Abstract
How axons repair themselves after injury is a fundamental question in neurobiology. With its conserved genome, relatively simple nervous system, and transparent body, C. elegans has recently emerged as a productive model to uncover the cellular mechanisms that regulate and execute axon regeneration. In this review, we discuss the strengths and weaknesses of the C. elegans model of regeneration. We explore the technical advances that enable the use of C. elegans for in vivo regeneration studies, review findings in C. elegans that have contributed to our understanding of the regeneration response across species, discuss the potential of C. elegans research to provide insight into mechanisms that function in the injured mammalian nervous system, and present potential future directions of axon regeneration research using C. elegans.
GENERAL FEATURES OF THE C. ELEGANS AXON REGENERATION MODEL
The adult C. elegans hermaphrodite is a transparent cylinder approximately 1mm long that contains 302 neurons (Figure 1). The worm’s nervous system includes motor, sensory, interneuron, and polymodal neurons and can be divided into 118 distinct classes of neurons, including GABAergic, cholinergic, chemosensory, mechanosensory, oxygen sensing, osmoceptors and proprioceptors (White et al., 1986). The development and positions of individual neurons are invariant from worm to worm. Together with the transparent cuticle, the invariant development of the nervous system makes the worm a tractable model of axon regeneration. The transparent cuticle provides the ability to sever individual fluorescently labeled axons with a laser and monitor severed axons for regenerative ability in vivo (Figure 2). The lack of inter-worm variability allows the effects of genetic and chemical manipulations on axon regeneration of specific, individual neurons to be compared between worms.
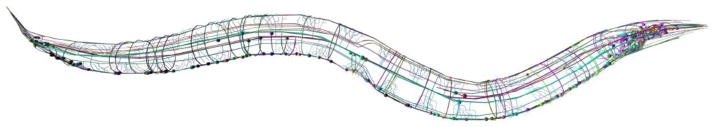
Figure 1. C. elegans nervous system.
The C. elegans nervous system consists of 302 neurons. Most cell bodies lie in the head (right) and tail (left) of the worm. GABA motor neuron cell bodies are located along the ventral nerve cord of the worm and extend commissures circumferentially to the dorsal nerve cord. Mechanosensory neurons extend axons along the lateral aspect of the worm. Image courtesy of OpenWorm.org and VirtualWorm.
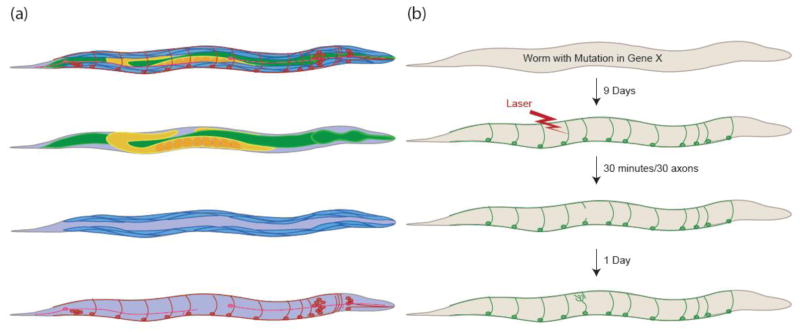
Figure 2. Axon Regeneration in C. elegans.
(a) Worms contain an alimentary system (green), reproductive system (yellow), muscle (blue), and nervous system (red and pink). Axon regeneration is mostly studied in the GABA motor neurons (red) and the mechanosensory neurons (pink). (b) Worms carrying a mutation in a gene of interest and expressing GFP in a specific type of neuron can be built and used to study axon regeneration in approximately 9 days. The diagram depicts a GABA axon severed with a laser and regenerating 24 hours later.
Most neurons are contained in ganglia located in the head and tail of the worm. In the head, the nerve ring is considered ‘the brain’ of the worm and consists of a neuropil of predominantly sensory neurons but also motor neurons and interneurons. Additional motor and mechanosensory neurons are located along the body of the worm. Among the various types of neurons, regenerative ability has been characterized for the GABA motor neurons, the mechanosensory neurons, and a few other types of neurons. The regenerative ability of most neuron types has not yet been studied.
The field of C. elegans axon regeneration began with the discovery that GFP-labeled GABA motor neurons regenerate after being severed with a femtosecond laser (Yanik et al., 2004). The GABA neuron cell bodies are positioned along the ventral nerve cord and extend commissures circumferentially towards the dorsal nerve cord (White et al., 1986; McIntire et al., 1993). They receive synaptic input from excitatory cholinergic neurons and make inhibitory en passant synaptic connections with body wall muscles. The commissural axons are accessible for laser surgery and imaging and have been used extensively for regeneration studies (Figure 2b). When a GABA motor axon commissure is cut at the midline, the two severed ends retract and the axon segment proximal to the cell body forms a growth cone 6–12 hours post-injury, then extends at 2–3 μm/hr towards the dorsal cord (Wu et al., 2007). Axon regeneration in mature animals is imprecise, as the axons that regenerate are often branched and occasionally grow around the distal stump rather than follow the same developmental path (Yanik et al., 2004; Wu et al., 2007) Not only do the axons regenerate, the regenerated axons are able to restore functional connections with the musculature (Yanik et al., 2004; El Bejjani and Hammarlund, 2012).
The mechanosensory or ‘touch’ neurons extend long axons along the length of the worm and regulate response to touch (Chalfie and Sulston, 1981). Like the GABA neuron commissures, the axons of the touch neurons are relatively easy to sever and study. Further, because these axons extend along the anterior-posterior axis, regenerative growth can be monitored over a much longer distance than the GABA neurons. Severed mechanosensory neurons regenerate more quickly, at a rate of approximately 7 μm/hr. However, in these neurons the majority of growth occurs between 24 and 48 hours (Wu et al., 2007). Unlike the GABA neurons, behavioral output dependent on regeneration of the touch neurons has not been described.
Because the nervous system is invariant between animals, the same single neuron can be studied across multiple animals. As such, C. elegans is an excellent model to analyze variability in regenerative ability in individual neurons. For example, although the GABA neurons can regenerate when severed with a femtosecond or pulsed UV laser, only 70% of injured axons form growth cones, while the remaining 30% do not (Wu et al., 2007; Hammarlund et al., 2009; Byrne et al., 2011). Variable regeneration is also observed in other neuron types, including the touch neurons (Yanik et al., 2006; Wu et al., 2007). One key determinant of regeneration success at the level of individual neurons is the rise in calcium observed after injury (Ghosh-Roy et al., 2010; Pinan-Lucarre et al., 2012; Yan and Jin, 2012). Axotomy of the PLM mechanosensory neurons causes an increase in localized calcium at the site of injury and the amount of calcium or cAMP determines regeneration success (discussed in more detail below) (Ghosh-Roy et al., 2010). It is likely that other cellular mechanisms besides calcium also contribute to the variability in regeneration.
The C. elegans nervous system also contains 56 glia. As in other invertebrate and early vertebrate systems, the glia wrap around nerve-nerve and nerve-muscle synapses, presumably to insulate signals from the surrounding environment (Ward et al., 1975; White et al., 1986). They also wrap around extending dendrites and axons to enhance extension and provide guidance. The role of glia in C. elegans axon regeneration has not been well characterized. In contrast to vertebrate glia, C. elegans glia do not produce myelin. The absence of myelin inhibition means that the neuronal mechanisms that regulate axon regeneration can be studied in the absence of this glial inhibitory pathway in C. elegans. However, other interactions between regenerating neurons and their supporting cells (including glia, but also other cell types) are undoubtedly important for regeneration. Further studies are necessary to address this important question.
Finally, C. elegans is a well-developed model organism, with a rapid life cycle and easy cultivation. The well-characterized genome facilitates the identification and investigation of the genetic and molecular determinants of axon regeneration (Box 1). Regulators of nervous system development and function are largely conserved between C. elegans and mammals. Approximately 40% of the worm’s genome is conserved with the mammalian genome (Shaye and Greenwald, 2011). In addition, the worm’s small size allows it to be easily cultured in large numbers on agar-filled petri plates containing non-pathogenic E. coli. The worm has a three-day life cycle and ten-day lifespan. The relatively short life cycle and lifespan facilitate quick construction of mutant strains and rapid analysis of the mechanisms that regulate regeneration at various ages, respectively. A strain containing a mutation in a gene of interest and a fluorescent marker expressed in a specific population of neurons can be built in approximately two weeks. Since regeneration is typically assessed within 24–48 hours after injury, the role of a given gene in axon regeneration can be quickly investigated.
BOX 1. C. ELEGANS IS AN EFFICIENT GENETIC MODEL WITH A HIGHLY CONSERVED GENOME.
C. elegans is a conserved, tractable, and efficient genetic system. The worm genome consists of approximately 21,000 genes on six chromosomes. Nearly half of the worm genome and most major signaling pathways, including Notch, Wnt, Insulin, and TGF-B, are conserved with mammals(Consortium, 1998; Shaye and Greenwald, 2011). Manipulating the genome and characterizing resulting phenotypes is relatively straightforward in C. elegans. Molecular mechanisms that control many processes, such as development, reproduction, and aging, have been investigated using genetic approaches in C. elegans. Knowledge gained from these studies can be applied to axon regeneration.
The conservation of function between C. elegans and mammalian genes predicts that genes found to regulate axon regeneration in C. elegans may have a similar function in mammalian axon regeneration. The genetic conservation also suggests the function of genes found to regulate axon regeneration in mammals may be further characterized in C. elegans. Indeed, multiple recently identified regulators of axon regeneration, including PTEN and DLK, have conserved function in C. elegans and in mammals (Park et al., 2008; Hammarlund et al., 2009; Itoh et al., 2009; Yan et al., 2009; Shin et al., 2012; Watkins et al., 2013; Byrne et al., 2014).
C. elegans is particularly amenable to genetic and genomic approaches, both classic and modern. Worms exist as hermaphrodites capable of self-fertilization and as males. Therefore, an isogenic strain of hermaphroditic worms can be maintained without organized mating and genotyping. Males are used to introduce a desired genetic mutation or fluorescent reporter into a hermaphroditic strain. In most regeneration experiments, a mating is used to create progeny who express GFP in a specific set of neurons and have a specific genetic mutation. The worm’s three-day reproductive cycle makes creating such progeny relatively quick: a neuron type-specific GFP reporter can be crossed into a strain containing a genetic mutation to produce a homozygous, double mutant (reporter; mutation), isogenic line in as few as nine days. Recently, modern CRISPR/Cas9 technology has added greatly to the ability to disrupt, express and characterize C. elegans genes in a spatial and temporal manner (Friedland et al., 2013). As with classic genetic approaches, the short C. elegans reproductive cycle makes modern genetic manipulations extremely fast compared to mammalian genetic manipulations.
The efficiency of C. elegans as a genetic model extends beyond time saving. In addition to a short life cycle, the worm has a relatively large brood size. A wild type hermaphroditic worm gives birth to approximately 250 isogenic progeny. As such, many identical animals are readily available for axotomy and analysis from a single parental worm. Worms also survive long-term freezing at extreme temperatures. The ability to freeze worms allows new and valuable worm strains to be maintained indefinitely with minimal effort or cost. Moreover, candidate gene analysis is aided by the availability of a large number of mutants generated by the worm community and C. elegans Gene Knockout Consortium, which are kept as frozen stocks at the ‘Caenorhabditis Genetics Center’ and at the ‘National Bioresource Project for the Experimental Animal C. elegans’.
Together, the conservation, tractability, and efficiency of C. elegans genetics facilitate the relatively quick and relevant investigation of conserved biological processes such as axon regeneration.
TECHNIQUES FOR AXON INJURY: LASERS AND GENETICS
Traditional mechanical techniques to injure axons, such as crushing nerves with forceps or severing nerves with a scalpel, have not been applied in C. elegans, due to the small size of the worm. Instead, optical or genetic techniques are used to sever axons. The first investigations of axon regeneration in C. elegans were carried out on axons that had been severed with amplified Ti-sapphire lasers that produce femtosecond pulses of near infrared light (780–800 nanometers) (Yanik et al., 2004). These lasers supply 10–40 nanojoules of energy with 200 femtosecond pulses at 1 kHz and vaporize 1–2 um of axon. Subsequently, unamplified femtosecond lasers that supply an order of magnitude less energy (1–3 nanojoules) with approximately 150 femtosecond pulses at a higher pulse rate (80–90 MHz) have been used to sever axons (Wu et al., 2007). The amount of energy supplied is the main determinant of the size of the injury; lower amounts of energy create a smaller area of damage (Bourgeois and Ben-Yakar, 2008). However, pulse rate also influences the specificity of injury; pulse rates in the kHz range create less damage and more specific injuries than pulse rates in the MHz range (Wu et al., 2007). Contrary to expectations, high trains of low energy pulses increase regeneration frequency compared to low trains of pulses of the same energy, indicating the total amount of energy used to injure the axon is not the sole determinant of regeneration success (Bourgeois and Ben-Yakar, 2008). Together, these experiments indicate femtosecond laser ablation using high trains of low energy pulses delivered at kHz pulse rates severs axons with minimal physical or thermal damage to the surrounding tissue.
Besides Ti-sapphire lasers, many axon regeneration studies use other types of pulsed lasers, such as nitrogen-pumped dye lasers (historically used for cell ablations) or solid-state lasers (Rao et al., 2008; Williams et al., 2011). These lasers are a fraction of the price of Ti-sapphire lasers. However, since they produce longer pulses, they may create a larger area of damage than Ti-sapphire lasers. In addition, these lasers may have difficulty severing axons deep within the animal. Therefore, they are excellent for cutting widely-spaced axons located just under the skin, such as the GABA and touch neurons.
In general, laser axotomy is a widely applicable technique, with the significant experimental advantage that the time and location of injury can be precisely controlled and limited to individual axons of interest. Although laser axotomy proceeds one neuron at a time, dedicated application of laser techniques can result in the study of a large number of genes (Chen et al., 2011; Nix et al., 2014).
For experimental questions that require even higher numbers of injured axons, a genetic mutation in the gene encoding β-spectrin (unc-70) has been used. Lack of β-spectrin causes axons to break when the animal moves (Hammarlund et al., 2007), likely due to defects in the spectrin-based membrane cytoskeleton (Xu et al., 2013; He et al., 2016). Loss of unc-70 function has no obvious effect on the development of motor axons. However, after the axons develop, the resulting fragile axons break as the worm moves. In this genetic background, axons successively break and regenerate without the use of laser axotomy. Thus, with this mutant, very large numbers of axons can be injured (at the expense of temporal and spatial precision), facilitating genetic and genomic screening (Hammarlund et al., 2009; Rohde et al., 2009; Nix et al., 2014). An alternative approach is to automate laser surgery, potentially increasing the number of axons that can be injured and analyzed (Guo et al., 2008; Gokce et al., 2014). The availability of diverse axon injury models in C. elegans enables researchers to select the optimal approach for a given experiment.
GENETIC SCREENS IDENTIFY REGULATORS OF AXON REGENERATION
A prominent benefit of the C. elegans regeneration model is the ability to conduct forward screens to identify genes involved in axon regeneration. To date, multiple screens of various types and scales have been undertaken, including RNAi screens, mutant screens, automated chemical screens, and candidate screens. These screens have made significant contributions to our understanding of axon regeneration, and have generated large amounts of data that awaits detailed analysis.
A large screen for regulators of axon regeneration in C. elegans took advantage of spontaneously broken axons caused by mutation of the unc-70 (β-spectrin) gene (Hammarlund et al., 2007; Hammarlund et al., 2009; Nix et al., 2014). Mutant unc-70 worms with GFP-labeled GABA motor neurons were screened against an RNAi library that targeted 5076 homologs of human genes (Nix et al., 2014). To induce RNAi in C. elegans, worms are fed bacteria that express double-stranded RNA corresponding to a particular gene (Fire et al., 1998; Timmons and Fire, 1998). Multiple independent investigations have found approximately 70% of GABA motor neurons regenerate in wild type animals (Yanik et al., 2004; Hammarlund et al., 2007; Wu et al., 2007; Byrne et al., 2011; Nix et al., 2011; Byrne et al., 2014). In contrast, when fed bacteria expressing a double stranded RNA with homology to a gene required for axon regeneration, few axons regenerate. In the first round of the screen, RNAi was used to characterize all genes in the library; in the second round, positive hits and other candidates were retested using laser axotomy in the corresponding genetic mutant background. In total, the screen identified more than 50 conserved genes that function in GABA neuron axon regeneration. Of these 50 genes, detailed analysis has been performed on the DLK-1 MAP Kinase pathway and its parallel MLK-1 MAP Kinase pathway, along with syndecan and the tRNA splicing ligase RTCB-1 have been characterized as regulators of axon regeneration (Hammarlund et al., 2009; Nix et al., 2011; Li et al., 2012; Edwards and Hammarlund, 2014; Kosmaczewski et al., 2015). The screen also led to the finding that Notch signaling inhibits axon regeneration (El Bejjani and Hammarlund, 2012). The remainder of these genes await further characterization.
Despite the positive results from this screen, it is not expected to identify all genes involved in axon regeneration. For example, not all genes were screened, and genes with functions essential to survival could not be tested for a role in axon regeneration. Further, RNAi does not always eliminate gene function in the C. elegans nervous system, even in a sensitized background (Kamath et al., 2001; Timmons et al., 2001; Sieburth et al., 2005; Wang et al., 2005). Therefore, other regulators of regeneration remain to be uncovered in the C. elegans genome.
A different type of screen was performed using laser axotomy. In this screen, 654 strains of worms with mutations in conserved genes were tested for their role in axon regeneration of PLM mechanosensory neurons (Chen et al., 2011). The screen identified 149 genes as regulators of axon regeneration. Most of the identified genes are required for axon regeneration and only 16 inhibit regeneration. The identified genes have diverse functions including, but not restricted to, synaptic vesicle endocytosis, neurotransmission, formation of the extracellular matrix, and axon guidance. Interestingly, the functions of many of the identified genes are specific to axon regeneration and not development, as the PLM axons have mostly wild type morphology pre-injury. One of the characterized hits is the conserved Arf Guanine nucleotide Exchange Factor efa-6, which inhibits axon regeneration by regulating microtubule dynamics (Chen et al., 2015). Interestingly, with some notable exceptions (e.g. the DLK pathway, syndecan), there is relatively little overlap between regeneration genes identified by these two screens, suggesting that axon regeneration in these two different neurons types (one motor, one sensory) is mediated in part by different cellular mechanisms.
In vivo, whole animal, chemical screens hold significant promise to identify regulators of axon regeneration. A semi-automated chemical screen of approximately 100 small molecules was carried out using microfluidics and a femtosecond laser (Samara et al., 2010). Chemicals that targeted the cytoskeleton and protein kinases affected regeneration while chemicals that targeted HDACs and vesicle trafficking did not. In particular, the screen identified three PKC (protein kinase C) inhibitors that inhibited regeneration and a PKC activator that enhanced axon regeneration. Expanded chemical screening has the potential to identify and characterize additional regulators of axon regeneration and reveal chemical approaches that may be translated to treat injury in humans.
CURRENT AREAS OF INVESTIGATION
Current investigations focus on the regulation and role of injury sensing, signal transduction, cytoskeletal dynamics, aging, and axon fusion in axon regeneration in C. elegans. In many cases, the investigations are motivated by the identification and characterization of genes in the screens described above. The emerging understanding of axon regeneration in C. elegans is that, as in the mammalian system, axon regeneration is a complex, orchestrated process regulated by the concerted function of multiple aspects of neuronal cell biology (Figure 3, Figure 4, and Table 1).
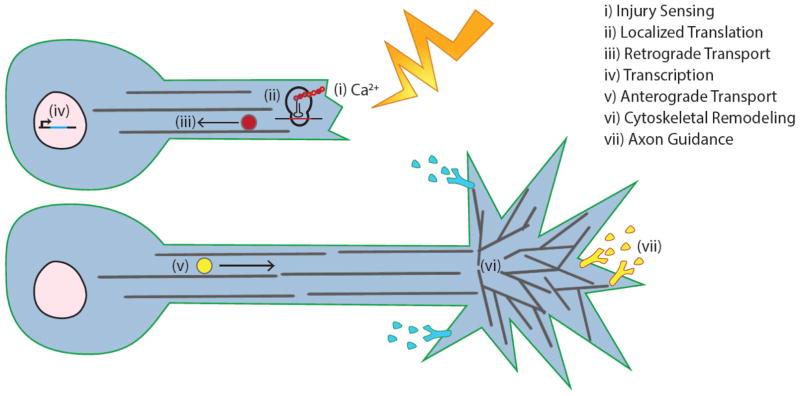
Figure 3. Current investigations focus on multiple components of axon regeneration in C. elegans.
A severed C. elegans axon (i) senses and transduces an injury signal mediated by calcium signaling, (ii) upregulates localized translation of genes, (iii) retrogradely transports vesicles along microtubules to the nucleus, (iv) upregulates transcription, (v) anterogradely transports vesicles along microtubules to the growth cone, (vi) remodels the cytoskeleton to drive new growth, (vi) and responds to guidance cues to steer itself towards its target. The precise order of these events is not understood as they likely overlap with one another.
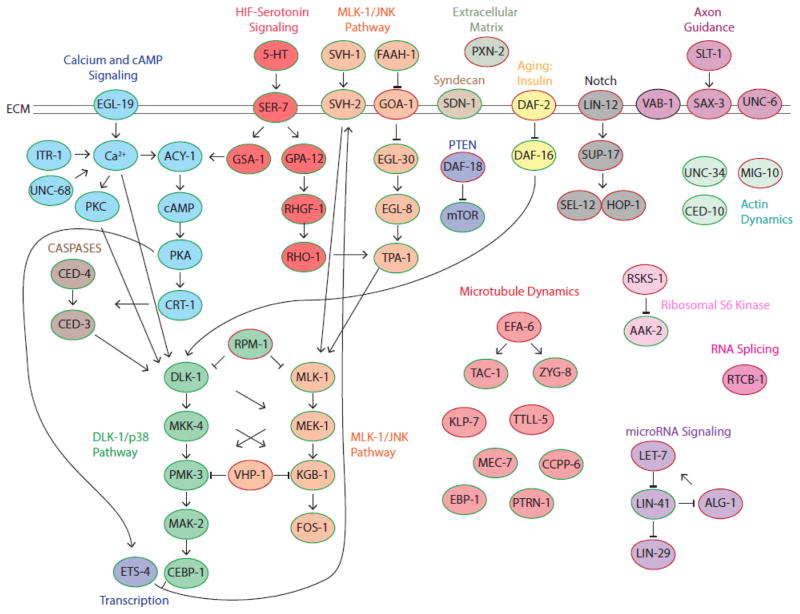
Figure 4. Regulators of Axon Regeneration in C. elegans.
Characterized components of pathways found to regulate axon regeneration in C. elegans are depicted in this model. Inhibitors of axon regeneration are outlined in red, while required components of axon regeneration are outlined in green. Note, many regulators of axon regeneration have been identified in C. elegans. Due to the resulting complexity of interactions between them, the model does not include all identified regulators.
Table 1. Regulators of Axon Regeneration in C. elegans.
Regulators of C. elegans axon regeneration are listed, along with their role in axon regeneration. For simplicity, the table does not include all identified regulators.
| Gene | Description | Neuron | Role in Regeneration | Reference | |
|---|---|---|---|---|---|
| Calcium and cAMP Signaling | egl-19 | Voltage-gated calcium channel alpha1 subunit | PLM | Required | Ghosh-Roy et al., 2010 |
| acy-1 | Adenylyl cyclase | PLM | Required | Ghosh-Roy et al., 2010 | |
| cAMP | cyclic AMP | PLM | Required | Ghosh-Roy et al., 2010 | |
| kin-2 | PKA subunit | PLM | Inhibitor | Ghosh-Roy et al., 2010; Chen et al., 2011 | |
| itr-1 | Inositol triphosphate receptor | PLM | Required | Ghosh-Roy et al., 2010 | |
| pde-4 | cAMP phosphodiesterase | PLM | Inhibitor | Ghosh-Roy et al., 2010 | |
| gsa-1 | Heterotrimeric G protein alpha subunit Gs | PLM | Required | Ghosh-Roy et al., 2010 | |
| jun-1 | bZIP transcription factor | PLM | Required | Ghosh-Roy et al., 2010 | |
| crt-1 | Calreticulin | ALM | Required | Pinan-Lucarre et al., 2012 | |
| unc-68 | Ryanodine receptor | ALM | Required | Sun et al., 2014 | |
| MLK Signaling | vhp-1 | MAPK phosphatase | GABA | Inhibitor | Nix et al., 2011 |
| svh-1 | HGF, MSP, and plasminogen family | GABA | Required | Li et al., 2012 | |
| svh-2 | Receptor tyrosine kinase | GABA | Required | Li et al., 2012 | |
| faah-1 | Fatty acid amide hydrolase | GABA | Required | Pastuhov et al., 2012 | |
| ets-4 | ETS transcription factor | GABA | Required | Li et al., 2015 | |
| mlk-1 | MAPKKK | GABA | Required | Hammarlund et al., 2009; Nix et al., 2011 | |
| mek-1 | MAPKK | GABA | Required | Hammarlund et al., 2009; Nix et al., 2011 | |
| kgb-1 | MAPK | GABA | Required | Nix et al., 2011 | |
| fos-1 | bZIP transcription factor | GABA | Required | Nix et al., 2014 | |
| goa-1 | Heterotrimeric G protein alpha subunit Go Heterotrimeric G protein alpha subunit Go Heterotrimeric G protein alpha subunit Go Heterotrimeric G protein alpha subunit Go Heterotrimeric G protein alpha subunit Go | GABA | Inhibitor | Pastuhov et al., 2012 | |
| egl-30 | Heterotrimeric G protein alpha subunit Gq Heterotrimeric G protein alpha subunit Gq Heterotrimeric G protein alpha subunit Gq Heterotrimeric G protein alpha subunit Gq Heterotrimeric G protein alpha subunit Gq | GABA | Required | Pastuhov et al., 2012 | |
| egl-8 | Phospholipase C beta | GABA | Required | Pastuhov et al., 2012 | |
| tpa-1 | Protein kinase C | GABA | Required | Pastuhov et al., 2012 | |
| DLK Signaling | rpm-1 | E3 ubiquitin ligase | GABA | Inhibitor | Hammarlund et al., 2009; Yan et al., 2009 |
| dlk-1 | MAPKKK | GABA, PLM | Required | Hammarlund et al., 2009; Yan et al., 2009 | |
| mkk-4 | MAPKK | GABA, PLM | Required | Hammarlund et al., 2009; Yan et al., 2009 | |
| pmk-3 | MAPK | GABA, PLM | Required | Hammarlund et al., 2009; Yan et al., 2009 | |
| mak-2 | MAPKAP kinase | PLM | Required | Yan et al., 2009 | |
| cebp-1 | bZIP CCAAT/enhancer-binding protein | PLM | Required | Yan et al., 2009 | |
| Cytoskeletal Dynamics | mek-7 | β-TUBULIN | ALM, PLM | Required | Kirszenblat et al., 2013 |
| efa-6 | Arf Guanine Nucleotide Exchange Factor | ALM, PLM | Chen et al., 2011; Chen et al., 2015 | ||
| klp-7 | Kinesin-13 family member | PLM | Inhibitor | Ghosh-Roy et al., 2012 | |
| ccpp-6 | Cytosolic carboxypeptidase | PLM | Required | Ghosh-Roy et al., 2012 | |
| ttll-5 | Putative tubulin polyglutamylase | PLM | Inhibitor | Ghosh-Roy et al., 2012 | |
| zyg-8 | Doublecortin-like-kinase | ALM, PLM | Required | Chen et al., 2015 | |
| tac-1 | Transforming-acidic-coiled-coil protein | ALM, PLM | Required | Chen et al., 2015 | |
| ptrn-1 | Microtubule-binding protein | PLM | Required | Chuang et al., 2014 | |
| ebp-1 | Microtubule End Binding Protein | PLM | Chen, et al., 2011 | ||
| Axon Guidance | sax-3 | Robo receptor | PLM | Inhibitor | Chen, et al., 2011 |
| slt-1 | Slit ligand | AVM, PLM | Inhibitor | Gabel, 2008, Chen, 2011 | |
| vab-1 | Eph Receptor Tyrosine Kinase | PLM | Inhibitor | Wu et al., 2007 | |
| unc-6 | Netrin | AVM | Inhibitor | Gabel et al., 2008 | |
| cwn-2 | Wnt ligand | PLM | Required | Chen, et al., 2011 | |
| Actin Dynamics | unc-34 | EVH1 domain-containing protein | AVM | Required | Gabel et al., 2008 |
| mig-10 | Lamellopodin homolog | AVM | Inhibitor | Gabel et al., 2008 | |
| ced-10 | Rac | AVM | Required | Gabel et al., 2008 | |
| Ageing | daf-2 | Insulin receptor | GABA | Inhibitor | Byrne et al., 2014 |
| daf-16 | FOXO transcription factor | GABA | Required | Byrne et al., 2014 | |
| PTEN Signaling | daf-18 | Phosphatase and tensin homolog | GABA | Inhibitor | Byrne et al., 2014 |
| Developmental Switch | let-7 | microRNA | AVM | Inhibitor | Zou et al., 2013 |
| lin-41 | Ring finger-B box-Coiled coil | AVM | Required | Zou et al., 2013 | |
| lin-29 | Zinc finger transcription factor | AVM | Inhibitor | Zou et al., 2013 | |
| alg-1 | Argonaute | AVM | Inhibitor | Zou et al., 2013 | |
| Notch Signaling | lin-12 | Notch | GABA | Inhibitor | el Bejjani, et al., 2012 |
| sup-17 | ADAM protein | GABA | Inhibitor | el Bejjani, et al., 2012 | |
| sel-12 | Presenilin | GABA | Inhibitor | el Bejjani, et al., 2012 | |
| hop-1 | Presenilin | GABA | Inhibitor | el Bejjani, et al., 2012 | |
| Extracellular Matrix | sdn-1 | Syndecan | Gaba | Required | Edwards et al., 2011 |
| pxn-2 | Peroxidasin | PLM, ALM | Inhibitor | Gotenstein et al., 2010 | |
| Caspases | ced-3 | Caspase | ALM | Required | Pinan-Lucarre, et al., 2012 |
| ced-4 | Caspase activator | ALM | Required | Pinan-Lucarre, et al., 2012 | |
| RNA ligase | rtcb-1 | RNA ligase | GABA | Inhibitor | Kosmaczewski et al., 2015 |
| S6 Kinase Signaling | rsks-1 | S6 kinase | PLM, ALM | Inhibitor | Hubert et al., 2014 |
| aak-2 | AMP kinase | PLM | Required | Hubert et al., 2014 | |
| Serotonin Signaling | tph-1 | tryptophan hydroxylase | PLM, GABA | Required | Alam et al., 2016 |
| hif-1 | hypoxia-induced factor | GABA, PLM | Required | Alam et al., 2016 | |
| ser-7 | 5-HT receptor | GABA | Required | Alam et al., 2016 | |
| gpa-12 | G protein alpha subunit | GABA | Required | Alam et al., 2016 | |
| rhgf-1 | RhoGEF | GABA | Required | Alam et al., 2016 | |
| rho-1 | RhoGTPase | GABA | Required | Alam et al., 2016 | |
| rga-5 | RhoGAP | GABA | Inhibitor | Alam et al., 2016 | |
| DLK-Independent Regeneration | egl-19 | Voltage gated calcium channel subunit | ASJ | Inhibitor | Chung et al., 2016 |
| tax-4 | Cyclic nucleotide-gated channel subunit | ASJ | Inhibitor | Chung et al., 2016 | |
| tax-2 | Cyclic nucleotide-gated channel beta subunit | ASJ | Inhibitor | Chung et al., 2016 | |
| osm-6 | Intraflagellar transport particle component | ASJ | Inhibitor | Chung et al., 2016 | |
| unc-36 | Alpha2/delta subunit of a voltage-gated calcium channel | ASJ | Inhibitor | Chung et al., 2016 | |
| daf-11 | Transmembrane guanylate cyclase | ASJ | Inhibitor | Chung et al., 2016 | |
| sax-1 | Ndr kinase | ASJ | Inhibitor | Chung et al., 2016 | |
| unc-43 | Type II calcium/calmodulin-dependent protein kinase | ASJ, ALM | Inhibitor | Chung et al., 2016 |
DUAL LEUCINE ZIPPER KINASE-1
The MAP kinase kinase kinase dlk-1 (dual leucine zipper kinase) is the best characterized intrinsic regulator of axon regeneration in C. elegans (Nakata et al., 2005; Hammarlund et al., 2009; Yan et al., 2009). In dlk-1 loss of function mutants, axon regeneration is critically compromised, and in animals that overexpress dlk-1, axon regeneration is improved compared to axon regeneration in wild type animals. dlk-1’s function in axon growth is specific to injury; dlk-1 is not required for developmental axon outgrowth. In addition to its function in the C. elegans neuronal injury response, dlk-1 also mediates regeneration in Drosophila and mice (Itoh et al., 2009; Xiong et al., 2010; Shin et al., 2012; Watkins et al., 2013) and also mediates other types of neuronal injury responses including Wallerian degeneration and cell death (Miller et al., 2009; Ghosh et al., 2011; Xiong and Collins, 2012; Xiong et al., 2012; Huntwork-Rodriguez et al., 2013; Pozniak et al., 2013).
In C. elegans, DLK-1 functions in injured axons at the time of injury to mediate regeneration (Hammarlund et al., 2009; Yan et al., 2009). DLK-1 is activated in response to an increase in localized calcium (Yan and Jin, 2012). Injury triggers an influx of calcium via a voltage-gated calcium channel (egl-19) (Ghosh-Roy et al., 2010) and triggers a release of calcium from stores in the endoplasmic reticulum in response to activated ryanodine receptor channels (Pinan-Lucarre et al., 2012; Sun et al., 2014). Calcium signaling triggers dissociation of an inhibitory DLK-1 isoform to activate DLK-1 function (Yan and Jin, 2012). In addition to calcium signaling, DLK-1 is activated by colchicine-induced microtubule disassembly via RHGF-1 (PDZ-RhoGEF) (Bounoutas et al., 2011; Chen et al., 2014), and DLK-1 function requires palmitoylation (Holland et al., 2016).
DLK-1 activation initiates a mitogen-activated protein kinase (MAPK) signaling cascade that includes the downstream map kinases mkk-4 (MAPKK) and pmk-3 (MAPK), along with mak-2, a MAPKAP kinase (Nakata et al., 2005; Hammarlund et al., 2009; Yan et al., 2009). The signaling cascade stabilizes and increases translation of the mRNA encoding the CCAAT/enhancer binding protein cebp-1 (Yan et al., 2009). cebp-1 regulates axon regeneration by regulating transcription of at least one component of the regeneration response, the receptor tyrosine kinase svh-2 (Li et al., 2012; Li et al., 2015). Other relevant targets of the DLK pathway downstream of cebp-1 have not been identified, but presumably have an essential function in axon regeneration.
Loss of DLK function not only blocks regeneration in otherwise normal animals, it also blocks regeneration in some backgrounds with improved regeneration. For example, manipulating Notch and Insulin signaling increases regeneration, but does not restore regeneration to dlk-1 mutants. By contrast, manipulations that affect axonal microtubules are able to produce regeneration even in mutants that lack DLK signaling (Ghosh-Roy et al., 2012; Chen et al., 2015). These data suggest a major output of DLK signaling is cytoskeletal remodeling (the role of the microtubule cytoskeleton in C. elegans axon regeneration is discussed below). A second form of DLK-independent regeneration is observed in axons of sensory neurons, involving reduced sensory activity (Chung et al., 2016). Thus, although DLK is a critical regeneration factor, some forms of enhanced regeneration can act without DLK. Further, at least three C. elegans neurons, ALM ASJ, and ASH, can regenerate even in the absence of DLK signals (Pinan-Lucarre et al., 2012; Chung et al., 2016).
One mechanism that could account for DLK-independent regeneration, such as observed in the ALM neuron, is activation of an alternative MAP kinase pathway. The MLK-1/JNK MAP kinase pathway has been identified as regulating axon regeneration, and can exhibit cross talk with the DLK pathway (Raivich et al., 2004; Hammarlund et al., 2009; Itoh et al., 2009; Nix et al., 2011). The parallel pathway consists of JNK (c-Jun N-terminal kinase), MLK-1 (MLK-type MAPKKK), MEK-1 (MKK7-type MAPKK), and KGB-1 (JNK-type MAPK) (Nix et al., 2011) (Figure 4).
Interestingly, in Drosophila melanogaster, the DLK and JNK pathways have converged: the fruit fly’s dlk-1 homolog, Wallenda, regulates axon regeneration via c-Jun N-terminal kinase (JNK) signaling (Xiong et al., 2010). In both C. elegans and D. melanogaster, the downstream JNK target Fos, but not Jun, regulates axon regeneration (Xiong et al., 2010; Nix et al., 2014). As in worms, mammals have parallel p38 and JNK pathways. In the murine model, DLK is required for retrograde transport of p-STAT3 and p-cJun (Shin et al., 2012). These data indicate although the DLK and MLK pathways have diverged slightly throughout evolution, they have maintained a shared function in regeneration.
The C. elegans MAP Kinase phosphatase VHP-1, identified in the large RNAi screen described above (Nix et al., 2011; Nix et al., 2014), negatively regulates both the DLK-1/p38 and the MLK-1/JNK MAP kinase pathways. Ten downstream regulators of VHP-1 were subsequently identified by their ability to suppress lethality caused by loss of vhp-1 gene function (Li et al., 2012). At least five suppressors of vhp-1 (svh) genes regulate regeneration via regulation of the MLK-1 pathway (Li et al., 2012; Pastuhov et al., 2012; Li et al., 2015). The SVH-1 growth factor is homologous to the hepatocyte growth factor (HGF), macrophage stimulating protein (MSP), and plasminogen family (Li et al., 2012). SVH-1’s receptor tyrosine kinase, SVH-2, is homologous to the hepatocyte growth factor receptor Met and the macrophage stimulating protein receptor Ron (Li et al., 2012). Both HGF and MSP have conserved roles in axon regeneration in rat optic nerves (Tonges 2011, Yin, 2003). Other characterized suppressors of VHP-1 include SVH-3 and SVH-5. SVH-3 encodes a fatty acid amide hydrolase required for regeneration (Pastuhov et al., 2012). SVH-5 encodes the ets-1 transcription factor, whose paralog ets-4, is required for axon regeneration (Pastuhov et al., 2012). These data suggest that expanding regeneration studies to additional neuron types will help identify key regeneration mechanisms that function together with or in parallel to DLK.
CYTOSKELETAL DYNAMICS
In addition to affecting calcium signaling, MAP kinase signaling, and gene transcription, injury also affects cytoskeletal dynamics. A primary component of the cytoskeleton is the microtubule, which regulates cell shape, axonal transport, and axon growth. Microtubules are relatively dynamic in the growing or regenerating axon and stable in the mature axon. Upon injury, neurons must destabilize their microtubules in order to regenerate. In injured C. elegans mechanosensory neurons, DLK-1 signaling regulates microtubule dynamics via a microtubule depolymerase KLP-7 (kinesin-13) and via a cytosolic carboxypeptidase CCPP-6 (Ghosh-Roy et al., 2012). KLP-7 inhibits growth of uninjured axons by stabilizing microtubules and CCPP-6 promotes axon growth by modifying alpha-tubulin post-translationally. Upon injury, KLP-7 is downregulated, which results in an increased number of growing microtubules. CCPP-6 then promotes microtubule growth. Thus, regulating microtubule dynamics in a timely manner upon injury allows the axon to regenerate.
The Arf6 guanine exchange factor (GEF) EFA-6 inhibits axon regeneration by binding to the doublecortin-like kinase ZYG-8 and the transforming acidic coiled-coil protein TAC-1, which are required for axon regeneration (Chen et al., 2011; Chen et al., 2015). Interestingly, EFA-6 and TAC-1 colocalize at the minus ends of microtubules with PTRN-1, which also functions in axon regeneration (Chuang et al., 2014; Chen et al., 2015). Manipulation of efa-6 and ptrn-1 can enhance regeneration, even in the absence of DLK signaling (Chen et al., 2011; Chuang et al., 2014). Therefore, as in the mammalian system, microtubule dynamics are an important component of the regeneration response and represent one way the injured axon reorganizes its subcellular resources to enable regrowth. The role of microtubules in axon regeneration has been recently reviewed (Tang and Chisholm, 2016).
AGE VS. AXON REGENERATION
As in the maturing mammalian central and peripheral nervous systems, C. elegans axons lose their ability to regenerate as they age (Pestronk et al., 1980; Tanaka et al., 1992; Verdu et al., 1995; Verdu et al., 2000; Wu et al., 2007; Gabel et al., 2008; Hammarlund et al., 2009; Nix et al., 2011; Zou et al., 2013; Byrne et al., 2014). In C. elegans, the loss of regenerative ability occurs in largely two phases: the first phase of decline occurs as the worm develops to adulthood (Wu et al., 2007; Gabel et al., 2008) and the second phase of decline occurs as adults age (Hammarlund et al., 2009; Nix et al., 2011; Byrne et al., 2014). Decline in regenerative ability is differentially regulated in the two phases.
The developmental decline in the number of severed mechanosensory neurons that reach their targets is partly attributable to errors in axon guidance (Wu et al., 2007; Gabel et al., 2008; Zou et al., 2013). However, the heterochronic genes let-7 (encodes a microRNA), lin-41 (encodes a tripartite motif protein) and alg-1 (encodes an argonaute protein) also contribute to the developmental decline in regenerative ability of mechanosensory neurons (Zou et al., 2013). The let-7 and lin-41 genes interact reciprocally: in young neurons, let-41, which promotes axon regeneration, inhibits let-7 expression via alg-1. However, let-7 microRNA expression is upregulated throughout development and eventually inhibits lin-41 via its 3′UTR. The conservation of let-7 microRNA in the mammalian genome suggests that inhibiting let-7 may be a strategy to enhance regeneration of injured mature neurons.
Axon regeneration also declines throughout adult age in worms and in mammals (Pestronk et al., 1980; Tanaka et al., 1992; Verdu et al., 1995; Verdu et al., 2000; Hammarlund et al., 2009; Nix et al., 2011; Byrne et al., 2014). The loss of intrinsic regenerative ability in aging adult neurons is not merely a secondary consequence of a decrepit animal, but is a regulated process occurring specifically in neurons (Byrne et al., 2014). Thus, adult decline of axon regeneration is an early effect of neuronal aging, occurring before other effects such as aberrant axon branching, defects in synaptic transmission, and decreased kinesin function (Pan et al., 2011; Tank et al., 2011; Toth et al., 2012; Liu et al., 2013; Li et al., 2016).
In aged GABA neurons, the loss of regenerative ability is regulated by the insulin signaling pathway (Byrne et al., 2014). This pathway is best known for its conserved role in regulating lifespan (Antebi, 2007; Kenyon, 2010). Briefly, the insulin receptor DAF-2/INSR/IGF1R promotes aging by inhibiting translocation of the transcription factor DAF-16/FOXO to the nucleus. In daf-2 loss of function mutants, DAF-16 translocates to the nucleus and regulates transcription of downstream genes, resulting in two-fold lifespan extension (Kenyon et al., 1993). In aged daf-2 loss of function mutants, axon regeneration is improved, but the regeneration effect can be uncoupled from lifespan extension and depends specifically on DAF-16 activity in neurons. Neuron-specific ChIP-seq analysis indicates that DAF-16 regulates a number of unique genes in neurons, including the critical regeneration factor DLK-1 (Byrne et al., 2014). These results were recently verified in a study using RNAseq (Kaletsky et al., 2015).
Further evidence supporting independent regulation of aging and regeneration is the finding that loss of function mutations in DAF-18/PTEN, a member of the canonical insulin pathway, increase axon regeneration despite shortening lifespan (Byrne et al., 2014). This result is surprising given DAF-18 functions antagonistically to DAF-2 in regulating lifespan. However, DAF-18/PTEN regulates axon regeneration via TOR (target of rapamycin), similar to its role in mice, and this function is independent of age and insulin signaling (Park et al., 2008; Byrne et al., 2014).
The above findings establish multiple molecular programs that regulate an aging neuron’s response to injury. In so doing, they demonstrate that C. elegans is an excellent system to identify and characterize novel regulators of adult axon regeneration. The conservation of let-7, daf-2, daf-16, dlk-1, daf-18, and TOR pathways with the mammalian genome suggest the knowledge gained from these and from future studies may inform strategies to induce regeneration of mammalian axons that do not regenerate because of their age.
AXON FUSION
An active area of C. elegans regeneration focuses on a fusion event that joins a regenerating axon to the distal segment of the severed axon. When mechanosensory axons are severed, the regenerating axon can fuse with the severed distal axon segment (Gabel et al., 2008; Neumann et al., 2011; Neumann et al., 2015). Reconnecting the severed axon segment to the cell body prevents degeneration of the severed axon (Yanik et al., 2006; Ghosh-Roy et al., 2010; Neumann et al., 2011). After injury, phosphatidylserine (PS) is externalized on the separated axon fragments, where it interacts with its receptor PSR-1 and with the transthyretin protein TTR-52, components of parallel conserved cell-corpse engulfment pathways. These signaling events are proposed to act as a ‘save me’ signal that mediates recognition of separated axonal fragments (Neumann et al., 2015). The fusogen EFF-1 then critically regulates fusion of the separated ends (Ghosh-Roy et al. 2010, Neumann et al. 2011, Neumann et al. 2015).
Mutation of ced-3 caspase, the core apoptotic executioner, or its activator, ced-4, delays fusion of severed ALM mechanosensory axons, likely as a result of inefficient axon regeneration (Pinan-Lucarre et al., 2012). CED-4 regulates regeneration independently of its canonical upstream regulators. Rather, CED-4 functions downstream of injury-induced calcium signaling and upstream of DLK-1. It is not yet known how activation of CED-3 by CED-4 results in regeneration rather than apoptosis.
Fusion may involve some form of self-recognition, as severed neurons fuse preferentially with the severed segment to which they were originally attached (Neumann et al., 2011). Fusion is an exciting method of neuronal repair as it eliminates the need for severed axons to regenerate over long-distances and form nascent connections with target cells. Although fusion of severed axons occurs in multiple organisms, currently fusion has not been identified in vertebrates. The involvement of conserved apoptotic molecules in C. elegans axonal fusion suggests that ectopic induction of these mechanisms in the mammalian system may be an approach to treat injury in the future.
ENVIRONMENT
Although most research in C. elegans to date has focused on intrinsic mechanisms that mediate regeneration, axon regeneration in the worm is also affected by extracellular cues. For example, as in the mammalian nervous system, axon guidance pathways including netrin, ephrin, WNT, and SLT/ROBO play an important role in the ability of severed axons to regenerate towards their target (Benson et al., 2005; Fabes et al., 2007; Wu et al., 2007; Gabel et al., 2008; Liu et al., 2008; Low et al., 2008; Giger et al., 2010; Zhang et al., 2010; Chen et al., 2011). C. elegans lacks some environmental features of mammalian axon regeneration, such as myelin, chondroitin sulfate proteoglycans, and macrophages. The simpler environment in C. elegans may facilitate the investigation of conserved extrinsic mechanisms that regulate axon regeneration in addition to myelin.
FUTURE AREAS OF INVESTIGATION
Research in the next decade is likely to yield great advances in understanding axon regeneration, as synergistic findings are emerging from multiple diverse approaches. With its strong knowledge base and robust research community, as well as its particular experimental advantages discussed above, C. elegans is positioned to make important contributions. Conservation of function between molecules that regulate axon regeneration in C. elegans and those that regulate axon regeneration in mammals, including DLK and PTEN, along with the conserved roles and regulators of calcium signaling, microtubule dynamics, axon guidance, and neuronal aging in axon regeneration, suggest findings in C. elegans will inform our understanding of mammalian axon regeneration (Neumann et al., 2002; Filbin, 2003; Raivich et al., 2004; Spencer and Filbin, 2004; Benson et al., 2005; Erturk et al., 2007; Fabes et al., 2007; Wu et al., 2007; Gabel et al., 2008; Liu et al., 2008; Low et al., 2008; Hammarlund et al., 2009; Itoh et al., 2009; Montolio et al., 2009; Yan et al., 2009; Ghosh-Roy et al., 2010; Giger et al., 2010; Zhang et al., 2010; Chen et al., 2011; Nix et al., 2011; Ghosh-Roy et al., 2012; Shin et al., 2012; Watkins et al., 2013; Byrne et al., 2014; Tang and Chisholm, 2016).
A particular strength of C. elegans is the ability to study cell biology in vivo. Currently, we are far from having a complete cellular understanding of axon regeneration. To date, only a few key C. elegans regeneration mechanisms, such as the DLK pathway and regulation of microtubules, have been subjected to detailed cell-biological analysis. By contrast, genetic approaches have resulted in identification of hundreds of genes that affect regeneration. A major challenge and opportunity for the next decade is to connect this rich genetic information to enable a detailed understanding of the cell biology of axon regeneration.
As a clearer understanding of the cell biology of regeneration emerges, it will answer the general puzzle of just what axon regeneration really is. Is regeneration an independent and highly specialized neuronal function? The DLK pathway, which in C. elegans seems to only function in regeneration, suggests that this is the case. Or rather, is regeneration a reconfiguration of more general neuronal properties? For example, the role of the cell death caspase CED-3 and the engulfment genes PSR-1 and TTR-52 in axon regeneration and axon fusion supports this second model. These findings also raise the fascinating cell biological question of how such multipurpose pathways are canalized during regeneration.
In addition, certain specific questions emerge as particularly urgent. What are the downstream effectors of DLK signaling? What factors control successful circuit reconnection, including the building of new synapses? How does fusion after axotomy work, and how is specificity achieved in this process? How do local calcium and microtubule dynamics shape the response to injury? Can C. elegans be used as a drug discovery platform for compounds that improve regeneration in the mammalian CNS? As new researchers enter the field of axon regeneration, C. elegans will contribute to answering these and other questions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Romero MI, Lush ME, Lu QR, Henkemeyer M, Parada LF. Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc Natl Acad Sci U S A. 2005;102:10694–10699. doi: 10.1073/pnas.0504021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, Kratz J, Emtage L, Ma C, Nguyen KC, Chalfie M. Microtubule depolymerization in Caenorhabditis elegans touch receptor neurons reduces gene expression through a p38 MAPK pathway. Proc Natl Acad Sci U S A. 2011;108:3982–3987. doi: 10.1073/pnas.1101360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois F, Ben-Yakar A. Femtosecond laser nanoaxotomy properties and their effect on axonal recovery in C. elegans. Opt Express. 2008;16:5963. doi: 10.1364/oe.16.005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Edwards TJ, Hammarlund M. In vivo laser axotomy in C. elegans. J Vis Exp. 2011;51:e2707. doi: 10.3791/2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Walradt T, Gardner KE, Hubbert A, Reinke V, Hammarlund M. Insulin/IGF1 signaling inhibits age-dependent axon regeneration. Neuron. 2014;81:561–573. doi: 10.1016/j.neuron.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chen CH, Lee A, Liao CP, Liu YW, Pan CL. RHGF-1/PDZ-RhoGEF and retrograde DLK-1 signaling drive neuronal remodeling on microtubule disassembly. Proc Natl Acad Sci U S A. 2014;111:16568–16573. doi: 10.1073/pnas.1410263111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chuang M, Koorman T, Boxem M, Jin Y, Chisholm AD. Axon injury triggers EFA-6 mediated destabilization of axonal microtubules via TACC and doublecortin like kinase. Elife. 2015:4. doi: 10.7554/eLife.08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O’Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang M, Goncharov A, Wang S, Oegema K, Jin Y, Chisholm AD. The microtubule minus-end-binding protein patronin/PTRN-1 is required for axon regeneration in C. elegans. Cell Rep. 2014;9:874–883. doi: 10.1016/j.celrep.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Awal MR, Shay J, McLoed MM, Mazur E, Gabel CV. Novel DLK-independent neuronal regeneration in Caenorhabditis elegans shares links with activity-dependent ectopic outgrowth. Proc Natl Acad Sci U S A. 2016;113:E2852–2860. doi: 10.1073/pnas.1600564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium C.e.S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Edwards TJ, Hammarlund M. Syndecan promotes axon regeneration by stabilizing growth cone migration. Cell Rep. 2014;8:272–283. doi: 10.1016/j.celrep.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bejjani R, Hammarlund M. Notch signaling inhibits axon regeneration. Neuron. 2012;73:268–278. doi: 10.1016/j.neuron.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabes J, Anderson P, Brennan C, Bolsover S. Regeneration-enhancing effects of EphA4 blocking peptide following corticospinal tract injury in adult rat spinal cord. Eur J Neurosci. 2007;26:2496–2505. doi: 10.1111/j.1460-9568.2007.05859.x. [DOI] [PubMed] [Google Scholar]
- Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat Rev Neurosci. 2003;4:703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel CV, Antoine F, Chuang CF, Samuel AD, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- Ghosh AS, Wang B, Pozniak CD, Chen M, Watts RJ, Lewcock JW. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J Cell Biol. 2011;194:751–764. doi: 10.1083/jcb.201103153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Dev Cell. 2012;23:716–728. doi: 10.1016/j.devcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Wu Z, Goncharov A, Jin Y, Chisholm AD. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J Neurosci. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger RJ, Hollis ER, 2nd, Tuszynski MH. Guidance molecules in axon regeneration. Cold Spring Harb Perspect Biol. 2010;2:a001867. doi: 10.1101/cshperspect.a001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce SK, Guo SX, Ghorashian N, Everett WN, Jarrell T, Kottek A, Bovik AC, Ben-Yakar A. A fully automated microfluidic femtosecond laser axotomy platform for nerve regeneration studies in C. elegans. PLoS One. 2014;9:e113917. doi: 10.1371/journal.pone.0113917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SX, Bourgeois F, Chokshi T, Durr NJ, Hilliard MA, Chronis N, Ben-Yakar A. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat Methods. 2008;5:531–533. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Jorgensen EM, Bastiani MJ. Axons break in animals lacking beta-spectrin. J Cell Biol. 2007;176:269–275. doi: 10.1083/jcb.200611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhou R, Wu Z, Carrasco MA, Kurshan PT, Farley JE, Simon DJ, Wang G, Han B, Hao J, et al. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc Natl Acad Sci U S A. 2016;113:6029–6034. doi: 10.1073/pnas.1605707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM, Collura KM, Ketschek A, Noma K, Ferguson TA, Jin Y, Gallo G, Thomas GM. Palmitoylation controls DLK localization, interactions and activity to ensure effective axonal injury signaling. Proc Natl Acad Sci U S A. 2016;113:763–768. doi: 10.1073/pnas.1514123113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntwork-Rodriguez S, Wang B, Watkins T, Ghosh AS, Pozniak CD, Bustos D, Newton K, Kirkpatrick DS, Lewcock JW. JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J Cell Biol. 2013;202:747–763. doi: 10.1083/jcb.201303066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh A, Horiuchi M, Bannerman P, Pleasure D, Itoh T. Impaired regenerative response of primary sensory neurons in ZPK/DLK gene-trap mice. Biochem Biophys Res Commun. 2009;383:258–262. doi: 10.1016/j.bbrc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2015 doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kosmaczewski SG, Han SM, Han B, Irving Meyer B, Baig HS, Athar W, Lin-Moore AT, Koelle MR, Hammarlund M. RNA ligation in neurons by RtcB inhibits axon regeneration. Proc Natl Acad Sci U S A. 2015;112:8451–8456. doi: 10.1073/pnas.1502948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hisamoto N, Matsumoto K. Axon Regeneration Is Regulated by Ets-C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca2+ Signaling Pathways. PLoS Genet. 2015;11:e1005603. doi: 10.1371/journal.pgen.1005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hisamoto N, Nix P, Kanao S, Mizuno T, Bastiani M, Matsumoto K. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat Neurosci. 2012;15:551–557. doi: 10.1038/nn.3052. [DOI] [PubMed] [Google Scholar]
- Li LB, Lei H, Arey RN, Li P, Liu J, Murphy CT, Xu XZ, Shen K. The Neuronal Kinesin UNC-104/KIF1A Is a Key Regulator of Synaptic Aging and Insulin Signaling-Regulated Memory. Curr Biol. 2016;26:605–615. doi: 10.1016/j.cub.2015.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang B, Lei H, Feng Z, Liu J, Hsu AL, Xu XZ. Functional aging in the nervous system contributes to age-dependent motor activity decline in C. elegans. Cell Metab. 2013;18:392–402. doi: 10.1016/j.cmet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang X, Lu CC, Kerman R, Steward O, Xu XM, Zou Y. Repulsive Wnt signaling inhibits axon regeneration after CNS injury. J Neurosci. 2008;28:8376–8382. doi: 10.1523/JNEUROSCI.1939-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K, Culbertson M, Bradke F, Tessier-Lavigne M, Tuszynski MH. Netrin-1 is a novel myelin-associated inhibitor to axon growth. J Neurosci. 2008;28:1099–1108. doi: 10.1523/JNEUROSCI.4906-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montolio M, Messeguer J, Masip I, Guijarro P, Gavin R, Antonio Del Rio J, Messeguer A, Soriano E. A semaphorin 3A inhibitor blocks axonal chemorepulsion and enhances axon regeneration. Chem Biol. 2009;16:691–701. doi: 10.1016/j.chembiol.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Neumann B, Coakley S, Giordano-Santini R, Linton C, Lee ES, Nakagawa A, Xue D, Hilliard MA. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–222. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- Neumann B, Nguyen KC, Hall DH, Ben-Yakar A, Hilliard MA. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev Dyn. 2011;240:1365–1372. doi: 10.1002/dvdy.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Bradke F, Tessier-Lavigne M, Basbaum AI. Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron. 2002;34:885–893. doi: 10.1016/s0896-6273(02)00702-x. [DOI] [PubMed] [Google Scholar]
- Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, Bastiani M. Axon regeneration genes identified by RNAi screening in C. elegans. J Neurosci. 2014;34:629–645. doi: 10.1523/JNEUROSCI.3859-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix P, Hisamoto N, Matsumoto K, Bastiani M. Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc Natl Acad Sci U S A. 2011;108:10738–10743. doi: 10.1073/pnas.1104830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CL, Peng CY, Chen CH, McIntire S. Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc Natl Acad Sci U S A. 2011;108:9274–9279. doi: 10.1073/pnas.1011711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuhov SI, Fujiki K, Nix P, Kanao S, Bastiani M, Matsumoto K, Hisamoto N. Endocannabinoid-Goalpha signalling inhibits axon regeneration in Caenorhabditis elegans by antagonizing Gqalpha-PKC-JNK signalling. Nat Commun. 2012;3:1136. doi: 10.1038/ncomms2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestronk A, Drachman DB, Griffin JW. Effects of aging on nerve sprouting and regeneration. Exp Neurol. 1980;70:65–82. doi: 10.1016/0014-4886(80)90006-0. [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, Slone RD, Xue J, Qiao Y, Weisberg S, Roodhouse K, et al. The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001331. doi: 10.1371/journal.pbio.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak CD, Sengupta Ghosh A, Gogineni A, Hanson JE, Lee SH, Larson JL, Solanoy H, Bustos D, Li H, Ngu H, et al. Dual leucine zipper kinase is required for excitotoxicity-induced neuronal degeneration. J Exp Med. 2013;210:2553–2567. doi: 10.1084/jem.20122832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Rao GN, Kulkarni SS, Koushika SP, Rau KR. In vivo nanosecond laser axotomy: cavitation dynamics and vesicle transport. Opt Express. 2008;16:9884–9894. doi: 10.1364/oe.16.009884. [DOI] [PubMed] [Google Scholar]
- Rohde CB, Gilleland C, Samara C, Norton S, Haggarty S, Yanik MF. Microfluidic in vivo screen identifies compounds enhancing neuronal regeneration. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:5950–5952. doi: 10.1109/IEMBS.2009.5334771. [DOI] [PubMed] [Google Scholar]
- Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, Yanik MF. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci U S A. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Spencer T, Filbin MT. A role for cAMP in regeneration of the adult mammalian CNS. J Anat. 2004;204:49–55. doi: 10.1111/j.1469-7580.2004.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Shay J, McLoed M, Roodhouse K, Chung SH, Clark CM, Pirri JK, Alkema MJ, Gabel CV. Neuronal regeneration in C. elegans requires subcellular calcium release by ryanodine receptor channels and can be enhanced by optogenetic stimulation. J Neurosci. 2014;34:15947–15956. doi: 10.1523/JNEUROSCI.4238-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Zhang QL, Webster HD. Myelinated fiber regeneration after sciatic nerve crush: morphometric observations in young adult and aging mice and the effects of macrophage suppression and conditioning lesions. Exp Neurol. 1992;118:53–61. doi: 10.1016/0014-4886(92)90022-i. [DOI] [PubMed] [Google Scholar]
- Tang NH, Chisholm AD. Regulation of Microtubule Dynamics in Axon Regeneration: Insights from C. elegans. F1000Res. 2016:5. doi: 10.12688/f1000research.8197.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank EM, Rodgers KE, Kenyon C. Spontaneous age-related neurite branching in Caenorhabditis elegans. J Neurosci. 2011;31:9279–9288. doi: 10.1523/JNEUROSCI.6606-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Toth ML, Melentijevic I, Shah L, Bhatia A, Lu K, Talwar A, Naji H, Ibanez-Ventoso C, Ghose P, Jevince A, et al. Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J Neurosci. 2012;32:8778–8790. doi: 10.1523/JNEUROSCI.1494-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu E, Buti M, Navarro X. The effect of aging on efferent nerve fibers regeneration in mice. Brain Res. 1995;696:76–82. doi: 10.1016/0006-8993(95)00762-f. [DOI] [PubMed] [Google Scholar]
- Verdu E, Ceballos D, Vilches JJ, Navarro X. Influence of aging on peripheral nerve function and regeneration. J Peripher Nerv Syst. 2000;5:191–208. doi: 10.1046/j.1529-8027.2000.00026.x. [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Watkins TA, Wang B, Huntwork-Rodriguez S, Yang J, Jiang Z, Eastham-Anderson J, Modrusan Z, Kaminker JS, Tessier-Lavigne M, Lewcock JW. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc Natl Acad Sci U S A. 2013;110:4039–4044. doi: 10.1073/pnas.1211074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Williams W, Nix P, Bastiani M. Constructing a low-budget laser axotomy system to study axon regeneration in C. elegans. J Vis Exp. 2011 doi: 10.3791/3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y, Chisholm AD. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A. 2007;104:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Collins CA. A conditioning lesion protects axons from degeneration via the Wallenda/DLK MAP kinase signaling cascade. J Neurosci. 2012;32:610–615. doi: 10.1523/JNEUROSCI.3586-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Hao Y, Sun K, Li J, Li X, Mishra B, Soppina P, Wu C, Hume RI, Collins CA. The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 2012;10:e1001440. doi: 10.1371/journal.pbio.1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, Collins CA. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Jin Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron. 2012;76:534–548. doi: 10.1016/j.neuron.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Neurosurgery: functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- Yanik MF, Cinar H, Cinar HN, Gibby A, Chisholm AD, Jin Y, Ben-Yakar A. Nerve regeneration in Caenorhabditis elegans after femtosecond laser axotomy. IEEE Journal of selected topics in quantum electronics. 2006;12:1283. [Google Scholar]
- Zhang HY, Zheng LF, Yi XN, Chen ZB, He ZP, Zhao D, Zhang XF, Ma ZJ. Slit1 promotes regenerative neurite outgrowth of adult dorsal root ganglion neurons in vitro via binding to the Robo receptor. J Chem Neuroanat. 2010;39:256–261. doi: 10.1016/j.jchemneu.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang CF, Chang C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340:372–376. doi: 10.1126/science.1231321. [DOI] [PMC free article] [PubMed] [Google Scholar]