has a similar effect on integrin related signalling, which suggests that the ofCS epitope is indeed crucial for these events to take place. It is likely that targeting the ofCS modification rather than the protein component of PGs such as CSPG4 will present a more universal and efficacious treatment strategy.
We have presented evidence for a function of ofCS in cellular adhesion, migration, and invasion. These are all driver functions of metastatic disease. Indeed we found that ofCS was present in both the primary and the metastatic lesions of human pancreatic cancer.
Finally we wanted to see if targeting ofCS with rVAR2 would interfere with tumor settlement in vivo. For this purpose we established two animal models exploring two essential events of metastatic spread; cell settlement and tumor implementation. Pre-incubating Lewis lung carcinoma cells with rVAR2 strongly inhibited settlement in distant organs and significantly prolonged lifespan of the treated mice. The same was evident for treatment of subcutaneous B16 melanoma tumors at time of implementation. This aligns with our data showing that rVAR2 inhibits cellular attachment, migration, invasion and integrin function. It also confirms what others have shown in targeting specific CSPG components of cancer cells using monoclonal antibodies (15).
Taken together, these data demonstrate the involvement of ofCS chains in cancer cell growth and motility, promoting ofCS as a candidate target for therapy.
Supplementary Material
Acknowledgments
This work was funded by the European Research Council (ERC); US Department of Defense (DoD); SPORE in Pancreatic Cancer (Rapid Autopsy Pancreas program), CA127297; TMEN Tumor Microenvironment Network, U54 CA163120, NCI Cancer Center Support Grant P30 CA36727, EDRN U01CA111294; Prostate Cancer Canada proudly funded by Canada Safeway (Grant # RS2014-02). The authors would also like to thank Birita Kjærbæk and Elham Alijazaeri for their technical assistance in the above mentioned experiments.
Footnotes
CONTRIBUTIONS
T.M.C, M.D, A.S, P.H.S, and T.G.T. designed the research; T.M.C, M.A.P, M.Ø.A, N.S, H.Z.O, A.R.C, A.E, S.L, M.Ø.M, M.F, and M.R performed the experiments; P.H, P.M.G, J.L.G, and M.A.H provided useful reagents and helpful discussions; T.M.C., A.S, and M.D. wrote the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Esko JD, Kimata K, Lindahl U. Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2. Cold Spring Harbor; NY: 2009. [PubMed] [Google Scholar]
- 2.Lanctot PM, Gage FH, Varki AP. The glycans of stem cells. Current opinion in chemical biology. 2007;11:373–80. doi: 10.1016/j.cbpa.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalathas D, Theocharis DA, Bounias D, Kyriakopoulou D, Papageorgakopoulou N, Stavropoulos MS, et al. Alterations of glycosaminoglycan disaccharide content and composition in colorectal cancer: structural and expressional studies. Oncology reports. 2009;22:369–75. [PubMed] [Google Scholar]
- 4.Chandrasekaran EV, Davidson EA. Glycosaminoglycans of normal and malignant cultured human mammary cells. Cancer research. 1979;39:870–80. [PubMed] [Google Scholar]
- 5.De Klerk DP. The glycosaminoglycans of human bladder cancers of varying grade and stage. The Journal of urology. 1985;134:978–81. doi: 10.1016/s0022-5347(17)47570-7. [DOI] [PubMed] [Google Scholar]
- 6.De Klerk DP, Lee DV, Human HJ. Glycosaminoglycans of human prostatic cancer. The Journal of urology. 1984;131:1008–12. doi: 10.1016/s0022-5347(17)50750-8. [DOI] [PubMed] [Google Scholar]
- 7.Sweet MB, Thonar EM, Immelman AR. Glycosaminoglycans and proteoglycans of human chondrosarcoma. Biochimica et biophysica acta. 1976;437:71–86. doi: 10.1016/0304-4165(76)90348-2. [DOI] [PubMed] [Google Scholar]
- 8.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–25. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nature reviews Cancer. 2015;15:540–55. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 10.Salanti A, Clausen TM, Agerbaek MO, Al Nakouzi N, Dahlback M, Oo HZ, et al. Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein. Cancer cell. 2015;28:500–14. doi: 10.1016/j.ccell.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, et al. Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast cancer research : BCR. 2011;13:R58. doi: 10.1186/bcr2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denholm EM, Lin YQ, Silver PJ. Anti-tumor activities of chondroitinase AC and chondroitinase B: inhibition of angiogenesis, proliferation and invasion. European journal of pharmacology. 2001;416:213–21. doi: 10.1016/s0014-2999(01)00884-6. [DOI] [PubMed] [Google Scholar]
- 13.Fthenou E, Zong F, Zafiropoulos A, Dobra K, Hjerpe A, Tzanakakis GN. Chondroitin sulfate A regulates fibrosarcoma cell adhesion, motility and migration through JNK and tyrosine kinase signaling pathways. In Vivo. 2009;23:69–76. [PubMed] [Google Scholar]
- 14.Garusi E, Rossi S, Perris R. Antithetic roles of proteoglycans in cancer. Cell Mol Life Sci. 2012;69:553–79. doi: 10.1007/s00018-011-0816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. Journal of the National Cancer Institute. 2010;102:1496–512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Price MA, Li GY, Bar-Eli M, Salgia R, Jagedeeswaran R, et al. Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer research. 2009;69:7538–47. doi: 10.1158/0008-5472.CAN-08-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, et al. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A) The Biochemical journal. 2007;403:553–63. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iida J, Skubitz AP, Furcht LT, Wayner EA, McCarthy JB. Coordinate role for cell surface chondroitin sulfate proteoglycan and alpha 4 beta 1 integrin in mediating melanoma cell adhesion to fibronectin. The Journal of cell biology. 1992;118:431–44. doi: 10.1083/jcb.118.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MA, Colvin Wanshura LE, Yang J, Carlson J, Xiang B, Li G, et al. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment cell & melanoma research. 2011;24:1148–57. doi: 10.1111/j.1755-148X.2011.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiura N, Shioiri T, Chiba M, Sato T, Narimatsu H, Kimata K, et al. Construction of a chondroitin sulfate library with defined structures and analysis of molecular interactions. The Journal of biological chemistry. 2012;287:43390–400. doi: 10.1074/jbc.M112.412676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Dam GB, van de Westerlo EM, Purushothaman A, Stan RV, Bulten J, Sweep FC, et al. Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. The American journal of pathology. 2007;171:1324–33. doi: 10.2353/ajpath.2007.070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deepa SS, Umehara Y, Higashiyama S, Itoh N, Sugahara K. Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. The Journal of biological chemistry. 2002;277:43707–16. doi: 10.1074/jbc.M207105200. [DOI] [PubMed] [Google Scholar]
- 23.Nikitovic D, Assouti M, Sifaki M, Katonis P, Krasagakis K, Karamanos NK, et al. Chondroitin sulfate and heparan sulfate-containing proteoglycans are both partners and targets of basic fibroblast growth factor-mediated proliferation in human metastatic melanoma cell lines. The international journal of biochemistry & cell biology. 2008;40:72–83. doi: 10.1016/j.biocel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, et al. Glycosaminoglycans: key players in cancer cell biology and treatment. The FEBS journal. 2012;279:1177–97. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 25.Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nature chemical biology. 2006;2:467–73. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 26.Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–4. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 27.Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. The Journal of experimental medicine. 2004;200:1197–203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Molecular microbiology. 2003;49:179–91. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.Clausen TM, Christoffersen S, Dahlback M, Langkilde AE, Jensen KE, Resende M, et al. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. The Journal of biological chemistry. 2012;287:23332–45. doi: 10.1074/jbc.M112.348839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahlback M, Jorgensen LM, Nielsen MA, Clausen TM, Ditlev SB, Resende M, et al. The chondroitin sulfate A-binding site of the VAR2CSA protein involves multiple N-terminal domains. The Journal of biological chemistry. 2011;286:15908–17. doi: 10.1074/jbc.M110.191510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janik P, Briand P, Hartmann NR. The effect of estrone-progesterone treatment on cell proliferation kinetics of hormone-dependent GR mouse mammary tumors. Cancer research. 1975;35:3698–704. [PubMed] [Google Scholar]
- 32.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nature reviews Clinical oncology. 2009;6:587–95. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 33.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature cell biology. 2004;6:154–61. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 34.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nature biotechnology. 2002;20:473–7. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 35.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 36.Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, et al. CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5352–63. doi: 10.1158/1078-0432.CCR-12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, Planaguma J, et al. Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PloS one. 2011;6:e23062. doi: 10.1371/journal.pone.0023062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyano JV, Carnemolla B, Albar JP, Leprini A, Gaggero B, Zardi L, et al. Cooperative role for activated alpha4 beta1 integrin and chondroitin sulfate proteoglycans in cell adhesion to the heparin III domain of fibronectin. Identification of a novel heparin and cell binding sequence in repeat III5. The Journal of biological chemistry. 1999;274:135–42. doi: 10.1074/jbc.274.1.135. [DOI] [PubMed] [Google Scholar]
- 39.Verfaillie CM, Benis A, Iida J, McGlave PB, McCarthy JB. Adhesion of committed human hematopoietic progenitors to synthetic peptides from the C-terminal heparin-binding domain of fibronectin: cooperation between the integrin alpha 4 beta 1 and the CD44 adhesion receptor. Blood. 1994;84:1802–11. [PubMed] [Google Scholar]
- 40.Iida J, Meijne AM, Spiro RC, Roos E, Furcht LT, McCarthy JB. Spreading and focal contact formation of human melanoma cells in response to the stimulation of both melanoma-associated proteoglycan (NG2) and alpha 4 beta 1 integrin. Cancer research. 1995;55:2177–85. [PubMed] [Google Scholar]
- 41.Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5985–8. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hynes RO. Integrins: a family of cell surface receptors. Cell. 1987;48:549–54. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 43.Iida J, Meijne AM, Oegema TR, Jr, Yednock TA, Kovach NL, Furcht LT, et al. A role of chondroitin sulfate glycosaminoglycan binding site in alpha4beta1 integrin-mediated melanoma cell adhesion. The Journal of biological chemistry. 1998;273:5955–62. doi: 10.1074/jbc.273.10.5955. [DOI] [PubMed] [Google Scholar]
- 44.Kamarajan P, Garcia-Pardo A, D’Silva NJ, Kapila YL. The CS1 segment of fibronectin is involved in human OSCC pathogenesis by mediating OSCC cell spreading, migration, and invasion. BMC cancer. 2010;10:330. doi: 10.1186/1471-2407-10-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Wang Y, Yu L, Sakakura K, Visus C, Schwab JH, et al. CSPG4 in cancer: multiple roles. Current molecular medicine. 2010;10:419–29. doi: 10.2174/156652410791316977. [DOI] [PubMed] [Google Scholar]
- 46.Korpetinou A, Skandalis SS, Moustakas A, Happonen KE, Tveit H, Prydz K, et al. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PloS one. 2013;8:e78157. doi: 10.1371/journal.pone.0078157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labropoulou VT, Theocharis AD, Ravazoula P, Perimenis P, Hjerpe A, Karamanos NK, et al. Versican but not decorin accumulation is related to metastatic potential and neovascularization in testicular germ cell tumours. Histopathology. 2006;49:582–93. doi: 10.1111/j.1365-2559.2006.02558.x. [DOI] [PubMed] [Google Scholar]
- 48.Stallcup WB, Huang FJ. A role for the NG2 proteoglycan in glioma progression. Cell adhesion & migration. 2008;2:192–201. doi: 10.4161/cam.2.3.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wegrowski Y, Maquart FX. Chondroitin sulfate proteoglycans in tumor progression. Advances in pharmacology. 2006;53:297–321. doi: 10.1016/S1054-3589(05)53014-X. [DOI] [PubMed] [Google Scholar]
- 50.Theocharis AD, Tsolakis I, Tzanakakis GN, Karamanos NK. Chondroitin sulfate as a key molecule in the development of atherosclerosis and cancer progression. Adv Pharmacol. 2006;53:281–95. doi: 10.1016/S1054-3589(05)53013-8. [DOI] [PubMed] [Google Scholar]
- 51.Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ. The biological role and regulation of versican levels in cancer. Cancer metastasis reviews. 2009;28:233–45. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 52.Woods A, Couchman JR. Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Molecular biology of the cell. 1994;5:183–92. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theocharis AD, Seidel C, Borset M, Dobra K, Baykov V, Labropoulou V, et al. Serglycin constitutively secreted by myeloma plasma cells is a potent inhibitor of bone mineralization in vitro. The Journal of biological chemistry. 2006;281:35116–28. doi: 10.1074/jbc.M601061200. [DOI] [PubMed] [Google Scholar]
- 54.Woods A, McCarthy JB, Furcht LT, Couchman JR. A synthetic peptide from the COOH-terminal heparin-binding domain of fibronectin promotes focal adhesion formation. Molecular biology of the cell. 1993;4:605–13. doi: 10.1091/mbc.4.6.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veiga SS, Elias M, Gremski W, Porcionatto MA, da Silva R, Nader HB, et al. Post-translational modifications of alpha5beta1 integrin by glycosaminoglycan chains. The alpha5beta1 integrin is a facultative proteoglycan. The Journal of biological chemistry. 1997;272:12529–35. doi: 10.1074/jbc.272.19.12529. [DOI] [PubMed] [Google Scholar]
- 56.Franco CR, Trindade ES, Rocha HA, da Silveira RB, Paludo KS, Chammas R, et al. Glycosaminoglycan chains from alpha5beta1 integrin are involved in fibronectin-dependent cell migration. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:677–86. doi: 10.1139/o09-047. [DOI] [PubMed] [Google Scholar]
- 57.Yu L, Favoino E, Wang Y, Ma Y, Deng X, Wang X. The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunologic research. 2011;50:294–302. doi: 10.1007/s12026-011-8232-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.