Abstract
Despite the developments for faster liquid chromatographic and mass spectral detection techniques, the standard in-solution protein digestion for proteomic analyses has remained relatively unchanged. The typical in-solution trypsin protein digestion is usually the slowest part of the workflow, albeit one of the most important. The development of a highly efficient immobilized enzyme reactor (IMER) with rapid performance for on-line protein digestion would greatly decrease the analysis time involved in a proteomic workflow. Presented here is the development of a silica based IMER for on-line protein digestion, which produced rapid digestions in the presence of organic mobile phase for both model proteins and a complex sample consisting of the insoluble portion of a yeast cell lysate. Protein sequence coverage and identifications evaluated between the IMER and in-solution digestions were comparable. Overall, for a yeast cell lysate with only a 10 sec volumetric residence time on-column, the IMER identified 507 proteins while the in-solution digestion identified 490. There were no significant differences observed based on identified protein’s molecular weight or isoelectric point between the two digestion methods. Implementation of the IMER into the proteomic workflow provided similar protein identification results, automation for sample analysis, and reduced the analysis time by 15 hr.
Keywords: IMER, immobilized enzyme, trypsin, digestion, membrane proteins
Introduction
Due to the importance of proteins in metabolic pathways, their identification, structure and function are a major focus of discovery [1, 2]. Proteomics encompasses a wide field, ranging from comprehensive studies which identify proteins within a complex mixture to more targeted approaches where specific proteins can be selected via liquid chromatographic (LC) and mass spectrometric (MS) techniques [2]. Regardless of the type of study, “shotgun” approaches are the most common, where proteins are first digested into peptides prior to MS analysis. This is accomplished through the addition of an enzyme to the sample. Traditionally high quality trypsin is used for this purpose since it hydrolyzes peptide bonds fairly reproducibly on the carboxy side of a lysine or an arginine residue, producing peptides within a desirable mass range for protein sequencing [3]. The digestion of the proteins into peptides is one of the most time consuming sample preparation steps, as it is typically performed as an overnight in-solution reaction. That being said, it is also the most important step for proper proteomic analysis [3–5]. Overall, the proteomic community is continuously demanding improved methods to increase sample throughput for the workflow while maintaining high accuracy digestions.
The instrumentation and hardware involved in LC separations and MS techniques continue to improve. Fast LC separation procedures and high resolution MS detection methods are commercially readily available. In addition, multi-dimensional LC can allow for an abundance of separation space for complex proteomic samples and electrospray ionization (ESI) has allowed for easy coupling of LC to MS for rapid protein/peptide identification. Although these LC and MS techniques have been greatly improved over the past 30 years, becoming faster and more efficient, the sample treatment for enzymatic digestion has remained relatively the same [6, 7]. A small amount of trypsin is added to the protein solution to reduce trypsin autolysis, however, this results in long incubation periods for complete protein digestion. The 5–24 h in-solution trypsin digestion is typically the slowest part in the sample workflow. One method to increase the speed and efficiency of the digestion is to immobilize the trypsin onto a substrate [4, 5]. Trypsin can be covalently bonded or physically adsorbed onto silica or organic materials such as styrene-, acrylamide-, and methacrylate-based copolymers [6]. This can provide immobilized trypsin on membranes [8–11], modified silica capillaries [12–14], microchips [15, 16], gel networks [17, 18], etc. [7]. Utilizing immobilized trypsin for a protein digestion provides advantages such as a high enzyme-to-substrate ratio, shorter digestion times, less trypsin autolysis, and low reagent consumption while maintaining highly efficient digestions [4–7, 12, 19]. While the high abundance of trypsin allows for less competition for catalytic sites, the decrease in trypsin autolysis provides less complex mass spectra for analysis. Bound to a substrate, trypsin can also be easily added and removed from the sample prior to LC-MS analysis [20] and reused [21]. Overall, immobilization has been found to provide quick proteolysis of protein samples which are easily adapted into LC-MS/MS workflows [5, 7].
Many different configurations for immobilized trypsin for a variety of applications have been discussed in the literature [5]. Off-line configurations include trypsin membrane cartridges, magnetic or glass beads, agarose gel beads, spin-columns, etc. while on-line approaches tend to immobilize trypsin in either porous monolithic materials [20, 22, 23] or particle supports packed into LC columns or capillary electrophoresis (CE) setups [5, 24–26]. One promising approach to increasing the proteomic workflow is to create a flow-through enzyme reactor, where the proteins pass through the reactor and are digested into peptides immediately on-column. This idea removes many laborious manual steps involved with the off-line digestion process, potentially eliminating sources of error. Although the idea of immobilized enzyme reactors (IMERs) is not new [27], recent developments in their ease of manufacture and potential for increasing the speed of the workflow have gained in popularity. The majority of works published for IMER digestion either place the IMER before a separation method (LC or CE) followed by mass spectrometric analysis in a true on-line fashion [21, 28, 29], or use the IMER to digest peptides followed by fraction collection prior to peptide analysis [30–32]. Although many studies focus on model proteins, IMER systems need to be tested with large sample amounts and more realistic samples to determine whether they are a promising tool for proteomic studies [3, 21].
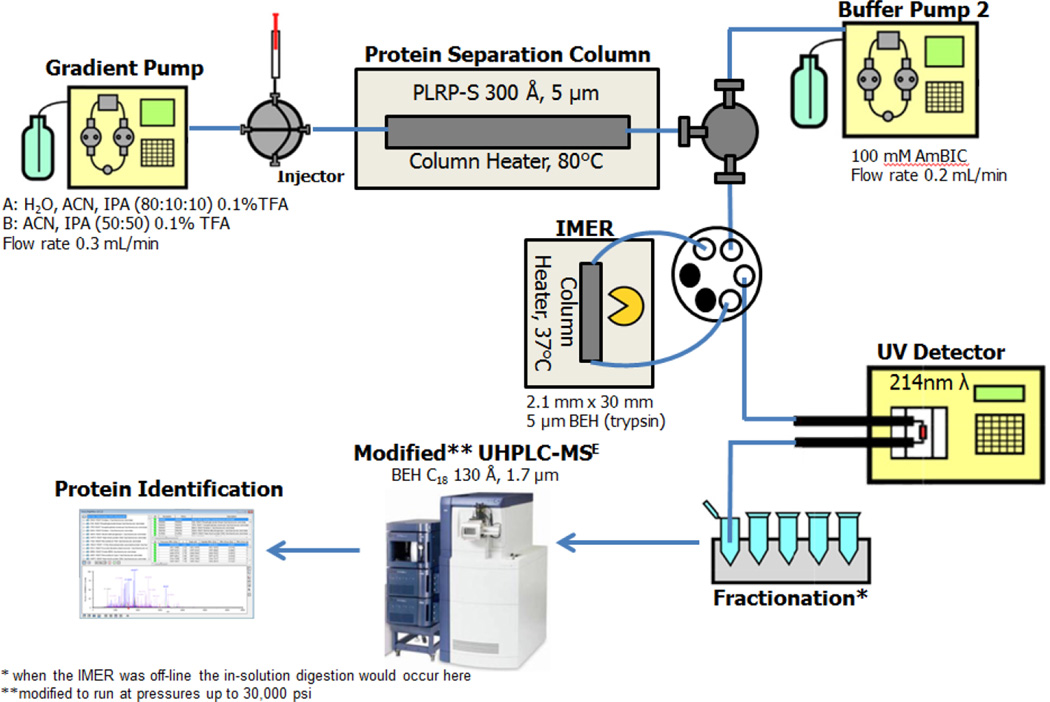
When working with a complex proteomic sample, there are many strategies for increasing the number of protein identifications. While shotgun analyses are most common [33], incorporating a multidimensional LC approach with a prefractionation step allows an increase in peak capacity [34] and protein identifications [35–38]. This is especially important when utilizing lower resolution mass spectrometers for protein identifications. For the multidimensional prefractionation scheme implemented in our lab, proteins are first separated on a first dimenison reverse phase column, fractions are collected and digested, then the resulting peptides are separated on a second dimension reverse phase column. This set up originally employed the 15 h in-solution protein digestion used by most laboratories; however, it is a prime candidate for the implementation of an IMER. Therefore, the focus of this work was to develop and characterize a highly active trypsin IMER that can be placed after the first dimension reverse phase separation column (Figure 1). This would result in the eluting proteins immediately digested into peptides and would not require the 15 h digestion step in the workflow. To our knowledge this is the first report of placing an IMER after a reverse phase separation. Although other types of separations can be utilized for the first dimension (such as the commonly utilized ion exchange chromatography), prior results from our lab reported more protein identifications when reverse phase was employed in the first dimension [39]. For the purpose of robustness, our work focused on using a silica substrate for the IMER, which has higher mechanical strength [40]. To validate IMER implementation into the workflow, it is widely accepted to compare the results from the IMER scheme to the in-solution counterpart. Both model proteins and complex yeast (Saccharomyces cerevisiae) cell lysate were digested using the IMER setup and compared to our previous method utilizing an in-solution digestion. For analysis, both protein sequence coverage and number of protein identifications will be compared to determine whether the IMER can provide an on-line digestion equivalent or superior to the traditional overnight protocol.
Figure 1.
Experimental workflow. The IMER was placed after the first dimension protein separation column in a fashion so that both experiments (on-line IMER digestion and IMER off-line with in-solution digestion) could be easily performed. Protein/peptide elution was monitored with UV detection and fractions were collected. Fractions were recombined and analyzed via a second dimension reverse phase separation with mass spectral detection.
Experimental
Immobilization Protocol
The immobilization protocol was adapted from Ahn, et al. [40]. Approximately 0.3 g of non-bonded bridged ethyl hybrid (BEH) silica particles (5 µm, 300 Å) from Waters Corporation (Milford, MA) were added to a 5 mL round bottom flask and kept under a constant flow of nitrogen. Then 110 µL of triethoxysilylbutyraldehyde (Tech-90, Gelest Inc., Morrisville, PA) in 2 mL of ethanol (Fisher Scientific, Pittsburgh, PA) was added. This was allowed to rotate for 2 hr at room temperature. The particles were rinsed with pH 9, 50 mM ammonium bicarbonate (Sigma Aldrich, St. Louis, MO) to remove excess linker and 80 mg of trypsin (Sigma Aldrich, TPCK treated, P/N T1426) in 2 mL 50 mM ammonium bicarbonate was added. The presence of a reversible trypsin inhibitor (such as benzamidine hydrochloride) during this step was deemed unnecessary, as preliminary results confirmed similar trypsin coverage and activity were obtained with or without the presence of the inhibitor. The ALD coupling solution (1 mL of 1 M NaCNBH3, Sterogene Bioseparations, Carlsbad, CA) was added and the solution was allowed to rotate for 2 hr at room temperature. Afterwards, 1 mL of 1 M ethanolamine (Sigma Aldrich) was added to quench the reaction and stirred for 30 min at room temperature. The particles were washed with 50 mM ammonium bicarbonate to remove excess trypsin and ethanolamine then stored in water with 0.08% trifluoroacetic acid (TFA, Sigma Aldrich) at 5°C. To determine activity of the immobilized trypsin, standard trypsin substrate BAEE (Nα-benzoyl-L-arginine ethyl ester hydrochloride, Sigma Aldrich, protocol EC 3.4.21.4) was used. In short, a known amount of immobilized particle slurry and BAEE solution were added to a cuvette. In the presence of trypsin BAEE is hydrolyzed to BA (Nα-benzoyl-L-arginine) and this conversion can be monitored at 253 nm. Following the published protocol, the change in absorbance was monitored for 5 min. This was compared to trypsin in solution and an estimated activity was calculated. To determine the amount of protein on the surface of the particles a BCA assay was used according to the manufacturer's instructions (Pierce™ BCA Protein Assay Kit, ThermoFisher Scientific, Waltham, MA).
Packing the Trypsin IMER
Once the particles were determined to be active, the above immobilization protocol was scaled up by a factor of 2 (0.6 g of particles) to pack into a chromatographic column. An Isco D-Series syringe pump module (Model 260D, Teledyne ISCO, Lincoln, NE) was used to pack the particles into an empty HPLC column assembly (30 mm × 2.1 mm × ¼” OD, Cat#25118, Restek, Bellefonte, PA). The syringe pump was connected to a pressure vessel which held the particles in a slurry (60 mg/mL in 50 mM ammonium bicarbonate, pH 8). The column was placed on top of the vessel with the outlet frit in place. The entire apparatus was rinsed with 50 mM ammonium bicarbonate prior to column packing. The column was packed at 3,000 psi for 30 min and the pressure was increased to 5,000 psi for 30 min to ensure proper packing for a range of HPLC pressures. When not in use the column was rinsed with water (pH adjusted to 4 with trifluoroacetic or formic acid) and stored at 5 °C. At this pH and temperature the trypsin activity can be conserved. Columns packed in this manner were used for up to 6 months without signs of degradation monitored via trypsin digestion efficiency.
Determining IMER Activity
The IMER was tested at a variety of flow rates. These flow rates were converted to a “volumetric residence time.” The volumetric residence time is defined as the column volume divided by the flow rate. Therefore it is the amount of time eluent is exposed to the IMER’s packed bed based on the calculated column volume and designated flow rate. To monitor the activity of the IMER under various conditions, BAEE was again used as the substrate. The IMER was placed before an analytical column (PLRP-S, 300 Å, 5 µm, 250 × 4.6 mm, Agilent Technologies, Santa Clara, CA) and 20 µL injections of 2.5 mM of BAEE were made. The mobile phase for the IMER was 50 mM ammonium bicarbonate, pH 8. The effluent from the IMER was collected directly onto the reverse phase PLRP-S column after passing through a valve. The valve allowed the eluting substrates of the IMER (either BAEE or BA) to preconcentrate onto the analytical column, then switched position for the subsequent BAEE/BA separation by gradient elution. Components eluted were detected via UV absorption at 253 nm. The gradient for the PLRP-S column consisted of water and acetonitrile, both with 0.1 % TFA. The gradient method is outlined in Table 1. Flow rates through the IMER from 10 µL/min up to 1 mL/min were tested, corresponding to 8 min and 5 sec volumetric residence times for digestion, respectively. For all experiments, the IMER was held at 37°C. Previously, room temperature (~22°C) and 60°C digestions were attempted with the IMER; however, higher coverage and number of digest peptides were obtained when the IMER was placed at 37°C. Therefore all experiments with the IMER presented in this study were conducted at 37°C.
Table 1.
Gradient Method on PLRP-S column for BAEE analysis and BSA elution.
| t (min) | % B |
|---|---|
| 0 | 5 |
| 5 | 5 |
| 20 | 21 |
| 25 | 85 |
| 27 | 85 |
| 30 | 5 |
| 35 | 5 |
Although the IMER appeared to produce adequate digestions, the goal of the IMER is to ultimately be placed following the analytical column (Figure 1), digesting the eluting proteins over a gradient run. Therefore, the IMER would be exposed to varying degrees of organic content combined with the 50 mM ammonium bicarbonate throughout the first dimension run. To determine the effect of organic mobile phase on the IMER’s digestion, a combination of mobile phases ranging between 5–85% B were combined in a 2:1 ratio with the 50 mM ammonium bicarbonate, pH 8. Mobile phase A consists of 80% water, 10% isopropanol, 10% acetonitrile, and 0.1% TFA while mobile phase B was 50:50 acetonitrile:isopropanol with 0.1% TFA. This was tested since it is the standard mobile phase used for elution of proteins from a yeast cell lysate within our lab. This experiment had to be performed in an off-line fashion since the resulting elutant required dilution (1:10) with water prior to injection onto the PLRP-S column for proper preconcentration of BA/BAEE.
Prefractionation workflow
The prefractionation workflow established for protein and peptide separation utilized by our lab is an off-line two dimensional system. For the first dimension, proteins are placed in 80% formic acid (MS grade, Fluka Analytical, Sigma Aldrich) and injected onto a PLRP-S (300 Å, 5 µm, 250 × 4.6 mm) polymeric reverse phase column at 80°C. The formic acid is added to aid in solubilization and avoid clogging the LC column. The sample is not allowed to sit idle for more than 2 min in formic acid to avoid protein formylation (+28 Da) [41] prior to separation. The IMER was implemented after the PLRP-S column as depicted in Figure 1. To accommodate the IMER, a buffer pump (Waters 600E pump consisting of 100mM ammonium bicarbonate pH 8) was added to preserve the IMER at pH ~7–8 during the entire gradient run. As depicted in Figure 1, the buffer pump was added after the PLRP-S column and prior to the trypsin column. Therefore a combination of flow rates from the gradient pump (HP 1050) and the buffer pump control the volumetric residence time through the IMER. The IMER itself was connected in a way that it could be easily taken off-line with the presence of a valve. This made the comparison with and without the IMER easier and allowed the IMER to avoid exposure to the formic acid used to solubilize the sample onto the PLRP-S column. After the IMER, the eluting peptides were collected into fractions. When the IMER was taken off-line with the control valve then the eluting proteins were collected into fractions. The progress of peptide/protein elution was monitored via UV detection (Waters CapLC2487) at 214 nm and 1 mL fractions were collected. Once the fractions were collected they were flash frozen and lyophilized to concentrate the sample and remove MS-incompatible solvents. These fractions were recombined post lyophilization to allow for a more manageable number of samples for the second dimension analysis. When the IMER was taken off-line and proteins were fractionated, the resulting recombined fractions were digested in-solution overnight following the standard digestion protocol [42] for our lab utilizing free (unbound) trypsin.
For the second dimension, all resulting peptides were analyzed and identified using a modified UHPLC coupled to MS [43]. In this modified high pressure separation system, the LC gradient and sample are placed into a gradient storage loop using a commercial nanoAcquity™ UPLC (Waters Corporation). The ultra-high pressure (30,000 psi) is achieved by pushing the gradient and sample from the loop using a pneumatic amplifier (DSHF-300, Haskel International LLC, Burbank, CA). The peptides were separated on a 75 µm I.D. × 87 cm (manufactured in-house) capillary column packed with 1.7 µm 130 Å BEH C18 particles (Waters Corporation). The column was held at 65°C and the eluting peptides were detected using a QToFPremier™ mass spectrometer (Waters Corp). Mobile phases of the second dimension consisted of water and acetonitrile with 0.1 % formic acid (optima grade, Fisher Scientific) and a gradient of 4 – 40% organic was run over 110 min. The column was connected to a silica nanospray emitter with a 20 µm I.D. and a 10 µm tip (New Objective, Woburn, MA). The mass spectrometer operated in MSE mode [44] performing parent ion scans from 50 to 1990 m/z over 0.6 sec at 5.0 V. The collision energy ramped from 15 to 40 V over 0.6 sec. Data was collected using MassLynx V4.1 SCN 872 and processed via ProteinLynx Global Server 2.5 (PLGS, Waters Corporation) set to a 4% false positive rate with a 1 × randomized yeast proteome database obtained from Uni-Prot protein knowledgebase (www.uniprot.org) on 2/3/2011. If the same proteins were identified in multiple fractions within a sample set, only the fraction with the highest identification score was counted. Therefore, a protein was only counted once despite whether the protein was split into multiple fractions during the first dimension separation. Each fraction was processed 3 times in the second dimension and a protein was required to be identified in at least 2 of the 3 replicate runs to be counted.
Analysis of model proteins
First a solution of bovine serum albumin (BSA, Sigma Aldrich, P/N A0281) was reduced and alkylated following the standard protocol [42]. One hundred microliters of a 0.070 µg/µL solution (~1 µM) was injected onto the PLRP-S column followed by on-line IMER digestion. The gradient is outlined in Table 1 and consisted of water and acetonitrile with 0.1% TFA. The flow rate was varied to test a range of analyte residence times (5 sec to 8 min) corresponding to trypsin exposure but the ratio of the gradient mobile phase flow rate to buffer flow rate was kept consistent at 2:1 (gradient:buffer) to guarantee the IMER remained at pH ~8 despite the desired volumetric residence time on column. Fractions were collected over the UV peak (at 214 nm) observed for BSA, lyophilized, and recombined into one sample for UHPLC analysis. For comparison, the same amount of BSA was injected onto the PLRP-S column without the IMER present (off-line) following the same procedure. Fractions containing the BSA peak were lyophilized, recombined, and digested in-solution following the standard digestion protocol [42]. Both samples, collected with and without the IMER, were processed on the previously described UHPLC system.
To determine the actual protein residence time and the effect of protein concentration on the IMER, model proteins BSA, myoglobin, cytochrome c, and RNAaseA (Sigma Aldrich, P/N A0281, M-1882, C-2037, and R5500, respectively) were injected directly onto the IMER (at 37°C) at a flow rate of 0.5 mL/min and eluting peptides analyzed via MS. Eluting peptides were collected and analyzed using an XBridge™ BEH (300 Å, C18, 3.5 µm, 4.6 × 50 mm) coupled to a standard flow ESI on the QTofPremier™ with the same mass spectrometer settings as previously described. The gradient for this analysis began at 4% acetonitrile and ramped to 50% acetonitrile in fifteen minutes. This was followed by a wash at 15.5 min of 80% acetonitrile which held for half a minute before re-equilibration of the column at 4% acetonitrile for 17–20 min. Concentrations from 10 nM to 1 µM were analyzed for BSA and myoglobin at various flow rates and in the presence of organic mobile phase during trypsin digestion.
Analysis of a yeast cell lysate
Saccharomyces cerevisiae (strain BY4741) was grown on glucose media until stationary phase was achieved (O.D. > 2). Cells were washed, resuspended in 50 mM ammonium bicarbonate pH 8, and protease inhibitors (Pierce Protease Inhibitor Mini Tablets, P/N 88665) were added and prepared to the manufacturer’s recommendations. Pressure assisted cell lysis utilized a French press cell, where the entire 25 mL sample was passed dropwise at 20,000 psi 3 times through the cell. The cell itself was chilled (4°C) and elutant was kept on ice throughout the process. The sample was centrifuged at ~1,200 g for 10 min at 4°C to remove unbroken cells (Beckman, L8–70 Ultracentrifuge). The supernatant was isolated and underwent ultracentrifugation at ~120,000 g (38,000 rpm Beckman 60Ti rotor) for 90 min at 4°C twice before the pellets (insoluble portions) were collected. To determine the amount of protein present a Bradford assay [45] (Coomassie Protein Assay Kit, Thermo Scientific) was performed once the pelleted proteins were resuspended in 50 mM ammonium bicarbonate buffer pH 8. Two milliliters of insoluble yeast cell lysate were removed (~1.5 mg of protein) and diluted to perform the reduction and aklyation step according to protocol [42]. Once the entire sample was reduced and alkylated, 550 µL of protein solution (~ 1.2 mg protein) was injected onto the prefractionation setup (Figure 1) with and without the IMER on-line. Mobile phase A consisted of 80% water, 10% isopropanol, 10% acetonitrile, and 0.1% TFA. Mobile phase B consisted of 50% isopropanol, 50% acetonitrile, and 0.1% TFA. Beginning at 0% B, the gradient ramped to 60% B in 100 min followed by a further increase to 90% at 110 min where it held for 10 min before returning and stabilizing at 0% B. The total run time was 150 min, allowing for formic acid elution for the first 10 min prior to gradient start time and ending with 20 min column re-equilibration. During the gradient, 100mM of ammonium bicarbonate, pH 8, was pumped into the fluidics after the PLRP-S column and prior to the trypsin column. The flow rate of the pump controlling the mobile phase gradient was 0.3 mL/min and the buffer pump was set to 0.2 mL/min. This resulted in a flow rate of 0.5 mL/min through the IMER (at 37°C) for a volumetric residence time for digestion of ~10 sec. Two minute wide (1 mL) fractions for each setup (with and without the IMER on-line) were collected, lyophilized, and recombined into 15 equivalent time fractions. Although 15 fractions were recombined, only fractions 1–9 were ultimately compared due to the lack of protein present in fractions 10–15. Once the fractions were recombined, the on-line IMER digested fractions were immediately ready for UHPLC peptide separation and protein identification. However, the 15 fractions collected without the IMER present were digested (with trypsin 1:30) in-solution according to protocol [42] with free (unbound) trypsin. Both fraction sets, collected with and without the IMER, were processed on the previously described UHPLC system.
Results and Discussion
Immobilization of trypsin
After the trypsin was immobilized onto the 5 µm BEH particles, 100 µL of slurry (consisting of 60 mg of trypsin immobilized particles in 10 milliliters of water with 0.08% TFA) was reacted with BAEE in solution and the absorbance monitored over 5 min at 253 nm. The slope of the change in absorbance per minute was used to calculate an activity of ~350,000 BAEE units/gram of particle. Since activity was confirmed, a larger batch of immobilized trypsin particles (0.6 g of particles) were produced and packed into a 30 mm × 2.1 mm empty column resulting in the IMER. Depending on the concentration of particles within an IMER, (assuming ~0.2 g per column) the resulting IMER would have an activity of ~70,000 BAEE units/column. In reality, however, the activity is potentially greater than this value since the interaction of a substrate with trypsin in a packed column would be greater than with trypsin immobilized particles a slurry (such as with the BAEE assay). The amount of total trypsin present within an IMER, determined by the BCA assay, was ~15 mg trypsin per column. Although this assay determined the total trypsin present, the amount of active trypsin (determined by the BAEE assay) is essential to the performance of the column itself.
IMER characterization
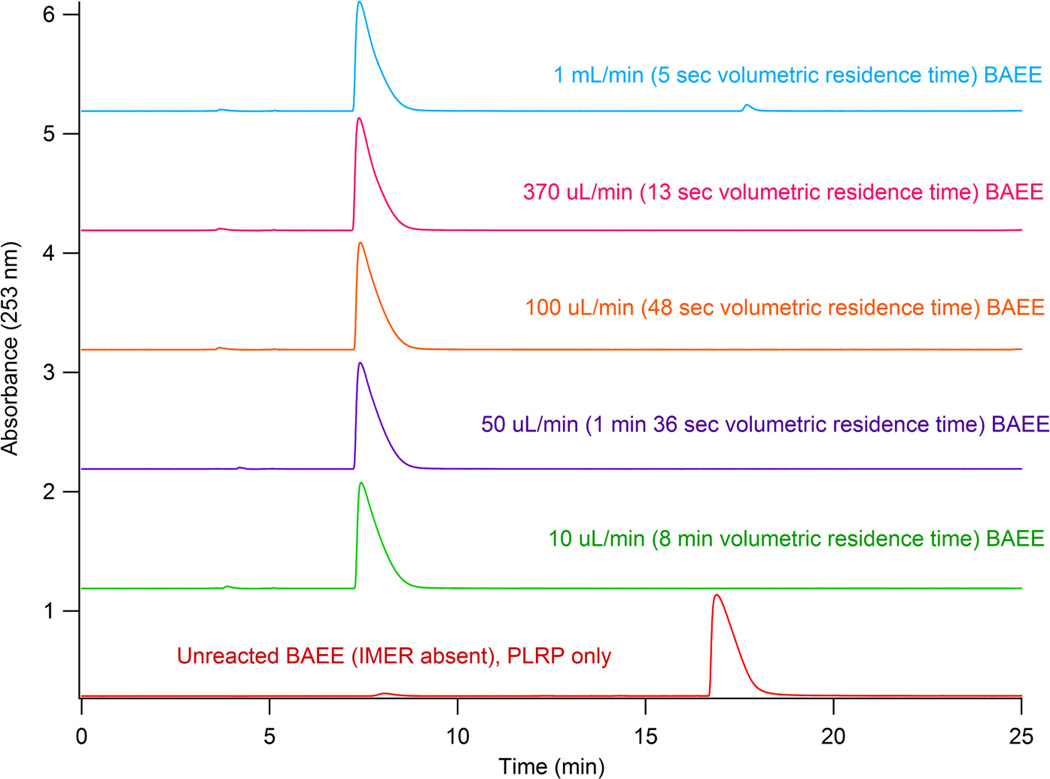
Once the IMER was packed it was initially tested with BAEE at multiple flow rates to test digestion speed. This was performed by placing the IMER before the PLRP-S analytical column so the reverse phase column could separate BA from BAEE. Between the IMER and the PLRP-S column a valve was installed to direct the effluent from the IMER, switching the analytes quickly and reproducibly onto the PLRP-S column. Buffer was allowed to flow through the IMER from 10 µL/min to 1 mL/min corresponding to a volumetric residence time for digestion between 8 min and 5 sec respectively. As shown in Figure 2, only the fastest speed tested (1 mL/min) resulted in a small amount of BAEE present. For all other digestion speeds tested no BAEE was observed.
Figure 2.
BAEE injected onto the IMER with variable buffer flow rates. The effulent of the IMER was collected onto a loop and analyzed for the presence of BA by separation on the PLRP-S column and UV detection. As shown, only a small amount of BAEE was observed at 1 mL/min.
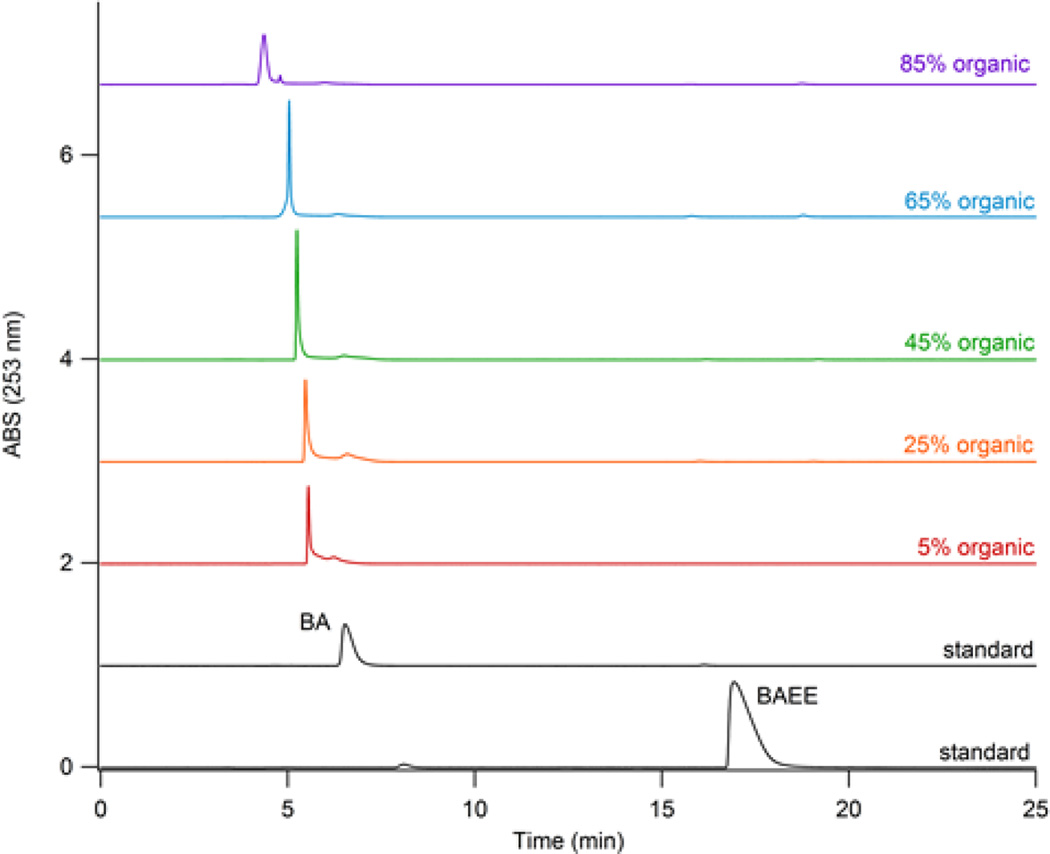
The ultimate objective for the workflow configuration (Figure 1) would implement the IMER after the protein separation column and it would experience the gradient as the proteins elute. This means that the digestion efficiency is required to remain constant despite the presence of organic solvents. To test the effect of organic solvents on the IMER digestion, varied amounts of mobile phase (5–85% B) were combined with the buffer flowing through the IMER at a flow rate of 1 mL/min. The resulting effluent was collected in a vial and underwent a 1:10 dilution. This was required for preconcentration of the sample onto the PLRP-S column for BA/BAEE analysis. As shown in Figure 3, no unreacted BAEE was observed regardless of the level of organic solvent present in the buffered mobile phase. Peak placement for BA and BAEE was confirmed with standards prepared in various organic mobile phase compositions. Despite the level of organic, BAEE remained at ~17 min, which supports that the results from the IMER (Figure 3) successfully converted BAEE to BA despite the level of organic mobile phase present.
Figure 3.
Comparison of organic content on IMER digestion efficiency. BAEE elutes ~ 17 min and BA ~ 6 min. The volumetric residence time for all IMER BAEE digestions was 1 mL/min.
Model protein results
Initial protein tests were performed using BSA as a model protein. The workflow outlined in Figure 1 was implemented and 100 nM BSA was injected (200 µL) with and without the IMER on-line. The fractions collected were combined into one sample and analyzed via the UHPLC. Flow rates of 10 µL/min (8 min IMER volumetric residence time), 100 µL/min (48 sec IMER volumetric residence time), 0.5 mL/min (10 sec IMER volumetric residence time), and 1 mL/min (5 sec IMER volumetric residence time) were tested. As shown in Table 2, high protein sequence coverage and number of digest peptides was obtained with on-line IMER digestion, especially considering the reduction in digestion time compared to in-solution. For example, a protein sequence coverage of 79% was obtained with an 8 minute volumetric residence time on column compared to a lengthy 15 hr in-solution digestion, which resulted in a coverage of 85%.
Table 2.
Analysis of 100 nM BSA on pre-fractionation workflow.
| Flow Rate | Volumetric Residence Time |
Coverage (%) |
Digest Peptides |
Trypsin Autolysis Digest Peptides |
Missed Cleavages |
|---|---|---|---|---|---|
| 10 µL/min | 8 min | 79% | 52 | 11 | 10 |
| 100 µL/min | 48 sec | 76% | 54 | 10 | 8 |
| 0.5 mL/min | 10 sec | 66% | 38 | 5 | 19 |
| 1 mL/min | 5 sec | 53% | 27 | 4 | 3 |
| In-solution | 15 hr | 85% | 72 | 14 | 33 (± 6) |
Since missed cleavages have been reported to be common with accelerated trypsin digestion via the use of immobilized enzyme [7], this was also investigated. Triplicate in-solution 15 hr digestions of BSA resulted in 33 (± 6) missed cleavages. With the flow rates discussed above, individual IMER digestions at 10 µL/min (8 min IMER volumetric residence time) had 10 missed cleavages, the 100 µL/min (48 sec IMER volumetric residence time) had 8, the 0.5 mL/min (10 sec IMER volumetric residence time) had 19, and the 1 mL/min (5 sec IMER volumetric residence time) had 3. Therefore, by utilizing the trypsin IMER a decrease in missed cleavages was observed for the flow rates tested compared to the in-solution digestion. Also, all three flow rates had a reduced number of trypsin autolysis peptides (Table 2).
In order to track the actual residence time of the model proteins on the IMER, four standard proteins (BSA, RNAase A, cytochrome C, and myoglobin) were individually injected onto the IMER and five 1-minute wide fractions were collected. These fractions were each injected onto a peptide separation column (Xbridge™, 4.6 × 50 mm, 3.5 µm, 300 Å BEH, Waters Corp.) with the gradient outlined in Table 1. When injected, the three smaller proteins (myoglobin, cytochrome c, and RNAase A) eluted from the column completely in 2 min. While the majority of BSA had eluted within this same time frame it was identified in subsequent fractions at a fraction of the intensity. The volumetric residence time within the IMER column (based on the flow rate and column volume) was 10 sec for 0.5 mL/min. However, it is not surprising that proteins are delayed due to their extensive interaction with trypsin on the column.
The concentration of the protein injected influences the amount of residence time delay on the IMER and the number of digest peptides/protein sequence coverage observed. When a large amount of protein is injected onto the IMER, the longer the elution time and the higher the number of digest peptides and protein sequence coverage. Therefore a balance is required to achieve high coverage while avoiding overloading the column and spreading proteins into multiple fractions. To investigate this further, model proteins were injected directly onto the IMER under a variety of flow rates and mobile phase conditions, and peptides collected directly onto a RPLC column (XBridge™). Concentrations as low as 10 nM were detected post-IMER injection for model proteins for 200 µL on-column injections. It is possible, with a more sensitive mass spectrometer, less than 10 nM concentrations could be detected. As the proteins approached >1 µM, longer elution times were required to remove the protein from the IMER. However, 1 µM injections provided high protein sequence coverage (> 75%) and excellent reproducibility for both protein sequence coverage and number of digest peptides without carryover effects. For identical injections (same injected concentration, flow rate, and organic content in the IMER’s mobile phase) the variation in number of digest peptides identified and protein sequence coverage between IMER digestions were ± 5 and ± 15%, respectively.
Similar to in-solution digestions, when longer exposure times result in higher coverage/digest peptides, the flow rate through the IMER could be simply reduced (increasing trypsin exposure time) allowing for more protein to be digested. Unlike the prior BAEE results, when organic content was present during the IMER digestion, the number of digest peptides and protein sequence coverage for model proteins did decrease slightly. Overall, the IMER produced rapid digestions while retaining high protein sequence coverage compared with literature studies [4, 13–15, 21–23, 30, 31], where typical digestion residence times are between 1.5 – 30 min. When shorter digestion times (< 1 min) were reported in the literature [18, 20, 24, 29, 30], the resulting protein sequence coverage was relatively low (30 – 55% for BSA) compared to the IMER presented in this study (53–79% for BSA depending on flow rate and concentration).
Reproducibility
Although only a single IMER was used for the comparison of the insoluble portion of the yeast cell lysate, three IMERs total were packed and tested with BAEE and model protein BSA. All three IMERs developed according to the proposed protocol completely hydrolyzed BAEE to BE for flow rates up to 1 mL/min and produced similar percent coverage (75–85%) for model protein BSA. Analysis of the slurry concentrations for all three immobilization batches resulted in 60–70,000 BAEE units/column and 10–15 mg trypsin/column.
Yeast cell lysate comparison
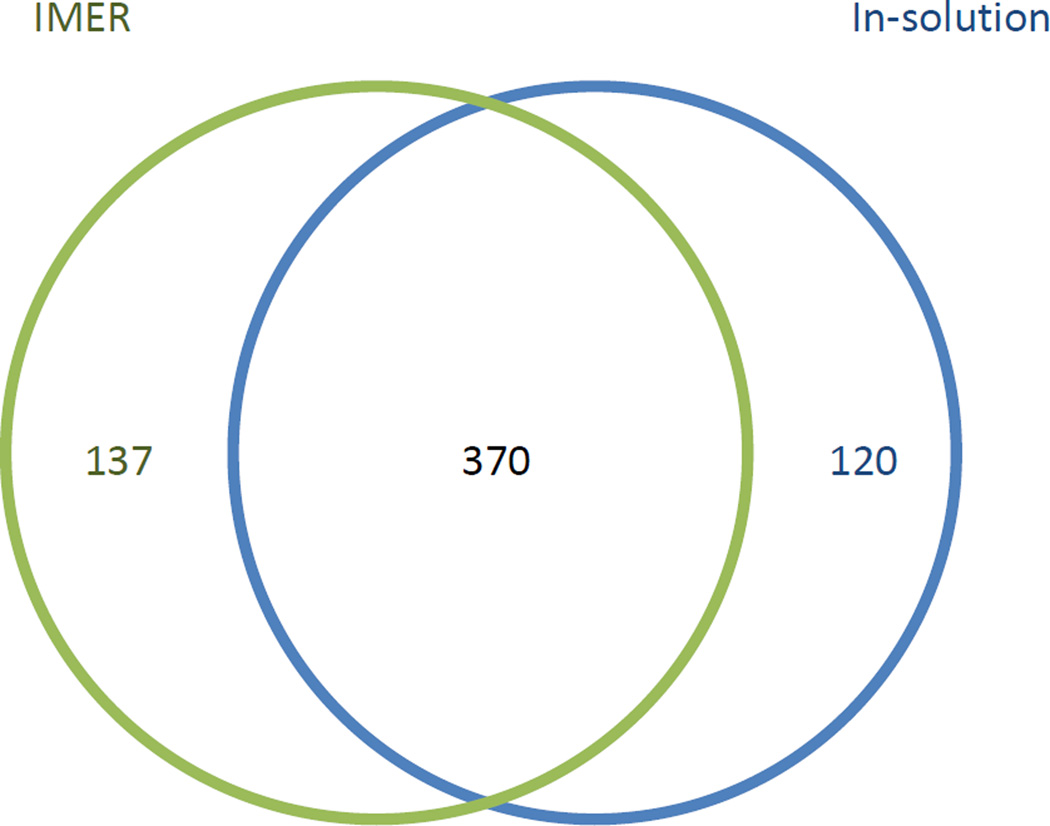
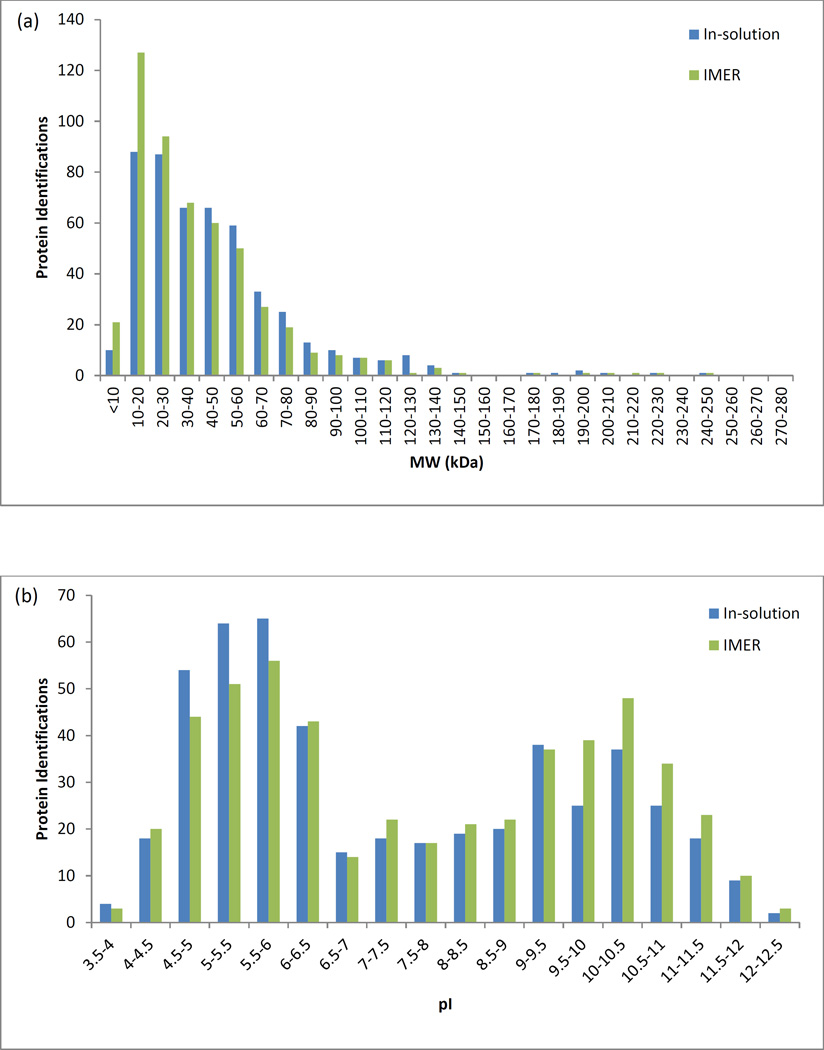
There are few reports in the literature of complex sample analysis (such as cell lysates) via on-line IMER digestion [13, 21, 26, 32]. Therefore, insoluble portion of a yeast cell lysate was processed using the workflow outlined in Figure 1. This was performed both with and without the IMER on-line. The process was monitored by UV and fractions were combined into 15 equal time fractions. These fractions were then processed in the second dimension using the UHPLC system. It was quickly determined that no proteins were present after fraction 9, so only fractions 1–9 were analyzed for both the IMER and in-solution digestion experiments. As shown in Figure 4, both experiments had roughly the same number of protein identifications. With the IMER on-line, the system identified 507 proteins. With the IMER removed, and applying in-solution digestion instead, the system identified 490 proteins. Therefore, both types of digestion produced similar results. It is important to note that if a more advanced mass spectrometer could be utilized, higher protein identifications can be expected. The similarities are more apparent when the protein identifications are plotted according to molecular weight (Figure 5a) and isoelectric point (Figure 5b). As shown in Figure 5, there was no serious bias for digestion method based on molecular weight or isoelectric point. Possibly the IMER resulted in more identifications of lower molecular weight proteins, but this parameter could be easily tuned based on the particle pore size. Utilizing particle substrates with larger pores could better accommodate larger molecular weight proteins.
Figure 4.
Venn diagram comparing the proteins identified with (green) and without (blue) the IMER on-line. Sample consisted of an insoluble portion of yeast (Saccharomyces cerevisiae) cell lysate.
Figure 5.
Proteins identified by both the IMER (green) and in-solution (blue) digestions plotted (a) by molecular weight and (b) by isoelectric point. Sample consisted of an insoluble portion of yeast (Saccharomyces cerevisiae) cell lysate.
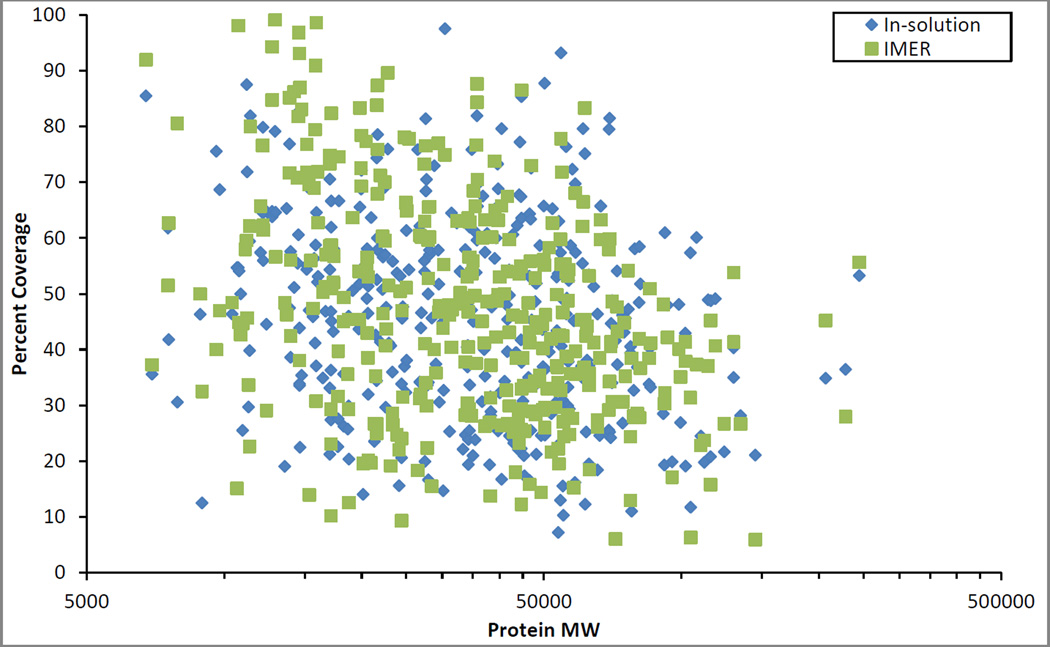
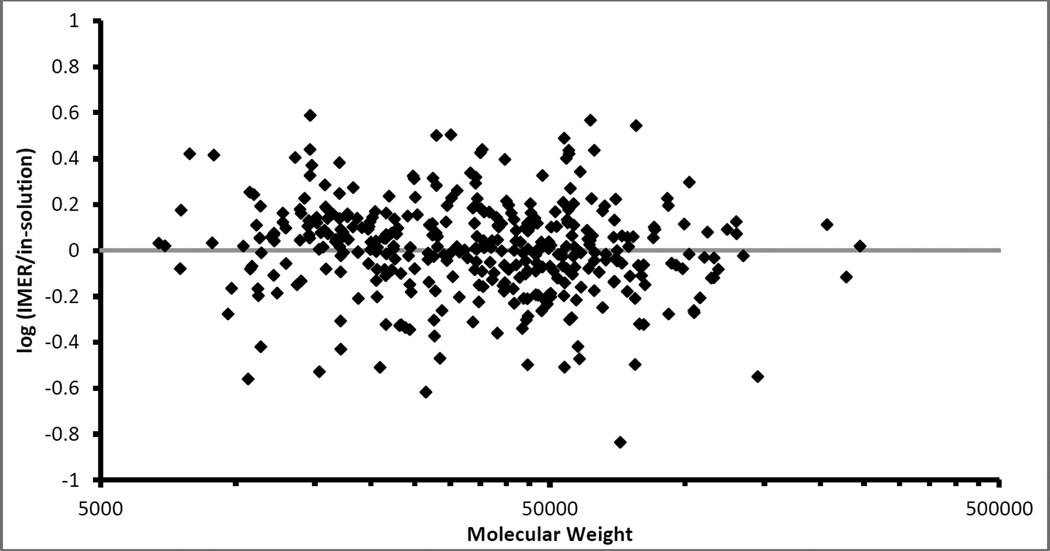
Examining the 370 proteins identified in both the IMER and in-solution digestion, protein sequence coverage was compared (Figures 6 and 7). In Figure 6, the proteins are plotted based on molecular weight versus protein sequence coverage. Overall, the IMER and the in-solution digestion had relatively similar coverage and this is further reflected in the average and median protein sequence coverage values. The IMER had an average protein sequence coverage of 47.5% and a median of 45.4% while the in-solution digestion had an average protein sequence coverage of 46.2% and a median of 46.4%. Taking the log of the ratio of the individual protein sequence coverage values for each of the 370 proteins identified in both methods (Figure 7), further displayed the similarity between the digestion methods. As shown in Figure 7, points appear evenly distributed above and below the axis, suggesting similar protein sequence coverage values were obtained for the IMER and in-solution digestions. Summarizing Figure 7, the in-solution digestion had 169 negative values where it produced higher coverage of the proteins. The IMER had 200 positive values where it produced higher coverage and there was one instance where the coverage was identical resulting in a value of zero. The IMER digestion presented in this study was faster than previous reports for on-line yeast cell lysate digestion [21, 46, 47] and identified approximately the same number of yeast proteins (541 reported in Feng, et al. [21] compared to 507 found in this study). It is important to note that the sample used in this study was the insoluble portion of a yeast cell lysate, containing approximately 60% proteins that are identified as membrane-associated. Additionally, the IMER manufactured for this study was used for digestions over a period of 6 months without signs of degradation or decrease in digestion efficiency.
Figure 6.
The protein sequence coverage for the 370 proteins identified by both the IMER (green) and in-solution (blue) digestions plotted against molecular weight. Sample consisted of an insoluble portion of yeast (Saccharomyces cerevisiae) cell lysate.
Figure 7.
The ratio of protein sequence coverage found in the 370 proteins identified by both the IMER and in-solution digestions. Sample consisted of an insoluble portion of yeast (Saccharomyces cerevisiae) cell lysate. Since the log(% coverage IMER/% coverage in-solution), the positive values indicate the IMER digestion had a higher protein sequence coverage than the in-solution digestion and negative values indicate the in-solution protein sequence coverage was higher.
Conclusions
The development of a highly efficient IMER with rapid performance for an on-line protein digestion was successful. Initially the IMER demonstrated promise through the hydrolysis of substrate BAEE for flow rates up to 1 mL/min (5 sec IMER volumetric residence time) and in the presence of high organic (85%) mobile phase. Initial studies with BSA provided protein sequence coverage that were comparable to in-solution trypsin digestion. The final comparison utilized a complex sample consisting of the insoluble portion of a yeast cell lysate. With only a 10 sec IMER volumetric residence time, the IMER identified 507 proteins while the in-solution digestion identified 490. There were no significant differences observed based on protein molecular weight or pI between the two digestion methods. In addition, the protein sequence coverage achieved using both methods were essentially equivalent. Therefore, the implementation of the IMER into the proteomic workflow provided similar protein identification results, automation for sample analysis, and reduced the analysis time by 15 hr. Future directions include fully implementing the IMER into a two dimensional on-line workflow. In this new workflow, the eluting peptides from the IMER will flow directly onto a second dimension LC column for immediate MS analysis. This will eliminate the use of fractionation, lyophilization, and further decrease analysis time.
Highlights.
Development of a highly efficient IMER for multidimensional proteomics.
IMER decreased analysis time without sacrificing digestion efficiency.
First known description of a RPLC→IMER→RPLC proteomic workflow.
IMER identified an equivalent number of proteins to the in-solution digestion.
IMER digested with high efficiency in the presence of organic mobile phase.
Acknowledgments
The authors would like thank Waters Corporation for their support, specifically Martin Gilar, Beatrice Muriithi, and Oksana Tchoul. This work was supported by the National Institute of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants #5U24DK097153 and #1R01DK101473-01A1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson N, Anderson N. Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P. Proteomics retrenches. Nat. Biotechnol. 2010;28:665–670. doi: 10.1038/nbt0710-665. [DOI] [PubMed] [Google Scholar]
- 3.Hustoft H, Malerod H, Wilson S, Reubsaet L, Lundanes E, Greibrokk T. A critical review of trypsin digestion for LC-MS based proteomics. In: Leung H, editor. Integrative Proteomics. 2012. pp. 73–92. InTech: On-line. [Google Scholar]
- 4.Freije J, Mulder P, Werkman W, Rieux L, Niederlander H, Verpoorte E, Bischoff R. Chemically modified, immobilized trypsin reactor with improved digestion efficiency. J. Proteome Res. 2005;4:1805–1813. doi: 10.1021/pr050142y. [DOI] [PubMed] [Google Scholar]
- 5.Ma J, Zhang L, Liang Z, Shan Y, Zhang Y. Immobilized enzyme reactors in proteomics. Trends Anal. Chem. 2011;30:691–702. [Google Scholar]
- 6.Ma J, Zhang L, Liang Z, Zhang W, Zhang Y. Recent advances in immobilized enzymatic reactors and their applications in proteome analysis. Anal. Chim. Acta. 2009;632:1–8. doi: 10.1016/j.aca.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 7.Regnier F, Kim J. Accelerating trypsin digestion: The immobilized enzyme reactor. Bioanalysis. 2014;6:2685–2698. doi: 10.4155/bio.14.216. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, Xu J, Locascio L, Lee C. Integrated microfluidic system enabling protein digestion, peptide separation, and protein identification. Anal. Chem. 2001;73:2648–2655. doi: 10.1021/ac001126h. [DOI] [PubMed] [Google Scholar]
- 9.Cooper J, Lee C. Integrated and ultrasensitive gel protein identification. Anal. Chem. 2004;76:2196–2202. doi: 10.1021/ac035318z. [DOI] [PubMed] [Google Scholar]
- 10.Cooper J, Chen J, Li Y, Lee C. Membrane-based nanoscale proteolytic reactor enabling protein digestion, peptide separation, and protein identification using mass spectrometry. Anal. Chem. 2003;75:1067–1074. doi: 10.1021/ac025768b. [DOI] [PubMed] [Google Scholar]
- 11.Luckarift H, Johnson G, Spain J. Silica-immobilized enzyme reactors. J. Liq. Chromatogr. RT. 2008;31:1568–1592. doi: 10.1016/j.jchromb.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Foo H, Smith N, Stanley S. Fabrication of an on-line enzyme micro-reactor coupled to liquid chromatography-tandem mass spectrometry for the digestion of recombinant human erythropoietin. Talanta. 2015;135:18–22. doi: 10.1016/j.talanta.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 13.Hustoft H, Vehus T, Brandtzaeg O, Krauss S, Greibrokk T, Wilson S, Lundanes E. Open tubular lab-on-column/mass spectrometry for targeted proteomics of nanogram sample amounts. PLOS One. 2014;9:1–10. doi: 10.1371/journal.pone.0106881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hustoft H, Brandtzaeg O, Rogeberg M, Misaghian D, Torsetnes S, Greibrokk T, Reubsaet L, Wilson S, Lundanes E. Integrated enzyme reactor and high resolving chromatography in "sub-chip" dimensions for sensitive protein mass spectrometry. Scientific Reports. 2013;3:1–7. doi: 10.1038/srep03511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Oleschuk R, Ouchen F, Li J, Thibault P, Harrison D. Integration of immobilized trypsin bead beds for protein digestion within a microfluidic chip incorporating capillary electrophoresis separations and an electrospray mass spectrometry interface. Rapid Commun. Mass SP. 2000;14:1377–1383. doi: 10.1002/1097-0231(20000815)14:15<1377::AID-RCM31>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Ekstrom S, Onnerfjord P, Nilsson J, Bengtsson M, Laurell T, Marko-Varga G. Integrated microanalytical technology enabling rapid and automated protein identification. Anal. Chem. 2000;72:286–293. doi: 10.1021/ac990731l. [DOI] [PubMed] [Google Scholar]
- 17.Kato M, Inuzuka K, Sakai-Kato K, Toyo'oka T. Monolithic bioreactor immobilizing trypsin for high-throughput analysis. Anal. Chem. 2005;77:1813–1818. doi: 10.1021/ac048388u. [DOI] [PubMed] [Google Scholar]
- 18.Qu H, Wang H, Huang Y, Zhong W, Lu H, Kong J, Yang P, Liu B. Stable microstructured network for protein patterning on a plastic microfluidic channel: strategy and characterization of on-chip enzyme microreactors. Anal. Chem. 2004;76:6426–6433. doi: 10.1021/ac049466g. [DOI] [PubMed] [Google Scholar]
- 19.Capelo J, Carreira R, Diniz M, Fernandes L, Galesio M, Lodeiro C, Santos H, Vale G. Overview on modern approaches to speed up protein identification workflows relying on enzymatic cleavage and mass spectrometry-based techniques. Anal. Chim. Acta. 2009;650:151–159. doi: 10.1016/j.aca.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Duan J, Sun L, Liang Z, Zhang J, Wang H, Zhang L, Zhang W, Zhang Y. Rapid protein digestion and identification using monolithic enzymatic microreactor coupled with nano-liquid chromatography-electrospray ionization mass spectrometry. J. Chrom. A. 2006;1106:165–174. doi: 10.1016/j.chroma.2005.11.102. [DOI] [PubMed] [Google Scholar]
- 21.Feng S, Ye M, Jiang X, Jin W, Zou H. Coupling the immobilized trypsin microreactor of monolithic capillary with µRPLC-MS/MS for shotgun proteome analysis. J. Proteome. Res. 2006;5:422–428. doi: 10.1021/pr0502727. [DOI] [PubMed] [Google Scholar]
- 22.Hwang H, Cho K, Kim J, Kim Y, Oh H. Protein analysis using a combination of an on-line monolithic trypsin immobilized enzyme reactor and collisionally-activated dissociation/electron transfer dissociation dual tandem mass spectrometry. B. Kor. Chem.. Soc. 2012;33:3233–3240. [Google Scholar]
- 23.Krenkova J, Lacher N, Svec F. Highly efficient enzyme reactors containing trypsin and endoproteinase LysC immobilized on porous polymer monolith coupled to MS suitable for analysis of antibodies. Anal. Chem. 2009;81:2004–2012. doi: 10.1021/ac8026564. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Zhang B, Zhang Q, Shi Y, Guo L, Yang L. Capillary electrophoresis-based immobilized enzyme reactor using particle-packing technique. J. Chrom. A. 2014;1352:80–86. doi: 10.1016/j.chroma.2014.05.058. [DOI] [PubMed] [Google Scholar]
- 25.Slysz G, Lewis D, Schriemer D. Detection and identification of sub-nanogram levels of protein in a nanoLC-trypsin-MS system. J. Proteome. Res. 2006;5:1959–1966. doi: 10.1021/pr060142d. [DOI] [PubMed] [Google Scholar]
- 26.Xia S, Tao D, Yuan H, Zhou Y, Liang Z, Zhang L, Zhang Y. Nano-flow multidimensional liquid chromatography platform integrated with combination of protein and peptide separation for proteome analysis. J. Sep. Sci. 2012;35:1764–1770. doi: 10.1002/jssc.201200052. [DOI] [PubMed] [Google Scholar]
- 27.Lilly M, Dunnill P. Immobilized-enzyme reactors. Methods in Enzymology. 1976;44:717–738. doi: 10.1016/s0076-6879(76)44051-x. Elsevier Science & Technology: On-line. [DOI] [PubMed] [Google Scholar]
- 28.Yuan H, Zhang L, Hou C, Zhu G, Tao D, Liang Z, Zhang Y. Integrated platform for proteome analysis with combination of protein and peptide separation via on-line digestion. Anal. Chem. 2009;81:8708–8714. doi: 10.1021/ac900310y. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Yuan H, Zhao B, Zhou Y, Jiang H, Zhang L, Liang Z, Zhang Y. Integrated platform with a combination of on-line digestion and 18O labeling for proteome quantification via an immobilized trypsin microreactor. Analyst. 2015;15:5227–5234. doi: 10.1039/c5an00887e. [DOI] [PubMed] [Google Scholar]
- 30.Spross J, Sinz A. A capillary monolithic trypsin reactor for efficient protein digestion in on-line and offline coupling to ESI and MALDI mass spectrometry. Anal. Chem. 2010;82:1434–1443. doi: 10.1021/ac9025362. [DOI] [PubMed] [Google Scholar]
- 31.Ruan G, Wei M, Chen Z, Su R, Du F, Zheng Y. Novel regenerative large-volume immobilized enzyme reactor: preparation, characterization and application. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014;967:13–20. doi: 10.1016/j.jchromb.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Rivera-Burgos D, Regnier F. Native protein proteolysis in an immobilized enzyme reactor as a function of temperature. Anal. Chem. 2012;84:7021–7028. doi: 10.1021/ac301114m. [DOI] [PubMed] [Google Scholar]
- 33.Vaisar T. Proteomic analysis of lipid-protein complexes. J. Lipid. Res. 2009;50:781–786. doi: 10.1194/jlr.R900005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malerod H, Lundanes E, Greibrokk T. Recent advances in on-line multidimensional liquid chromatography. Anal. Methods. 2010;2:110–122. [Google Scholar]
- 35.Stobaugh J, Fague K, Jorgenson J. Prefractionation of intact proteins by reversed-phase and anion-exchange chromatography for the differential proteomic analysis of Saccharomyces cerevisiae. J. Proteome. Res. 2013;12:626–636. doi: 10.1021/pr300701x. [DOI] [PubMed] [Google Scholar]
- 36.Puangpila C, Mayadunne E, El Rassi Z. Liquid phase based separation systems for depletion, prefractionation, and enrichment of proteins in biological fluids and matrices for in-depth proteomics analysis - an update covering the period 2011–2014. Electrophoresis. 2015;36:238–252. doi: 10.1002/elps.201400434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan C, Reardon K, Wright P. Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics. 2005;5:2468–2478. doi: 10.1002/pmic.200401266. [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald A, Walsh B. New method for prefractionation of plasma for proteomic analysis. Electrophoresis. 2010;31:3580–3585. doi: 10.1002/elps.201000298. [DOI] [PubMed] [Google Scholar]
- 39.Ahn J, Jung M, Wyndham K, Yu Y, Engen J. Pepsin immobilized on high-strength hybrid particles for continuous flow on-line digestion at 10,000 psi. Anal. Chem. 2012;84:7256–7262. doi: 10.1021/ac301749h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitelegge J, Halgand F, Souda P, Zabrouskov V. Top-down mass spectrometry of integral membrane proteins. Expert. Rev. Proteomics. 2006;3:585–596. doi: 10.1586/14789450.3.6.585. [DOI] [PubMed] [Google Scholar]
- 41.Duke Proteomics Core Facility. In-solution tryptic digestion protocol. Durham, NC: [Google Scholar]
- 42.Fague K. The University of North Carolina at Chapel Hill Doctoral Dissertation; 2014. Multidimensional separations with ultrahigh pressure liquid chromatography-mass spectrometry for the proteomics analysis of Saccharomyces cerevisiae. [Google Scholar]
- 43.Geromanos S, Vissers J, Silva J, Dorschel C, Li G, Gorenstein M, Bateman R, Langridge J. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 44.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Inerowicz D, Hedrick V, Regnier F. Integrated sample preparation methodology for proteomics: analysis of native proteins. Anal. Chem. 2013;85:8039–8045. doi: 10.1021/ac401477w. [DOI] [PubMed] [Google Scholar]
- 46.Jiang S, Zhang Z, Li L. A one-step preparation method of monolithic enzyme reactor for highly efficient sample preparation coupled to mass spectrometry-based proteomics studies. J. Chrom. A. 2015;1412:75–81. doi: 10.1016/j.chroma.2015.07.121. [DOI] [PMC free article] [PubMed] [Google Scholar]