Abstract
The development of new anti-androgens such as enzalutamide or androgen synthesis inhibitors like abiraterone have improved patient outcomes in the treatment of advanced prostate cancer (PCa). However, due to the development of drug resistance and tumor cell survival, the majority of these patients progress to the refractory state of castration-resistant prostate cancer (CRPC). Thus, newer therapeutic agents and a better understanding of their mode of action are needed for treating these CRPC patients. We demonstrated previously that targeting the Binding Function 3 (BF3) pocket of the androgen receptor (AR) has great potential for treating patients with CRPC. Here we explore the functional activity of this site by using an advanced BF3-specific small molecule (VPC-13566) that was previously reported to effectively inhibit AR transcriptional activity and to displace the BAG1L peptide from the BF3 pocket. We show that VPC-13566 inhibits the growth of various PCa cell lines including an enzalutamide-resistant cell line and reduces the growth of AR-dependant PCa xenograft tumors in mice. Importantly we have used this AR BF3 binder as a chemical probe and identified a co-chaperone, small glutamine-rich tetratricopeptide repeat (TPR)-containing protein alpha (SGTA), as an important AR-BF3 interacting partner. Furthermore, we employed this AR BF3-directed small molecule to demonstrate that inhibition of AR activity through the BF3 functionality can block translocation of the receptor into the nucleus. These findings suggest that targeting the BF3 site has potential clinical importance especially in the treatment of CRPC and provide novel insights on the functional role of the BF3 pocket.
Keywords: Androgen receptor, castration-resistant prostate cancer, Binding Function 3, nuclear translocation, co-chaperone binding site
INTRODUCTION
Prostate cancer (PCa) is the most commonly diagnosed type of carcinoma in men in western countries (1). Many studies have shown that PCa growth and progression are sustained by androgen receptor (AR; AR gene) signaling and activity (2). Hence, androgen depletion therapy (ADT) is the standard of care for advanced PCa (3–5). Despite initial clinical benefits, ADT is not curative, and the majority of patients will develop a refractory form of the disease known as castration-resistant PCa (CRPC). Many aberrant mechanisms underlie the disease progression to the CRPC stage such as AR mutations, overexpression of the receptor, and expression of constitutively active AR splice-variants lacking the ABS entirely (6).
All current anti-androgens used for PCa treatment, including the first generation drugs flutamide, bicalutamide, nilutamide and the second generation enzalutamide (7, 8), inhibit AR by competing with endogenous steroids for binding to the androgen binding site (ABS) and share a common molecular scaffold that increases the likelihood of cross-resistance when gain-of-function mutations occur in the ABS. Because of the emergence of resistance and the side effects of conventional and experimental anti-androgens, new anti-AR therapeutics are needed, such as drugs that would disrupt the interaction between the AR and its co-activator proteins.
Recently we described the preliminary development of such small molecule inhibitors targeting the binding function 3 (BF3) pocket of the AR (9–12) - a surface site identified by Estebanez-Perpina et al. (13) and suggested to be important for co-regulator recruitment, AR transactivation and N/C interactions (14). Indeed, Jehle et al. (15) demonstrated that a duplicated GARRPR motif at the N-terminal region of the co-chaperone Bag-1L functions through the BF3 mediation and is very important for fine-tuning of AR function. In addition, this surface site has been implicated in FKBP52-dependent AR regulation (16).
In this work, we describe the characterization of a greatly improved BF3-specific small molecule (VPC-13566) that is effective in inhibiting AR transcriptional activity in vitro as well as the growth of AR-dependent PCa cell lines, including an enzalutamide-resistant cell line. Importantly, these potent and selective AR BF3 binders can be used as a chemical probe to help identify previously unknown AR partners and, hence, provide additional insights into AR biology. Indeed, using VPC-13566 as a probe, we were able to discover that the small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), known to regulate AR activity and to affect AR subcellular distribution (17), is in fact an AR BF3 interacting partner. Equally important, we used VPC-13566 to demonstrate that the inhibition of AR activity through the BF3 site can functionally block translocation of the receptor into the nucleus and lead to growth reduction of PCa xenograft tumors in mice.
MATERIALS AND METHODS
eGFP AR Transcription Assay and PSA Assay
The inhibition effects of compounds were assessed as described previously (11).
Cell Viability Assay
This assay was performed as previously reported in (11). PC3 cells lacking the AR and LNCaP cells were authenticated by IDEXX Laboratories (Maine, USA) in August 2014 and were maintained in RPMI 1640 media (Life Technologies) and 5 % FBS (Hyclone ThermoFisher Scientific) at 37 °C and 5 % CO2. Cultures were routinely monitored for mycoplasma contamination.
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations of VPC-13566, GARRPR and PARTPP motifs were performed starting from their docking poses in the BF3 pocket of the LBD crystal structure (4HLW) as predicted by Glide (18). All MD simulations were performed with the CUDA accelerated Amber 14 program. AR LBD force field parameters were obtained from the ff14SB force field and the ligand (VPC-13566 and DHT) parameters came from generalized amber force field with charges derived from a RESP fit using an HF/6-31G* electrostatic potential calculated using the Gaussian 09 program. MD simulations were carried out within AMBER 14 on WestGrid facilities from Compute Calculation Canada (https.//www.westgrid.ca). The production MD simulation was conducted for 50 or 100 ns without any restraints under the NPT ensemble condition at a temperature of 298 K and pressure of 1atm.
AR-LBD expression and purification
The AR LBD in fusion with an N-terminal (His)6 –avidin (GLNDIFEAQKIEWHE) tag was expressed in E. Coli BL21-DE3 cells. Cells were lysed in buffer containing 500mM Li2SO4, 50mM HEPES pH7.2, 5% Glycerol, 5mM β-mercaptoethanol, 10mM imidazole, 20μM DHT and 0.1mM phenylmethanesulfonyl fluoride. After sonication and centrifugation, the samples were loaded onto a Ni-NTA affinity column and subsequently eluted with 300 mM imidazole. After an overnight dialysis against 150mM Li2SO4, 50mM HEPES pH7.5, 10% Glycerol, 20μM of DHT and 0.5mM tris(2-carboxyethyl)phosphine, the protein was further purified by size-exclusion chromatography equilibrated in the dialysis buffer.
Biolayer interferometry (BLI) assay
The direct reversible interactions between VPC-13566 or SGTA peptides and the AR LBD were measured as previously described (19).
Proximity Ligation Assay (PLA) and Immunofluorescence (IF)
LNCaP cells were starved in phenol red free RPMI supplemented with 5% charcoal-stripped serum (CSS). After 4 days, cells were seeded into BD Falcon Culture slides (Fisher Scientific) (10,000 cells/well in 300μL volume). After 24h, cells were treated with 100 μL of 4X VPC-13566, enzalutamide or the vehicle DMSO. For the siRNA control, cells were treated with 10nM (3.6 pmol) siAR-4 or siSCR control for 24 h using Lipofectamine RNAiMax (Life Technologies) as per manufacturer’s instructions. The next day after treatment, cells were induced by adding 1nM R1881 for 24h, then fixed in 4% PFA. For PLA analysis, LNCaP cells were blocked with 3% BSA and incubated with 1/200 dilution of AR antibody (441) (sc-7305, Santa Cruz Biotechnology) and Bag1L antibody (16148-1-AP, Proteintech). Protein-protein interactions were analyzed using Duolink-based in situ PLA (Sigma-Aldrich), according to manufacturer’s instructions. Signals were quantified using a confocal microscope and the Duolink ImageTool. For IF, AR antibody (441) at 1/1000 dilution was used.
Lanthascreen TR-FRET displacement assay
The displacement of a fluorescein isothiocyanate (FITC) labelled SGTA peptide (FITC-MPQDLRSPARTPPSEEDSAEA) from the BF3 pocket of the LBD by the compounds VPC-13566 and VPC-14449 was assessed using a Time-Resolved Fluorescence Energy Transfer (TR-FRET) as previously described (20, 21).
Combination effect of VPC-13566 and enzalutamide
The LNCaP eGFP cells (1) were starved in phenol-red-free RPMI 1640 supplemented with 5% CSS for 5 days, followed by plating into 96-well plates at 20,000 cells/well. After 4h, cells were stimulated with 0.1nM R1881 and treated with either the vehicle (DMSO), enzalutamide (IC25 = 40nM), VPC-13566 (IC25 = 50nM) or combination of enzalutamide and VPC-13566 (IC25 of each). The level of PSA excreted into the media was measured after 3 days. Four independent experiments with 4 replicate wells each were done and the average and SEM were plotted.
Mammalian 2-hybrid assay
For the mammalian two-hybrid assay, full length AR and SGTA (residues 1–123) were cloned in pACT and pBIND vectors (CheckMate™, Promega), respectively. PC3 cells in phenol-red-free RPMI 1640 supplemented with 5% CSS were seeded in 96-well plates at 5000 cells/well. After 24h, cells were transfected with 8.5 ng of wild-type or mutated pACT-AR, 34 ng of pBIND-SGTA and 25 ng of the reporter plasmid PG5-luciferase. After 48h, cells were stimulated with 0.1 nM of R1881. Cells were lysed the next day and the luminescence signal was measured after adding 50 μL of luciferase assay reagent (Promega). Each measurement was done in 8 replicates with a biological replicates of n=3.
Tumor Xenografts
6–8 weeks old male Hsd:Athymic Nude-Foxn1nu mice (Harlan Laboratories) weighing 25–35 g were implanted by subcutaneous injection with LNCaP cells (2 million cells in BD Matrigel diluted 1:2 in DMEM media) at the posterior dorsal site. Tumor volume and serum PSA levels were measured weekly. When serum PSA levels reached more than 25 ng/ml, mice were surgically castrated. When PSA recovered to pre-castration levels, mice were placed into treatment groups by cyclical enrollment as follows: vehicle (5 % dmso, 10 % (2-Hdroxypropyl)-b-cyclodextrin (332593-25G; ALDRICH), 85 % water), 100 mg/kg BID of VPC-13566 (single dose of 200 mg/kg once per day for 2 days per week), or 10 mg/kg enzalutamide via intraperitoneal injection for 4 weeks. Mice were weighed weekly and monitored daily for signs of toxicity. All experiments were performed in accordance with protocols approved by the University of British Columbia Animal Care Committee.
RESULTS
VPC-13566 reduces AR transcriptional activity and PSA expression
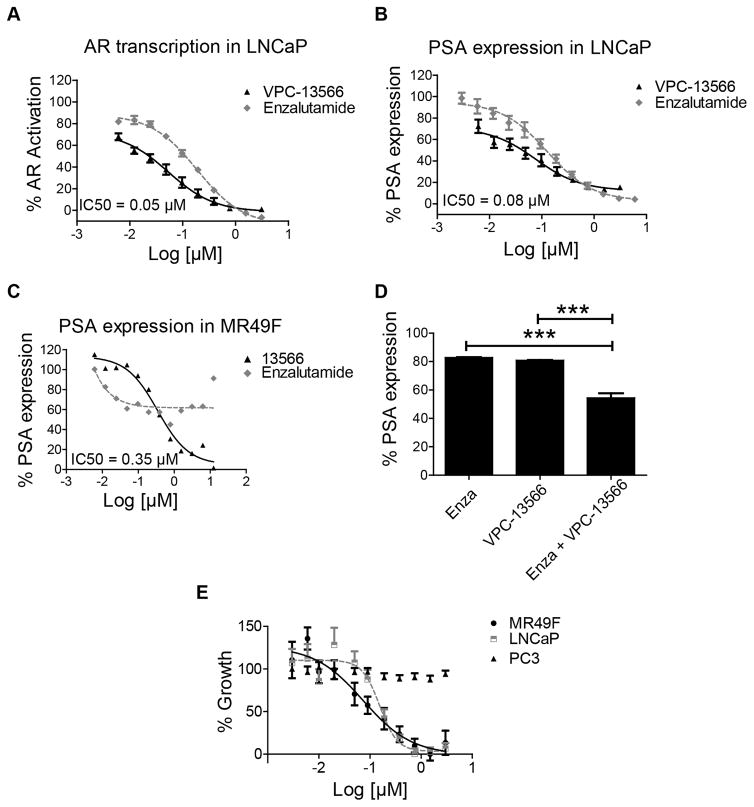
Previously we reported a BF3-specific small-molecule inhibitor “compound 23” that demonstrated some promising AR inhibition in androgen sensitive LNCaP and enzalutamide-resistant MR49F cells in vitro and in vivo (12). We have used “compound 23” as a template to develop further improved BF3 inhibitors that demonstrate enhanced affinity and drug-like properties (21). In this work the compound VPC-13566 was tested for its ability to inhibit AR transcriptional activity in LNCaP cells using an enhanced green fluorescent protein (eGFP) AR transcriptional assay (22), where the expression of eGFP is under the direct control of an androgen responsive probasin-derived promoter. Using a concentration-dependent titration, we established that the IC50 values are in the range of 0.05 μM and 0.19 μM for VPC-13566 and for enzalutamide, respectively (Figure 1A). This data indicates a 6-fold improvement in the potency of VPC-13566 in comparison with the parental compound 23 (IC50 = 0.31 μM) (12). To further validate VPC-13566 as an AR inhibitor, we tested its activity by quantifying its effect on the production of endogenous prostate specific antigen (PSA) in PCa cell lines. PSA is an AR-regulated serine protease and is widely used as a biomarker for PCa (23). As expected, VPC-13566 induced a significant decrease in secreted PSA levels in LNCaP cells with a corresponding IC50 value established at 0.08 μM (Figure 1B). VPC-13566 was further evaluated in the enzalutamide-resistant PCa cell line, MR49F (24). As anticipated, enzalutamide was confirmed to be ineffective against MR49F cells, whereas compound VPC-13566 caused a significant reduction in PSA expression (IC50 = 0.35 μM; see Figure 1C). These results provide additional assurance as to the effectiveness of our lead BF3 inhibitor and its applicability for targeting enzalutamide-resistant PCa cells.
Figure 1. Effect of VPC-13566 on AR transcriptional activity and cell growth.
A- Dose-response inhibition of AR transcriptional activity by VPC-13566 and enzalutamide in LNCaP cells. Data points represent the mean ± SEM of three independent experiments performed in triplicate. B–C Effect of VPC-13566 in comparison to enzalutamide on PSA expression in LNCaP cells (B) and enzalutamide resistant cells (MR49F) (C). D- The combination of VPC-13566 (IC25 = 50 nM) and enzalutamide (IC25 = 40 nM) decreases the AR regulated PSA expression in LNCaP cells. E- The effect of VPC-13566 on cell viability in LNCaP, enzalutamide resistant cell line (MR49F) and PC3 cells. % cell viability is plotted in dose dependent manner. Data points represent the mean ± SEM of three independent experiments performed in triplicate.
Combination treatment of VPC-13566 and enzalutamide in LNCaP cells
As enzalutamide and VPC-13566 target two different mechanisms of AR signaling, we decided to evaluate the effect of the combination of these two inhibitors on AR transcriptional activity. LNCaP cells were treated with a single dose of Enzalutamide alone or in combination with VPC-13566 at concentrations corresponding to their respective IC25 values. As a combination, enzalutamide and VPC-13566 inhibited PSA expression by 46% (Figure 1D). These results suggest that the combined use of enzalutamide and VPC-13566 additively improved the inhibition of PSA transcription in LNCaP cells.
VPC-13566 potently inhibits androgen-stimulated proliferation of LNCaP and enzalutamide-resistant PCa cell lines
To determine the efficacy of VPC-13566 on the viability of various PCa cell lines, its activity was assessed in LNCaP, MR49F and PC3 cells. MTS assay was performed after 4 days of treatment with VPC-13566 in a dose dependent manner. VPC-13566 is very effective in inhibiting the growth of both LNCaP and enzalutamide-resistant cells, achieving IC50 values of 0.15 and 0.07 μM, respectively and did not show any effect on AR-independent PC3 cells, confirming its AR-specific activity (Figure 1E). In addition, at a concentration of 5 μM, VPC-13566 did not affect the transcriptional activity of the Estrogen Receptor, showing the selectivity of this molecule towards AR (data not shown).
Docking of BF3 inhibitor VPC-13566 using molecular dynamics simulations
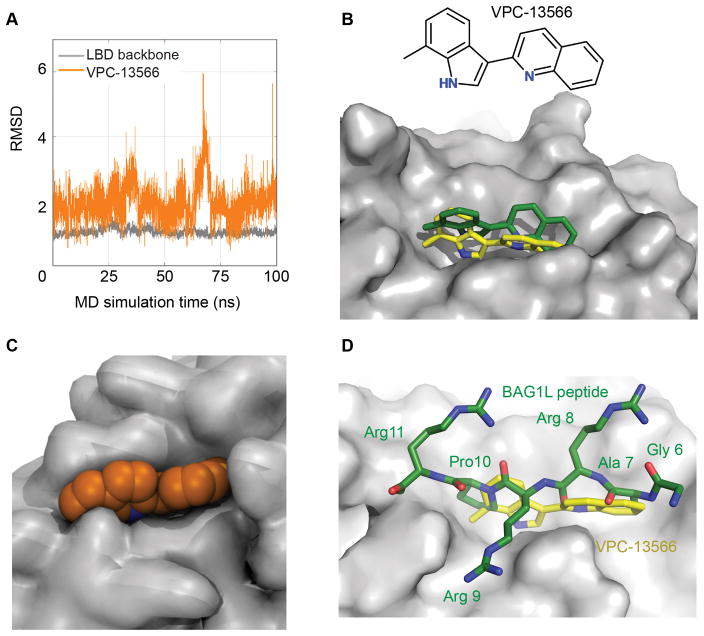
To better understand the interaction of the small molecule inhibitor with the BF3 site, we have conducted explicit solvent molecular dynamics (MD) simulations on the target BF3 site for 100ns with VPC-13566. The binding pose of novel compound VPC-13566 predicted by the Glide SP program (18) was used as a starting point for MD. The stability of the receptor and ligand conformations was then assessed by the root mean squared deviation (RMSD) between the initial conformation and each snapshot during MD simulations, and by the GB/SA binding free energy calculations.
The RMSD was calculated by using the initial complex conformation as reference. Over the 100ns run, the plateau of backbone RMSD indicates that the AR LBD was well equilibrated and the average RMSD of VPC-13566 motion was around 2Å (Figure 2A). This clearly demonstrates that the ligand fits into the BF3 site very well with a binding orientation similar to the initial conformation. We observed that the RMSD increases at around 67ns and this reflects the rotation of the quinoline group in the VPC-13566 ligand while its 7-methylindole group was keeping its binding pose.
Figure 2. Molecular dynamics simulations of VPC-13566 binding to the AR ligand binding domain.
A- The observed root mean square deviation (RMSD) values for VPC-13566 (orange) and backbone atoms of AR LBD residues (grey) during 100 ns MD simulations. B- Initial docked conformation (green) and representative docking pose of VPC-13566 obtained from MD simulations (yellow). C- Surface representation of BF3 pocket (grey) and VPC-13566 (orange) showing that the ligand fits well into the pocket. D- Superimposition of Bag1L peptide, GARRPR (green) and VPC-13566 (yellow) into the BF3 pocket (grey).
In order to identify the most frequently occurring binding pose of VPC-13566 during MD simulations, we applied cluster analysis to the VPC-13566 conformations. The clustering was conducted on the basis of RMSD of the ligand’s non-hydrogen atoms by using the hierarchical agglomerative clustering approach and five clusters were obtained. About 45% of the ligand conformations were assigned to the first ranked cluster and its centroid conformation in the cluster was found to be similar to its input conformation (Figure 2B). To determine the most stable binding pose, we proceeded to the GB/SA binding free energy calculation for all the protein-ligand complex conformations in the cluster and defined the conformation that gave the most stable binding free energy. The binding free energy of the representative binding pose, where VPC-13566 was well surrounded by BF3 residues, is −33.6kcal/mol (Figure 2C).
As mentioned above, it was previously demonstrated that the GARRPR motif from the co-chaperone BAG1L interacts with the BF3 pocket of AR (15). Next, we verified whether this binding model for VPC-13566 in the BF3 site would also work with the GARRPR motif. Indeed, the superimposition of the binding orientations of VPC-13566 and GARRPR motif shows that VPC-13566 perfectly occupies the binding region of the hexapeptide motif inside the BF3 pocket (Figure 2D).
VPC-13566 interacts with the BF3 pocket and hinders co-chaperone binding
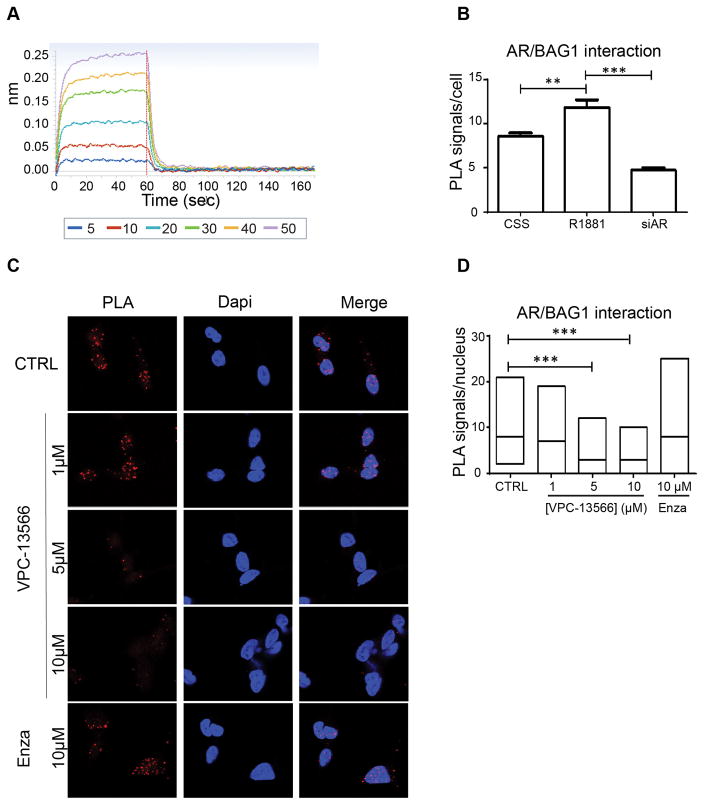
The superimposition of VPC-13566 and GARRPR motif predicted by MD was then tested experimentally. Direct binding of VPC-13566 to the ligand binding domain (LBD) of the AR was measured using Bio-Layer Interferometry (BLI) technology. VPC-13566 showed a reversible and dose-dependent interaction with purified LBD protein (Figure 3A). To demonstrate that this interaction is occurring through the BF3 site, we used in situ proximity ligation assay (PLA) (25) to evaluate the effect of VPC-13566 on BF3-interacting BAG1L protein in LNCaP cells. As BAG1L protein is mainly nuclear, the interaction between AR and BAG1L was highest in the presence of R1881 (Figure 3B). On the other hand, when cells were treated with a siRNA against the AR-LBD (siAR4) (26), the fluorescence signal was decreased as expected (Figure 3B). To evaluate the effect of VPC-13566 on AR/BAG1L interaction, we only considered the nuclear signal of PLA. Treatment with VPC-13566 reduced the interaction between BAG1L and AR in a dose responsive manner, which suggests that this compound disrupt the binding to BF3. Indeed, the ABS binder enzalutamide did not affect this interaction (Figure 3C–D).
Figure 3. The binding of VPC-13566 to the BF3 pocket of AR.
A- BLI dose-response curves (5–50 μM) reflecting the direct binding of the VPC-13566 to the purified ligand binding domain of AR. The specific binding to the BF3 site was confirmed by the decrease of the endogenous AR/BAG1L interaction in LNCaP cells as seen by proximity ligation assay (B–D). Statistical significance was set at P < 0.01 (**) and P < 0.001 (***).
Identification of the co-chaperone SGTA as a BF3 binder
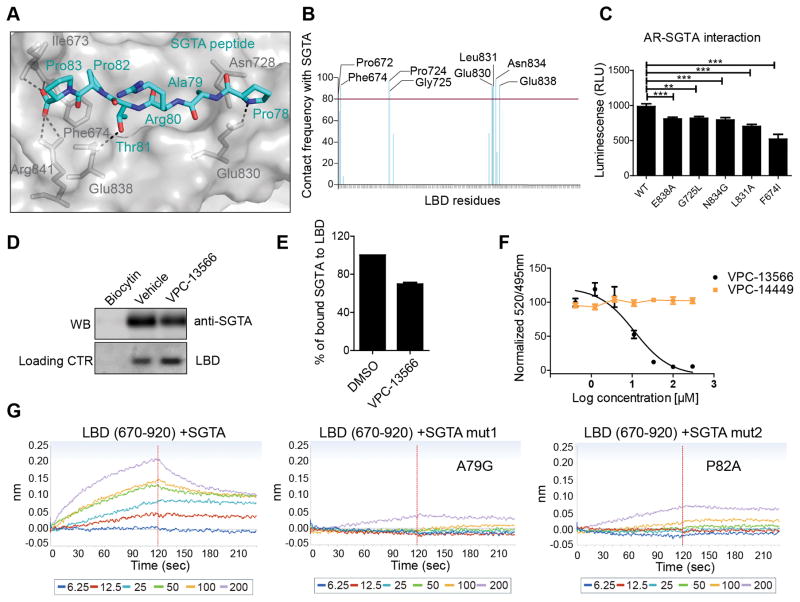
The co-chaperone small glutamine–rich tetratricopeptide repeat (TPR) containing protein alpha (SGTA) has been reported to play a role in AR maturation in the cytoplasm and to prevent its inappropriate translocation into the nucleus in the absence of the specific ligand. Upon androgen binding, SGTA is exchanged by another TPR containing protein such as the 52 kDa FK506 binding protein (FKBP52), an essential step for AR nuclear shuttling (17, 27, 28). It has been previously shown that the N-terminal region of SGTA (residues 21–40) is important for interaction with the AR hinge region (residues 630–645) (17, 29). We identified a PARTPP motif in the SGTA sequence (residues 78–83) that is very similar to the sequences of FKPB52 (GSAGSPP) and BAG1L (GARRPR), shown to interact with the BF3 region of AR (15, 16, 30). Therefore, we evaluated the possibility of SGTA’s binding to the BF3 surface using MD simulation and molecular docking. These in silico experiments predicted that the SGTA peptide (PARTPP) is able to bind stably to the BF3 surface and establish strong interactions with residues in this pocket (Figure 4A). The SGTA peptide makes a network of H-bond interactions with Ile673, Phe674, Arg841, Glu838 and Glu830. Importantly, several residues had the contact with the peptide over 80% of total experimental time frame of the MD simulations (see figure 4B).
Figure 4. The binding of SGTA to the BF3 pocket of AR.
A- The most common MD simulations binding conformation of SGTA peptide (cyan) into the BF3 pocket (grey). Critical BF3 residues that make H-bond interactions (Dotted lines) are highlighted (grey sticks). B- BF3 residues that interact with the PARTPP peptide more than 80% of the MD simulations time. C-Mutations in the BF3 pocket affect SGTA-AR interaction in mammalian two hybrid assay. D–E Pull down of SGTA protein by a purified AR-LBD protein (residues 670-920) and the effect of 100 μM VPC-13566 on LBD/SGTA interaction. F- Displacement of a FITC-SGTA peptide from the BF3 pocket by increasing concentrations of VPC-13566 using a TR-FRET assay. G- The direct binding of SGTA peptide containing the PARTPP motif to a purified LBD using BLI. Mutating A79 or P82 of this motif disrupted the LBD-peptide interaction.
The significance of these residues was further evaluated experimentally using a mammalian 2-hybrid assay where mutations affecting residues in the BF3 pocket significantly decreased the interaction between the AR receptor and SGTA (Figure 4C). In order to confirm the direct binding experimentally, we purified a biotinylated form of the isolated LBD domain of the AR (residues 670–920), immobilized it on streptavidin beads, and then incubated it with LNCaP cell lysate. We next checked by western blot if the LBD protein pulls down SGTA. Indeed a band corresponding to SGTA was detected with the AR-LBD but not with the biocytin control (Figure 4D). We then tested the effect of VPC-13566 (100 μM) on the LBD/SGTA binding and noticed a significant decrease in the binding (Figure 4D–E). In addition, VPC-13566 was able to displace a FITC-labeled SGTA peptide (FITC-MPQDLRSPARTPPSEEDSAEA) from the BF3 pocket of a purified LBD domain in a TR-FRET assay with a Kd = 11.4 ± 1.4 μM. As a negative control in this experiment, we used compound VPC-14449 which only targets the DNA Binding domain of the AR (31). As expected, there was no peptide displacement observed with this other compound (Figure 4F).
We next evaluated the direct interaction between biotinylated LBD and an unlabeled SGTA peptide (71MPQDLRSPARTPPSEEDSAEA91) using BLI. Indeed, the SGTA peptide showed a direct binding to the isolated LBD domain of the AR that lacks the hinge region. Mutating Alanine 79 or Proline 82 of SGTA disrupted the binding, indicating that similarly to BAG1L and FKBP52, those two residues are essential for LBD/SGTA interaction (Figure 4G). All these results suggest that there exists a direct interaction between the PARTPP motif of SGTA and the BF3 pocket of the AR.
Compound VPC-13566 reduces AR translocation to the nucleus
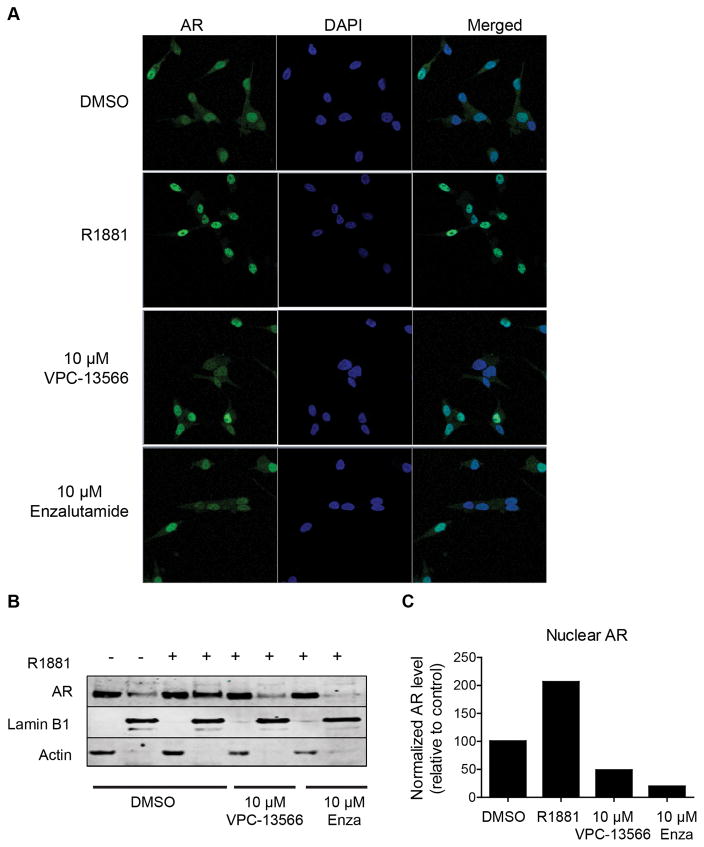
Next we investigated whether VPC-13566 could be used as a probe to better understand the role of the BF3 site in AR activity. It is well known that in the presence of androgen, the AR translocates into the nucleus and modulates the transcription of specific regulated genes. It has been previously reported that small molecules that are predicted to bind to the BF3 surface of AR blocks the 52 kDa FK506 binding protein (FKBP52) dependent AR nuclear translocation (16). Hence, we evaluated if VPC-13566 could affect androgen-induced translocation of AR from the cytoplasm to the nucleus. Using confocal microscopy, the immunofluorescence staining of AR in LNCaP cells showed an increase in the nuclear fraction of the receptor when cells were stimulated with R1881 (Figure 5A). In contrast, this androgen-specific effect was reduced when cells were additionally treated with 10 μM of VPC-13566 (Figure 5A). A similar effect was observed with enzalutamide, which is known to block AR nuclear translocation (32). To confirm this finding, cell fractionation was performed on LNCaP cells stimulated with R1881 alone or with VPC-13566. Cytoplasmic and nuclear proteins were isolated and probed with an AR antibody. In the absence of R1881 stimulation, AR was mainly cytoplasmic. However, in the presence of androgen the nuclear fraction of AR increased. Treatment with VPC-13566 in the presence of R1881 clearly reduced nuclear localization of the receptor (Figure 5B–C).
Figure 5. VPC-13566 blocks AR nuclear translocation.
A- LNCaP cells were stimulated with R1881 and treated with 10 μM of either VPC-13566 or enzalutamide (Enza). The cellular localization of endogenous AR, cell nucleus (DAPI) and the merged images of AR and DAPI were assessed using confocal microscopy. Enzalutamide and VPC-13566 reduced the nuclear localization of AR. The blockage of AR nuclear translocation by VPC-13566 was confirmed by western blot after nuclear/cytoplasmic fractionation (B–C).
VPC-13566 inhibits AR-dependent growth of xenograft tumors in vivo
The in vivo effect of VPC-13566 was evaluated with a xenograft model of castration-resistant prostate cancer. The pharmacokinetic analysis of VPC-13566 and its metabolites following intravenous (IV) or intraperitoneal (IP) administration demonstrated moderate metabolic stability of VPC-13566, with one primary and two medium level glucuronides established as dominant biotransformation products. The IV and IP serum profiles of VPC-13566 suggested that it would be best administered IP with substantial retention up to 24h. For IV and IP, Cmax was established at approximately 14 and 1 μM and half-lives were estimated as 2.3 and 2.9 hours respectively, and a very long terminal half-life for IP only. Based on the initial in vitro data (AR eGFP IC50 = 0.05 μM) it was to be expected that the plasma concentration of the drug should remain within the predicted therapeutic window for at least 24 hours when delivered IP.
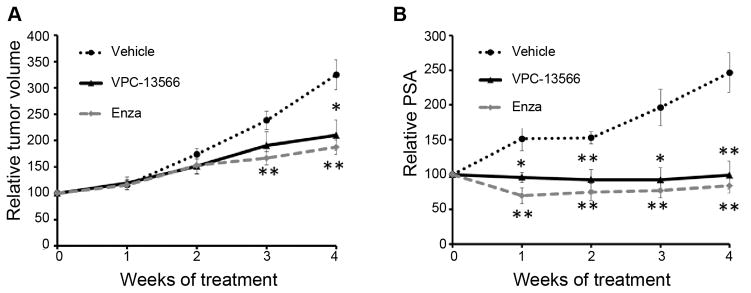
A dose of 100 mg/kg administered IP twice a day for 5 days and followed by a single dose of 200 mg/kg once per day for 2 days per week, for 4 weeks, was chosen with no visible signs of toxicity seen in the mice. The in vivo screening for tumor growth was performed using the castration-resistant LNCaP xenograft model (26, 33–35). Mice were implanted with LNCaP tumor cells and castrated upon serum PSA reaching 25 ng/ml. When tumor regrowth was observed and serum PSA returned to pre-castration levels, the mice were treated with VPC-13566, vehicle control, or enzalutamide. Tumor growth was effectively suppressed by VPC-13566 (p < 0.05) and enzalutamide (P < 0.01) (Figure 6A). Moreover, VPC-13566 significantly decreased serum PSA to a comparable degree as enzalutamide (p < 0.01, Figure 6B). Together, these results indicate that this class of AR inhibitor has the potential to yield an AR targeting drug that could have clinical utility in the treatment of patients with castration-resistant tumors.
Figure 6. In vivo effect of VPC-13566 in LNCaP castration resistant xenograft model.
A-The in vivo effect of VPC-13566 on the tumor volume. Data are presented as mean ± SEM of % tumor volume at time of treatment initiation (each group, n=5). B- The in vivo effect of VPC-13566 on PSA level. Data are presented as mean ± SEM of % PSA level at time of treatment initiation (each group, n=5). A p value < 0.05 was considered significant (*) and < 0.01 very significant (**) compared to vehicle control (two-tailed T-Test).
DISCUSSION
The human AR is the main driver of CRPC mainly due to the acquisition of new growth-promoting functions during PCa development and progression through genetic and epigenetic mechanisms. In order to overcome drug resistance associated with traditional anti-androgens that block steroids from binding to the ABS, a new promising approach has emerged by inhibiting the receptor through alternatives sites such as its BF3 functional region. The BF3 site is a pocket located on the LBD surface and is formed by residues from helices 1, 3′ and 9 and the loop connecting H3 and H3′ (36). This pocket is important for co-chaperone recruitment, AR transcriptional activation and allosteric regulation of the adjacent activation function 2 (AF2) site (14–16). In this study, we report on the development of a novel BF3 inhibitor with 6-fold improved efficacy compared to the parental prototype compound previously reported by our group (12). According to the MD simulation experiments, VPC-13566 tightly occupies the binding regions of an hexapeptide motif and disrupts key H-bond interactions and Van der Waals contacts formed between BAG1L 7ARRP10 peptide and BF3 residues (Figure 2D). Experimentally, VPC-13566 was able to compete with the BAG1L co-chaperone for binding to the BF3 pocket (Figure 3C–D).
It has been previously demonstrated that alanine and proline residues in the GSAGSPP and GARRPR sequences of FKBP52 and BAG1L respectively, are essential for the binding to the AR BF3 pocket (15, 30). Since the BF3 pocket is considered a co-chaperone binding site and is at the boundary with the hinge region, we suspected that a similar motif could exist in the SGTA protein, a co-chaperone that interacts with the AR hinge and plays a role in receptor cytoplasmic regulation (28, 29). Indeed, analysis of the SGTA protein sequence has identified a PARTPP motif (residues 78–83) that had the potential to be recognized by the BF3 site. Using pull-down and TR-FRET assays, we were able to demonstrate that SGTA is able to bind to a purified AR LBD protein lacking the hinge region (residues 670–920) and this interaction was reduced in presence of the BF3 inhibitor VPC-13566 (Figure 4D–F). Critical BF3 residues engaged into the SGTA binding were identified using MD simulations and then confirmed by site directed mutagenesis (Figure 4A–C). In addition, the importance of SGTA residues alanine 79 and proline 82 was validated by the BLI technique (Figure 4G). This combination of in silico predictions and experimental data suggested that SGTA is a BF3 interacting partner. To our knowledge, this is the first report of such a role for SGTA.
In addition, it has been reported that the BF3 site of the AR plays a role in the receptor shuttling into the nucleus (16). Indeed, we have established that VPC-13566 blocked the androgen-dependent AR translocation into the nucleus most probably through interfering with the exchange between the TRP containing proteins SGTA and FKBP52, a necessary step for nuclear shuttling. It is intriguing to note that the BF3 site was important for both the interaction with cytoplasmic (SGTA) or nuclear (BAG1L) cofactors. This suggests that VPC-13566 can act on both translocation pathways of nuclear transcription (Figures 3 and 4), and thus could complicate the development of resistance against this drug by the cell.
VPC-13566 may have clinical benefits, as it is able to inhibit in vitro the growth and PSA expression of enzalutamide-resistant MR49F cells (Figure 1C–E). Additionally, VPC-13566 demonstrated significant inhibition of tumor growth as well as serum PSA levels in the LNCaP xenograft model (Figure 6). Interestingly, the combination of enzalutamide and VPC-13566 showed an additive effect on the inhibition of PSA expression in LNCaP cells, indicating that two binding sites on the AR could be targeted simultaneously without mutual interference from these two drugs (Figure 1E). In addition, in another study we demonstrated that compound VPC-13566 was capable of inhibiting the transcriptional activity of several resistance-associated AR mutants (21), showing that targeting alternative sites on the AR might help overcoming the resistance frequently seen with clinically used drugs that exclusively target the ABS.
It has been reported that full length AR heterodimerizes, in the absence of androgens, with splice variants lacking the LBD and thus the BF3 site. This heterodimerization is suggested to play a role in AR nuclear translocation under castration conditions and therefore in the activation of AR target genes (37). In our hands, we did not see any effect of this compound on the cell growth of 22Rv1 in the absence of androgens where the growth is solely driven by AR splice variants (data not shown), however this does not exclude an effect on the transcriptional activity of certain genes regulated by the heterodimer.
In conclusion, we identified a new, very potent AR inhibitor VPC-13566 that targets the AR BF3 site, a functional surface pocket distant from the mutations-prone ABS area of the androgen receptor. Using this inhibitor, we demonstrated that the BF3 site is involved with AR nuclear translocation and transcriptional regulation through interacting with cytoplasmic and nuclear co-chaperones, such as FKBP52, BAG1L and the newly identified SGTA. Thus, VPC-13566 may possess an important clinical potential for the treatment of drug-resistant forms of PCa and could be used concurrently or simultaneously with the current anti-androgen treatments such as enzalutamide.
Acknowledgments
Financial support: This work was supported by Prostate Cancer Canada with generous support from Canada Safeway Grant SP2013-02 (P.S Rennie). This research was also supported by the Department of Defense (Prostate Cancer Research Program) under award number W81XWH-12-1-0401 (A. Cherkasov). The authors acknowledge financial support from the Canadian Institutes of Health Research (operating grant #272111; A. Cherkasov) and from Canadian Cancer Society Research Institute (Grant# 701585; A. Cherkasov). The salary of N. Lallous was supported by Award Number P50CA097186 from the National Cancer Institute. R. S.N. Munuganti had a graduate fellowship from the Prostate Cancer Foundation-BC.
The authors thank Bonny Chow, Alisse Saunders, Ronnie Tse, Josef Murillo, Clement Yau, Shaghayegh Esfandnia, Lauren Leachman and Justin Lardizabal for helping with this study.
Abbreviations
- BF3
Binding Function 3
- AR
Androgen receptor
- PCa
Prostate Cancer
- CRPC
Castration resistant prostate cancer
- LBD
Ligand Binding Domain
- PLA
Proximity ligation assay
- IF
Immunofluorescence
- BLI
Biolayer interferometry
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA: a cancer journal for clinicians. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. The lancet oncology. 2009;10:981–91. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. The lancet oncology. 2012;13:983–92. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 5.Toren PJ, Kim S, Pham S, Mangalji A, Adomat H, Guns ES, et al. Anticancer activity of a novel selective CYP17A1 inhibitor in preclinical models of castrate-resistant prostate cancer. Molecular cancer therapeutics. 2015;14:59–69. doi: 10.1158/1535-7163.MCT-14-0521. [DOI] [PubMed] [Google Scholar]
- 6.Wyatt AW, Gleave ME. Targeting the adaptive molecular landscape of castration-resistant prostate cancer. EMBO molecular medicine. 2015;7:878–94. doi: 10.15252/emmm.201303701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman-Censits J, Kelly WK. Enzalutamide: a novel anti-androgen for patients with castrate resistant prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:1335–9. doi: 10.1158/1078-0432.CCR-12-2910. [DOI] [PubMed] [Google Scholar]
- 8.Pal SK, Stein CA, Sartor O. Enzalutamide for the treatment of prostate cancer. Expert opinion on pharmacotherapy. 2013;14:679–85. doi: 10.1517/14656566.2013.775251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban F, Leblanc E, Li H, Munuganti RS, Frewin K, Rennie PS, et al. Discovery of 1H-indole-2-carboxamides as novel inhibitors of the androgen receptor binding function 3 (BF3) Journal of medicinal chemistry. 2014;57:6867–72. doi: 10.1021/jm500684r. [DOI] [PubMed] [Google Scholar]
- 10.Lack NA, Axerio-Cilies P, Tavassoli P, Han FQ, Chan KH, Feau C, et al. Targeting the binding function 3 (BF3) site of the human androgen receptor through virtual screening. Journal of medicinal chemistry. 2011;54:8563–73. doi: 10.1021/jm201098n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munuganti RS, Leblanc E, Axerio-Cilies P, Labriere C, Frewin K, Singh K, et al. Targeting the Binding Function 3 (BF3) Site of the Androgen Receptor Through Virtual Screening. 2. Development of 2-((2-phenoxyethyl) thio)-1H-benzimidazole derivatives. Journal of medicinal chemistry. 2013;56:1136–48. doi: 10.1021/jm3015712. [DOI] [PubMed] [Google Scholar]
- 12.Munuganti Ravi SN, Hassona Mohamed DH, Leblanc E, Frewin K, Singh K, Ma D, et al. Identification of a Potent Antiandrogen that Targets the BF3 Site of the Androgen Receptor and Inhibits Enzalutamide-Resistant Prostate Cancer. Chemistry & Biology. 2014;21:1476–85. doi: 10.1016/j.chembiol.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16074–9. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosdidier S, Carbo LR, Buzon V, Brooke G, Nguyen P, Baxter JD, et al. Allosteric conversation in the androgen receptor ligand-binding domain surfaces. Mol Endocrinol. 2012;26:1078–90. doi: 10.1210/me.2011-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jehle K, Cato L, Neeb A, Muhle-Goll C, Jung N, Smith EW, et al. Coregulator Control of Androgen Receptor Action by a Novel Nuclear Receptor-Binding Motif. The Journal of biological chemistry. 2014;289:8839–51. doi: 10.1074/jbc.M113.534859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Leon JT, Iwai A, Feau C, Garcia Y, Balsiger HA, Storer CL, et al. Targeting the regulation of androgen receptor signaling by the heat shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11878–83. doi: 10.1073/pnas.1105160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchanan G, Ricciardelli C, Harris JM, Prescott J, Yu ZC, Jia L, et al. Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer research. 2007;67:10087–96. doi: 10.1158/0008-5472.CAN-07-1646. [DOI] [PubMed] [Google Scholar]
- 18.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of medicinal chemistry. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 19.Axerio-Cilies P, Lack NA, Nayana MR, Chan KH, Yeung A, Leblanc E, et al. Inhibitors of androgen receptor activation function-2 (AF2) site identified through virtual screening. Journal of medicinal chemistry. 2011;54:6197–205. doi: 10.1021/jm200532b. [DOI] [PubMed] [Google Scholar]
- 20.Singh K, Munuganti RS, Leblanc E, Lin YL, Leung E, Lallous N, et al. In silico discovery and validation of potent small-molecule inhibitors targeting the activation function 2 site of human oestrogen receptor alpha. Breast cancer research : BCR. 2015;17:27. doi: 10.1186/s13058-015-0529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lallous N, Volik SV, Awrey S, Leblanc E, Tse R, Murillo J, et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 2016;17:10. doi: 10.1186/s13059-015-0864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavassoli P, Snoek R, Ray M, Rao LG, Rennie PS. Rapid, non-destructive, cell-based screening assays for agents that modulate growth, death, and androgen receptor activation in prostate cancer cells. The Prostate. 2007;67:416–26. doi: 10.1002/pros.20532. [DOI] [PubMed] [Google Scholar]
- 23.Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:383–91. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 24.Kuruma H, Matsumoto H, Shiota M, Bishop J, Lamoureux F, Thomas C, et al. A Novel Antiandrogen, Compound 30, Suppresses Castration-Resistant and MDV3100-Resistant Prostate Cancer Growth In Vitro and In Vivo. Molecular cancer therapeutics. 2013;12:567–76. doi: 10.1158/1535-7163.MCT-12-0798. [DOI] [PubMed] [Google Scholar]
- 25.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods (San Diego, Calif) 2008;45:227–32. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Snoek R, Ghaidi F, Cox ME, Rennie PS. Short hairpin RNA knockdown of the androgen receptor attenuates ligand-independent activation and delays tumor progression. Cancer research. 2006;66:10613–20. doi: 10.1158/0008-5472.CAN-06-0028. [DOI] [PubMed] [Google Scholar]
- 27.Cheung-Flynn J, Prapapanich V, Cox MB, Riggs DL, Suarez-Quian C, Smith DF. Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol Endocrinol. 2005;19:1654–66. doi: 10.1210/me.2005-0071. [DOI] [PubMed] [Google Scholar]
- 28.Philp LK, Butler MS, Hickey TE, Butler LM, Tilley WD, Day TK. SGTA: a new player in the molecular co-chaperone game. Hormones & cancer. 2013;4:343–57. doi: 10.1007/s12672-013-0151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trotta AP, Need EF, Butler LM, Selth LA, O’Loughlin MA, Coetzee GA, et al. Subdomain structure of the co-chaperone SGTA and activity of its androgen receptor client. Journal of molecular endocrinology. 2012;49:57–68. doi: 10.1530/JME-11-0152. [DOI] [PubMed] [Google Scholar]
- 30.Riggs DL, Cox MB, Tardif HL, Hessling M, Buchner J, Smith DF. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Molecular and cellular biology. 2007;27:8658–69. doi: 10.1128/MCB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalal KM, MR, Li H, Sharma A, Ban F, Leblanc E, Dehm S, Cherkasov A, Rennie PS. Selectively Targeting the DNA Binding Domain of the Androgen Receptor as a Prospective Therapy for Prostate Cancer. The Journal of biological chemistry. 2014;289:26417–29. doi: 10.1074/jbc.M114.553818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyake H, Nelson C, Rennie PS, Gleave ME. Acquisition of chemoresistant phenotype by overexpression of the antiapoptotic gene testosterone-repressed prostate message-2 in prostate cancer xenograft models. Cancer research. 2000;60:2547–54. [PubMed] [Google Scholar]
- 34.Sato N, Gleave ME, Bruchovsky N, Rennie PS, Goldenberg SL, Lange PH, et al. Intermittent androgen suppression delays progression to androgen-independent regulation of prostate-specific: Antigen gene in the LNCaP prostate tumour model. Journal of Steroid Biochemistry and Molecular Biology. 1996;58:139–46. doi: 10.1016/0960-0760(96)00018-0. [DOI] [PubMed] [Google Scholar]
- 35.Snoek R, Cheng H, Margiotti K, Wafa LA, Wong CA, Wong EC, et al. In vivo Knockdown of the Androgen Receptor Results in Growth Inhibition and Regression of Well-Established, Castration-Resistant Prostate Tumors. Clinical Cancer Research. 2009;15:39–47. doi: 10.1158/1078-0432.CCR-08-1726. [DOI] [PubMed] [Google Scholar]
- 36.Lallous N, Dalal K, Cherkasov A, Rennie PS. Targeting alternative sites on the androgen receptor to treat castration-resistant prostate cancer. Int J Mol Sci. 14:12496–519. doi: 10.3390/ijms140612496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer research. 75:3663–71. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]