Abstract
Repetitive DNA sequences, comprising up to 50% of the genome in all eukaryotes, play important roles in a wide range of cellular functions, such as transcriptional regulation, genome stability and cellular differentiation. However, due to technical difficulties in differentiating their sequences, DNA repeats remain one of the most mysterious parts of eukaryotic genomes. Key questions, such as how repetitive entities behave at individual level and how the internal architecture of these repeats is organized, are still poorly understood. Recent advances from our group reveal unexpected position-dependent variation within tandem DNA repeats in fission yeast. Despite sharing identical DNA sequences, the peri-centromeric repeats are organized into diverse epigenetic states and chromatin structures. We demonstrate that this position-dependent variation requires key heterochromatin factors and condensin. Our works further suggest that the peri-centromeric repeats are organized into distinct higher-order structures that ensure proper positioning of CENP-A, the centromere-specific histone H3 variant, to centromeres. These most recent developments offer insights into the mechanisms underlying the position effect within tandem DNA arrays, and have broad implications in the field of epigenetics and chromatin biology.
Keywords: DNA repeats, heterochromatin, position effect, centromere positioning, Fission yeast Schizosaccharomyces pombe, epigenetics, CENP-A
One of the most conserved features in eukaryotic organisms is the presence of many repetitive DNA sequences. The sequence complexity of these DNA repeats varies from complete genes (such as the ribosomal DNA) to simple sequences of a few base pairs. These repeats can be organized into long tandem arrays (such as peri-centromeric repeats), or widely dispersed in the genome (such as tRNA and transposon elements) (Richard et al., 2008; Sutherland and Richards, 1995). For a long time, researchers considered DNA repeats as “junk DNA”. However, recent advances have implicated DNA repeats in many important cellular events, including transcriptional control, disease development, evolution, cellular differentiation and genome stability (Armour 2006, Gemayel, et al. 2012). Despite their importance, DNA repeats remain the least understood structures in the genome (Gemayel et al., 2012; Martienssen et al., 2004; Pearson et al., 2005). It is largely due to the technical challenges of analyzing repetitive DNA.
Repetitive DNA sequences are hotspots for meiotic crossover and other recombination events, which may cause genome instability (Jeffreys et al., 1999a; Vader et al., 2011). To prevent this, DNA tandem arrays are often packaged into constitutive heterochromatin, such as centromeres and telomeres. Constitutive heterochromatin is the stable, compact chromatin structure containing low level of transcriptional activities (Bierhoff et al., 2014; Saksouk et al., 2015). Hypermethylation of histone 3 at lysine 9 (H3K9me) is a conserved hallmark of constitutive heterochromatin, and plays a key role in heterochromatin assembly. It is generally assumed that epigenetic state is evenly distributed among the tandem repeats in constitutive heterochromatin. Notably, ribosomal DNA (rDNA), which encodes rRNA, also comprises of tandem arrays of rDNA genes in eukaryotic genomes. Since rRNA, an important component of ribosome, is essential for protein translation in all eukaryotes, rDNA has to be transcriptionally active. Interestingly, it has been shown that some fractions of rDNA repeats can be silenced and form heterochromatin-like structure (Guetg and Santoro, 2012; Pasero and Marilley, 1993). Moreover, the heterochromatic state in rDNA region is not static, and can be influenced upon stress, cell cycle progression and other stimuli (Benoit et al., 2013; Hamperl et al., 2013). Whether similar position-dependent silencing takes place within tandem repeats in constitutive heterochromatin, such as peri-centromeres, has not been explored. If that also is the case in constitutive heterochromatin, how specific chromatin states are maintained and regulated at the individual repeat units? More importantly, what is the functional significance of this interesting epigenetic phenomenon?
Fission yeast Schizosaccharomyces pombe is a superb model to study constitutive heterochromatin. S. pombe is a single-cell, genetic-tractable eukaryotic organism that is relatively easy to manipulate. Like many other eukaryotes, peri-centromeric regions in S. pombe contain tandem DNA repeats that are not found in the peri-centromeres in budding yeast Saccharomyces cerevisiae (He et al., 2014). Key mechanisms involved in heterochromatin regulation, such as methylation of histone H3 lysine 9 (H3K9me) and the RNAi interference (RNAi) machinery, are absent in budding yeast but preserved in S. pombe (Nakayama et al., 2001; Verdel et al., 2004). Heterochromatin of peri-centromeric repeats has been implicated in centromere positioning and chromosome segregation (Bernard et al., 2001; Folco et al., 2008; Gonzalez and Li, 2012), providing an excellent opportunity to examine the functional relevance of position-dependent epigenetic events in the tandem arrays.
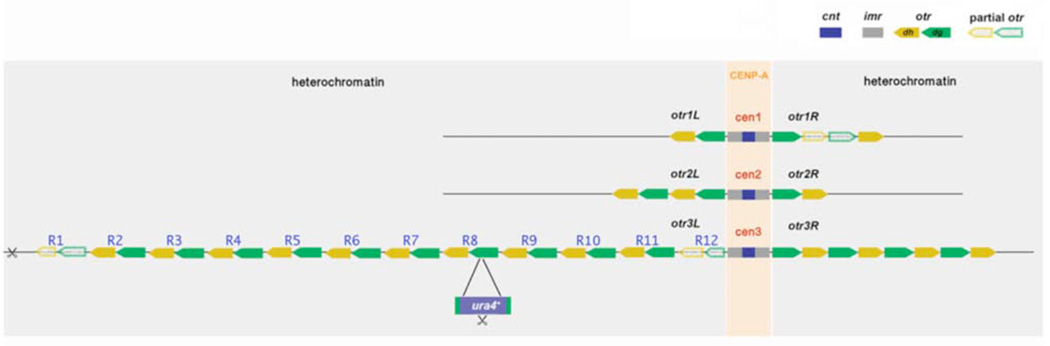
Fission yeast has three chromosomes, each chromosome harboring a single centromere surrounded by peri-centromeric tandem repeats. Like higher eukaryotic organisms including humans, fission yeast has large regional centromere that is epigenetically defined. Specifically, fission yeast centromere contains a core region cnt (centromere core domain), which is flanked with imperfect repeat regions imr (innermost repeats). Both cnt and imr regions are marked with CENP-Acnp1, the centromeric specific H3 variant. Beyond imr regions there are tandem arrays of otr (outermost repeat regions) repeats, 6.7kb each that consists of alternating dg and dh repeats. otr regions are heterochromatic, and enriched with H3K9me (He et al., 2014; Takahashi et al., 2000). It has been challenging to assemble the exact copy numbers of otr repeats on each chromosome. By estimation, Chromosome I and II have 1~2 repeats on either side of cnt, while Chromosome III has 7~11 repeats on the left arm (otr3L) and 4 repeats on the right arm (otr3R). The considerable size of otr3L repeats makes it a favorable region to study heterochromatin structures in tandem array (Figure 1) (Wood et al., 2002). However, powerful genetic tools such as sequencing, chromatin immunoprecipitation (ChIP) and chromosome conformation capture (3C), which completely rely on DNA sequence specificities, fail to assemble large tandem arrays or differentiate among individual repeats bearing identical sequences.
Figure 1. Schematic representation of centromeres in fission yeast and the development of repeat-specific reporter.
Central core regions (cnt) are flanked by innermost repeats (imr) and outermost repeats (otr). Each of otr repeats consists of dg and dh repeat units as indicated. CENP-Acnp1 is enriched in cnt and imr regions, while otr repeats are organized into peri-centromeric heterochromatin. The ura4+ reporter flanked with part of dg sequence was randomly inserted into otr repeats by homologues recombination. The location of ura4+ insertion is mapped by estimating the distance between two unique restriction sites in ura4+ and a heterochromatin boundary region, respectively (indicated by two scissors).
A novel approach to study tandem repeats at individual leve
To address how peri-centromeric repeats are regulated at the individual level, He et al. recently developed a collection of strains that mark individual repeats in peri-centromeric tandem array in S. pombe via the insertion of the ura4+ reporter gene (He et al., 2016). The exogenous ura4+ sets apart the repeat unit containing the reporter from other identical repeats. Therefore, for the first time, it is feasible to dissect structural and functional nature of heterochromatin states within peri-centromeric repeats at individual level.
To mark the individual repeat, the ura4+ reporter flanked with a section of dg repeats was constructed and transformed into wild-type S. pombe cells. The reporter was randomly inserted into the peri-centromeric repeats via homologous recombination. Strains that survive on the minimum medium without uracil were selected. The exact location of ura4+ insertion within the peri-centromeric repeats was mapped based on the distance between unique restriction enzyme sites in ura4+ and the boundary region of the repeat array by Southern blot analysis. This experimental approach was adapted from a previous report in which a repeat-specific reporter was developed for rDNA tandem array in budding yeast (Vader et al., 2011). Mapping results by He et al. indicated that otr3L in S. pombe contains a total of 12 repeats. While the first repeat 1 (R1), which is distal to core centromeres, and the last repeat (R12) in otr3L have only partial repeat sequence, the rest of otr3L tandem array contains full-length 6.7kb repeat DNA (Figure 1). He et al. generated a collection of ura4+ insertion for every otr3L repeat with exception of R6, R9, and R12.
Not all repeats are created equal
Upon analyzing the transcriptional level of ura4+ by growth assay and Northern blotting, an apparent position-dependent effect is identified within the otr3L tandem array. Transcription of ura4+ is relatively active in repeats close to core centromere region, whereas ura4+ in distal repeats are more silenced. Taking advantage of repeat-specific ura4+ insertions, H3K9me enrichment in individual repeat unit can be assayed by ChIP. Repeats distal to the core centromere are enriched with H3K9me, while repeats close to the core centromere have less H3K9me. The distribution of H3K9 methylation is well correlated with the position dependent gradient of transcriptional activity.
In fission yeast, H3K9me is mediated by Clr4 complex (ClrC), which contains Cul4, Rik1, Dos1, Dos2, Lid2, and the catalytic subunit histone methyltransferase Clr4 (Hong et al., 2005; Horn et al., 2005; Jia et al., 2005; Li et al., 2005; Li et al., 2008; Thon et al., 2005). Loss of any of these subunits results in significant reduction of silencing in peri-centromeric heterochromatin. In fission yeast, assembly of pericentromeric heterochromatin is also regulated by RNAi, a phenomenon found in many other organisms (Lejeune and Allshire, 2011; Volpe and Martienssen, 2011). Noncoding RNAs (ncRNAs) within peri-centromeric repeats are transcribed by RNA polymerase II (RNAPII) and later processed into siRNA by Dicer, RITS (RNA-induced transcriptional silencing), and RDRC (RNA-directed RNA polymerase complex). RITS complex, containing Ago1, Tas3 and the chromo-domain protein Chp1, is directed to peri-centromeric repeats via base-pairing interactions between Ago1-bound siRNAs and ncRNAs. This siRNA-containing complex in turn recruits ClrC for heterochromatin assembly(Kato et al., 2005; Motamedi et al., 2004). Similar to human HP1 (heterochromatin protein 1), the chromo domain-containing Chp1 specifically binds to H3K9me (Schalch et al., 2009). Correspondingly, He et al. found that Chp1 distribution among otr3L repeats is correlated with the distribution of H3K9me, as Chp1 is enriched in repeats distal to centromeres, but not in repeats close to centromeres that are less silenced.
The collection of strains with the repeat-specific reporter also provides an opportunity for investigating the replication timing between different repeats. It is generally believed that euchromatin replicates earlier than heterochromatin (Gilbert, 2002). However, peri-centromeric heterochromatin in fission yeast replicate at the early S phase. Similar phenomenon also was found in mammals (Kim et al., 2003). Using BrdU-ChIP, He et al. revealed that the replicating timing of the individual repeats in otr3L tandem array can vary considerably. In general, the repeats exhibiting strong silencing in otr3L tandem array appears to replicate earlier. The reason for the early replication of these repeats remains unclear.
These results indicate that although the DNA repeats with a tandem array contain same sequence, the function and epigenetic state for each repeat can be very different, suggesting that each individual repeat unit can be organized into distinct higher-order architecture. Consistent with this idea, siRNAs generated from an ura4+ hairpin can silence the ura4+ reporter inserted within otr3L tandem array in trans in a position-dependent manner. Overexpression of the ura4+ hairpin promotes heterochromatin formation in the otr repeats exhibiting weaker silencing, but has little effect on the repeats containing strong silencing.
Mechanisms regulating position effect in tandem repeats
How this position-dependent heterochromatin differentiation is achieved in the repeats despite sharing identical sequences? Heterochromatin assembly required machineries such as ClrC and RITS. Disruptions of these factors also lead to the total loss of the position effect within otr3L repeats, indicating that they are the key effectors for the position effect. He et al. also found that Cut3, a condensin subunit, is required for the position effect in the peri-centromeric region. An ancient but highly conserved family of proteins, condensin is important for chromosome compaction and organization. It is well established that condensin is recruited to peri-centromeric and rDNA tandem arrays, underscoring its role in higher order chromatin structures across repetitive sequences (Bloom, 2014; Hirano, 2016). He et al found that disruption of Cut3 alters the profile of position-dependent transcription in otr3L tandem array without total loss of silencing, suggesting that Cut3 functions as an upstream regulator of this position effect. Wang et al. also showed that condensin is essential for the position effect in rDNA tandem repeat in budding yeast, indicating that the role of condensin in regulating the organization of DNA repeats is conserved (Wang et al., 2016).
He et al. further revealed that Cut3 physically interacts with RITS complex, and is required for the association of the RITS with individual repeats. We propose that condensin acts as the upstream instructor to recruit the proper amount of silencing effectors, such as the RITS complex, to individual repeats, which in turn establish the specific epigenetic state in these repeats. However, among individual otr3L repeats there is no apparent bias in Cut3 distribution, suggesting additional factors may be required to establish position-dependent heterochromatin organizations. Cohesins and other chromatin remodeling complexes are possible candidates to control the position effect within the peri-centromeric repeat array (Mizuguchi et al., 2014; Robellet et al., 2014).
Biological significance of position effect in peri-centromeric repeats
Centromeres play essential role in equal chromosome segregation during mitosis and meiosis. In most eukaryotes, the underlying DNA sequence in centromere is not sufficient to organize the assembly of centromeres, indicating that epigenetic mechanism is important for centromere specification. CENP-A, the histone H3 variant, is considered as the epigenetic mark to define the centromeres (Allshire and Karpen, 2008; Black and Cleveland, 2011; Gonzalez et al., 2014). Multiple proteins, including HJURP and Mis16 complex, have been identified to be the key CENP-A loading factors (Gonzalez et al., 2013; Hayashi et al., 2004; Sanchez-Pulido et al., 2009; Takahashi et al., 2005). However, how CENP-A is precisely incorporated into centromeres remains unclear. The position effect in peri-centromereic otr repeats in S. pombe revealed by He et al. suggests that these repeats is organized into unique three-dimensional architecture. He et al. further showed that disruption of the position effect led to the dissociation of CENP-A from centromeres and mis-targeting of CENP-A to ectopic regions. These results indicate that the higher-order structure of peri-centromeric repeats may promote CENP-A positioning to centromeres. Consistent with this, peri-centromeric heterochromatin has previously been implicated in centromeric localization of CENP-A in multiple organisms, including Neurospora crassa, S. pombe, and mouse cell lines (Boyarchuk et al., 2014; Folco et al., 2008; Smith et al., 2011).
Although heterochromatin is generally considered to be transcriptional inactive, transcription at peri-centromeric repeats was found in wide ranges of organisms and under various conditions, such as differentiation, heat-shock response and tumorigenesis (Enukashvily and Ponomartsev, 2013; Eymery et al., 2009; Hall et al., 2012; Saksouk et al., 2015). Paradoxically, the transcription in these regions can play a critical role in heterochromatin assembly. In fission yeast, peri-centromeric transcription peaks in early S phase, and subsequently processed into siRNAs. This cell-cycle dependent transcription in peri-centromeric repeats has been proposed to explain the establishment of heterochromatin during S phase (Chen et al., 2008; Gonzalez and Li, 2012; Kloc et al., 2008). The finding of position effect in otr3L indicated that under basal conditions, some repeats (R10 and R11) are relative open and susceptible to gene transcription. It is possible that siRNAs generated from these less condensed repeats may facilitate the establishment and maintenance of heterochromatin states in other otr repeats, in same or other chromosomes, throughout the cell cycle. Supporting this idea, as mentioned above, siRNAs generated from artificial introduced ura4+ hairpin is able to induce peri-centromeric ura4+ silencing in trans. Further experiments need to be performed to address this hypothesis. In addition, tandem DNA repeats are hotspot for aberrant replication, repair and recombination, which can lead to genome instability and diseases (Armour, 2006; Jeffreys et al., 1999b; Murray et al., 1999). The position effect in tandem DNA repeats may also reflect the higher order structure of these repeats that can be used to resolve the potential dysfunction of these DNA metabolic processes.
Concluding remarks
Understanding tandem DNA repeats has been challenging due to its repetitive nature. Recent advances in the repeat-specific reporter approach allow us to study the behavior of tandem repeats at the individual level, revealing unexpected position effect within tandem repeat arrays. Using the peri-centromeric repeats as a model, we found that the position effect within the repeat array is regulated by key heterochromatin factors, such as RITS and CLRC complexes, and condensin. Out study further indicates that the internal position effect within tandem repeats may be involved in many aspects of array biology, including gene expression, chromatin structure and DNA replication. Particularly, our works suggest that the pericentromeric repeats are organized into specific higher-order structures to ensure proper targeting of CENP-A to centromeres. However, many important questions remain open. One of key questions is what are the factors responsible for regulating the position effects within tandem DNA repeat? Also, how are the tandems repeats spatially organized? Is there important cross talk between individual repeats? It will also be interesting to know how the position effect in tandem repeats is regulated through the cell cycle and in response to environmental stresses. Answers to these questions will help us understand this uncharted territory in eukaryotic genomes.
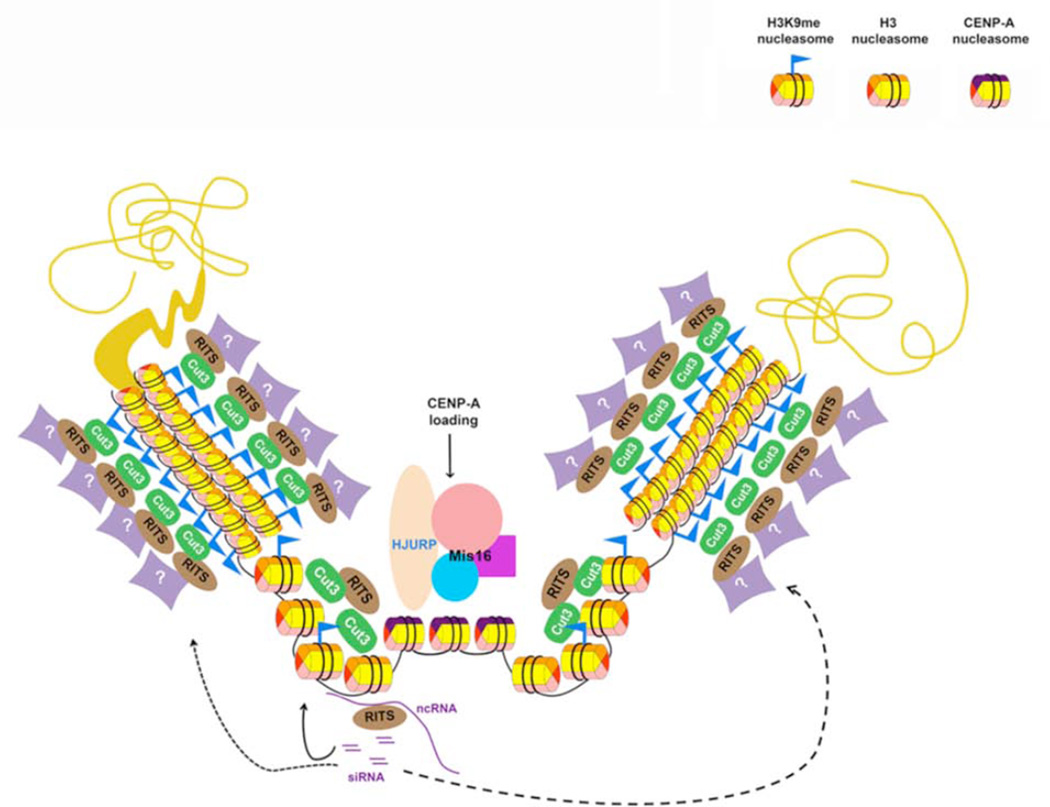
Figure 2. A proposed model for the position effect in peri-centromeric tandem repeats.
The condensin subunit Cut3 and other unknown upstream factors recruit key silencing effectors, such as RITS complex, to pericentromeric repeat arrays, and mediate the level of the silencing at individual repeats, leading to the position-dependent silencing. The less silenced repeats may generate siRNAs that in turn facilitate heterochromatin assembly in other repeats in trans. Position effects in pericentromeric repeat arrays indicates that these repeats are organized into distinct three-dimensional higher-order structure; such spatial arrangement helps to define and position the CENP-Acnp1 enriched centromere core regions.
Acknowledgments
We thank members of the Li laboratory, particularly Drs. Qianhua Dong, David Aristizabal-Corrales, and Haijin He for reading the manuscript. F. L. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. The Li laboratory is supported by funding from NIH grant 1R01 GM106037 and NSF grant MCB-1330557.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour JA. Tandemly repeated DNA: why should anyone care? Mutation research. 2006;598:6–14. doi: 10.1016/j.mrfmmm.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Benoit M, Layat E, Tourmente S, Probst AV. Heterochromatin dynamics during developmental transitions in Arabidopsis - a focus on ribosomal DNA loci. Gene. 2013;526:39–45. doi: 10.1016/j.gene.2013.01.060. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bierhoff H, Postepska-Igielska A, Grummt I. Noisy silence: non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics. 2014;9:53–61. doi: 10.4161/epi.26485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom KS. Centromeric heterochromatin: the primordial segregation machine. Annual review of genetics. 2014;48:457–484. doi: 10.1146/annurev-genet-120213-092033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk E, Filipescu D, Vassias I, Cantaloube S, Almouzni G. The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during the cell cycle. J Cell Sci. 2014;127:3347–3359. doi: 10.1242/jcs.148189. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Enukashvily NI, Ponomartsev NV. Mammalian satellite DNA: a speaking dumb. Advances in protein chemistry and structural biology. 2013;90:31–65. doi: 10.1016/B978-0-12-410523-2.00002-X. [DOI] [PubMed] [Google Scholar]
- Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, Van den Broeck A, Brambilla E, Fournier A, Callanan M, et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic acids research. 2009;37:6340–6354. doi: 10.1093/nar/gkp639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Cho J, Boeynaems S, Verstrepen KJ. Beyond Junk-Variable Tandem Repeats as Facilitators of Rapid Evolution of Regulatory and Coding Sequences. Genes-Basel. 2012;3:461–480. doi: 10.3390/genes3030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Current opinion in cell biology. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, He H, Dong Q, Sun S, Li F. Ectopic Centromere Nucleation by CENP-A in Fission Yeast. Genetics. 2014 doi: 10.1534/genetics.114.171173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, He H, Sun S, Li C, Li F. Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proc Natl Acad Sci U S A. 2013;110:606–611. doi: 10.1073/pnas.1214874110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Li F. DNA replication, RNAi and epigenetic inheritance. Epigenetics. 2012;7:14–19. doi: 10.4161/epi.7.1.18545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LE, Mitchell SE, O'Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2012;20:535–546. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- Hamperl S, Wittner M, Babl V, Perez-Fernandez J, Tschochner H, Griesenbeck J. Chromatin states at ribosomal DNA loci. Bba-Gene Regul Mech. 2013;1829:405–417. doi: 10.1016/j.bbagrm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- He H, Gonzalez M, Zhang F, Li F. DNA replication components as regulators of epigenetic inheritance--lesson from fission yeast centromere. Protein Cell. 2014;5:411–419. doi: 10.1007/s13238-014-0049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zhang S, Wang D, Hochwagen A, Li F. Condensin Promotes Position Effects within Tandem DNA Repeats via the RITS Complex. Cell reports. 2016;14:1018–1024. doi: 10.1016/j.celrep.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes & Development. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A, Barber R, Bois P, Buard J, Dubrova YE, Grant G, Hollies CRH, May CA, Neumann R, Panayi M, et al. Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis. 1999a;20:1665–1675. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1665::AID-ELPS1665>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Barber R, Bois P, Buard J, Dubrova YE, Grant G, Hollies CR, May CA, Neumann R, Panayi M, et al. Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis. 1999b;20:1665–1675. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1665::AID-ELPS1665>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Current Biology. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Current Biology. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 Is Required for Coordinating H3K4 and H3K9 Methylation of Heterochromatin and Euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R, Lippman Z, May B, Ronemus M, Vaughn M. Transposons, tandem repeats, and the silencing of imprinted genes. Cold Spring Harb Sym. 2004;69:371–379. doi: 10.1101/sqb.2004.69.371. [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Fudenberg G, Mehta S, Belton JM, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Murray J, Buard J, Neil DL, Yeramian E, Tamaki K, Hollies C, Jeffreys AJ. Comparative sequence analysis of human minisatellites showing meiotic repeat instability. Genome research. 1999;9:130–136. [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Pasero P, Marilley M. Size Variation of Rdna Clusters in the Yeasts Saccharomyces-Cerevisiae and Schizosaccharomyces-Pombe. Mol Gen Genet. 1993;236:448–452. doi: 10.1007/BF00277147. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Edamura KN, Cleary JD. Repeat instability: Mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative Genomics and Molecular Dynamics of DNA Repeats in Eukaryotes. Microbiol Mol Biol R. 2008;72:686-+. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X, Fauque L, Legros P, Mollereau E, Janczarski S, Parrinello H, Desvignes JP, Thevenin M, Bernard P. A genetic screen for functional partners of condensin in fission yeast. G3. 2014;4:373–381. doi: 10.1534/g3.113.009621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Simboeck E, Dejardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics & chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland GR, Richards RI. Simple tandem DNA repeats and human genetic disease. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3636–3641. doi: 10.1073/pnas.92.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2005;360:595–606. doi: 10.1098/rstb.2004.1614. discussion 606-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klart AJS. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477:115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Martienssen RA. RNA interference and heterochromatin assembly. Cold Spring Harb Perspect Biol. 2011;3:a003731. doi: 10.1101/cshperspect.a003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Mansisidor A, Prabhakar G, Hochwagen A. Condensin and Hmo1 Mediate a Starvation-Induced Transcriptional Position Effect within the Ribosomal DNA Array. Cell reports. 2016;14:1010–1017. doi: 10.1016/j.celrep.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]