Abstract
Long non-coding RNA (lncRNA) may contribute to the initiation and progression of tumor. In this study, we first systematically compared lncRNA and mRNA expression between GBM and paired normal brain tissues using microarray data. We found 27 lncRNA and 82 mRNA significantly up-regulated in GBM, as well as 198 lncRNA and 285 mRNA significantly down-regulated in GBM. We identified 138 co-expressed lncRNA-mRNA pairs from these differentially expressed lncRNA and genes. Subsequent pathway analysis of the lncRNA-paired genes indicated that EphrinB-EPHB, p75-mediated signaling, TNF alpha/NF-κB, and ErbB2/ErbB3 signaling pathways might be altered in GBM. Specifically, lncRNA RAMP2-AS1 had significant decrease of expression in GBM tissues and showed co-expressional relationship with NOTCH3, an important tumor promoter in many neoplastic diseases. Our follow up experiment indicated that (1) an overexpression of RAMP2-AS1 reduced GBM cell proliferation in vitro and also reduced GBM xenograft tumors in vivo; (2) NOTCH3 and RAMP2-AS1 co-expression rescued the inhibitory action of RAMP2-AS1 in glioblastoma cells; and (3) RNA pull-down assay revealed a direct interaction of RAMP2-AS1 with DHC10, which may consequently inhibit, as we hypothesize, the expression of NOTCH3 and its downstream signaling molecule HES1 in GBM. Taken together, our data revealed that lncRNA expression profile in GBM tissue was significantly altered; and RAMP2-AS1 might play a tumor suppressive role in GBM through an indirect inhibition of NOTCH3. Our results provided some insights into understanding the key roles of lncRNA-mRNA co-regulation in human GBM and the mechanisms responsible for GBM progression and pathogenesis.
Keywords: Glioblastoma, Glioma stem cells, LncRNA-mRNA co-expression network, lncRNA-RAMP2-AS1, NOTCH3
Introduction
Gliomas account for approximately 30% of primary tumors in the brain (1), and can be categorized into four grades (I-IV) by the histopathological classification according to The World Health Organization (WHO) (2). Among the four grades, glioblastoma (GBM, grade IV) has the most dismal prognosis with an overall survival of less than 14 months (1). Despite multimodal and aggressive treatments that include surgical resection, local radiotherapy and systemic chemotherapy, the outcome of GBM patients remains poor (3, 4). To improve treatment efficacy, a better understanding of glioma pathogenesis at the genetic and molecular levels is urgently needed.
Molecular profiling of normal and tumor tissues has revealed that long non-coding RNA (lncRNA) is dysregulated in many human malignancies, including prostate (5), colorectal (6), breast (7), bladder (8), liver (9) and brain cancers (10). Multiple lncRNA genes are postulated to function as oncogenes and tumor suppressors and regulate many hallmarks of cancer. In particular, the regulatory roles of lncRNA in expression, activity and localization of protein-coding genes have attracted much attention (11).
Recent evidence has indicated that lncRNA may play important roles in glioma pathogenesis (12, 13). For example, it has been reported that lncRNA may regulate biological processes in glioma, such as cellular proliferation and apoptosis, which contribute to tumorigenesis (14). Aberrant expression of lncRNA has also been implicated for clinical phenotypes (15) and patient prognosis of GBM (16), which could be further exploited as potential diagnostic and therapeutic targets (17).
An increasing number of lncRNAs have been characterized in cancer including GBM, lncRNA H19 has been shown to promote glioma angiogenesis and invasion (18, 19). Another study reported that knockdown of lncRNA XIST exerted tumor-suppressive functions in human glioblastoma stem cells by up-regulating the microRNA -152 (20). lncRNA HOTAIR was found to be a cell cycle-regulator and essential for proliferation in human glioblastoma (21, 22). Although these lncRNAs were found their roles in some glioma paradigm, systematic investigation of lncRNA in GBM has not been reported yet (23). In this study, we performed a systematic analysis of lncRNAs with their paired mRNAs in GBM, from which the we pinpointed several lncRNAs whose aberrant expression in GBM specimens potentially altered GBM-specific pathways. Specifically, we studied the function of a lncRNA, RAMP2-AS1, in GBM both in vitro and in vivo. Our data indicated that RAMP2-AS1 might contribute to GBM through targeting the NOTCH3 signaling pathway. Our study provides some important insights of lncRNAs into the mechanisms responsible for GBM pathogenesis.
Materials and Methods
Acquisition of clinical specimens and ethical standards
Glioblastoma specimens were obtained from 20 GBM patients who underwent surgical treatment at Navy General Hospital, China, from January 2011 to December 2013 (Table S1). Glioma was diagnosed according to the 2007 WHO Classification of Tumors of the Central Nervous System. Normal samples were obtained from the normal brain tissues around the tumors (approximately 2-3 cm from the tumor border) of the same GBM patients. Written informed consent of the patients was provided by their legal surrogates to permit surgical procedures and use of resected tissues. This study was approved by the Specialty Committee on Ethics of Biomedicine Research, Navy General Hospital of PLA, China (permission number: 0506-2006).
Microarray expression profiles for lncRNA and mRNA
The Agilent human lncRNA and mRNA array V4.0 was designed with four identical arrays per slide (4 × 180K format), with each array containing probes interrogating approximately 41,000 and 34,000 human lncRNAs and mRNAs, respectively. These lncRNA and mRNA target sequences were merged from multiple databases, such as 23,898 from GENCODE (V19) (24), 14,353 from Human LincRNA Catalog (25), 7760 from RefSeq, 5627 from UCSC (http://genome.ucsc.edu/), 13,701 from NRED (ncRNA Expression Database, http://nred.matticklab.com/cgi-bin/ncrnadb.pl), 21,488 from LNCipedia (http://www.lncipedia.org/), 1038 from H-InvDB (http://www.h-invitational.jp/hinv/ahg-db/news.jsp), 3019 from lncRNAs-a (Enhancer-like) (25), 1053 from Antisense ncRNA pipeline (26), 407 Hox ncRNAs, 962 UCRs (27), and 848 from Chen Ruisheng lab (Institute of Biophysics, Chinese Academy of Science, http://www.ibp.cas.cn/nRNA). Each RNA sequence was detected by probes and repeated twice. The array also contained 4974 control probes from Agilent. Data normalization of the 2 channel ratios was achieved using an intensity-dependent “Lowess” module implemented in the R programming language. Differential expression (DE) of lncRNA/gene was defined according to the following criteria: > 2.0 fold-change and P <0.05. We have submitted the data of microArray to GEO. The GEO accession number is GSE77452 (Differentially expressed lncRNAs and genes in GBM compared to matched normal brain samples).
RNA extraction, labeling and hybridization
Total RNA was extracted from 3 pairs of snap-frozen GBM specimens and matched noncancerous tissues using Trizol (Invitrogen, California), and purified with mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to manufacturer's protocol. The amplified cRNA was purified using the RNA Clean-up Kit (MN). The cDNA, labeled with a fluorescent dye (Cy5 and Cy3-dCTP), was produced by Eberwine's linear RNA amplification method and subsequent enzymatic reaction. Labeled controls and test samples, labeled with Cy5-dCTP and Cy3-dCTP, were dissolved in 80 μL hybridization solution containing 3×SSC, 0.2% SDS, 5×Denhardt's solution and 25% formamide. Arrays hybridization was performed in an Agilent Hybridization Oven overnight at a rotation speed of 20 rpm and a temperature of 42°C, then washed with two consecutive solutions (0.2% SDS and 2× SSC) at 42°C and room temperature for 5 minutes.
lncRNA-gene co-expression network construction
Pre-processed RNA-Seq data of 19 different human normal tissues including brain was downloaded from lncRNA function database (28). The downloaded expression profiling data consisted of 13,249 lncRNA and 20,447 protein-coding genes. We excluded lncRNA or genes that had low variance (belonged to lowest quartile). Furthermore, an lncRNA or mRNA gene was excluded from the analysis if >25% samples had missing values. We computed Pearson correlation coefficient for all possible lncRNA-gene, lncRNA-lncRNA, and gene-gene pairs. A pair was considered as significantly co-expressed if the absolute correlation score was >0.75 and correlation P<0.05 (P values adjusted by Benjamini-Hochberg multiple test correction method) (29).
Quantitative real-time polymerase chain reaction (qRT-PCR) validation
Total RNA was extracted from 3 GBM and 3 matched normal brain samples using TRIzol reagent (Invitrogen Life Technologies), and then reverse-transcribed using Fermentas RT reagent Kit (Perfect Real Time) according to the manufacturer's instructions. Two microgram of total RNA was converted to cDNA according to the manufacturer's protocol. Expression of lncRNA was measured by real-time RT-PCR using SYBR Premix Ex Taq on MX3000 instrument. The primers used in this study are shown in Table S2. PCR was performed in a total reaction volume of 8 μl, including 5 μl 2×PCR master mix (Superarray), 0.5 μl of PCR Forward Primer (10μM), 2 μl of cDNA, and diluted to 8 μl with double distilled water. The quantitative real-time RT-PCR reaction was set at an initial denaturation step of 10 minutes at 95°C; and 95°C (10 seconds), 60°C (60 seconds), 95°C (10 seconds) in a total 40 cycles, with a final step heating slowly from 60 to 99°C. All samples normalized to GAPDH to calculate relative lncRNA concentrations.
Cell lines and cell culture
The human glioblastoma cell lines U87 and U251 were obtained from the American Type Culture Collection. These cell lines were purchased between 2006 and 2010 and authenticated by morphologic and growth curve analysis before this study beginning. U87 and U251 cell lines were derived from human glioblastoma specimens and cultured in D/F12 medium supplemented with 10% fetal bovine serum (FBS), (Hyclone, USA).
Lentiviral infection and gene transfection
Lentivirus (LV) containing RAMP2-AS1 (P34516) and NOTCH3 full-length sequence were obtained from Shanghai GenePharma Co., Ltd. Viral particles were harvested 48 hours after cotransfection of the lentiviral vector (or the control LV vector) and the packaging vectors into HEK293T cells using Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, CA, USA). U87 and U251 cells were then infected with the LV (LV-RAMP2-AS1 or LV-NOTCH3), or the control virus (NC). The infection ratio was determined by fluorescence labeling and real-time RT-PCR.
Cell cycle distribution
U87 and U251 cells (1×105 cells) were plated in 60-mm culture plates, and the cells were infected with LV-RAMP2-AS1, LV- NOTCH3 and control virus respectively. After 96h, the cells were trpsinized, fixed in 70% ethanol, washed once with PBS, and then labeled with propidium iodide (Sigma-Aldrich, USA) in the presence of RNase A (Sigma-Aldrich) for 30 min in the dark (50g/mL). Samples were run on a FACScan flow cytometer (Becton-Dickinson, USA), and the percentages of cells within each phase of the cell cycle were analyzed using Cell Quest software.
Cell proliferation analysis
Glioblastoma cells (2×103 cells per well) were placed into 96-well plates. Cells were infected with LV-RAMP2-AS1 and LV controls for 24, 48 and 72 hours, respectively. Thereafter, CCK-8 reagent was added to the cells. The cells were further cultured for two hours, and the optical density (OD) at 450 nm was measured by a microplate reader according to the manufacturer's instructions (Thermo Fisher Scientific, USA).
Immunofluorescent staining
Cells were washed three times with ice-cold PBS and fixed with 4% paraformaldehyde-PBS. Cell were then incubated with 0.1% Triton-PBS for 15 minutes, 1% bovine serum albumin-PBS for 10 minutes, and then treated with goat anti-human Ki67 monoclonal antibody (1:1000, Santa Cruze, Texas, USA at 4°C overnight. FITC labeled mouse anti-goat IgG (1:1000, Santa Cruz, Texas, USA) was used for visualization.
RNA pull-down assay
Biotin-labeled RNAs were in vitro transcribed using the Biotin RNA Labeling Mix and T7 RNA polymerase (Ambion, USA), and purified with the RNeasy Mini Kit (QIAGEN, Germany) on-column digestion of DNA. To prepare the glioblastoma cell-U87 nuclear extract, frozen U87 cells were homogenized using a dounce homogenizer with 15-20 strokes in nuclear isolation buffer (250 mM sucrose, 10mM Tris-HCL [PH 7.5], 1mM EDTA with protease inhibitors). Nuclear pellets were collected by centrifugation at 1,000 × g for 10 min, resuspended in 1 ml RNA immunoprecipitation buffer (150mM Nacl, 20 mM Tris[PH7.4], 1mM EDTA, 0.5% Triton X-100 with protease inhibitors and RNaseOUT). The lysates were mechanically sheared again using a dounce homogenizer with 15–20 strokes. Nuclear membrane and other debris were pelleted by centrifugation at 12,000 rpm for 10 min. The folded sense or anti-sense RNAs (1 ug) were added into 2 mg pre-cleared nuclear lysates (supplemented with 0.2 mg/ml heparin, 0.2 mg/ml yeast tRNA and 1 mM DTT) and incubated at 4_C for 1 hr. Sixty microliters of washed Streptavidin-coupled Dynabeads (Invitrogen, USA) were added to each binding reaction and further incubated at 4°C for 1 hr. Beads were washed briefly five times with RIP buffer and heated at 70°C for 10 min in 13 LDS loading buffer, and the retrieved proteins were visualized by SDS-PAGE and silver staining. The unique protein bands shown in the sense RNA pull-down were identified by Mass Spectrometry.
Western blot
Protein lysates were prepared as previously described (30). The protein samples were resolved by SDS-PAGE and transferred onto PVDF membranes (Roche, Basel, Switzerland). The membranes were then incubated with the following antibodies: Rabbit anti-NOTCH3 (Cell Signaling Technology, USA), mouse anti-HES1 (R&D, USA), mouse anti-DHC10 (R&D, USA) and mouse anti-P16/P21/P27 (Santa Cruz, USA). Chemiluminescence antibody-labeled protein bands were detected using a G:BOX F3 (Syngene, Cambridge, UK).
Xenografts
U87 and U251 (3×108 cells) infected with LV-RAMP2-AS1 or LV control were injected (subcutaneous) into the upper right flanks of nude mice. Thirty days after injection, the xenograft were harvested and volumes measured according to the formula: V (mm3) = (a)×(b2/2), where a is the largest diameter of tumor and b is the smallest diameter of tumor. For conducting intracerebral xenografted tumors, nude mice were anesthetized and stereotaxically inoculated in the right striatum (Bregmaanteroposterior: −0.5mm, mediolateral: +2mm, dorsoventral: −3mm) with 1×108 tumor cells derived from the subcutaneous tumors. After 3 weeks of intracerebral implantation, the mice were anesthetized deeply and perfused with saline that contained 100 U/ml of heparin (Sigma-Aldrich, USA). They were then fixed with 4% of poly-formaldehyde that was prepared in PBS. Next, the paraffin sections (4μm) of the xenografted tumors were analyzed by H&E staining. Experiments involving mice were performed in accordance with an approved Institutional Animal Care and Use Committee protocol (permission number: 0308-2013).
Statistical analysis
The difference in results between two groups was examined by two-tailed Student t-test, a P value <0.05 was considered statistically significant. The mean ± S.D. values were displayed in the figures. All statistical analyses were performed and graphs were generated using the SPSS software (version 12.0; Windows platform), or R software package (version 3.2.2). Detail survival analysis using a cohort of 20 GBM patient samples (in-house data) or 150 GBM patient samples (extracted from The Cancer Genome Atlas [TCGA] project) was provided in Supplemental data 1.
Results
Systematic identification of lncRNA in GBM
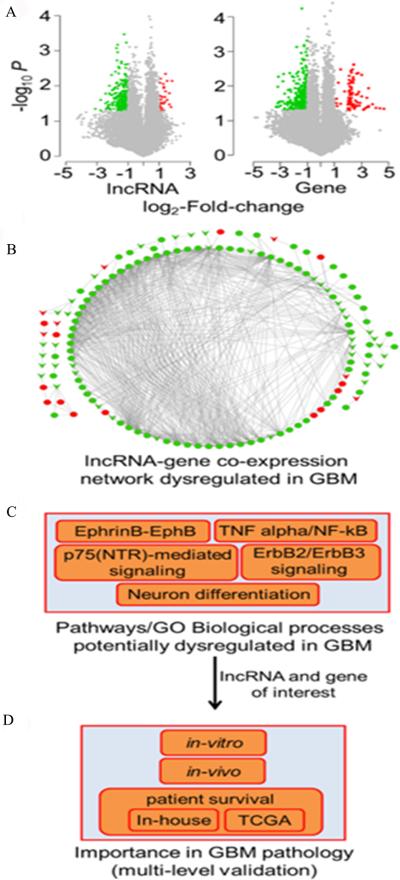
We first performed microarray expression profiling of lncRNA and mRNA in 3 human GBM and matched normal brain tissue samples. These analyses yielded 27 up- and 198 down-regulated lncRNA, as well as 82 up- and 285 down-regulated protein-coding genes in GBM compared to normal brain tissues of the same patients that passed the criteria of >2 fold-change and P <0.05 (Fig. 1A). Next, we mapped the aberrantly expressed lncRNA and mRNA onto an lncRNA-mRNA co-expression network, which was constructed based on significant expression correlation (absolute Pearson correlation coefficient >0.75, P <0.05 after adjusted by Benjamini-Hochberg [BH] multiple testing correction method) (31) among all the lncRNA and protein-coding genes across 19 human normal tissues, including brain (32). In the network, there were three types of co-expression links: between a coding RNA and a coding RNA, between a non-coding RNA and a coding RNA, and between a non-coding RNA and a non-coding RNA (Fig. 1B). For each type of co-expression link, we summarized dysregulation patterns of associated molecules, number of dysregulated links, lncRNA, and genes in Supplemental Table S3 (33).
Figure 1.
Flowchart for deciphering functional lncRNA(s) in GBM. The framework has the following major steps. A, Identification of differentially expressed (DE) lncRNAs and genes in GBM compared to matched normal samples. Volcano plots show DE probes for lncRNAs and mRNA genes. X-axis and Y-axis show fold-change and −log(P values), respectively. Red, green, and grey dots represent up-, down-, and no significant expression change, respectively. B, Construction of lncRNA-gene co-expression network. Red and green circle nodes denote up- and down-regulated genes, respectively. Red and green v-shaped nodes denote up- and down-regulated lncRNAs, respectively. Up- or down-regulation was measured in GBM patient samples compared to the matched normal brain tissues. Edges denote significant expression correlation between nodes. C, Detection of GBM-specific pathways and Gene Ontology biological process terms enriched in lncRNA-gene co-expression network. D, Selection of lncRNAs and genes of interest to perform follow-up experiments in-vitro and in-vivo to determine lncRNA's pathogenic potential in GBM. Additionally, Kaplan-Meier analysis assessed the ability of the selected lncRNA to predict survival of GBM patients using an in-house as well as a large cohort of GBM patient samples available in The Cancer Genome Atlas (TCGA) project.
To illustrate functional importance of the dysregulated lncRNA, we performed pathway and Gene Ontology (GO) term enrichment analyses of their co-expressed genes that were also differentially expressed in GBM. This approach facilitated prioritizing lncRNA whose aberrant expression potentially alters the expression of genes that belong to GBM specific pathways and biological processes (BPs). We identified 10 significantly enriched pathways (adjusted P <0.05, hypergeometric test followed by BH multiple test correction) using the Pathway Commons database as the pathway annotation dataset (34), which is embedded into the software WebGestalt (28, 35) (Supplementary Table S4). Intriguingly, most of the inferred pathways are linked to functions related to GBM pathogenesis, such as EphrinB-EPHB pathway, p75(NTR)-mediated signaling, TNF alpha/NF-kB, Neurotrophic factor-mediated Trk receptor signaling, and ErbB2/ErbB3 signaling events (Fig. 1C). The GO functional annotations were also informative in the context of GBM development. Specifically, we obtained 11 BP terms (non-redundant terms that belong to level 5 or more), among which most are closely linked to the biology of neurogenesis, neuron regeneration and differentiation. The result suggested that these relevant biological functions might be altered in the origin and development of GBM (Supplementary Table S5).
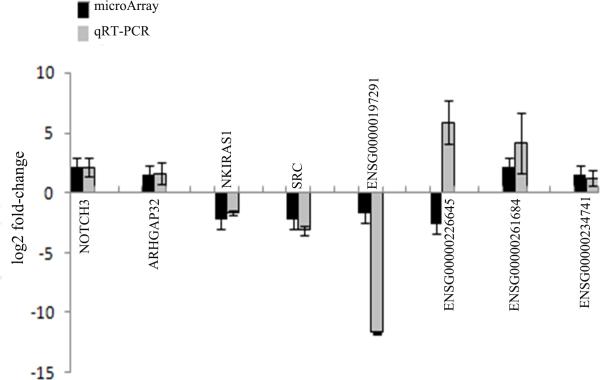
From the GBM relevant pathways and GO BP terms, we pinpointed 4 protein-coding genes and their co-expressed lncRNAs (ENSG00000197291-NOTCH3, ENSG00000226645-ARHGAP32, ENSG00000261684-NKIRAS1, and ENSG00000234741-SRC) (Supplemental Table S6) and performed real-time RT-PCR using 3 pairs of snap-frozen GBM specimens and matched noncancerous tissues to verify whether these important pathway-linked molecules were indeed dysregulated in GBM. Our real-time RT-PCR confirmed aberrant expression patterns of all the lncRNA (except ENSG00000226645) and genes that were observed in the microarray data (Fig. 2), suggesting that these lncRNAs and genes were dysregulated in GBM patient samples and that they might have important roles in GBM pathology. For example, in the co-expression network, lncRNA ENSG00000197291 (RAMP2-AS1) was linked to NOTCH3, a key regulator of many signaling pathways involved in cancer stem cells (CSCs) differentiation and development of glioma (36-39). It has been shown that activation of NOTCH3 promotes invasive glioma in a tissue-specific manner (40). The results from both the microarray and real-time RT-PCR showed that, when compared with normal brain tissue, the expression level of RAMP2-AS1 in GBM significantly decreased (P<0.05), whereas the expression of NOTCH3 significantly increased (P<0.05, Fig. 2).
Figure 2.
Real-time RT-PCR verification of lncRNA and mRNA expression. Four lncRNAs and four mRNAs were selected to examine their expression between GBM and normal control by real-time RT-PCR.
Low RAMP2-AS1 expression correlates with poor prognosis in GBM
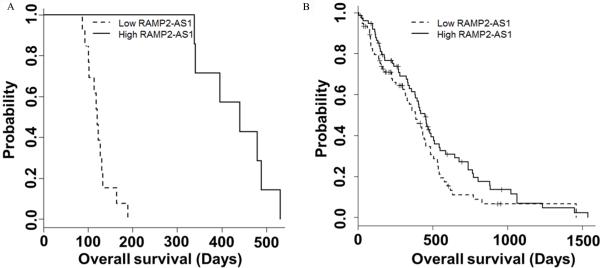
Considering the lower expression of RAMP2-AS1 in GBM than normal brain tissue, we further examined whether RAMP2-AS1 expression correlated with patient survival. We separated 20 GBM patient samples into two groups by using the median expression of RAMP2-AS1. Our analysis revealed that the patients with low expression of RAMP2-AS1 had significantly shorter survival times (P <0.001, log-rank test; Fig. 3A) than those with a high expression level of RAMP2-AS1 (detail procedure in Supplementary data1). We also assessed 5-year survival by dividing a larger cohort of 150 GBM patients extracted from TCGA project into two groups based on the median RAMP2-AS1 expression (Supplementary data1). The result showed a similar trend to what we observed in our patient samples, though statistical significance is moderate (P =0.094, log-rank test; Fig. 3B).
Figure 3.
Decreased expression of RAMP2-AS1 in GBM correlates with poor clinical outcome of GBM patients. A, Kaplan-Meier survival curves using the data from 20 GBM patients with respect to RAMP2-AS1 expression. B, A large cohort of 150 GBM patient samples from TCGA was evaluated by Kaplan-Meier analysis with respect to RAMP2-AS1 expression.
RAMP2-AS1 regulates the cell cycle progression mediated by NOTCH3
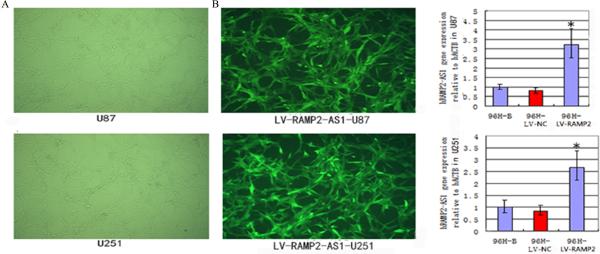
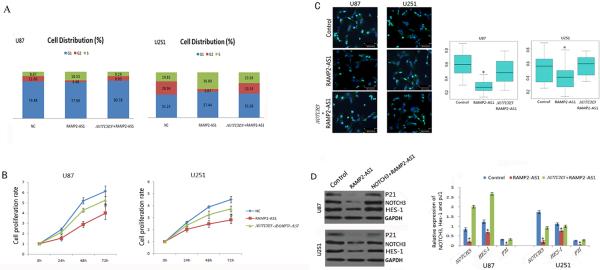
To determine the exact effects of RAMP2-AS1 on the growth of GBM cells, we generated GBM cell lines with over-expression of RAMP2-AS1 by infection with the LV-RAMP2-AS1. The immunofluorescence and qRT-PCR analysis showed that the infection efficiency of LV-RAMP2-AS1 in U251 and U87 were greater than 90% (Figs. 4A and 4B). U87 and U251 cells were infected with LV-RAMP2-AS1 only, or co-infected with LV-NOTCH3 at the same time. At 96h after treatment, flow cytometry was performed to examine the cell cycle. We found that RAMP2-AS1 overexpression could block the GBM cell cycle progress and NOTCH3 co-expression rescued the cell cycle arrest (Fig.5A). The proteins which are relative to cell cycle, such as P21, P27 and P16 were prepared for Western blot analysis. The result showed that RAMP2-AS1 overexpression decreased the expression of P21 significantly and NOTCH3 could restore P21 to basal levels (Fig.5D).
Figure4.
The infection effect of LV-RAMP2-AS1 on GBM cells. A, After 96h of LV-RAMP2-AS1(5×108TU/ml) infection, U87 and U251 cells were observed under fluorescence microscope. B, After 96h of LV-RAMP2-AS1 infection, qRT-PCR were performed to analyze the expression of RAMP2-AS1in U87 and U251 cells. Experiments were performed in triplicate in three times. The values shown are mean ± SD. *P <0.05 vs. control group.
Figure 5.
RAMP2-AS1 inhibits the growth of GBM cells. A, Flow cytometry was performed to examine the S arrest in U87 and U251 cells after treatment with control LV vector LV-NC, LV-RAMP2-AS1(RAMP2-AS1) and LV- NOTCH3 combined with LV-RAMP2-AS1 (Notch3+RAMP2-AS1) for 96h. B, U87 and U251 GBM cells which were infected with above vectors respectively and subjected to CCK-8 cell proliferation assays. C, Ki67 expression of U87 and U251 GBM cells which were infected with above vectors respectively were detected by immunofluorescence (left). The Ki67 data is presented as the ratio of Ki67 positive cells (light blue) to total DAPI positive cells (light blue U dark blue) (right). D, The expression of protein NOTCH3, HES-1 and P21 in U87 and U251 cells was determined by Western blot after 72h of above vectors infection. All above experiments were performed in triplicate in three times. The values shown are mean ± SD (n=5 in each group). *P <0.05 vs. control group.
RAMP2-AS1 inhibits GBM cell growth
Cell-counting kit-8 assays indicated that cell proliferation was reduced in both U87 and U251 cells when RAMP2-AS1 was over-expressed (P<0.05, Fig. 5B). Consistent with the decrease in cell proliferation, we observed a significant lower level of Ki67 expression (0.2808±0.091 and 0.3911±0.185236) in U87 and U251 cells, which had increased RAMP2-AS1 expression, than that of the control cells (0.59189±0.1734 and 0.5282±0.2050) (P<0.05, Fig. 5C).
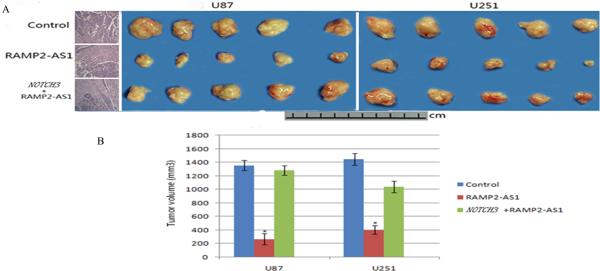
To further evaluate the effects of RAMP2-AS1 on GBM growth in vivo, RAMP2-AS1-overexpressing U87 and U251 cells and the negative control cells (those were infected with control virus) were injected into the flanks of nude mice. Tumors were allowed to grow for 30 days and then they were extracted and measured for mean volumes. The xenografts of U87 and U251 cells with overexpressed RAMP2-AS1(269±80.56mm3 and 403±62.17mm3) were significantly smaller than those GBM tumors formed from control cells (1355±76.32mm3 and 1447±85.95mm3) (P <0.05, Figure 6A, 6B). Furthermore, the H&E staining of intracranial implanted tumors showed that the subcutaneous tumors still remained the pathologic characters of GBM, such as cellular heterogeneity, rapid proliferation, angiogenesis and extensive invasion (Figure 6 A). The results above indicated that RAMP2-AS1 inhibited the growth of GBM cells both in vitro and in vivo, suggesting its tumor suppressive role in GBM.
Figure 6.
RAMP2-AS1 inhibits the growth of GBM in vivo. A, U87 and U251 GBM cells which were infected with LV-NC, LV-RAMP2-AS1 and LV- NOTCH3 combined with LV-RAMP2-AS1 were injected subcutaneously into nude mice. After 30 days tumors were removed and photographed and their volumes were measured. There were five mice in each group. *: P <0.05 each group vs. the NC group (right). The tumor cells derived from above subcutaneous xenografts were implanted into right striatum of nudes and analyzed by H&E staining (left). B, The volume of subcutaneous xenografts of the groups mentioned above. Bars are mean±SD. *p<0.05, n=5 per group.
RAMP2-AS1 may regulate the proliferation of GBM through NOTCH3 signaling
Our co-expression network analysis suggested that RAMP2-AS1 expression correlated with NOTCH3 expression (Fig. 1). To determine whether NOTCH3 is regulated by RAMP2-AS1, we measured the expression of NOTCH3 after over-expression of RAMP2-AS1. Expression of NOTCH3 protein and its downstream signaling molecule, HES1, decreased significantly in both U87 and U251 cells which were infected by LV-RAMP2-AS1 when we compared with those cells expressing control vector (P= 0.039738, Fig. 5D). To determine the specificity of NOTCH3 regulation, we overexpressed NOTCH3 together with RAMP2-AS1 in U87 and U251 cells, and did not observe the reduction of HES1 expression by Western blot analysis (Fig. 5D). Furthermore, we observed that the growth inhibition of U87 and U251, both in vitro and in vivo induced by RAMP2-AS1 over-expression, was rescued by NOTCH3 over-expression (Fig. 5B, Fig.5C and Fig. 6). These results suggested that the RAMP2-AS1-mediated down-regulation of NOTCH3 likely contributed to the reduction of GBM proliferation.
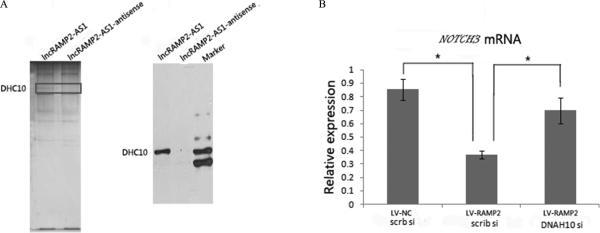
RAMP2-AS1 interacts with DHC10 to regulate NOTCH3 expression
We next investigated how RAMP2-AS1 regulates NOTCH3 expression. First, we performed an RNA pull-down experiment using nuclear extracts of U87 cells to identify the proteins interacting with RAMP2-AS1. Several additional bands were present in the SDS-PAGE silver staining analysis of the fraction precipitated with biotin-labeled RAMP2-AS1 compared to an antisense control (Fig. 7A). Proteins in these specific bands were identified by mass spectrometry, one of which was human dynein heavy chain 10 (DHC10), a known microtubule-associated protein (41). We confirmed the specific interaction between DHC10 and RAMP2-AS1 by immunoblotting (Fig. 7B). Next, qRT-PCR confirmed that DNAH10siRNA in combination with RAMP2-AS1 could restore NOTCH3 expression associated with RAMP2-AS1 depletion (Fig. 7C). Put together, our data suggested that RAMP2-AS1 might inhibit the expression of NOTCH3 mediated by DNAH10 in GBM pathology.
Figure7.
RAMP2-AS1 interacts with DHC10. A, Silver-stained SDS-PAGE gel analysis of proteins in nuclear extract of U87 cells that are bound to biotinylated RAMP2-AS1 or its antisense. The highlighted regions were analyzed by mass spectrometry, identifying DHC10 as a protein unique to RAMP2-AS1 (left). Immunoblotting analysis of proteins in nuclear extract of U87 cells that are bound to biotinylated RAMP2-AS1 or its antisense using an anti-DHC10 antibody (right). B, NOTCH3 expression in U87 cells receiving LV, LV-RAMP2-AS1, scramble siRNA(scrb si), or DHC10siRNA in combination as indicated (Error bars represent SD, *p<0.05).
Discussion
GBM is the most common and aggressive type of primary brain tumor in humans. It accounts for 52% of all parenchymal brain tumor cases and 20% of all intracranial tumors (42). Although the mechanisms of GBM occurrence and development have been extensively investigated during the past two decades, the pathogenesis of this disease is still ill defined, and the gene regulation involved in this disease remains largely unclear.
Increasing lines of evidence suggested that lncRNA may be important factors in controlling gene expression (43). Hence, we examined the expression profiles of both lncRNAs and mRNAs in GBM tissue and matched control samples (Fig.1A). Subsequently, we constructed lncRNA-mRNA co-expression network based on publicly available, matched lncRNA and mRNA expression profiling data and mapped the differently expressed lncRNAs and mRNAs in GBM into this unique reference network. This approach was taken to examine which lncRNA-mRNA pairs have been aberrantly expressed in GBM and have potential roles in the origination and development of GBM. Our results may better reflect the genetic variations of GBM due to the ruling out the difference at the individual level. Thus, this co-expression network analysis could generate useful hypotheses for further study of the functional roles of the differentially expressed lncRNAs in the development of GBM. Importantly, our systems biology approach found that the abnormally expressed lncRNAs were mostly involved in the pathways and biological functions that are related to GBM pathogenesis, indicating the effectiveness of our approach.
While the functional roles of most of the lncRNAs in the co-expression network have not been previously studied, the mRNAs that have co-expression relationship with those lncRNAs have been previously reported to have important roles in various tumors including prostate cancer, glioma, breast cancer and squamous cell carcinoma of the lung (39, 44-45). Therefore, according to the functional roles of these mRNAs, we could prioritize the lncRNAs that might play key roles in GBM. Our investigation of lncRNA-mRNA network revealed that NOTCH3, a key gene in the progression of GBM, had co-expression relationship with the lncRNA RAMP2-AS1. The expression of RAMP2-AS1 was significantly reduced in GBM. Notch signaling has a critical function in the specification, proliferation, and survival of stem/progenitor cells in a number of tissues, including the central and peripheral nervous systems (46). The pathway is widely implicated in neoplasia, and in most contexts it promotes neoplastic growth (47-52). It has been shown that activation of NOTCH3 pathway promotes murine T-acute lymphoblastic leukemia (T-All), similar to that has been seen in humans (50, 53, 54).
Notch is also thought to play important role in poorly differentiated tumor cells. Inhibition of this pathway may deplete “cancer stem cells”, which are resistant to radiation and standard chemotherapies (55-58). Small molecules targeting Notch have shown great promise in preclinical testing of several tumor models. Based on such studies, phase I clinical trials for leukemia and breast cancer have been initiated using gamma-secretase inhibitors that block the activation of Notch receptors (59, 60). Similar to leukemia and breast, Notch signaling pathway has also been shown to promote proliferation of glioma cells and play different roles in primary and secondary GBM (61, 62); however, the mechanisms of Notch regulation in GBM has not yet clear.
In our study, we found that RAMP2-AS1 expression was significantly reduced in primary GBM tissues compared with normal brain tissues and lower RAMP2-AS1 expression was correlated with poor overall survival of GBM patients (Fig. 3). We utilized a cohort of 150 GBM patient samples from TCGA and a cohort of 20 patient samples from our in house data. Although the survival analysis of a large cohort of 150 GBM patients extracted from TCGA showed that the patients with low expression of RAMP2-AS1 had only moderately shorter survival times than those with a high expression level of RAMP2-AS1 (P =0.094, log-rank test; Fig. 3B, Supplementary data 1), the survival analysis of our in-house 20 primary GBM revealed significant survival difference between the two groups. Several factors might contribute to the difference between the two datasets: specimens background (ethnic background, gender, age, etc.), sample heterogeneity and treatment (e.g., all the 20 in-house GBM samples were primary GBM and the patients had experienced the same treatment), and statistical power.
Recently, Pastori et al. applied single molecule sequencing to quantify the expression of lncRNAs in GBM. They reported the top 100 up- and down- regulated lncRNAs in GBM versus control samples, and RAMP2-AS1 was not found in that list (63). Pastori et al. used epilepsy samples as control, while we used matched normal brain tissue samples. This different selection of control tissues may partially explain why RAMP2-AS1 showed different result in the two studies, though further investigation is warranted. Our co-expression network analysis of lncRNA-mRNA genes revealed that RAMP2-AS1 was significantly correlated with NOTCH3, which was significantly up-regulated in GBM (Fig.2). This is consistent with many previous studies in which Notch signaling acts as a tumor promotor (45, 46). Our study also showed that over-expression of RAMP2-AS1 inhibited the proliferation of GBM cells in vitro and in vivo in part by down-regulating NOTCH3 and its downstream molecule, HES1(Fig.5, Fig.6). These results indicated that RAMP2-AS1 might contribute to GBM through its regulation of NOTCH3. Furthermore, although we did not detect the direct interaction between RAMP2-AS1 and NOTCH3, our RNA pull-down experiment indicated another protein, DHC10 had direct interaction with RAMP2-AS1. For all the results we had, we speculated that RAMP2-AS1 might interact with NOTCH3 mediated by DHC10 (Fig.7).
While our results are still preliminary, the findings are promising toward setting us future research direction focusing on lncRNA-mRNA synergistic regulation in the pathogenesis of GBM. It is our belief that the lncRNA-mRNA pairs highlighted in EphrinB-EPHB, p75-mediated signaling, TNF alpha/NF-κB and ErbB2/ErbB3 signaling pathways warrant future investigation in GBM or its related phenotype.
Supplementary Material
Acknowledgments
This work was supported by the National Nature Science Foundation of China (Grants No. 81172384 and No.30873029). Z.M. Zhao was partially supported by National Institutes of Health (NIH) grants (R01LM011177, R21CA196508 and R03CA167695). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the CapitalBio Corporation for performing the microarray detection of lncRNA & mRNA and Dr. Zheng Wang (Chinese Academy of Medical Sciences) for his assistance in establishing animal models. We thank the three reviewers' constructive comments which helped us substantially improve the quality of this work in revision.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 4.Taylor LP. Diagnosis, treatment, and prognosis of glioma: five new things. Neurology. 2010;75:S28–S32. doi: 10.1212/WNL.0b013e3181fb3661. [DOI] [PubMed] [Google Scholar]
- 5.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotesaggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marín-Béjar O, Marchese FP, Athie A, Sánchez Y, González J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Su M, Lu G, Wang J. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer. 2013;12:101. doi: 10.1186/1476-4598-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 11.Zong X, Tripathi V, Prasanth KV. RNA splicing control: Yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;6:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian EB, Li J, Xie YS, Zong G, Zhao B. LncRNAs: new players in gliomas, with special emphasis on the interaction of lncRNAs With EZH2. J Cell Physiol. 2014;230:496–503. doi: 10.1002/jcp.24549. [DOI] [PubMed] [Google Scholar]
- 13.Sun YZ, Wang Z, Zhou D. Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med Hypotheses. 2013;81:319–321. doi: 10.1016/j.mehy.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Amit D, Matouk IJ, Lavon I, Birman T, Galula J, Abu-Lail R, et al. Transcriptional targeting of glioblastoma by diphtheria toxin-A driven by both H19 and IGF2-P4 promoters. Int. J. Clin. Exp. Med. 2012;5:124–135. [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, et al. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis and stemness of glioblastoma cells. J Neurosurg. 2016;124:129–36. doi: 10.3171/2014.12.JNS1426. [DOI] [PubMed] [Google Scholar]
- 19.Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;38:359–69. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Yilong Y, Jun M, Yixue X, Ping W, Zhen L, Jing L, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer letters. 2015;359:75–85. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 21.Kailiang Z, Xiaotian S, Xuan Z, Lei H, Luyue C, Zhendong S, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2014;6:537–46. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiara P, Philipp K, Clara P, Veronica P, Claude-Henry V, Jann NS, et al. The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. PNAS. 2015;112:8326–31. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Chang HY. Molecular mechanisms of long non-coding RNAs. Mol Cell. 2011;6:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 25.Jiang Q, Ma R, Wang J, Wu X, Jin S, Peng J, et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC genomics. 2015;16(Suppl 3):S2. doi: 10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic acids research. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 30.Alqudah MA, Agarwal S, Al-Keilani MS, Sibenaller ZA, Ryken TC, Assem M. NOTCH3 is a prognostic factor that promotes glioma cell proliferation, migration and invasion via activation of CCND1 and EGFR. PloS One. 2013;8:e77299. doi: 10.1371/journal.pone.0077299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 32.Jiang Q, Ma R, Wang J, Wu X, Jin S, Peng J, et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC genomics. 2015;16(Suppl 3):S2. doi: 10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic acids research. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra R, Edmonds MD, Sun J, Zhao M, Yu H, Eischen CM, et al. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014;20:1356–68. doi: 10.1261/rna.042754.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 37.Alqudah MA, Agarwal S, Al-Keilani MS, Sibenaller ZA, Ryken TC, Assem M. NOTCH3 is a prognostic factor that promotes glioma cell proliferation, migration and invasion via activation of CCND1 and EGFR. PloS One. 2013;8:e77299. doi: 10.1371/journal.pone.0077299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito N, Fu J, Zheng S, Yao J, Wang S, Liu DD, et al. A high Notch pathway activation predicts response to γ secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells. 2014;32:301–312. doi: 10.1002/stem.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu P, Yu S, Jiang R, Kang C, Wang G, Jiang H, et al. Differential expression of Notch family members in astrocytomas and medulloblastomas. Pathol Oncol Res. 2009;15:703–710. doi: 10.1007/s12253-009-9173-x. [DOI] [PubMed] [Google Scholar]
- 40.Pierfelice TJ, Schreck KC, Dang L, Asnaghi L, Gaiano N, Eberhart CG. Notch3 activation promotes invasive glioma formation in a tissue site-specific manner. Cancer research. 2011;71:1115–1125. doi: 10.1158/0008-5472.CAN-10-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arai E, Gotoh M, Tian Y, Sakamoto H, Ono M, Matsuda A, et al. Alterations of the spindle checkpoint pathway in clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Int J Cancer. 2015;137:2589–606. doi: 10.1002/ijc.29630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wick W, Weller M, Weiler M, Batchelor T, Yung AW, Platten M. Pathway inhibition: emerging molecular targets for treating glioblastoma. Neuro Oncol. 2011;6:566–579. doi: 10.1093/neuonc/nor039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One. 2010;4:E10316. doi: 10.1371/journal.pone.0010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarma P, Bag I, Ramaiah MJ, Kamal A, Bhadra U, Pal Bhadra M. Bisindole-PBD regulates breast cancer cell proliferation via SIRT-p53 axis. Cancer Biol Ther. 2015;16:1486–1501. doi: 10.1080/15384047.2015.1071731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng G, Ju X, Meng Q, Yu ZJ, Ma LB. Emodin inhibits the proliferation of PC3 prostate cancer cells in vitro via the Notch signaling pathway. Mol Med Rep. 2015;12:4427–4433. doi: 10.3892/mmr.2015.3923. [DOI] [PubMed] [Google Scholar]
- 46.Kim JS, Kim ES, Liu D, Lee JJ, Behrens C, Lippman SM, et al. Activation of insulin-like growth factor 1 receptor in patients with non-small cell lung cancer. Oncotarget. 2015;6:16746–16756. doi: 10.18632/oncotarget.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierfelice TJ, Schreck KC, Eberhart CG, Gaiano N. Notch, neural stem cells, and brain tumors. Cold Spring Harb Symp Quant Biol. 2008;73:367–75. doi: 10.1101/sqb.2008.73.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 49.Sun J, Gong X, Purow B, Zhao Z. Uncovering miroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol. 2012;8(7):e1002488. doi: 10.1371/journal.pcbi.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 51.O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Li Y, Banerjee S, Sarkar FH. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer Res. 2008;28:3621–3630. [PubMed] [Google Scholar]
- 53.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 54.Lee SY, Kumano K, Masuda S, Hangaishi A, Takita J, Nakazaki K, et al. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 2005;19:1841–1843. doi: 10.1038/sj.leu.2403896. [DOI] [PubMed] [Google Scholar]
- 55.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 56.Wicha MS. Targeting breast cancer stem cells. Breast. 2009;18:S56–58. doi: 10.1016/S0960-9776(09)70274-7. [DOI] [PubMed] [Google Scholar]
- 57.Talora C, Campese AF, Bellavia D, Felli MP, Vacca A, Gulino A, et al. Notch signaling and diseases: an evolutionary journey from a simple beginning to complex outcomes. Biochim Biophys Acta. 2008;1782:489–497. doi: 10.1016/j.bbadis.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, et al. Notch Promotes Radioresistance of Glioma Stem Cells. Stem Cells. 2009;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kvinlaug BT, Huntly BJ. Targeting cancer stem cells. Expert Opin Ther Targets. 2007;11:915–927. doi: 10.1517/14728222.11.7.915. [DOI] [PubMed] [Google Scholar]
- 60.Papayannidis C, DeAngelo DJ, Stock W, Huang B, Shaik MN, Cesari R, et al. A phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015;5:e350. doi: 10.1038/bcj.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 62.Marie-Therese S, Karina K, Hans SP. The functional role of Notch signaling in human gliomas. Neuro-Oncology. 2010;12:199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alqudah MA, Agarwai S, AI-keilani MS, Sibenaller ZA, Ryken TC, Assem M. Notch3 is a prognostic factor that promotes glioma cell proliferation, migration and invasion via activation of CCND1 and EGFR. PLoS One. 2013;8(10):e77299. doi: 10.1371/journal.pone.0077299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.