Abstract
Focal adhesion kinase (FAK) is a major drug target in cancer and current inhibitors targeted to the ATP-binding pocket of the kinase domain have entered clinical trials. However, preliminary results have shown limited single-agent efficacy in patients. Despite these unfavorable data, the molecular mechanisms which drive intrinsic and acquired resistance to FAK-kinase inhibitors are largely unknown. We have demonstrated that receptor tyrosine kinases (RTKs) can directly bypass FAK-kinase inhibition in cancer cells through phosphorylation of FAK’s critical tyrosine 397 (Y397). We also showed that HER2 forms a direct protein-protein interaction with the FAK-FERM-F1 lobe, promoting direct phosphorylation of Y397. Additionally, FAK-kinase inhibition induced two forms of compensatory RTK reprogramming: 1) the rapid phosphorylation and activation of RTK signaling pathways in RTKHigh cells and 2) the long-term acquisition of RTKs novel to the parental cell line in RTKLow cells. Finally, HER2+ cancer cells displayed resistance to FAK-kinase inhibition in 3D–growth assays using a HER2 isogenic system and HER2+ cancer cell lines. Our data indicate a novel drug resistance mechanism to FAK-kinase inhibitors whereby HER2 and other RTKs can rescue and maintain FAK activation (pY397) even in the presence of FAK-kinase inhibition. These data may have important ramifications for existing clinical trials of FAK inhibitors and suggest that individual tumor stratification by RTK expression would be important to predict patient response to FAK-kinase inhibitors.
Keywords: FAK inhibitors, Drug Resistance, Growth factors and receptors, Molecular modeling, Novel mechanisms
Introduction
Focal adhesion kinase (FAK) is considered a major cancer drug target due to its vast overexpression in 80% of all solid tumors both at the protein and mRNA level (1–3). Additionally, it has been shown to play a critical role in multiple aspects of tumor progression such as proliferation, survival, invasion, metastasis, angiogenesis, cancer stem cell maintenance, and recently, immune cell suppression (4–11). Multiple studies using transgenic mouse models have shown a defect in tumor growth, invasion, and metastasis with specific FAK deletion (12–15). In further support, FAK overexpression has been clinically correlated with higher tumor stage, metastasis, and in some tumors (breast, lung, endometrium, liver, GI system, and esophagus) is associated with poor prognosis (16).
FAK functions as a dual kinase and scaffolding protein where it forms complexes with various oncogenic proteins such as receptor tyrosine kinases (RTKs), integrins, and tumor suppressor proteins (17–20). Upon extracellular stimuli, FAK is activated by autophosphorylation of critical residue tyrosine 397 (Y397) through the FAK-kinase domain (21). Phosphorylated Y397 subsequently serves as a canonical SH2-domain docking site for SRC-family kinases, PI3K, and GRB7 (22–24). These signaling molecules then lead to the full activation of FAK through phosphorylation of effector residues (Y861 & Y925) as well as the activation of downstream oncogenic pathways ERK and AKT (25–27).
In an attempt to inhibit FAK in cancer, numerous groups have developed FAK-kinase inhibitors which bind to the ATP-binding pocket of the FAK-kinase domain and block catalysis (28–31). Although very potent (low nM) and specific inhibitors of FAK-kinase activity have been discovered, limited efficacy has been observed in Phase I/II clinical trials with lack of mechanistic knowledge explaining this phenomenon (32–34). Furthermore, there are no current molecular markers available that successfully predict the response of FAK-kinase inhibitors in patients, nor are there biomarkers that would predict resistance to FAK-kinase inhibition. As there are 18 reported clinical trials involving FAK-kinase inhibitors (35), the ability to predict both response and resistance to FAK-kinase inhibitors is critical to clinical development.
Along with the canonical FAK activation pathway through autophosphorylation, it is also known that FAK can be activated independently of FAK catalytic activity (19). PDGF- and EGF-stimulated FAK phosphorylation and cell migration occurs independently of FAK-kinase activity in a genetically modified FAK kinase-dead (K454R) model system. However, the tyrosine 397 site via the Y397F mutation was shown to be indispensable for FAK-dependent migration. Despite these data, it is still unknown how Y397 is activated by RTKs and whether multiple oncogenic RTKs can directly phosphorylate FAK at Y397 as a resistance mechanism to FAK-kinase inhibitors.
In this report, we have investigated whether multiple oncogenic RTKs could re-activate FAK and therefore cause resistance to FAK-kinase inhibitors. We found that multiple classes of RTKs directly phosphorylated FAK at Y397 independently of FAK catalytic activity. FAK-kinase inhibitor treatment induced rapid RTK activation, leading to FAK reactivation as well as MAPK/AKT activation. Additionally, the initial presence of HER2 activity predicted resistance to FAK-kinase inhibitors in vitro. Together, these studies have identified a novel drug resistance mechanism to FAK-kinase inhibitors through transphosphorylation by RTKs. We call this “oncogenic protection” of FAK due to the variety of oncogenic RTKs that can phosphorylate Y397 to protect a cancer cell’s ability to maintain signaling through SH2 domain pathways. Furthermore, we believe that these results have identified a fragile point in FAK signaling pathways that can be exploited with precision-targeted therapeutics directed at FAK Y397 opposed to simply targeting the intrinsic FAK-kinase activity.
Materials and Methods
Cell Culture
Cell lines MDA-MB-231, MDA-MB-453, and MCF7 (courtesy of Katerina Gurova), FAK−/− MEFs (courtesy of Duško Ilić), SYF−/− MEFs (courtesy of Irwin Gelman), H292 (courtesy of Pamela Hershberger), MCF7/HER2 tet-off cells (courtesy of Andrei Bakin) were cultured at 37°C with 5% CO2. H292 cells were cultured in RPMI 1640 (GIBCO, Life Technologies), 10% HI FBS (GIBCO, Life Technologies), 1% Pen Strep (GIBCO, Life Technologies), MEM Nonessential Amino Acids (cellgro, Mediatech, Inc.), Sodium Pyruvate (cellgro, Mediatech, Inc.), and HEPES (GIBCO, Life Technologies), and 0.2% Normocin (InvivoGen). Remaining cell lines were cultured in DMEM (GIBCO, Life Technologies), 10% HI FBS (GIBCO, Life Technologies), 1% Pen Strep (GIBCO, Life Technologies), and 0.2% Normocin (InvivoGen). Cells lines were not submitted for cell authentication services however were instead validated for proper HER2, EGFR, and FAK expression via western blotting and were tested for mycoplasma infection via the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Creation of HER2/HER3 stably transduced FAK−/− MEFs
HER2 and HER3 retrovirus was prepared from 293T-Phoenix cells transfected with either pLXSN-HER2 or pLXSN-HER3 vector (courtesy of Dr. H. Shelton Earp III) similarly as described (36). Cultured FAK−/− MEFs were transduced with retrovirus and selected for HER2High/HER3High expression using FACS (Roswell Park Flow Cytometry Core). Antibodies used for selection were Alexa Fluor® 488 anti-human erbB2/HER-2 (cat # 324410, BioLegend) and APC-conjugated anti-hErbB3/Her3 (cat # FAB3481A, R&D Systems). Stable cells were subsequently transiently transfected with various HA-FAK constructs (WT, Y397F, K454R) using LipoD293™ (SignaGen® Laboratories) and processed for western blotting analysis.
Protein Purification
His-tagged avian FAK-FERM (AA 31-405) in modified pET vector (provided by Dr. Michael Eck) was expressed in BL21 (DE3) E. coli (Life Technologies) and purified on Ni-NTA resin (Thermo Scientific) using buffers as described (37). His-tagged human FAK-CD (AA 677-1052) was cloned into pET15b vector and similarly purified on Ni-NTA resin (Thermo Scientific). GST-FAK-NT (AA 1-415), GST-FAK-KD (AA 416-676), GST-FAK-CD (AA 677-1052), GST-FAK-NT1 (AA 1-126), GST-FAK-NT2 (AA 127-243), and GST-FAK-NT3 (244-415) protein in pGEX-4T1 vector were expressed in BL21 (DE3) E. coli and purified on Glutathione Sepharose 4B resin (GE Healthcare) as previously described (20, 38). GST-HER2-ICD (AA 676-1255), GST-HER2-ICD1 (AA 676-801), GST-HER2-ICD2 (AA 802-1029), and GST-HER2-ICD3 (AA 1030-1255) constructs were designed into the pGEX-4T1 vector, sequenced by the Roswell Park Sequencing Core, expressed in BL21 (DE3) E. coli, and purified on Glutathione Sepharose 4B resin (GE Healthcare). HER2-ECD protein (cat#BMS362) was purchased from eBiocience. Recombinant HER2-ICD (cat#PV3366), EGFR (cat#PV3872), Tie2 (cat#PV3628), EphA2 (cat#PV3688), and FGFR4 (cat#P3054) were purchased from Life Technologies.
In vitro kinase assay
Purified FAK-FERM (100ng) or FAK-CD (100ng) were incubated with purified HER2 (100ng) or additional RTK for 30 min in the presence or absence of ATP (10µM) in standard tyrosine kinase buffer: 20mM HEPES (pH=7.3), 5mM MgCl2, 5mM MnCl2, 2mM DTT, 0.1mg/mL BSA, and 0.1mM Na3VO4. Proteins were resolved on 4–20% gradient gels and probed for phosphorylated FAK (Y397), FAK (Y925), HER2 (Y1248), FAK-FERM, FAK-CD, and HER2/RTK using standard western blotting techniques.
Pull Down Assays
Purified GST-FAK or GST constructs were incubated in NP40 buffer plus 0.1% BSA with purified HER2-ECD and HER2-ICD. GST or GST-FAK constructs were pulled down using Glutathione Sepharose 4B (GE Healthcare Life Sciences) and washed three times with NP40 buffer. Proteins were eluted off of beads in 2X laemmli buffer (BioRad) and boiled. Samples were resolved on 4–20% gradient gels and probed for HER2-ECD or HER2-ICD using standard western blotting techniques and antibodies as described below. Secondary gels were run and stained with SimplyBlue™ SafeStain to confirm protein loading.
PathScan® RTK Signaling Antibody Array Kit (Chemiluminescent Readout)
Cell lysates were collected using NP40 lysis buffer containing protease (Roche) and phosphatase inhibitors (Roche) and were incubated on profiler slides according to manufacturer’s instructions (Cell Signaling) (map in Sup Methods). Slides were imaged via chemilumienescence on film and dot intensities were analyzed using ImageJ densitometry software and graphed in Graphpad Prism 6. Phosphorylation (relative to 0h) was quantified for each protein by subtracting the negative control dots within each panel, respectively. Subsequently, values were divided by signal obtained at 0h to obtain phosphorylation levels relative to 0h.
Matrigel-on-top 3D growth assay
Cells were plated at a density of 1,000 cells per well in a 1:50 solution of matrigel:complete DMEM culture media on top of a base matrix composed of 1:1 matrigel:DMEM media. Doxycycline (1µg/mL) was added for MCF7-HER2 Tet-Off cells. Defactinib was added at various concentrations the day after initial plating. Cell proliferation was evaluated after 5 days using CellTiter AQueous One Solution Cell Proliferation Assay (Promega) according to manufacturer’s instructions. Viability was plotted relative to DMSO control and IC50’s were calculated using Dose-response – Inhibition nonlinear regression algorithm (log(inhibitor) vs. response) in Graphpad Prism 6.
Molecular Modeling
Protein Data Bank files 2AL6 (crystal structure of FAK FERM domain), 3RCD (crystal structure of HER2 kinase domain), 4RIW (crystal structure of EGFR kinase domain), and 2J0J (crystal structure of FAK FERM-Kinase domains) were downloaded and utilized for all modeling experiments. “Active” surface residues determined from CPORT studies were utilized as input residues for restraints to drive the HADDOCK docking process (39). Restraints were only set to “active” residues that corresponded to FAK FERM F1 lobe and HER2 Kinase N-lobe sub-domains that were found to be required for HER2-FAK binding in experimental assays. EGFR restraints were solely based from CPORT results. The HADDOCK docking protocol was performed similarly as described (40). The top-ranking HADDOCK cluster (based on HADDOCK score and Z-score) was selected and visualized using PyMOL software (Incentive version). Images were ray-traced and saved for publication-quality purposes.
Statistical analysis
Comparisons between two groups were made using a Students t test (GraphPad Prism6). Data were considered significant when p<0.05. Two-way ANOVA and Tukey’s multiple comparison test were used to calculate significance when comparing multiple groups within the same experiment (GraphPad Prism6).
Results
Oncogenic Receptor Tyrosine Kinases HER2 and EGFR reactivate FAK in FAK-kinase inhibited cancer cells and activate AKT/ERK independent of FAK-kinase activity
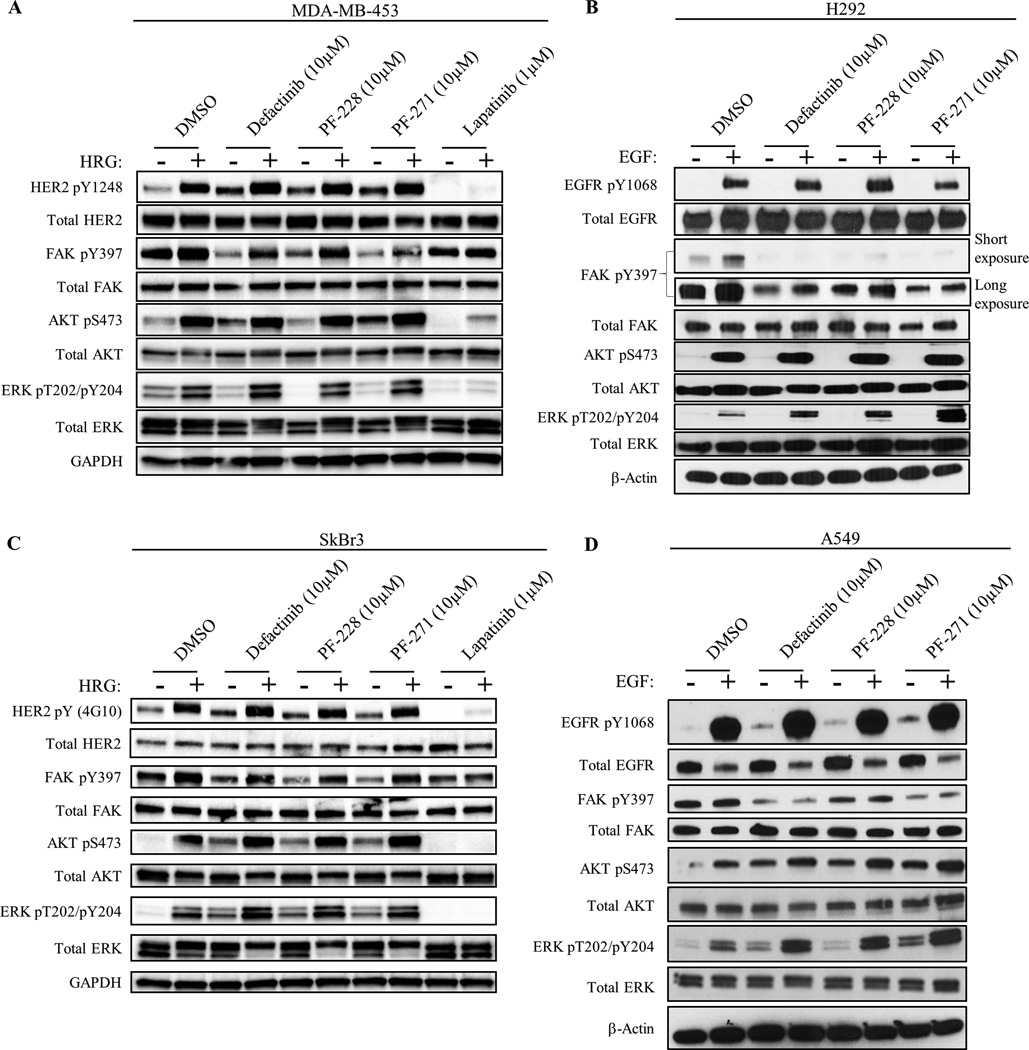
FAK has been shown to be primarily activated at Y397 by autophosphorylation through the FAK-kinase domain. Whereas some data suggest that RTKs can signal through FAK independently of FAK-kinase activity (19), it is still unclear how RTKs activate FAK and whether this alternative pathway plays a role in cancer cell resistance to FAK-kinase inhibitors. Thus, we hypothesized that oncogenic RTKs could directly transphosphorylate FAK Y397 as a drug resistance mechanism to bypass FAK-kinase inhibition. In order to test this, in vitro studies were carried out in HER2+ MDA-MB-453 and SkBr3 breast cancer cells as well as EGFR+ H292 and A549 lung cancer cells (Fig. 1, Supplementary Fig. S1, S2, and Table S1) treated with three different FAK-kinase inhibitors (defactinib, PF-228, and PF-271), two of which have been in clinical trials (28, 29, 32, 34, 41, 42). We used 10µM of FAK-kinase inhibitor treatment to ensure full inactivation of the FAK enzyme in a variety of cell lines (Supplementary Fig. S3 and S4). HER2 activation by Heregulin (HER2-activating ligand) reactivated FAK Y397 phosphorylation after treatment with all three FAK-kinase inhibitors in both MDA-MB-453 and SkBr3 cell lines, suggesting that FAK Y397 phosphorylation could be maintained independent of its intrinsic kinase activity. While the effects of EGF stimulation on FAK reactivation were not as robust as HRG stimulation, EGF stimulation slightly reactivated FAK pY397 after treatment with PF-228 in H292 cells. Nonetheless, growth factor stimulation activated both AKT and ERK pathways in the presence of all three FAK-kinase inhibitors in all four cell lines, indicating that FAK-kinase activity was dispensable for downstream pathway activation. Conversely, FAK-depletion using FAK-null MEFs or FAK siRNA in MDA-MB-453 cells partially diminished HER2-dependent activation of AKT/ERK, indicating that FAK total protein, however not kinase activity, could regulate downstream AKT/ERK pathways (Supplementary Fig. S5). We also noticed that even in the absence of HRG or EGF stimulation, all three FAK kinase inhibitors stimulated phosphorylation of HER2/AKT in MDA-MB-453 cells as well as HER2/AKT/ERK in SkBr3 cells and EGFR/AKT/ERK in A549 cells (Fig. 1). These data indicated a novel drug resistance pathway to FAK-kinase inhibitors through the reactivation of FAK Y397 and downstream AKT/ERK pathways by oncogenic RTKs.
Fig. 1. HER2 and EGFR phosphorylate FAK Y397 independently of FAK-kinase activity.
Immunoblots showing phosphorylation of FAK and downstream molecules in HER2+ breast cancer cells (A) MDA-MB-453 and (C) SkBr3, as well as EGFR+ lung cancer cells (B) H292 and (D) A549. Cancer cells were serum-starved overnight followed by drug treatment with FAK-kinase inhibitor (defactinib, PF-228, PF-271) or HER2/EGFR inhibitor (Lapatinib) for 1 hour and stimulated with either Heregulin-β1 (HRG) or Epidermal Growth Factor (EGF) for 30 min. Images shown are representative of three independent experiments. Densitometry and statistical analysis is found in Supplementary Fig. S1, S2, and Table S1.
To verify that the compensatory increases in HER2, AKT, and ERK after FAK-kinase inhibitor treatment were in fact due to FAK-kinase inhibition and not off-target kinase effects of the 10µM dose, we revisited our dose-titration experiments using clinical-stage FAK inhibitor, defactinib (Supplementary Fig. S3 and S4). In MDA-MB-453 and SkBr3 cells, once FAK pY397 levels were decreased by defactinib treatment (0.3–1.0µM and 0.004µM respectively), compensatory increases in pHER2, pAKT, and pERK were also observed. In MDA-MB-453 cells, maximal FAK inhibition (10–30µM) caused maximal compensatory increases in pAKT and pERK. Intriguingly, at higher doses of defactinib (3µM) in SkBr3 cells, pHER2 and pAKT levels were maximally increased and FAK pY397 levels were paradoxically restored. In H292 and A549 cells, compensatory increases in pEGFR and pERK were observed after low dose defactinib treatment (0.001–1µM). We also noticed that 0.01–0.3µM defactinib treatment in H292 cells induced paradoxical increases in FAK pY397 that were in alignment with increases in pEGFR, suggesting that FAK-kinase inhibition led to FAK hyperphosphorylation by EGFR. To further confirm the FAK-specificity of these compensatory increases, we performed FAK siRNA studies in SkBr3 cells using all three FAK-kinase inhibitors (Supplementary Fig. S6). FAK knockdown by siRNA itself induced the phosphorylation of AKT, ERK, and the reactivation of FAK pY397. In addition, FAK siRNA reduced the relative increase in pAKT and pERK after FAK-kinase inhibitor treatment. In all, these data supported that the compensatory increases in pHER2, pAKT, and pERK upon FAK-kinase inhibition were indeed due to the specific inhibition of FAK.
HER2 and EGFR phosphorylate kinase-dead FAK to maintain cell migration and invasion
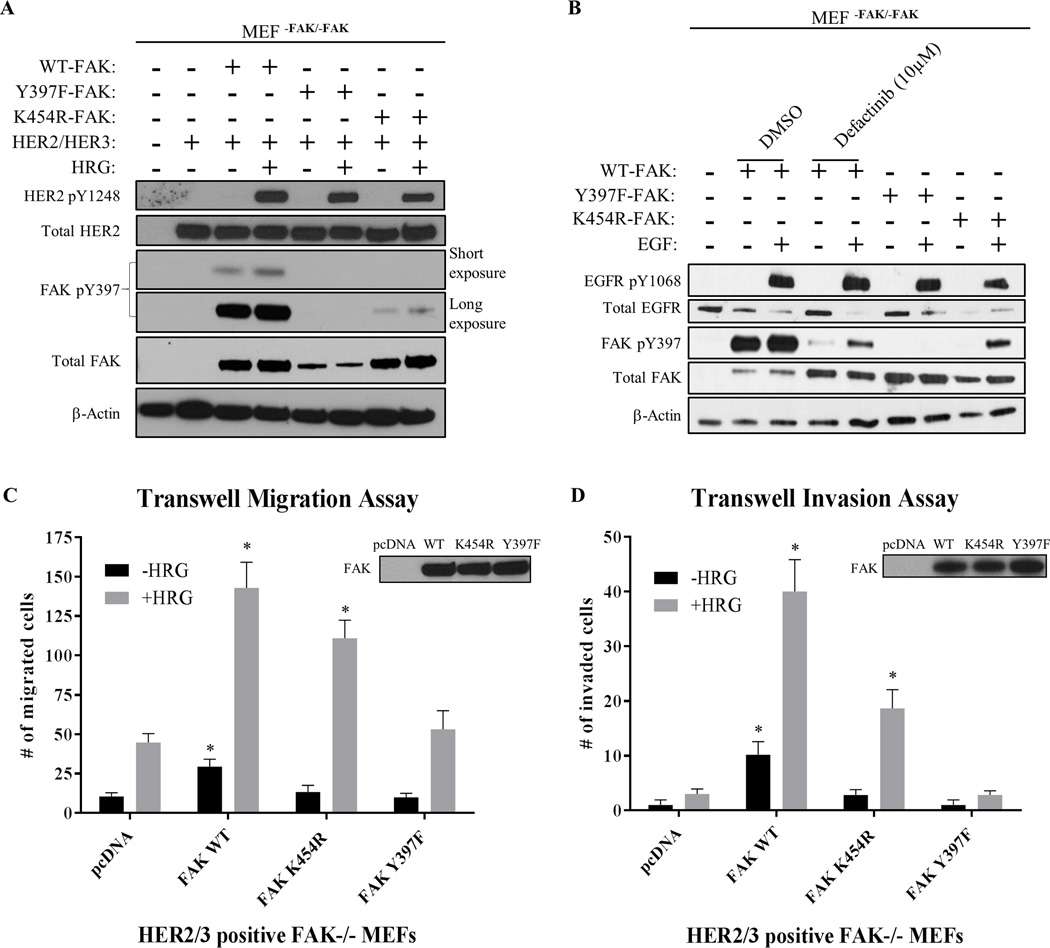
Next, we used genetic studies in FAK-null MEFs to confirm the phenomenon of RTK-driven Y397 phosphorylation (Fig. 2A, 2B, and Supplementary Fig. S7). Both WT-FAK and K454R-FAK (kinase-dead) were found to be re-phosphorylated at Y397 upon stimulation of both HER2/HER3-stably expressing and EGFR+ FAK-null MEFs. Additionally, studies in SRC/YES/FYN (SYF)-null MEFs confirmed FAK Y397 phosphorylation by HER2 independently of known SYF adaptor proteins (Supplementary Fig. S8). These data demonstrated that Y397 can still be phosphorylated by RTKs independently of both FAK and SRC activity, consistent with the RTK-dependent drug resistance we observed with all three FAK-kinase inhibitors.
Fig. 2. HER2 and EGFR phosphorylate kinase-dead FAK to maintain cell migration and invasion.
Immunoblots showing phosphorylation of FAK in (A) HER2+/HER3+ MEFFAK-/FAK-, and (B) EGFR+ MEFFAK-/FAK- cells. MEFs transfected with FAK constructs (WT, K454R-kinase dead, and Y397F) were serum-starved overnight followed by drug treatment with FAK-kinase inhibitor (defactinib) for 1 hour and stimulated with either Heregulin-β1 (HRG) or Epidermal Growth Factor (EGF) for 30 min. Both HER2 and EGFR activation partially reactivated kinase-dead FAK phosphorylation at Y397. Images shown are representative of three independent experiments. Densitometry and statistical analysis is found in Supplementary Fig. S7. (C) Transwell migration and (D) invasion assays performed in transfected HER2+/HER3+ MEFFAK-/FAK- cells. Transfected MEFs were serum-starved overnight and placed into upper chamber to allow chemotaxis towards BSA (-HRG) or HRG (+HRG) placed into the lower chamber. Assays were performed for 3 hours (migration) or 24 hours (invasion). Western blot panels in upper right demonstrate levels of transfected FAK. Two-way ANOVA statistical analysis was performed for results from three independent experiments. Asterisks represent multiple comparison corrected p-values < 0.05 in comparison to pcDNA.
Because HER2 reactivated kinase-dead FAK phosphorylation at Y397 in MEFs, we then hypothesized that HER2 could maintain FAK-dependent biological functions as well. To test this, we performed transwell migration and invasion assays utilizing HER2/HER3-stably expressing FAK-null MEFs that were transiently transfected with WT-FAK, K454R-FAK, or Y397F-FAK. As shown in Figure 2C, HRG-stimulated migration was enhanced by not only WT-FAK, but K454R-FAK as well. Conversely, only mutation of Y397 directly (Y397F-FAK) was found to completely block HRG-stimulated migration. Intriguingly, basal cell migration (-HRG) was fully inhibited by K454R mutation, suggesting that basal migration, but not HER2-stimulated migration, is regulated by FAK-kinase activity. These findings were confirmed in cell invasion assays as well (Figure 2D). Kinase-dead FAK (K454R) moderately reduced HRG-stimulated invasion compared to WT-FAK, however only Y397F-FAK completely abrogated FAK-enhanced cell invasion. These data showed that HER2 can maintain FAK-dependent biological functions (migration and invasion) under kinase inhibition as a result of the FAK transphosphorylation phenomenon.
HER2 directly binds to FAK as a structural mechanism to promote Y397 phosphorylation
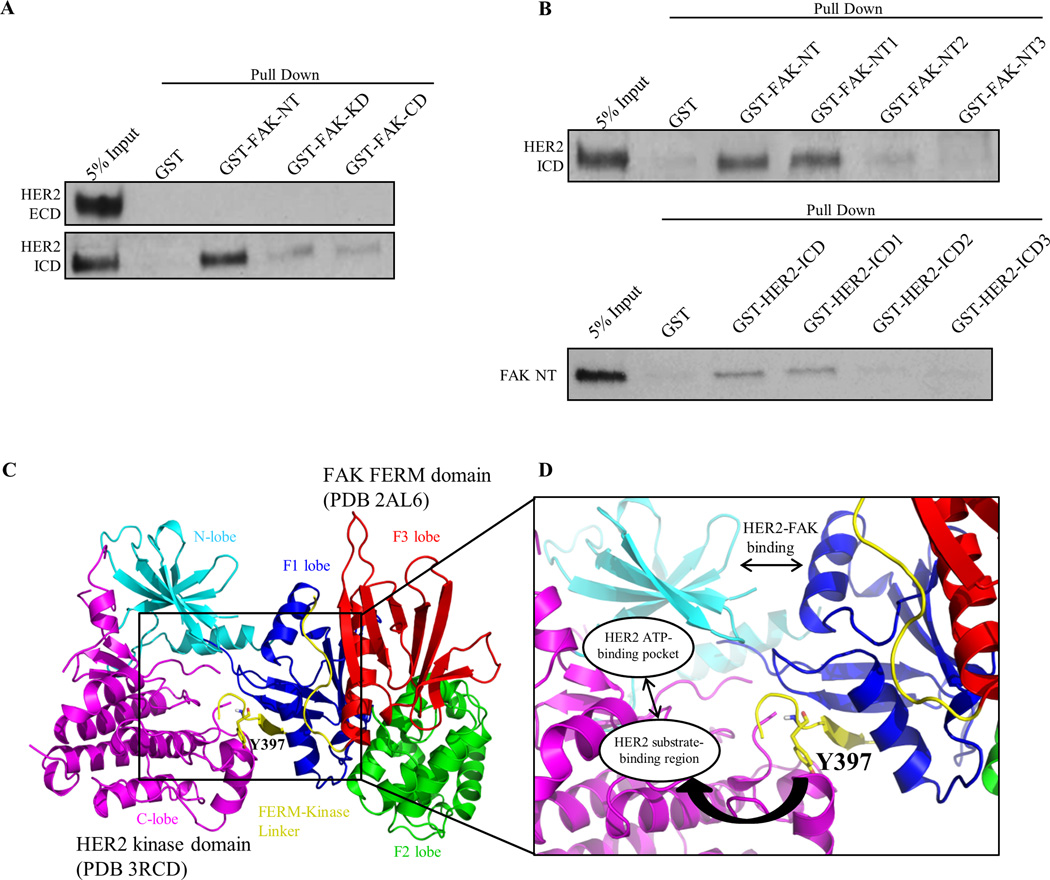
Because FAK was reactivated by RTKs independently of both FAK and SRC activity, we hypothesized that RTKs were directly binding to FAK to promote phosphorylation. Although HER2 has been previously shown to co-localize and form a complex with FAK in cells, it is still unknown whether HER2 and FAK form a direct protein-protein interaction (43, 44). To further confirm the mechanism of direct FAK activation by HER2, we performed a series of GST pull-down assays with purified recombinant proteins. GST-FAK constructs were cloned according to FAK-NT (N-terminus), -KD (Kinase Domain), or -CD (C-terminal Domain) regions and fusion-proteins were incubated with either HER2-ICD (Intracellular Domain) or -ECD (Extracellular Domain) proteins (Fig. 3A and Supplementary Fig. S9A). Significant binding was detected between GST-FAK-NT and HER2-ICD, but minimal binding was observed for FAK-KD and –CD regions. As expected, no binding was detected between GST-FAK constructs and HER2-ECD protein.
Fig. 3. HER2 directly binds FAK as a structural mechanism to promote Y397 phosphorylation.
(A) GST pull-down assays demonstrating direct binding of FAK-NT (N-terminal domain) to HER2-ICD (Intracellular domain). GST, GST-FAK-NT, GST-FAK-KD (Kinase domain), and GST-FAK-CD (C-terminal domain) proteins were incubated with HER2-ECD (Extracellular domain) or HER2 ICD (Intracellular domain) proteins and resulting immunoblots represent the bound fraction. (B) Upper panel: GST pull-down assays showing direct binding of FAK-NT1 (FERM-F1 lobe) to HER2-ICD. GST-FAK-NT1, GST-FAK-NT2, and GST-FAK-NT3 were cloned according to FAK-FERM-F1, -F2, and -F3 lobes, respectively. Lower panel: GST pull-down assays showing direct binding of HER2-ICD1 (kinase N-lobe) to FAK-NT (FERM). GST-HER2-ICD, -ICD1, -ICD2, and -ICD3 were cloned according to HER2-ICD, kinase N-lobe, kinase C-lobe, and c-terminal tail regions, respectively. Images shown are representative of three independent experiments. (C) Structural model of the HER2-FAK interaction as determined by HADDOCK docking studies. The N-lobe (cyan) and C-lobe (magenta) of the HER2 kinase domain (PDB 3RCD) as well as the F1 lobe (blue), F2 lobe (green), F3 lobe (red), and FERM-kinase linker (yellow) of the FAK FERM domain (PDB 2AL6) are depicted. The model shown represents the top-ranked cluster in HADDOCK. (D) Zoomed inset of the HER2-FAK model showing proximity of the binding interface to Y397 of the FERM-kinase linker region and orientation of Y397 within the HER2 substrate-binding region. All images were created in PyMOL
To further define the binding region between HER2 and FAK, we performed similar assays with GST-FAK-NT1, NT2, and NT3 proteins cloned according to the F1, F2, and F3 lobes of the FAK N-terminal FERM domain (Fig. 3B and Supplementary Fig. S9B). The FAK NT1 (or F1 lobe) region, but not NT2 or NT3, was found to bind to HER2-ICD. Additionally, we tested various HER2 segments with GST-HER2-ICD, ICD1, ICD2, and ICD3 proteins cloned according to whole ICD, kinase domain N-lobe, kinase domain C-lobe, and c-terminal tail regions of the HER2 ICD (Fig. 3B and Supplementary Fig. S9C). Both the HER2-ICD and ICD1 (or kinase N-lobe) regions, but not ICD2 or ICD3, were found to bind to FAK-NT. These data confirmed the direct interaction of FAK and HER2 between the FAK FERM F1 lobe and HER2 kinase N-lobe.
We then examined the x-ray crystal structure of the FAK-FERM domain (PDB 2AL6) and observed that Y397, located within the flexible FERM-Kinase linker region, binds back onto the structured FERM F1 lobe where HER2 interacts (Supplementary Fig. S10). Since Fl lobe binding could provide proximity of HER2 to Y397, we utilized HADDOCK protein-protein docking to develop a structural model for HER2-FAK binding and putative Y397 transphosphorylation (40). FAK FERM (PDB 2AL6) and HER2 kinase domain (PDB 3RCD) crystal structures were computationally analyzed for active surface residues likely to be involved in protein-protein interactions using the program CPORT (39). Subsequently, CPORT-predicted residues that overlapped with experimentally validated amino acids from GST pull-down assays were set as restraints to drive the HER2-FAK docking model. Intriguingly, HADDOCK produced a HER2-FAK structural model similar to a prototypical kinase-substrate interaction (Fig. 3C). As observed in Fig. 3D, the FAK FERM F1 lobe makes contact with the HER2 N-lobe to orient the flexible Y397 FERM-linker segment close to the HER2 substrate-binding region and ATP-binding pocket. HADDOCK docking was also performed similarly with the EGFR kinase domain (PDB 4RIW) to approximate a model between EGFR and FAK (Supplementary Fig. S11). In agreement with the HER2 model, the FAK FERM domain was predicted to bind EGFR through the F1 lobe, facilitating orientation of Y397 into the EGFR ATP-binding pocket. These data provided structural rationale for the HER2-FAK interaction, where binding of HER2 (and potentially other RTKs) to the F1 lobe promotes proximity to Y397, allowing HER2 to directly phosphorylate Y397.
HER2, EGFR, and additional RTKs directly phosphorylate FAK FERM at Y397
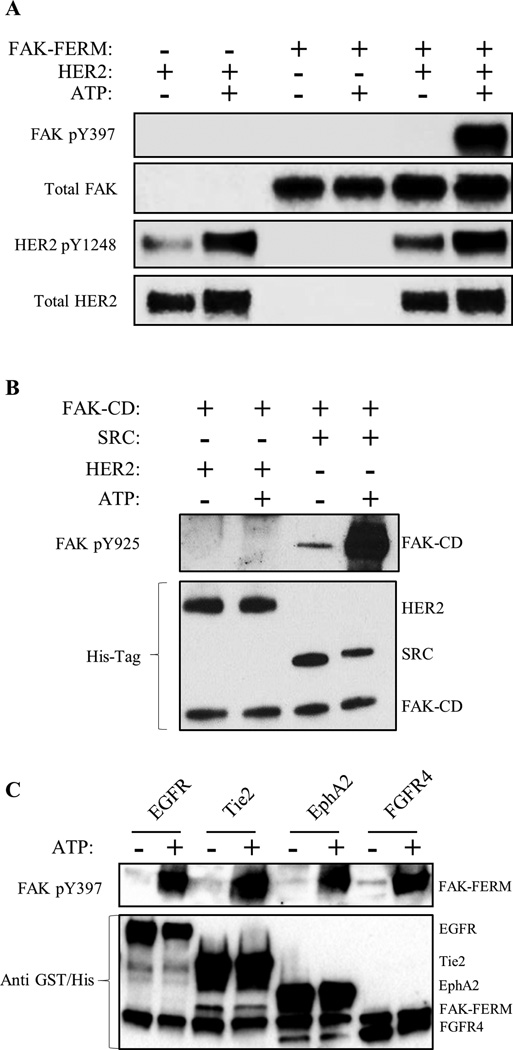
Direct HER2-FAK binding assays as well as our HADDOCK docking model suggested that HER2, and potentially other RTKs, could directly activate FAK at Y397 (Fig. 3). To evaluate direct phosphorylation and experimentally validate our HADDOCK model, we tested whether HER2 could directly phosphorylate FAK using an in vitro kinase assay with purified HER2-ICD, FAK-FERM, and FAK-CD proteins (Fig. 4A and 4B). In agreement with our HADDOCK model, HER2 directly phosphorylated FAK-FERM specifically at Y397. However, no phosphorylation was detected at FAK-CD Y925, confirming the specificity of the kinase reaction. Intriguingly, multiple RTKs (EGFR, Tie2, EphA2, FGFR4) were also found to directly phosphorylate FAK at Y397 (Fig. 4C), indicating a redundant FAK activation pathway by multiple RTKs. These data supported a direct Y397 phosphorylation mechanism by HER2 and other RTKs, where HER2 directs interacts with the FAK-FERM F1 lobe to phosphorylate Y397.
Fig. 4. HER2, EGFR and additional RTKs directly phosphorylate FAK at Y397.
(A) Direct in vitro kinase assay between HER2-ICD and FAK-FERM domain purified proteins showing direct phosphorylation of FAK Y397 by HER2 with the addition of ATP. (B) Direct in vitro kinase assay between HER2, SRC, and FAK-CD domain purified proteins showing the direct phosphorylation of Y925 only by SRC (positive control) but not HER2 with the addition of ATP. (C) Direct in vitro kinase assay between EGFR, Tie2, EphA2, and FGFR4 and FAK-FERM domain purified proteins showing direct phosphorylation of FAK Y397 by RTKs with the addition of ATP. Images shown are representative of three independent experiments.
Small molecule inhibition of FAK kinase activity induces compensatory RTK reprogramming
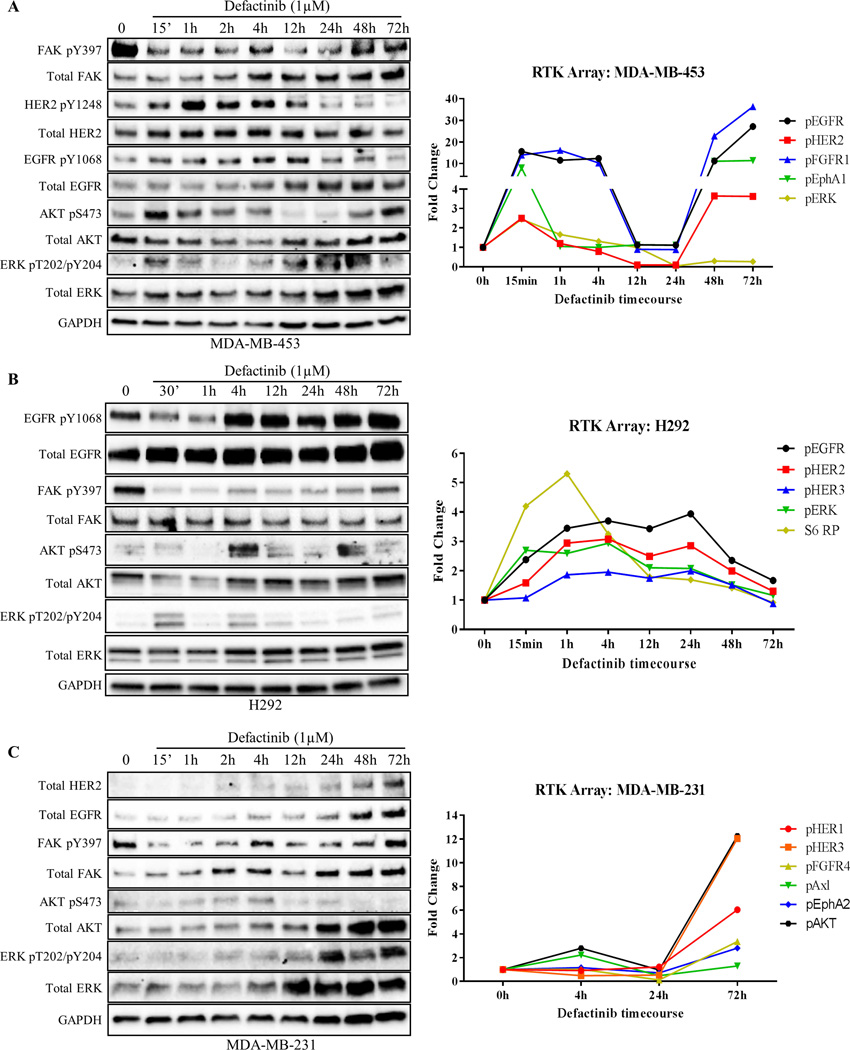
As mentioned previously, even in the absence of growth factor stimulation, all three FAK-kinase inhibitors stimulated the phosphorylation of RTK/AKT/ERK pathways as a drug resistance mechanism (Fig. 1). Given our data of FAK reactivation by RTKs, we hypothesized that cancer cells might induce novel RTK signatures (rapid vs. long-term depending on initial RTK expression) in response to FAK-kinase inhibitors as a mechanism to maintain FAK and AKT/ERK signaling. To test this, we performed time course experiments in RTKHigh cell lines (MDA-MB-453, SkBr3, H292) as well as RTKLow cell lines (MDA-MB-231, MDA-MB-468) and evaluated changes in phosphorylation patterns. We focused on defactinib due to its current involvement in multiple ongoing clinical trials and its recently failed Phase II clinical trial (34). Additionally, we used a dose of 1µM, which is in alignment with patient serum concentrations of defactinib achieved with the recommended phase II dose of 425 mg BID (42). In MDA-MB-453 cells, defactinib treatment induced rapid activation of HER2, EGFR, AKT, and ERK within 15 min, with maximal activation of HER2 and EGFR coming at 1 and 4 hours, respectively (Fig. 5A and Supplementary Fig. S12A). When FAK-kinase inhibition was lost and pY397 levels were restored at 48–72 hours, pHER2 and pEGFR levels also returned to basal levels.
Fig. 5. Cancer cells demonstrate rapid and long-term compensatory RTK reprogramming to defactinib.
(A) Timecourse experiment where HER2+ breast cancer cells (MDA-MB-453) were treated with FAK-kinase inhibitor (defactinib) for the indicated timepoints. Note; defactinib induced EGFR/HER2 and AKT/ERK phosphorylation within 15 min of treatment. RTK arrays were performed (right panel) with matched immunoblot confirming results. (B) Timecourse experiment where EGFR+ lung cancer cells (H292) were treated with defactinib showing rapid compensatory RTK reprogramming. RTK arrays (right panel) showed activation of EGFR, ERK, and S6RP within 30 min. (C) Timecourse experiment where triple-negative breast cancer cells (MDA-MB-231) were treated with defactinib showing long-term (72h) RTK reprogramming in MDA-MB-231 cells. Note, initially HER2− cells were found to have low levels of HER2 at 2–4 hours and high levels at 48–72 hours after defactinib treatment. RTK arrays (right panel) showed upregulation of additional RTKs, FGFR4 and EphA2, 72 hours after treatment. Immunoblot images shown are representative of three independent experiments. Densitometry and statistical analysis is found in Fig. S12. RTK array results represent results from one RTK array chip (Supplementary Table S2).
In addition, we utilized RTK arrays to assess a broader kinome response to FAK-kinase inhibition (Fig. 5, Supplementary Fig. S13, S14, and Table S2). In H292 cells, we found consistent rapid activation of RTK pathways as observed in MDA-MB-453 cells, with increases in EGFR, HER2, ERK, and AKT from 15 min to 4 hours post treatment. Immunoblots were used to confirm our findings of EGFR, AKT, and ERK activation after defactinib treatment (Fig. 5B and Supplementary Fig. S12B). Intriguingly, S6 ribosomal protein (S6 RP), a marker of increased translation of mRNA transcripts encoding for proteins regulating cell cycle progression, was rapidly activated at 15–30 minutes in two of three RTKHigh cell lines. These data showed that RTKHigh cancer cells are involved in a rapid feedback loop, where FAK-kinase inhibition induces corresponding RTK activation to compensate for loss of FAK Y397 phosphorylation. Conversely, in RTKLow cell lines, the RTK reprogramming in response to defactinib treatment occurred at longer timepoints (48 and 72 hours). In MDA-MB-231 triple-negative (ER/PR/HER2−) breast cancer cells, defactinib induced the expression of HER2 and EGFR as well as the activation of other RTKs such as FGFR4 and EphA2 at 72 hours. Immunoblots were used to confirm to total protein increases in both EGFR and HER2 after defactinib treatment (Fig. 5C and Supplementary Fig. 12C). These data demonstrated that RTKLow cancer cells require longer time points to undergo kinome changes, most likely due to the requirement to express new RTKs. Additionally, these data showed that the selective pressure of FAK-kinase inhibition is able to drive triple-negative breast cancer cells to express HER2.
RTK positivity predicts resistance to FAK kinase inhibitors
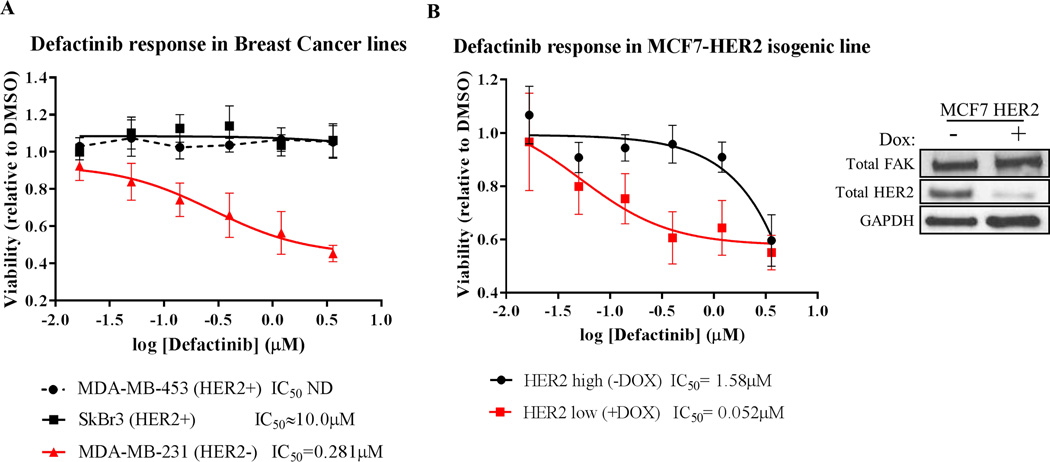
Based on our findings of rapid vs. long-term RTK reprogramming in RTKHigh vs. RTKLow cells, we predicted that cancer cells driven by oncogenic RTKs would be more resistant to FAK inhibition than those that are not driven by RTKs. We utilized 3D matrigel-on-top growth assays to measure of the efficacy of FAK kinase inhibitors, as the 3D-environment of these assays were shown to recapitulate the cellular requirement for FAK (45, 46). Indeed, HER2-addicted breast cancer cell lines MDA-MB-453 (IC50 ND) and SkBr3 (IC50 > 10µM) showed no changes in viability in response to defactinib treatment in 3D growth assays, whereas defactinib treatment decreased the viability of HER2− cell line MDA-MB-231 (IC50 = 0.281µM) in a dose-responsive manner (Fig. 6A). We confirmed these findings using the MCF7-HER2 Tet-Off system, where HER2 expression was modulated by the simple removal or addition of doxycycline in this isogenic system (Fig. 6B). HER2High (-Dox) cells showed minimal response to defactinib (IC50 = 1.58µM), whereas HER2Low (+Dox) cells were quite responsive to defactinib treatment (IC50 = 0.052µM). Together, these data showed that HER2 expression levels can distinguish cancer cells that may or may not respond to FAK-kinase inhibitors and demonstrated that RTKs such as HER2 can drive resistance to FAK-kinase inhibitors.
Fig. 6. HER2-positive cancer cells are resistant to FAK-kinase inhibition.
Matrigel on-top 3D growth assays with (A) a panel of breast cancer cell lines and (B) MCF7-HER2 isogenic line (Tet-Off) treated with titrating doses of defactinib. HER2 positivity confers resistance to defactinib treatment (IC50 values shown below graph). Viability values shown are averaged results from triplicate measurements in three independent experiments. Error bars shown represent 95% confidence intervals. The immunoblot right of (B) shows HER2 knockdown with 1µg/mL doxycycline treatment.
Discussion
The data described in this report have identified a novel mechanism of drug resistance that may explain some of the failures of FAK-kinase inhibitors in clinical trials. Here, we have shown that oncogenic RTKs, such as EGFR or HER2, directly phosphorylated FAK at Y397 to rescue kinase-inhibited FAK in cancer cells. Additionally, HER2 formed a direct protein-protein interaction complex with FAK nearby this critical residue Y397. We also have shown that RTKHigh cell lines displayed rapid resistance to FAK-kinase inhibitors due to upregulation of RTK signaling pathways while RTKLow cells were initially sensitive to the inhibitors. However, RTKLow cell lines, such as MDA-MB-231, started to express HER2 and EGFR protein after longer incubations with defactinib, explaining a possible acquired-resistance mechanism to FAK-kinase inhibitors. An overview of our findings is summarized in Supplementary Fig. S15. In all, we have identified a common resistance mechanism to FAK-kinase inhibitors, whereby RTKs can bypass FAK-kinase inhibition to re-phosphorylate FAK Y397.
In our analysis, we noticed that RTKHigh cell lines showed a trend whereby RTK activation led to increased FAK pY397, but we also observed that different RTKHigh cell lines had different capacities to re-phosphorylate FAK at Y397. Additionally, EGFR+ cells line, A549, showed minimal reduction of pY397 by FAK-kinase inhibitors and compensatory increases in pEGFR, suggesting an innate resistance to these inhibitors. Also observed was a temporal difference between cell lines in their ability to induce compensatory RTK reprogramming after defactinib treatment. To explain these results, we propose that the ability of RTKs to transphosphorylate FAK at Y397 is very cell line-dependent, where multiple variables (i.e. RTK levels, mutation status, adaptor protein levels) can affect both the levels and kinetics of Y397 phosphorylation. Future experiments will be designed to evaluate other cellular factors that may drive Y397 phosphorylation and FAK-kinase inhibitor resistance.
Of additional notice was the strong compensatory activation of HER2/AKT/ERK pathways as a drug resistance mechanism to FAK-kinase inhibitors. We acknowledge that high-dose kinase inhibitor treatment has the potential for off-target kinase effects. Nonetheless, our data with low concentrations of defactinib and FAK-specific siRNA suggest that the compensatory RTK reprogramming after FAK-kinase inhibitor treatment was FAK-specific. Kinase inhibitor off-target kinase effects are generally inhibitory (due to the conserved ATP-binding region) and not agonistic in nature. In addition, the chemical structures of all three FAK-kinase inhibitors (defactinib, PF-271, PF-228) are different and therefore display different off-target kinase effects as observed (28, 29, 41). However, all three FAK-kinase inhibitors still activated HER2/EGFR and AKT/ERK in a similar manner, supporting the notion that these compensatory increases are FAK-specific. Further, as mentioned previously, the recommended phase II dose of defactinib is 425 mg BID, and at steady state dosing (day 15) the average serum Cmax was greater than 1.0 ug/mL, or 1.95 uM, indicating that our experimental doses are not only appropriate for cell culture studies, but are also in line with circulating levels of drug in patients (42). Therefore, we believe our data describe a clinically relevant drug resistance mechanism to FAK-kinase inhibitors at concentrations utilized in current clinical trials.
Surprisingly, we found that multiple RTKs from different family-members could directly phosphorylate FAK FERM at Y397. Additionally, we found a wide array of upregulated RTKs in response to the FAK-kinase inhibitor in widespread clinical trials, defactinib. These findings suggest there are redundant transphosphorylation mechanisms where cancer cells can hijack multiple different RTK pathways (depending on cellular context and genomic accessibility) to maintain FAK Y397 pathway activation during FAK-kinase inhibition. Also, FAK depletion by siRNA in SkBr3 cells induced the hyperactivation of FAK Y397 despite lower levels of total FAK, implying cancer cell dependency on FAK Y397 phosphorylation. Therefore, we propose FAK Y397 phosphorylation as a fragile point in cancer cells, required to maintain cancer cell motility and proliferation. We have termed our observation “oncogenic protection of FAK” because of the ability of different oncogenic RTKs to phosphorylate and protect this fragile Y397 site. In fact, several other groups have shown the requirement of FAK for tumor initiation and progression (12, 13). Additionally, another group has implicated the FERM domain in binding of FAK to RTKs in cells, but did not show a direct interaction (19). We have demonstrated a direct interaction of FAK and HER2 and further hypothesize that multiple RTKs bind to the FAK-F1 lobe as a means to directly phosphorylate Y397. Although our direct kinase and cellular data support the redundant phosphorylation of FAK Y397 by multiple RTKs, we acknowledge the known promiscuity issue with in vitro kinase assays (47). Future cellular evaluation of novel RTK-FAK signaling complexes as well as in vivo drug resistance models will be helpful as we seek to use precision RTK biomarkers of a patient’s tumor to predict their response/resistance to FAK kinase inhibitors.
Several recent reports have characterized a novel drug resistance mechanism to kinase inhibitors involving rapid dynamic kinase signaling called kinome reprogramming (48–50). These groups have identified global changes in cancer cell kinome expression and phosphorylation events after kinase inhibitor treatment. Interestingly, MEK-inhibitor and HER2-inhibitor treatment induced global upregulation of redundant intracellular signaling nodes, RTKs, and MAPK/AKT signaling pathways. Our data describe a novel and previously uncharacterized form of compensatory RTK reprogramming to FAK-kinase inhibitors through the re-phosphorylation of FAK Y397 but are in agreement with these studies where other kinase inhibitors induced global changes in kinase expression/activity. It remains to be determined whether FAK-kinase inhibition induces both whole kinome as well as non-kinase forms of drug resistance.
At the time this manuscript was prepared, there were 18 reported clinical trials involving FAK inhibitors on clinicaltrials.gov (35). Although FAK is considered a promising drug target for cancer therapy, questions still remain on which function of FAK (kinase vs. scaffold) is optimal to target and under which molecular context (17). Several pre-clinical studies have shown that molecular markers, such as MerlinLow expression, can be utilized to stratify tumor sub-types which may be responsive to FAK-kinase inhibitors (46). Unfortunately, clinical trial results of the COMMAND (Control Of Mesothelioma with MAiNtenance Defactinib) trial have shown limited efficacy and no difference in control vs. defactinib-treated patients with MerlinLow tumors (34). As such, it is important to identify new molecular markers, such as HER2 and other RTKs that may have the ability to predict which population of patients will or will not respond to FAK-kinase inhibitor therapy.
Finally, the data presented here have several direct clinical implications regarding FAK-kinase inhibitors. First, we show that expression of RTKs in cancer cells, such as HER2, can cause resistance to FAK-kinase inhibitors due to re-phosphorylation of Y397. This suggests an opportunity to retrospectively analyze tumor samples from patients who have received FAK-kinase inhibitors to determine the levels of specific RTKs such as HER2 and EGFR and correlate them with clinical response. Second, our data suggest that RTKLow cancers will be initially sensitive to FAK-kinase inhibitors and that RTK expression can be induced as a mechanism of acquired resistance. This suggests the need to regularly monitor tumors for changes in their kinome that could predict acquired resistance to the FAK-kinase inhibitor. Third, these studies provide a molecular rationale for combination therapy of FAK-kinase inhibitors with RTK inhibitors such as lapatinib or erlotinib. However, we predict eventual resistance to combination therapy, due to the observed upregulation of diverse RTKs in RTK reprogramming assays. As an alternative, the discovery and development of FAK-scaffold inhibitors that directly inhibit FAK Y397 may provide a more durable response to FAK-based therapy (51) by blocking the fragile Y397 site to globally prevent its phosphorylation. As we further understand the dynamic nature that surrounds FAK and its complex interactome, we believe that more precision-targeted therapeutics will be elucidated.
Supplementary Material
Acknowledgments
We would like to thank Drs. Vita Golubovskaya, Elena Kurenova, and Shelton Earp for scientific discussions regarding the research topic, Mr. Thomas Mathers for pharmaceutical industry advice, the Roswell Park Flow Cytometry Core for performing FACS analysis, and Mr. George Clinton for creative influences.
Funding: This work was funded by the NCI (R01CA065910) awarded to W.G. Cance, a Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (T32CA009072) awarded to T.A. Marlowe, and a NCI Cancer Center Support Grant for Roswell Park Cancer Institute (P30CA016056).
Abbreviations
- FAK
Focal Adhesion Kinase
- RTKs
Receptor Tyrosine Kinases
- HER2
Human Epidermal Growth Factor Receptor 2
- EGFR
Human Epidermal Growth Factor Receptor
- FERM
4.1 Ezrin Radixin Moesin
Footnotes
Competing interests: We declare no financial conflicts of interest. W.G. Cance is Chief Scientific Officer of FAKnostics, LLC, a company focused on FAK biomarkers and therapeutics.
References
- 1.Weiner TM, Liu ET, Craven RJ, Cance WG. Expression of focal adhesion kinase gene and invasive cancer. Lancet. 1993;342:1024–1025. doi: 10.1016/0140-6736(93)92881-s. [DOI] [PubMed] [Google Scholar]
- 2.Owens LV, Xu L, Craven RJ, Dent GA, Weiner TM, Kornberg L, et al. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Research. 1995;55:2752–2755. [PubMed] [Google Scholar]
- 3.Lark AL, Livasy CA, Calvo B, Caskey L, Moore DT, Yang X, et al. Overexpression of focal adhesion kinase in primary colorectal carcinomas and colorectal liver metastases: immunohistochemistry and real-time PCR analyses. Clin Cancer Res. 2003;9:215–222. [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Schaller MD, Borgman CA, Cobb BS, Vines RR, Reynolds AB, Parsons JT. pp125fak a structurally distinctive protein-tyrosine kinase associated with focal adhesions. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5192–5196. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanks SK, Calalb MB, Harper MC, Patel SK. Focal adhesion protein-tyrosine kinase phosphorylated in response to cell attachment to fibronectin. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8487–8491. doi: 10.1073/pnas.89.18.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siesser PMF, Hanks SK. The Signaling and Biological Implications of FAK Overexpression in Cancer. Clin Cancer Res %R 101158/1078-0432CCR-06-0456. 2006;12:3233–3237. doi: 10.1158/1078-0432.CCR-06-0456. [DOI] [PubMed] [Google Scholar]
- 8.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Advanced drug delivery reviews. 2011;63:610–615. doi: 10.1016/j.addr.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S, et al. Mammary Epithelial-Specific Ablation of the Focal Adhesion Kinase Suppresses Mammary Tumorigenesis by Affecting Mammary Cancer Stem/Progenitor Cells. Cancer Res. 2009;69:466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serrels A, Lund T, Serrels B, Byron A, McPherson RC, von Kriegsheim A, et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell. 2015;163:160–173. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean GW, Komiyama NH, Serrels B, Asano H, Reynolds L, Conti F, et al. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 2004;18:2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci U S A. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provenzano PP, Inman DR, Eliceiri KW, Beggs HE, Keely PJ. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am J Pathol. 2008;173:1551–1565. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K–dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. The Journal of Clinical Investigation. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golubovskaya VM, Kweh FA, Cance WG. Focal adhesion kinase and cancer. Histol Histopathol. 2009;24:503–510. doi: 10.14670/HH-24.503. [DOI] [PubMed] [Google Scholar]
- 17.Cance WG, Kurenova E, Marlowe T, Golubovskaya V. Disrupting the scaffold to improve focal adhesion kinase-targeted cancer therapeutics. Science Signaling. 2013;6:e10. doi: 10.1126/scisignal.2004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zachary I, Rozengurt E. Focal adhesion kinase (p125FAK): a point of convergence in the action of neuropeptides, integrins, and oncogenes. Cell. 1992;71:891–894. doi: 10.1016/0092-8674(92)90385-p. [DOI] [PubMed] [Google Scholar]
- 19.Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 20.Golubovskaya VM, Finch R, Cance WG. Direct Interaction of the N-terminal domain of Focal Adhesion Kinase with the N-terminal transactivation domain of p53. J Biol Chem. 2005;280:25008–25021. doi: 10.1074/jbc.M414172200. [DOI] [PubMed] [Google Scholar]
- 21.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 22.Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Molecular & Cellular Biology. 1994;14:1680–1688. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HC, Appeddu PA, Isoda H, Guan JL. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. Journal of Biological Chemistry. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 24.Han DC, Guan JL. Association of focal adhesion kinase with Grb7 and its role in cell migration. J Biol Chem. 1999;274:24425–24430. doi: 10.1074/jbc.274.34.24425. [DOI] [PubMed] [Google Scholar]
- 25.Calalb MB, Zhang X, Polte TR, Hanks SK. Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochemical & Biophysical Research Communications. 1996;228:662–668. doi: 10.1006/bbrc.1996.1714. [DOI] [PubMed] [Google Scholar]
- 26.Schlaepfer DD, Hunter T. Evidence for in vivo phosphorylation of the Grb2 SH2-domain binding site on focal adhesion kinase by Src-family protein-tyrosine kinases. Molecular & Cellular Biology. 1996;16:5623–5633. doi: 10.1128/mcb.16.10.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia H, Nho RS, Kahm J, Kleidon J, Henke CA. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. J Biol Chem. 2004;279:33024–33034. doi: 10.1074/jbc.M313265200. [DOI] [PubMed] [Google Scholar]
- 28.Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- 29.Roberts WG, Ung E, Whalen P, Cooper B, Hulford C, Autry C, et al. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res. 2008;68:1935–1944. doi: 10.1158/0008-5472.CAN-07-5155. [DOI] [PubMed] [Google Scholar]
- 30.Tanjoni I, Walsh C, Uryu S, Tomar A, Nam JO, Mielgo A, et al. PND-1186 FAK inhibitor selectively promotes tumor cell apoptosis in three-dimensional environments. Cancer Biol Ther. 2010;9:764–777. doi: 10.4161/cbt.9.10.11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Q, Hjelmeland AB, Keir ST, Song L, Wickman S, Jackson D, et al. A novel low-molecular weight inhibitor of focal adhesion kinase, TAE226, inhibits glioma growth. Mol Carcinog. 2007;46:488–496. doi: 10.1002/mc.20297. [DOI] [PubMed] [Google Scholar]
- 32.Infante JR, Camidge DR, Mileshkin LR, Chen EX, Hicks RJ, Rischin D, et al. Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J Clin Oncol. 2012;30:1527–1533. doi: 10.1200/JCO.2011.38.9346. [DOI] [PubMed] [Google Scholar]
- 33.Gan H, Nebot N, Soria JC, Arkenau HT, Blagden S, Plummer R, et al. Pharmacokinetics/pharmacodynamics (PK/PD) of GSK2256098, a Focal Adhesion Kinase (FAK) Inhibitor, in Patients with Advanced Solid Tumors. 24th EORTC-NCI-AACR Symposium on ‘Molecular Targets and Cancer Therapeutics’; 6 to 9 November 2012; Dublin, Ireland. 2012. [Google Scholar]
- 34.Verastem, Inc. ClinicalTrials.gov [Internet] Bethesda MD: National Library of Medicine (US); 2000. [cited 2016 April 24]. Placebo Controlled Study of VS-6063 in Subjects With Malignant Pleural Mesothelioma (COMMAND) Available from: http://clinicaltrials.gov/ct2/show/record/NCT01870609 NLM Identifier: NCT01870609. [Google Scholar]
- 35.U.S. National Institutes of Health. ClinicalTrials.gov [Internet] Bethesda MD: National Library of Medicine (US); 2000-. [cited 2016 April 24]. Search: “FAK inhibitor”. Available from: http://clinicaltrials.gov/ct2/results?term=FAK+inhibitor&Search=Search. [Google Scholar]
- 36.Sartor CI, Zhou H, Kozlowska E, Guttridge K, Kawata E, Caskey L, et al. Her4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol Cell Biol. 2001;21:4265–4275. doi: 10.1128/MCB.21.13.4265-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem. 2006;281:252–259. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- 38.Zheng D, Kurenova E, Ucar D, Golubovskaya V, Magis A, Ostrov D, et al. Targeting of the protein interaction site between FAK and IGF-1R. Biochemical and Biophysical Research Communications. 2010;388:3015. doi: 10.1016/j.bbrc.2009.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Vries SJ, Bonvin AM. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 41.National Center for Biotechnology Information. [accessed Aug. 9, 2016];PubChem Compound Database; CID=25117126. https://pubchem.ncbi.nlm.nih.gov/compound/25117126.
- 42.Jones SF, Siu LL, Bendell JC, Cleary JM, Razak AR, Infante JR, et al. A phase I study of VS-6063, a second-generation focal adhesion kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs. 2015;33:1100–1107. doi: 10.1007/s10637-015-0282-y. [DOI] [PubMed] [Google Scholar]
- 43.Benlimame N, He Q, Jie S, Xiao D, Xu YJ, Loignon M, et al. FAK signaling is critical for ErbB-2/ErbB-3 receptor cooperation for oncogenic transformation and invasion. J Cell Biol. 2005;171:505–516. doi: 10.1083/jcb.200504124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang SE, Xiang B, Zent R, Quaranta V, Pozzi A, Arteaga CL. Transforming growth factor beta induces clustering of HER2 and integrins by activating Src-focal adhesion kinase and receptor association to the cytoskeleton. Cancer Res. 2009;69:475–482. doi: 10.1158/0008-5472.CAN-08-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2012;2:706–721. doi: 10.1158/2159-8290.CD-11-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro IM, Kolev VN, Vidal CM, Kadariya Y, Ring JE, Wright Q, et al. Merlin deficiency predicts FAK inhibitor sensitivity: a synthetic lethal relationship. Science translational medicine. 2014;6 doi: 10.1126/scitranslmed.3008639. 237ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning BD, Cantley LC. Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002:e49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 48.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015;11:390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kampen KR, Ter Elst A, Mahmud H, Scherpen FJ, Diks SH, Peppelenbosch MP, et al. Insights in dynamic kinome reprogramming as a consequence of MEK inhibition in MLL-rearranged AML. Leukemia. 2014;28:589–599. doi: 10.1038/leu.2013.342. [DOI] [PubMed] [Google Scholar]
- 51.Golubovskaya VM, Nyberg C, Zheng M, Kweh F, Magis A, Ostrov D, et al. A Small molecule Inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the Y397 site of Focal Adhesion Kinase decreases tumor growth. J Med Chem. 2008;51:7405–7416. doi: 10.1021/jm800483v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.