Abstract
Glioblastoma (GBM) is a primary brain cancer that is resistant to all treatment modalities. This resistance is due, in large part, to invasive cancer cells that disperse from the main tumor site, escape surgical resection, and contribute to recurrent secondary lesions. The adhesion and signaling mechanisms that drive GBM cell invasion remain enigmatic, and as a result there are no effective anti-invasive clinical therapies. Here we have characterized a novel adhesion and signaling pathway comprised of the integrin αvβ8 and its intracellular binding partner, Spinophilin (Spn), which regulates GBM cell invasion in the brain microenvironment. We show for the first time that Spn binds directly to the cytoplasmic domain of β8 integrin in GBM cells. Genetically targeting Spn leads to enhanced invasive cell growth in pre-clinical models of GBM. Spn regulates GBM cell invasion by modulating the formation and dissolution of invadopodia. Spn-regulated invadopodia dynamics are dependent, in part, on proper spatiotemporal activation of the Rac1 GTPase. GBM cells that lack Spn showed diminished Rac1 activities, increased numbers of invadopodia and enhanced extracellular matrix (ECM) degradation. Collectively, these data identify Spn as a critical adhesion and signaling protein that is essential for modulating GBM cell invasion in the brain microenvironment.
Keywords: extracellular matrix, vascular basement membrane, itgb8, ppp1r9b, pp1
Introduction
GBM is a rapidly progressive and highly invasive primary brain cancer that has a median survival time of less than two years after diagnosis (1). While a great deal is known about genes and pathways that promote GBM initiation and growth, relatively little is understood about mechanisms that promote tumor cell invasion. (2) GBM cells often exploit blood vessels and their ECM-rich basement membranes for selective growth and invasion in the brain microenvironment (3). Most cells interact with the ECM via integrins, a family of cell surface receptors consisting of α and β subunits (4). Integrin-ECM affinity is modulated via “inside-out” signaling mechanisms involving proteins such as talins and kindlins that bind to β integrin cytoplasmic domains and induce conformational changes in extracellular regions (5). ECM engagement subsequently triggers “outside-in” signaling pathways via adhesion protein complexes and other effectors, leading to changes in the cytoskeletal network and alterations in gene expression (6).
The five members of the αv integrin subfamily recognize arginine-glycine-aspartic acid (RGD) peptide motifs in many ECM ligands, most of which are abundantly expressed in the brain (7). β8 integrin is a 100 kDa protein that heterodimerizes exclusively with the 130 kDa αv subunit (8). αvβ8 integrin binds mainly to latent TGFβs, which are produced by cells as inactive ECM-bound complexes (9). Cell type-specific gene knockouts in mice have revealed that glial-expressed αvβ8 integrin promotes latent TGFβ activation and signaling in vascular endothelial cells to control CNS angiogenesis and development of the blood-brain barrier (10–15). αvβ8 integrin protein expression levels are also altered in human nervous system malignancies, including GBM (16,17) and brain metastases (18).
While αvβ8 integrin adhesion to latent TGFβs plays important roles in development and disease, we understand surprisingly little about its control of intracellular signal transduction. Alignment of the five β integrin subunits that pair with αv reveals that the β8 cytoplasmic domain (β8cyto) shares no sequence homology with β1A, β3A, β5, and β6. Using genetic screens we identified Band 4.1B and the Rho GDP Dissociation Inhibitor 1 (RhoGDI1) as proteins that binds to β8 integrin (19). There are at least three members of the RhoGDI family, and these proteins function to extract GDP-bound Rhos from membranes and sequester them in the cytoplasm (20). Thus, RhoGDIs act in concert with guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) to balance Rho protein activation. While other integrins stimulate Rho GTPases via interactions with GEFs, αvβ8 integrin forms complexes with RhoGDI1 to suppress activation of Rho GTPases (21). More recently, we have discovered that αvβ8 integrin also forms complexes with the cytoplasmic tyrosine phosphatase PTP-PEST, and this complex dynamically regulates the phosphorylation status of RhoGDI1 in migrating cells (22).
Spinophilin (Spn) is a 130 kDa scaffolding protein that contains protein phosphatase 1 (PP1), PDZ, and actin binding domains (23) that is highly expressed in the brain where it regulates various signaling events to promote dendritic spine formation (24). Spn also modulates cytoskeletal dynamics, in part, via direct interactions with F-actin as well as α-actinin, a focal adhesion protein and actin regulatory factor (25). Spn is a regulatory subunit for PP1 and controls PP! substrate specificity by sub-cellular localization (26). For example, in developing axons Spn is reported to interact with doublecortin, leading to its dephosphorylation by PP1 and alterations in cytoskeletal dynamics (27). Recent reports have shown that down-regulation of Spn gene expression promotes tumor cell growth and enhances metastasis in cancers of the colon, lung and breast (28), although the exact signaling mechanisms that control these processes remain unclear.
Here, we have used biochemical and genetic strategies to identify Spn in GBM cells as a binding partner for the β8 integrin cytoplasmic tail. We show that Spn is essential for the negative regulation of perivascular GBM cell migration and invasion in the brain microenvironment. Genetic inhibition of Spn expression in mouse astrocytoma cells or human GBM cells leads to significantly larger brain tumors due to enhanced invasive growth patterns. Lastly, we show that Spn suppresses brain tumor cell invasion, in part, via control of Rac1 GTPase activities and invadopodia disassembly. Collectively, these results reveal new and important functions for Spn in regulating tumor cell invasive growth in the brain via modulation of invadopodia dissolution.
Materials and Methods
Proteomics experiments
Detergent-soluble lysates (1 mg/ml) from primary human GBM cells were used for immunoprecipitation experiments with control rabbit IgG or a rabbit polyclonal antibody directed against the β8 integrin cytoplasmic domain that has been described elsewhere (19). Antibody complexes were immobilized using Protein G-Sepharose. Immunoprecipitated proteins were resolved on an SDS polyacrylamide gel under reducing conditions. Control and experimental bands were cut from the gel, and subjected to in-gel digestion with Trypsin. Peptides were extracted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as follows. Peptides were resuspended in 2.5% formic acid (FA), 2.5% acetonitrile (MeCN) and were loaded using a Micro AS autosampler (Thermo Electron) onto a microcapillary column of 100 μm inner diameter packed with 12 cm of reversed-phase Magic C18 packing material (5 μm, 200 Å; Michrom Bioresources, Inc., Auburn, CA, USA). SEQUEST matches were filtered by XCorr scores to a less than 1% false discovery rate when the control proteins were eliminated and protein matches were required to have three spectral matches, no identifiable false positive peptides remained.
Experimental mice
Generation of Spn+/− mice used in this study has been detailed elsewhere (24). Spn+/− mice, maintained on a mixed C57Bl6/129S4/FVB mixed genetic background, were interbred to generate Spn+/+, Spn+/− and Spn−/− littermates for experimental analyses. Genotypes were confirmed using genomic DNA isolated from ear tissue and PCR-based methodologies. Primer sequences used for genomic PCR reactions are as follows: 5′-GGAGACCACTCCCATTCCC -3′, 5′-GGCGTCAGATATAGACAGAGG -3′and 5′-AGGTGAGATGACAGGAGATC-3′. PCR amplifies a wild-type product of 500 base pairs and mutant (Spn−/−) product of 750 base pairs, with both products being amplified in heterozygous (Spn+/−) animals. NCR-nu/nu mice were purchased from Jackson Laboratories and used for all experiments involving intracranial injections of mouse astrocytoma cells and human GBM cells. For brain isolation, adult mice were anesthetized and brains were fixed by cardiac perfusion with ice-cold 4% PFA/PBS.
Isolation and transformation of mouse astroglial cells
Astroglial progenitors were cultured from cerebral cortices of wild type or Spn−/− littermates (P0) and propagated on laminin-coated dishes, as described previously (McCarty et al., 2005). To generate retroviral stocks, we transfected 293T Phoenix cells with pLXSP-puro-E6/E7 and pLXSNneo-G12VH-Ras (29). Primary astroglial cells were transduced with the E6/E7 retroviral supernatant and cells were selected in growth media containing 1 mg/ml puromycin for 5 days. Puromycin-resistant clones were pooled and transduced with G12VH-Ras retrovirus supernatant, and selected for 10 days in G418 (900 μg/ml).
Human GBM tissue samples and cultured cells
Approval for the use of human specimens was obtained from the Institutional Review Board (IRB) at the University of Texas MD Anderson Cancer Center. LN229 GBM cells were purchased from American Tissue Culture Collection (ATCC). Cells were cultured in DMEM high glucose sodium pyruvate supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum, at 37 °C in an atmosphere with 5% CO2. Primary human GBM cells used for original identification of β8 integrin-Spn interactions were cultured from patient samples. Briefly, tumor tissue was dissociated by trituration with a polished glass pipette and cells were cultured in the following growth media: DMEM-F12 (Mediatech), 20 ng/ml EGF and bFGF (Biosource), B27 supplement (Gibco) and one unit per ml penicillin-streptomycin (Sigma-Aldrich).
Antibodies and plasmids
The anti-β8 integrin antibody used for immunoblotting has been described previously (McCarty et al., 2004, 2005). The following antibodies were purchased: rabbit anti-Spn and anti-PP1 antibodies (Santa Cruz Biotech, Santa Cruz, CA, USA), chicken anti-Nestin IgY (Neuromics, Edina, MN, USA), rat anti-mouse CD31 (BD Biosciences, San Jose, CA, USA), rabbit anti-β-actin pAb (Sigma, St Louis, MO, USA), rabbit anti-GFAP pAb (Dako, Carpinteria, CA, USA), and anti-Ras and anti-E7 antibodies (Santa Cruz Biotech, Santa Cruz, CA, USA). The anti-myc mAb was purchased form Clontech Laboratories (Mountanview, CA, USA). Secondary antibodies used for imaging were goat anti-rabbit, goat anti-chicken, goat anti-rat and goat anti-mouse, all conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Carlsbad, CA, USA). Most sections were analyzed using a Zeiss Axio Imager.Z1 microscope. For Far western blotting experiments detergent-soluble lysates from LN229 cells expressing non-targeting control shRNAs or shRNAs targeting Spn were resolved pn a 7.5% SDS gel and transferred to nitrocellulose membranes. Purified GST or GST-β8 integrin probes (5 μg/ml) were added to the membranes for 12 hours. Membranes were probed with an anti-GST antibody conjugated with HRP (Amersham) and membranes were analyzed by chemiluminescence.
pGIPZ lentiviral plasmids expressing various shRNAs (Dharmacon) were obtained from an institutional core facility via Dharmacon. The full-length human Spn cDNA was provided by Dr. Amacio Carnero. Myc-tagged human Spn cDNAs were generated by standard PCR-based methods. Constructs were cloned into the pcDNA3.1A plasmid for mammalian expression. For the Crispr/Cas9 experiments we cloned gDNAs into the LentiCRISPR (pXPR-001) expression vector purchased from Addgene. Infected cells were selected in puromycin and Spn targeting was confirmed by immunoblotting and genomic analyses. gDNA sequences were selected using an open source design tool (http://crispr.mit.edu). The Spn gDNA sequences are as follows: 5′-ACTTTTGATGCGGTGGACGT-3′, 5′-GTTCGAACGGAGCGCCCCAG-3′, 5′-GGATTACGATCGTCGCAACG-3′, and 5′-GCGACGATCGTAATCCTCGT-3′.
Intracranial tumor cell implantation
All animal procedures were conducted under Institutional Animal Care and Use Committee–approved protocols. Nude mice were anesthetized and an automated micropump (Stoelting Instruments, Wood Dale, IL, USA) was used to dispense cells in phosphate-buffered saline. Astroglial progenitors cultured for less than four passages after transformation were used for the experiments. Nude mice were stereotactically injected with 1 × 105 Spn+/+ or Spn−/− transformed astroglial cells. Xenograft tumors were generated by stereotactically injecting 2×105 LN229 GBM cells into immunocompromised mice.
Invadopodia assays and Rac1 activity FRET
LN229 control cells or cells expressing Spn gDNAs were plated on acid-washed coverslips coated with fluorescent gelatin and incubated for 6 hours. Subsequently, cells were fixed in 4% paraformaldehyde and stained for F-actin and cortactin to visualize invadopodia. LN229 cells expressing Cas9 or Spn gDNA/Cas9 and the Raichu-Rac1 single chain FRET probe (4×104 cells) were plated on the fibronectin-coated (10 μg/mL) MatTek dishes with phenol red-free complete growth media. Cells were incubated at 37 °C in an atmosphere with 5% CO2 for 20 hours prior to imaging. Time-lapse series images were acquired every minute for 30 minutes using a Zeiss LSM780 confocal microscope equipped with a Plan-Apo 40X/1.40NA oil objective. Ratio maps for visualization of Rac1 activity were generated using ImageJ. Briefly, stack of CFP and FRET images were first processed using a 3 × 3 median filter to remove background. Then, the median filtered stacks were used to create a ratio stack the FRET channel divided by the CFP channel.
Statistical analysis
Student’s t test was performed to determine statistically significant differences between groups. The Wilcoxon rank sum test was used for analysis of Kaplan-Meier survival results. Excel (Microsoft, Redmond, WA) was used to calculate statistics.
Results
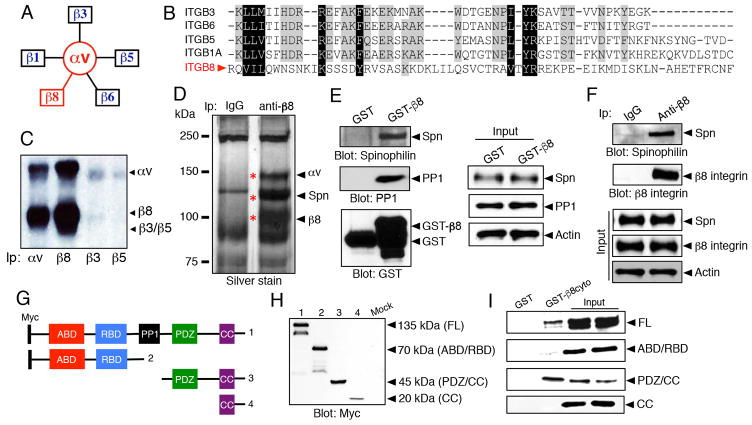
β8 integrin, which heterodimerizes exclusively with the αv integrin subunit (Figure 1A), contains a cytoplasmic tail that is distinct from other integrin subunits. Alignment of the primary amino acid sequence reveals lack of conserved motifs, e.g., NPXY motifs and juxtamembrane sequences, which are present in the other β subunits that heterodimerize with αv integrin. (Figure 1B). We analyzed levels of cell surface αv-containing integrins using primary human GBM cells and biotinylation/immunoprecipitation strategies. As shown in Figure 1C, αvβ8 integrin is the major αv-containing integrin, with significantly lower levels of αvβ3 and αvβ5 integrin detected on the tumor cell surface. To identify intracellular signaling proteins that interact with the β8 integrin cytoplasmic tail we performed co-immunoprecipitation experiments with anti-β8 integrin antibodies and primary human GBM cells followed by mass spectrometry-based analyses. As shown in Figure 1D and Supplemental Table 1, the major proteins that co-immunoprecipitate with β8 integrin include αv integrin and the scaffolding protein, Spinophilin/PPP1R9B. Interactions between β8 integrin and Spn were confirmed in pull-down assays using a GST-tagged fusion protein containing the entire cytoplasmic tail of β8 integrin (Figure 1E). In addition, Spn-β8 integrin protein interactions were verified by co-immunoprecipitation in LN229 GBM cells (Figure 1F). In addition to the biochemical interactions in cell lysates, we used Far western blotting approaches to show direct protein-protein interactions between Spn and the β8 integrin cytoplasmic domain in vitro (Supplemental Figure 1A). Spn and β8 integrin also co-localized in cultured LN229 GBM cells, as revealed by immunofluorescence (Supplemental Figure 1B).
Figure 1. Spinophilin binds to the cytoplasmic tail of β8 integrin.
(A); The αv sub-family of integrins consists of five members. Note that β8 integrin dimerizes exclusively with the αv subunit. (B); Alignment of primary amino acid sequences from the five β integrin subunits that pair with αv integrin. Note that the β8 integrin cytoplasmic tail does not share common motifs with other integrin subunits. (C); αvβ8 integrin is the major αv-containing integrin on the surface of primary GBM cells, as revealed by cell surface biotinylation and immunoprecipitation. Antibodies used for immunoprecipitation are indicated at the bottom of the image. (D); Primary human GBM cells were immunoprecipitated with control IgG or an anti-β8 integrin antibody and gels were silver stained. Trypsin-digested bands were identified by mass spectrometry. Note that αv integrin and Spn are the major proteins co-immunoprecipitate with β8 integrin. (E); GST or GST fused to the cytoplasmic domain of β8 integrin (GST-β8cyto) were purified from bacteria and used as probes in GSC lysates to confirm interactions between GST-β8cyto and Spn. (F); Spn and β8 integrin interact in LN229 GBM cell lysates as determined by co-immunoprecipitation. (G, H); Schematic diagram (G) and immunoblot (H) showing domain structure and full-length myc-tagged Spn protein and various deletion constructs expressed in HEK-293T cells. (I); GST or GST-β8cyto proteins were purified from bacteria and used as probes in lysates from Spn−/− mouse astrocytes forcibly expressing full-length Spn or various deletion constructs. Note that full-length Spn and the deletion construct containing the PDZ/CC region interacts with β8 integrin. Abbreviations: ABD; actin binding domain, RBD; receptor binding domain, PP1; protein phosphatase 1 binding domain, CC; coiled coil domain.
To identify the region of the Spn protein that interacts with β8 integrin we generated and expressed various recombinant myc-tagged protein constructs lacking select N-terminal and C-terminal regions (Figure 1G–H and Supplemental Figure 1C–F). The GST-β8 integrin fusion protein was used to test for interactions with myc-tagged Spn constructs expressed in Spn−/− astrocytes. As shown in Figure 1I, The N-terminal region of Spn containing the receptor binding domain (RBD) and the actin binding domain (ABD) did not interact with β8 integrin. In contrast, the C-terminus of Spn protein, consisting of the PDZ domain and the coiled coil (CC) domain, is required for interactions with β8 integrin. The isolated CC domain did not show an interaction, identifying a requirement for the Spn PDZ domain, possibly in conjunction with the CC region, in mediating interactions with the β8 integrin cytoplasmic tail.
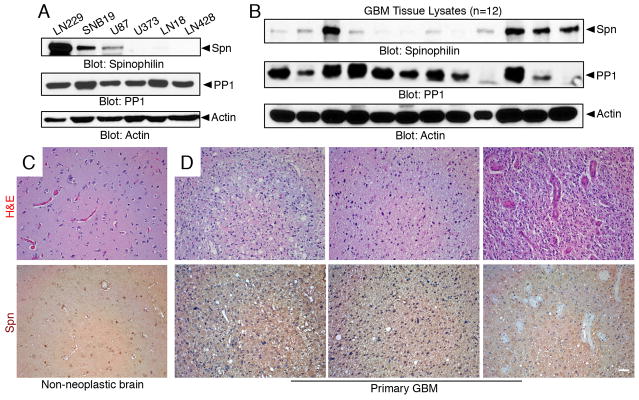
We next analyzed Spn protein expression levels in human GBM cells and tissue samples. Anti-Spn immunoblots were performed with detergent-soluble lysates from six different human GBM cell lines, revealing robust Spn expression in LN229 cells (Figure 2A). Lower levels of Spn protein were detected in SNB19 and U87 GBM cells, and undetectable Spn levels were found in LN18, U373 and LN428 GBM cells. In addition, anti-Spn antibodies were used to immunoblot lysates from 12 different freshly resected human GBM samples. Robust levels of Spn were detected in some tumor samples and low or undetectable levels were present in other tumors (Figure 2B). Given that Spn is a regulatory subunit for PP1, we also monitored PP1 protein levels in cell lines and tumor lysates. As shown in Figure 2A–B, PP1 was detected in all GBM cells and most tumor lysates analyzed by anti-PP1 immunoblotting. PP1 can function independently of Spn; hence, this likely explains why PP1 and Spn expression does not entirely overlap in the various GBM samples. To analyze spatial patterns of Spn protein in tissue samples we next used immunohistochemistry with anti-Spn antibodies. As shown in Figure 2C, Spn protein was diffusely expressed in neural cells and blood vessels in the normal brain. In human GBM sections, Spn was expressed mostly in tumor cells as well as in some intratumoral capillaries, although glomeruloid-like blood vessels, which develop via microvascular cell hyperproliferation and are a defining feature of GBM, were largely negative for Spn expression (Figure 2D). In addition to the primary GBM specimens, we analyzed Spn expression by immunofluorescence in recurrent GBM samples taken from patients after radiation and chemotherapy. Spn protein expression was detected at robust levels primarily within GBM cells of these recurrent tumor samples (Supplemental Figure 2).
Figure 2. Spn is differentially expressed in GBM cell lines and human tumor samples.
(A, B); Lysates from six different human GBM cell lines (A) and 12 different resected primary human GBM samples (B) were immunoblotted with anti-Spn, anti-PP1 or anti-Actin antibodies. (C, D); Formalin fixed paraffin embedded sections from non-neoplastic human brain (C) or three different GBM samples (D) were stained with H&E (upper panels) or anti-Spn antibodies (lower panels). Note that Spn protein is diffusely expressed in normal brain tissue. Spn is also expressed in GBM cells, but is largely absent in blood vessels within the tumor microenvironment. Scale bar, 50 μm.
Querying the open source The Cancer Genome Atlas (TCGA) database for human GBM (http://csbi.ltdk.helsinki.fi/anduril/tcga-gbm) revealed that Spn/PPP1R9B mRNA expression in tumors was diminished by approximately 50%, with a median expression fold-change of 0.527 in GBM samples versus non-cancerous control brain tissue (data not shown). Analysis of data in the Ivy GBM Atlas Project (GAP) (http://glioblastoma.alleninstitute.org/rnaseq/search/index.html) revealed that Spn mRNA was heterogeneously expressed in different tumor regions. Most notably, Spn mRNA is enriched as well as at the margins of tumor samples and in some infiltrative tumor regions, but was expressed at significantly lower levels in core tumor regions and in intratumoral regions containing microvascular proliferation (Supplemental Figure 3A), which is consistent with the immunohistochemistry results in Figure 2. In GBM there are at least four molecular sub-types based on genomic alterations and gene expression profiles (30). However, querying the Ivy GAP database revealed Spn mRNA was differentially expressed in tumor regions, regardless of classification by GBM molecular sub-type (Supplemental Figure 3B), although Spn levels were notably higher in classical and neural sub-types. Segregating tumor samples based on relative Spn mRNA expression levels (Spnhigh versus Spnlow) did not reveal statistically significant differences in overall patient survival (Supplemental Figure 3C).
Next, to determine roles for Spn in brain tumor initiation, growth and invasion in vivo we developed mosaic mouse models of GBM. Retroviral-delivered oncogenes were used to sequentially immortalize and transform primary mouse astroglial cells as we have previously reported (16) (Supplemental Figure 4A–B). Cells were immortalized with E6/E7 proteins and transformed using a H-Ras-V12G oncogene. E6/E7 proteins inhibit the tumor suppressors p53 and Rb, which are commonly inactivated in GBM. Hyperactivation of the Ras pathway via EGFR amplification or mutation is linked to GBM progression, and expression of oncogenic Ras in mice leads to spontaneous brain tumorigenesis (31). Transformed astrocytes generate intracranial tumors in vivo that displayed many histological features of grade III anaplastic astrocytoma (Supplemental Figure 4C), and some tumors progress to GBM-like status (data not shown).
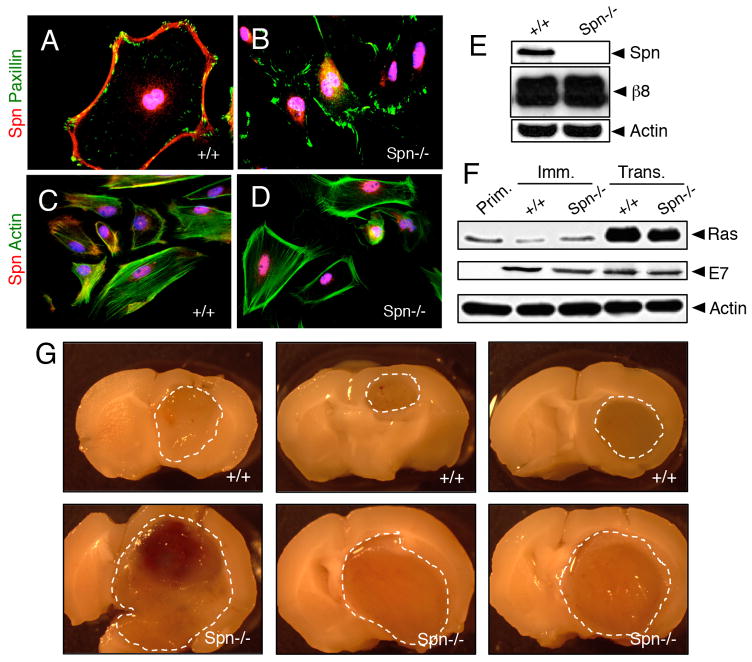
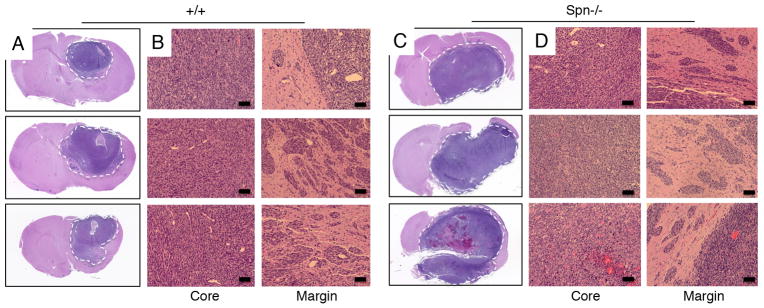
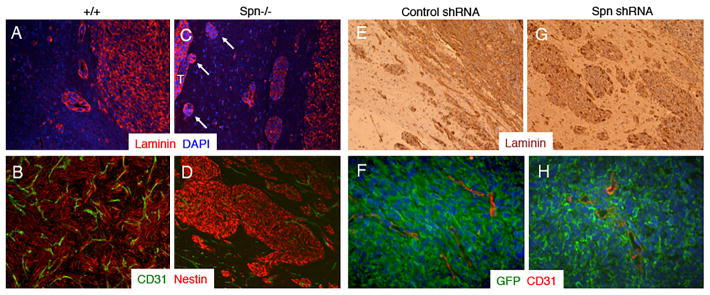
Analysis of Spn protein expression in primary mouse astroglial cells isolated from wild type mouse pups revealed enrichment at the cell membrane and co-localization with F-actin, whereas Spn protein was absent in Spn−/− cells owing to gene ablation (Figure 3A–E). Expression of retroviral-delivered oncoproteins was confirmed in wild type and Spn−/− mouse astrocytes (Figure 3F). We intracranially injected wild type and Spn−/− transformed mouse astrocytes into immunocompromised adult mice (n=8 mice per cell type). Although all mice were sacrificed at two months post-injection, animals injected with Spn−/− cells showed more obvious tumor-related neurological deficits including a hunched posture, ataxia and seizures (data not shown). Brain tumor growth and invasion were analyzed in coronal brain slices. As shown in Figure 3G, in comparison to wild type transformed astrocytes, Spn−/− transformed cells generated significantly larger intracranial tumors. Microscopic analysis of H&E-stained brain sections confirmed that Spn−/− tumors were significantly larger in relation to tumors derived from wild type cells (Figure 4A–D). Spn−/− astrocytes generated tumors with increased invasive properties., and clusters of tumor cells often showed perivascular dispersal patterns away from the primary mass with secondary lesions forming at the pial surface of the brain (Figure 4C–D). Immunofluorescence staining revealed that wild type and Spn−/− transformed astrocytes expressed laminin and nestin within the brain microenvironment, and tumors were well vascularized, as revealed by anti-CD31 immunofluorescence (Figure 5A–D).
Figure 3. Genetically targeting Spn in a mosaic model of brain cancer enhances invasive tumor cell growth.
(A–D); Primary astrocytes were cultured from the cerebral cortices of wild type (A, C) or Spn−/− (B, D) neonatal mice. Adherent cells were immunolabeled with anti-Spn (red) combined with anti-paxillin (green) to visualize focal adhesions (A, B) or phalloidin-FITC (green) to visualize the actin cytoskeleton (C, D). (E); Lysates from primary astrocytes isolated from wild type or Spn−/− pups were immunoblotted with anti-Spn. (F); Lysates from primary, immortalized and transformed mouse brain astrocytes were immunoblotted with anti-Ras or anti-E7 antibodies. (G); Images of three different brains harboring tumors derived from wild type (top) or Spn−/− transformed mouse astrocytes (bottom). Note that Spn−/− tumors are significantly larger than control tumors, and indicated by dashed lines.
Figure 4. Spn−/− mosaic mouse models display enhanced invasive growth in vivo.
(A–D); Brains of mice harboring tumors generated from wild type (A, B) or Spn−/− (C, D) transformed mouse astrocytes were sliced coronally, embedded in paraffin, and sections were stained with H&E. Note that tumors derived from Spn−/− transformed cells are significantly larger than wild type tumors. Spn−/− cells show more robust invasion away from the primary mass, often reaching the pial surface of the brain (arrows in D).
Figure 5. Spn negatively regulates tumor cell invasion in mouse models of brain cancer.
(A–D); Coronal sections through brain tumors derived from wild type (A, B) or Spn−/− transformed mouse astrocytes (C, D) were immunolabeled with anti-laminin (A, C) or anti-CD31 combined with anti-Nestin (B, D). Note that Spn−/− astrocytoma cells express laminin and nestin and display enhanced perivascular invasion with invasive cells reaching in distal pial regions (arrows in C). (E–H); Sections from control shRNA (E, F) or Spn shRNA (G, H) LN229 xenografts were subjected to either immunohistochemistry analysis (E, G) with an anti-laminin antibody or immunofluorescence analysis using anti-GFP and anti-CD31 antibodies combined with DAPI.
In addition to the mosaic mouse models, we analyzed functions for Spn in LN229 human GBM cells, which express high levels of endogenous Spn protein (Figure 2A). Lentiviral-delivered shRNAs were used to target the Spn gene, which led to diminished expression of Spn mRNA (data not shown) and protein (Supplemental Figure 5A). Intracranial implantation of LN229 cells expressing non-targeting control shRNAs or shRNAs targeting Spn revealed important roles in tumor growth and invasion in the brain. In comparison to animals injected with LN229 cells expressing control shRNAs, mice injected with cells expressing Spn shRNAs developed earlier and more obvious tumor-related neurological deficits (data not shown). Microscopic analysis revealed that LN229 tumors lacking Spn were significantly larger (Supplemental Figure 5B–E), and often grew to comprise nearly the entire brain hemisphere. Microscopic analysis of H&E stained samples confirmed larger tumor volumes and enhanced invasion in LN229 tumors expressing Spn shRNAs, as compared to tumors derived from cells expressing control shRNAs (Supplemental Figure 6). We confirmed continued stable down-regulation of Spn protein expression in vivo by immunofluorescently labeling LN229 brain tumor sections with anti-Spn antibodies (data not shown). Immunofluorescence and immunohistochemical microscopic analyses revealed enhanced invasion in tumors formed from LN229 cells expressing Spn shRNAs (Figure 5E–H and Supplemental Figure 7). Collectively, data from both the mosaic mouse models and LN229 xenograft models reveal important and novel functions for Spn in negatively regulating tumor cell growth and invasion in the brain microenvironment.
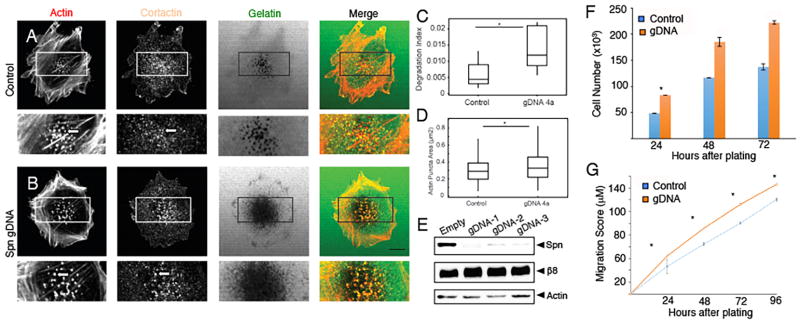
A critical step in the invasion process is the formation of invadopodia, which are membrane protrusions enriched in F-actin, the actin binding protein cortactin, and other signaling effectors (32). Invadopodia are utilized by cancer cells to survey the surrounding microenvironment in order to initiate and sustain pro-invasive signaling cascades. Therefore, we analyzed Spn-dependent invadopodia formation in GBM cells on FITC-gelatin (Supplemental Figure 8). Since the lentiviral-delivered shRNAs targeting Spn did not yield a complete inhibition of gene expression (Supplemental Figure 5A), we utilized Crispr/Cas9 gene editing methodologies to more effectively block Spn gene expression. In comparison to control cells, LN229 cells that lack Spn contain increased numbers of actin-expressing and cortactin-expressing invadopodia (Figure 6A–B). Cells expressing Spn gDNAs also displayed a significant increase in the degradation of FITC-gelatin (Figure 6C) and showed diminished peripheral actin levels, likely owing to increased actin in invadopodia (Figure 6D). As shown in Figure 6E, we obtained a near complete loss of Spn protein expression using lentiviruses expressing Cas9 and different gDNAs targeting various regions of the Spn gene. Loss of Spn gene expression in LN229 cells also led to increased proliferation (Figure 6F) and migration in scratch-wound assays (Figure 6G).
Figure 6. Spn regulates invadopodia dynamics to promote GBM cell invasive growth.
(A, B); Control LN229 cells (A) or cells expressing Spn gDNAs (B) were analyzed on coverslips coated with FITC-gelatin for invadopodia formation. Note that cells lacking Spn contain increased numbers of invadopodia, as evidenced by actin and cortactin expression/localization and FITC-gelatin degradation. (C, D); Quantitation of Spn-dependent FITC-gelatin degradation (C) and actin puncta (D) in LN229 cells. The degradation index was calculated as the ratio of degradation area/actin area per field (n = 8–10 fields/condition), *p<0.05. Box and whisker plots show distribution of actin in invadopodia defined as actin co-localized with cortactin. The results summarize at least 320 actin-rich puncta per condition, *p<0.05. (E); Crispr/Cas9-mediated gene editing was used to target Spn in LN229 cells. Note the absence of Spn protein as revealed by immunblotting detergent-soluble lysates. (F, G); LN229 cells expressing Cas9 and control gDNAs or Cas9 and gDNAs targeting Spn were assayed for proliferation (F) and migration (G). In comparison to control cells, LN229 cells expressing Cas9 and gDNAs targeting Spn display enhanced migration in vitro in scratch-wound assays.
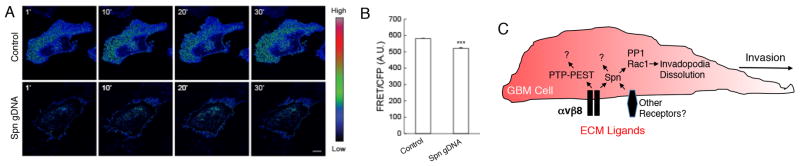
The results in Figure 6 reveal that while Spn is dispensable for the formation of invadopodia in GBM cells, it likely has important roles in the dynamic maintenance and/or dissolution of invadopodia structures. Various reports have shown that adhesion-mediated activation of members of the Rho family of small GTPases, comprised of RhoA, Cdc42 and Rac1, is essential for invadopodia dynamics and cell invasion (33). In particular, proper spatiotemporal regulation of Rac1 by the TrioGEF promotes invadopodia disassembly and cell invasion (34). Spn interacts with various GEFs (35) to control F-actin dynamics in the nervous system. To determine if Spn regulates Rac1 activation, we utilized the Raichu-Rac1 fluorescent biosensor (36) to monitor levels of Rac1 activities modulated by GEFs and GAPs. Raichu-Rac1 consists of truncated Rho sequences fused to the Rac-interactive binding domain (CRIB) of Pak1 flanked by YFP at the N-terminus and CFP at the C-terminus. Intramolecular interactions between GTP-bound Rac1 and CRIB juxtapose YFP and CFP, leading to Förster resonance energy transfer (FRET) from CFP to YFP. As shown in Figure 7A–B, significantly lower levels of GTP-bound Rac1 protein are detected in LN229 cells that lack Spn. Time lapse imaging studies confirmed diminished levels of Raichu-Rac1 FRET in LN229 GBM cells lacking Spn expression (Supplemental Videos 1 and 2). Based on these data, we propose a model in which Spn recruits GEFs to locally active Rac1 (and possibly other Rho GTPases) to promote invadopodia maintenance and disassembly. Tumor cells lacking Spn form invadopodia, but do not normally maintain and disassemble these structures, leading to increased numbers of invadopodia per cell, enhanced ECM degradation, and more robust cell invasion in the brain microenvironment (Figure 7C).
Figure 7. Spn promotes Rac1 GTPase activation in GBM cells.
(A); LN229 cells infected with control lentivirus or lentivirus expressing gDNAs targeting Spn were transiently transfected with a plasmid expressing the Rac1-Raichu FRET biosensor. Shown are representative FRET/CFP ratio images taken from a confocal time-lapse series. Images are presented using an intensity modulated display to associate intensity (activity) with color and hue (red = high activity; blue = low activity). Scale bar = 10 μm. (B); Quantitative analysis of FRET activity (mean +/− s.e.m., *** p-value <0.001) in arbitrary units. FRET ratios were collected for each cell in 2–4 fields (2–4 cells per field) for every time point for analysis (n=1147). Experiments were done in triplicate. (C); A model for Spn regulation of invadopodia dynamics in GBM cells. Spn normally suppresses GBM cell invasion by promoting Rac1 activation, which leads to dissolution of invadopodia and diminished invasion. Loss of Spn leads to increased numbers of invadopodia and enhanced tumor cell invasion, which is supported by our in vitro and in vivo data. αvβ8 integrin interacts directly with Spn, which likely regulates invadopodia formation and disassembly. Other cell surface receptors and intracellular proteins also bind to Spn and likely control its functions independently of αvβ8 integrin. Furthermore, in addition to Rac1 Spn interacts with additional signaling effectors, such as PP1, to modulate invadopodia dynamics.
Discussion
Here, we have characterized mechanisms by which the intracellular scaffolding protein Spn regulates invasive cell growth in the malignant brain cancer GBM. We report the following novel findings: (i) the C-terminus of Spn binds directly to the β8 integrin cytoplasmic tail (Figure 1), (ii) Spn is expressed in human GBM cells in vitro and in tumor samples in situ (Figure 2), (iii) Spn suppresses perivascular tumor cell invasion in pre-clinical mouse models of GBM (Figures 3–5), (iv) Spn modulates invadopodia dynamics and disassembly in GBM cells (Figure 6), and (v) Spn promotes the activation status of the Rac1 GTPase to regulate invadopodia dynamics (Figure 7). Collectively, these data identify Spn as a critical intracellular scaffolding protein that controls invasion by negatively regulating GBM cell invadopodia dynamics.
In studies of cancers outside of the brain, diminished levels of Spn have been reported to correlate with enhanced malignancy. For example, targeting Spn gene expression in breast tumor cells using RNAi-mediated methodologies increases self-renewal of stem cell populations and enhances migration and metastasis to the lung (37,38). Similar findings have been reported in primary tumors of the colon (39,40). Reflecting the multi-domain composition of Spn and its scaffolding and signaling roles, the mechanisms by which Spn loss promotes malignancy are complex. For example, down-regulation of Spn leads to differential gene expression in breast cancer cells and alters PP1 sub-cellular localization and specificity for substrates including retinoblastoma and other proteins (39,41). TCGA data reveal that Spn expression is diminished in GBM versus non-cancerous brain samples, and our experimental results show that Spn down-regulation in GBM cells causes enhanced invasive growth in the brain microenvironment. However, GBM patients with relative low versus high levels of Spn expression do not show differences in overall survival, revealing that Spn alone is not an accurate biomarker to predict outcome. It will be interesting to determine if combining Spn and additional genes to generate an invasive “gene signature” will better predict patient prognosis or response to therapy.
The C-terminal domain of Spn, which contains the CC and PDZ domains, interacts with the β8 integrin cytoplasmic tail, suggesting that Spn oligomerization, which is mediated by the CC domain (42), is important for integrin binding and/or signaling. The Spn C-terminus contains a type 5 PDZ domain, which interacts with short class 1 and class 2 PDZ binding peptide sequences (43). Interestingly, amino acids 713–716 within the cytoplasmic tail of human β8 integrin comprise an SNKI peptide sequence that is similar to SVKI, which is the class 2 PDZ peptide-binding motif. Hence, we postulate that the PDZ domain mediates interactions with β8 integrin cytoplasmic sequence, enabling the N-terminal regions of Spn to interact with additional effector proteins, including F-actin and PP1. Subsequent ECM engagement and outside-in signaling events likely dependent on Spn oligomerization via the CC domain. Given its roles in mediating activation of latent TGFβs, it is likely that these are involved in β8 integrin signaling via Spn. It is also possible that Spn may be involved in inside-out activation of αvβ8 integrin, thus modulating integrin affinity for ECM protein ligands. Inside-out integrin signaling commonly occurs via interactions with intracellular scaffolding proteins such as talins and kindlins (44). However, β8 integrin lacks cytoplasmic tail NPXY binding motifs found in β1A, β2, β3A, β5, and β6 integrins that commonly recruit FERM domain-containing effector proteins that promote activation and signaling. In addition, the intracellular juxtamembrane region of β8 integrin is divergent from other integrin subunits. This region is involved in a “handshake” with the adjacent α integrin subunit to maintain the extracellular region in an inactive state (45). This juxtamembrane handshake is disrupted upon interactions between the β subunit and talins and kindlins. Furthermore, the β8 integrin extracellular region lacks a “deadbolt” domain that facilitates inside-out activation in other integrins (45), suggesting constitutive activation and ECM ligand engagement independently of classical integrin “inside-out” activation mechanisms.
A prior report used proteomics-based methods to identify multiple proteins that co-immunoprecipitate with Spn in the rodent brain (25), but in that study β8 integrin was not reported as a Spn-associated protein. Here, we have utilized an antibody directed against the β8 integrin cytoplasmic domain and a GST fusion protein containing the β8 integrin cytoplasmic tail to identify and validate Spn interactions in GBM cells. Interestingly, anti-Spn antibodies we have tested did not co-immunoprecipitate β8 integrin from cells, suggesting that the Spn protein pool that interacts with β8 integrin is not recognized by these antibodies, or that the β8 integrin-Spn interaction masks the anti-Spn antibody epitopes. In the proteomics screen mentioned above, α-actinin (Actn1) was identified as a major Spn-interacting protein in the mouse brain. Indeed, Actn1 and Actn4 were identified in our proteomic experiments as β8 integrin-interacting proteins, but we have not detected interactions between Actn1/4 and β8 integrin by Far western blotting (data not shown), suggesting that its interaction with β8 integrin is likely the result of Spn associations. Indeed, we have confirmed interactions between Spn and Actn1 by co-immunoprecipitation and co-localization at GBM cell-ECM contacts, likely nascent and/or focal adhesions (data not shown). Genetically deleting β8 integrin expression, or truncating its cytoplasmic signaling domain to abrogate signaling, leads defects in GBM cell migration and invasion (17,21). However, in this report we show that tumor cells that lack Spn, a β8 integrin effector protein, display enhanced invasiveness. Therefore, we propose that β8 integrin balances pro-invasive and anti-invasive signal transduction pathways. In the case of the Spn signaling cascade, β8 integrin serves to localize Spn to the cell’s leading edge to promote invadopodia disassembly and negatively regulate invasive signaling events. In the absence of Spn this balance is perturbed, and β8 integrin continues to signal via other pro-invasive effectors, such as the tyrosine phosphatase PTP-PEST. In addition, Spn in part likely modulates some intracellular signaling events independently of β8 integrin that impact GBM ell growth and invasion.
Lastly, understanding the signaling mechanisms that drive perivascular invasion in GBM is not only fundamentally important but also clinically relevant. Brain tumor cells that disperse into healthy brain tissue escape surgical resection, remain largely resistant to chemotherapy and radiation, and invariably give rise to recurrent lesions (46). The significance of GBM cell invasion has also been underscored by recent clinical trials with the vascular endothelial growth factor (VEGF) neutralizing antibody Bevacizumab (47). Patients treated with Bevacizumab showed short-term improvements in progression-free survival; however, overall survival rates have not improved due to inevitable tumor recurrence. Unexpectedly, about 40% of patients display unusually robust patterns of invasive growth and lethal secondary lesions (48). Furthermore, in pre-clinical models of breast cancer and pancreatic cancer small molecule inhibitors of VEGF receptors have also been reported to enhance invasion and metastasis (49). Tumor recurrence and enhanced invasion following Bevacizumab therapy has been linked, in part, to deregulation of the c-Met signaling pathway (50), although other intracellular signaling events are likely involved. Based on the data that we report here, it is intriguing to speculate that anti-VEGF therapies may lead to increased invasive growth in GBM, or enhanced metastasis in non-brain tumors, via modulating Spn expression ore related signaling effectors in cancer cells.
Implications.
Tumor cell invasion is a major clinical obstacle in glioblastoma and this study identifies a new signaling pathway regulated by Spinophilin in invasive glioblastoma.
Acknowledgments
This work was supported by grant funding to J.H.M. from NIH/NINDS (R01NS087635, R01NS078402 and R21NS085688), and from the Cancer Prevention and Research Institute of Texas (RP140411). Funding to J.H.M. was also provided, in part, by an NIH/NCI SPORE in Brain Cancer (P50CA127001). The mass spectrometry analyses were funded by the Vermont Genetics Network through an NIH/NIGMS grant (8P20GM103449) from the IDeA Network for Biomedical Research Excellence (INBRE) INBRE program.
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Annual review of pathology. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Anderson JC, Gladson CL. The role of the extracellular matrix in angiogenesis in malignant glioma tumors. Brain Pathol. 2005;15(4):318–26. doi: 10.1111/j.1750-3639.2005.tb00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8(8):610–22. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 4.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326(5957):1216–9. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C, Ye F, Ginsberg MH. Regulation of Integrin Activation. Annual review of cell and developmental biology. 2011 doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 6.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11(9):633–43. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weis SM, Cheresh DA. alphaV integrins in angiogenesis and cancer. Cold Spring Harb Perspect Med. 2011;1(1):a006478. doi: 10.1101/cshperspect.a006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura SL, Sheppard D, Pytela R. Integrin alpha v beta 8. Interaction with vitronectin and functional divergence of the beta 8 cytoplasmic domain J Biol Chem. 1994;269(46):28708–15. [PubMed] [Google Scholar]
- 9.Worthington JJ, Klementowicz JE, Travis MA. TGFbeta: a sleeping giant awoken by integrins. Trends Biochem Sci. 2010;36(1):47–54. doi: 10.1016/j.tibs.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Allinson K, Lee H, Fruttiger M, McCarty J, Arthur H. Endothelial Expression of TGFβ Type II Receptor Is Required to Maintain Vascular Integrity during Postnatal Development of the Central Nervous System. PLos One. 2012 doi: 10.1371/journal.pone.0039336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold TD, Ferrero GM, Qiu H, Phan IT, Akhurst RJ, Huang EJ, et al. Defective Retinal Vascular Endothelial Cell Development As a Consequence of Impaired Integrin alphaVbeta8-Mediated Activation of Transforming Growth Factor-beta. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32(4):1197–206. doi: 10.1523/JNEUROSCI.5648-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarty JH, Lacy-Hulbert A, Charest A, Bronson RT, Crowley D, Housman D, et al. Selective ablation of alphav integrins in the central nervous system leads to cerebral hemorrhage, seizures, axonal degeneration and premature death. Development. 2005;132(1):165–76. doi: 10.1242/dev.01551. [DOI] [PubMed] [Google Scholar]
- 13.Mobley AK, Tchaicha JH, Shin J, Hossain MG, McCarty JH. {beta}8 integrin regulates neurogenesis and neurovascular homeostasis in the adult brain. J Cell Sci. 2009;122(Pt 11):1842–51. doi: 10.1242/jcs.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen HL, Lee YJ, Shin J, Lee E, Park SO, McCarty JH, et al. TGF-beta signaling in endothelial cells, but not neuroepithelial cells, is essential for cerebral vascular development. Lab Invest. 2011;91(11):1554–63. doi: 10.1038/labinvest.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proctor JM, Zang K, Wang D, Wang R, Reichardt LF. Vascular development of the brain requires beta8 integrin expression in the neuroepithelium. J Neurosci. 2005;25(43):9940–8. doi: 10.1523/JNEUROSCI.3467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchaicha JH, Mobley AK, Hossain MG, Aldape KD, McCarty JH. A mosaic mouse model of astrocytoma identifies alphavbeta8 integrin as a negative regulator of tumor angiogenesis. Oncogene. 2010;29(31):4460–72. doi: 10.1038/onc.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchaicha JH, Reyes SB, Shin J, Hossain MG, Lang FF, McCarty JH. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by beta8 integrin. Cancer Res. 2011;71(20):6371–81. doi: 10.1158/0008-5472.CAN-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schittenhelm J, Klein A, Tatagiba MS, Meyermann R, Fend F, Goodman SL, et al. Comparing the expression of integrins alphavbeta3, alphavbeta5, alphavbeta6, alphavbeta8, fibronectin and fibrinogen in human brain metastases and their corresponding primary tumors. International journal of clinical and experimental pathology. 2013;6(12):2719–32. [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty JH, Cook AA, Hynes RO. An interaction between {alpha}v{beta}8 integrin and Band 4.1B via a highly conserved region of the Band 4. 1 C-terminal domain. Proc Natl Acad Sci U S A. 2005;102(38):13479–83. doi: 10.1073/pnas.0506068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12(8):493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyes SB, Narayanan AS, Lee HS, Tchaicha JH, Aldape KD, Lang FF, et al. alphavbeta8 integrin interacts with RhoGDI1 to regulate Rac1 and Cdc42 activation and drive glioblastoma cell invasion. Molecular biology of the cell. 2013;24(4):474–82. doi: 10.1091/mbc.E12-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HS, Cheerathodi M, Chaki SP, Reyes SB, Zheng Y, Lu Z, et al. Protein tyrosine phosphatase-PEST and beta8 integrin regulate spatiotemporal patterns of RhoGDI1 activation in migrating cells. Mol Cell Biol. 2015;35(8):1401–13. doi: 10.1128/MCB.00112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarrouilhe D, di Tommaso A, Metaye T, Ladeveze V. Spinophilin: from partners to functions. Biochimie. 2006;88(9):1099–113. doi: 10.1016/j.biochi.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Feng J, Yan Z, Ferreira A, Tomizawa K, Liauw JA, Zhuo M, et al. Spinophilin regulates the formation and function of dendritic spines. Proc Natl Acad Sci U S A. 2000;97(16):9287–92. doi: 10.1073/pnas.97.16.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baucum AJ, 2nd, Jalan-Sakrikar N, Jiao Y, Gustin RM, Carmody LC, Tabb DL, et al. Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Molecular & cellular proteomics: MCP. 2010;9(6):1243–59. doi: 10.1074/mcp.M900387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. The FEBS journal. 2013;280(2):596–611. doi: 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, et al. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129(3):579–91. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carnero A. Spinophilin: a new tumor suppressor at 17q21. Current molecular medicine. 2012;12(5):528–35. doi: 10.2174/156652412800619987. [DOI] [PubMed] [Google Scholar]
- 29.Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61(18):6674–8. [PubMed] [Google Scholar]
- 30.Verhaak RG, Valk PJ. Genes predictive of outcome and novel molecular classification schemes in adult acute myeloid leukemia. Cancer treatment and research. 2010;145:67–83. doi: 10.1007/978-0-387-69259-3_5. [DOI] [PubMed] [Google Scholar]
- 31.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nature genetics. 2000;25(1):55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 32.Seano G, Primo L. Podosomes and invadopodia: tools to breach vascular basement membrane. Cell cycle. 2015;14(9):1370–4. doi: 10.1080/15384101.2015.1026523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Destaing O, Block MR, Planus E, Albiges-Rizo C. Invadosome regulation by adhesion signaling. Curr Opin Cell Biol. 2011;23(5):597–606. doi: 10.1016/j.ceb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nature cell biology. 2014;16(6):574–86. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan XP, Alldritt J, Svenningsson P, Allen PB, Wu GY, Nairn AC, et al. The Rho-specific GEF Lfc interacts with neurabin and spinophilin to regulate dendritic spine morphology. Neuron. 2005;47(1):85–100. doi: 10.1016/j.neuron.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat Protoc. 2009;4(11):1623–31. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- 37.Ferrer I, Verdugo-Sivianes EM, Castilla MA, Melendez R, Marin JJ, Munoz-Galvan S, et al. Loss of the tumor suppressor spinophilin (PPP1R9B) increases the cancer stem cell population in breast tumors. Oncogene. 2016;35(21):2777–88. doi: 10.1038/onc.2015.341. [DOI] [PubMed] [Google Scholar]
- 38.Schwarzenbacher D, Stiegelbauer V, Deutsch A, Ress AL, Aigelsreiter A, Schauer S, et al. Low spinophilin expression enhances aggressive biological behavior of breast cancer. Oncotarget. 2015;6(13):11191–202. doi: 10.18632/oncotarget.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estevez-Garcia P, Lopez-Calderero I, Molina-Pinelo S, Munoz-Galvan S, Salinas A, Gomez-Izquierdo L, et al. Spinophilin loss correlates with poor patient prognosis in advanced stages of colon carcinoma. Clin Cancer Res. 2013;19(14):3925–35. doi: 10.1158/1078-0432.CCR-13-0057. [DOI] [PubMed] [Google Scholar]
- 40.Ress AL, Stiegelbauer V, Schwarzenbacher D, Deutsch A, Perakis S, Ling H, et al. Spinophilin expression determines cellular growth, cancer stemness and 5-flourouracil resistance in colorectal cancer. Oncotarget. 2014;5(18):8492–502. doi: 10.18632/oncotarget.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrer I, Blanco-Aparicio C, Peregrina S, Canamero M, Fominaya J, Cecilia Y, et al. Spinophilin acts as a tumor suppressor by regulating Rb phosphorylation. Cell cycle. 2011;10(16):2751–62. doi: 10.4161/cc.10.16.16422. [DOI] [PubMed] [Google Scholar]
- 42.Stephens DJ, Banting G. In vivo dynamics of the F-actin-binding protein neurabin-II. The Biochemical journal. 2000;345(Pt 2):185–94. [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell communication and signaling: CCS. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annual review of cell and developmental biology. 2011;27:321–45. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 45.Jannuzi AL, Bunch TA, West RF, Brower DL. Identification of integrin beta subunit mutations that alter heterodimer function in situ. Mol Biol Cell. 2004;15(8):3829–40. doi: 10.1091/mbc.E04-02-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drappatz J, Norden AD, Wen PY. Therapeutic strategies for inhibiting invasion in glioblastoma. Expert review of neurotherapeutics. 2009;9(4):519–34. doi: 10.1586/ern.09.10. [DOI] [PubMed] [Google Scholar]
- 47.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3(5):391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 48.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nature reviews Cancer. 2008;8(8):592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15(3):220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarty JH. Glioblastoma resistance to anti-VEGF therapy: has the challenge been MET? Clin Cancer Res. 2013;19(7):1631–3. doi: 10.1158/1078-0432.CCR-13-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]