Abstract
Alkylating agents are a commonly used cytotoxic class of anticancer drugs. Understanding the mechanisms whereby cells respond to these drugs is key to identify means to improve therapy while reducing toxicity. By integrating genome-wide gene expression profiling, protein analysis and functional cell validation, we herein demonstrated a direct relationship between NRF2 and Endoplasmic Reticulum (ER) stress pathways in response to alkylating agents, which is coordinated by the availability of glutathione (GSH) pools. GSH is essential for both drug detoxification and protein thiol homeostasis within the ER, thus inhibiting ER stress induction and promoting survival; an effect independent of its antioxidant role. NRF2 accumulation induced by alkylating agents resulted in increased GSH synthesis via GCLC/GCLM enzyme, and interfering with this NRF2 response by either NRF2 knockdown or GCLC/GCLM inhibition with buthionine sulfoximine (BSO) caused accumulation of damaged proteins within the ER, leading to PERK-dependent apoptosis. Conversely, upregulation of NRF2, through KEAP1 depletion or NRF2-myc overexpression, or increasing GSH levels with N-acetylcysteine (NAC) or glutathione-ethyl-ester (GSH-E), decreased ER stress and abrogated alkylating agents-induced cell death. Based on these results, we identified a subset of lung and head-and-neck carcinomas with mutations in either KEAP1 or NRF2/NFE2L2 genes that correlate with NRF2 targets overexpression and poor survival. In KEAP1 mutant cancer cells, NRF2 knockdown and GSH depletion increased cell sensitivity via ER stress induction in a mechanism specific to alkylating drugs. Overall, we show that the NRF2-GSH influence on ER homeostasis implicates defects in NRF2-GSH or ER stress machineries as affecting alkylating therapy toxicity.
Keywords: NRF2, glutathione, ER stress, alkylating agents, cancer
Introduction
Alkylating chemotherapeutics, such as cyclophosphamide, carmustin and temozolomide, and alkylating-like compounds, such as cisplatin (CDDP)/platinum compounds, modify nucleotides, inducing DNA crosslinks and strand breaks which impede DNA replication, activate DNA repayir and, in case of inefficient cell recovery, trigger cell death cascades (1–5). Even though the alkyl group is expected to attach to the N7 nitrogen atom of the purine ring in DNA, alkylating agents also may react with RNA and proteins (6–8) as well as increase reactive oxygen species (ROS), which may lead to cell damage via oxidative stress (9, 10). While the fast proliferating phenotype of cancer cells makes them more sensitive to DNA damage, off-target effects to both proliferative and non-proliferative normal tissues are well documented in alkylating therapies (1–5). Thus, understanding the mechanisms whereby cells respond to these drugs is key to identify means of improving therapy or, at least, minimize the unintended damage to normal tissues.
NRF2 (Nuclear factor (erythroid-derived 2)-like 2; NFE2L2) is a transcription factor directly related to resistance to a variety of xenobiotics and oxidative stress (11, 12). Under unstressed/normal conditions, NRF2 is a short-lived (T1/2: 20 min) protein that interacts in the cytoplasm with the adaptor Kelch like-ECH-associated protein 1 (KEAP1) which helps Cullin-3 to rapidly ubiquitinate and target NRF2 for degradation. Oxidative or electrophilic stresses alter critical cysteine residues in KEAP1, disrupting its interaction with NRF2 and allowing NRF2 to accumulate and translocate into the nucleus. Nuclear NRF2 binds to the antioxidant response element (ARE) in the upstream promoter region of genes such as thioredoxins (TXNRD1), sulfiredoxin (SRXN1), hemoxygenase-1 (HMOX1), NAD(P)H quinone oxidoreductase 1 (NQO1), glutamate cysteine ligase catalytic/modifier (GCLC/GCLM) and Glutathione-S-transferases (GSTs) among others, thus improving the antioxidant and detoxification machinery of cells (11, 12).
ER stress is a well-conserved proteotoxicity response mechanism that can promote cell survival as well as apoptosis depending on the duration of the stimulus and which ER sensor is activated (13, 14). While ATF6 and IRE1α are first responders to unfolded cargo that promote chaperone production and ER biogenesis to enhance ER folding capacity (15, 16), PERK responds to high and persistent unfolded cargo by inhibiting global mRNA translation via eIF2α phosphorylation and promoting apoptosis via CHOP (17, 18). Because the folding of nascent proteins within the ER lumen requires a more pro-oxidant environment compared to other cellular compartments, the ER is highly sensitive to variations in redox homeostasis, especially imbalances in protein thiol (protein-SH) and GSH pools (14, 19–21). However, there is limited evidence for how antioxidant-associated transcription factors such as NRF2 affect ER homeostasis. Our prior work revealed that NRF2-GSH synthesis genes and unfolded protein responses are key to cell survival in a MMS survival RNAi screen performed in D. melanogaster cells (22). Further, we showed that these processes appear to be conserved across fly and mammalian systems suggesting a potential relationship (22).
In this study, we performed genome-wide gene expression profiling of cancer cells (MDA-MB231 and U2OS) exposed to methyl methanesulfonate (MMS), a prototypical alkylating agent that does not require bioactivation (23). Protein level analysis, metabolite quantifications and functional cell assays were used to validate the predicted activation of NRF2 and ER stress pathways. We delineated the coordination between NRF2 and ER stress, which involves a NRF2-dependent GSH synthesis necessary to maintain ER protein-SH homeostasis and inhibit ER stress-mediated apoptosis via PERK. Throughout this study, the phenotypes observed with MMS were extended to clinically relevant alkylating agents such as 4-hydroperoxycyclophosphamide (4-HC) and the alkylating-like agent CDDP
Materials and Methods
Cell culture and treatments
MDA-MB231 and MCF7 breast cancers were obtained from ATCC in 2012 and 2014, respectively. U2OS (osteosarcoma) cells were obtained from collaborators in 2011. MDA-MB231 and U2OS cells were authenticated by examining RNA sequencing data produced in 2014 for this project and comparing against mutations known to be present in each cell line. The A549 (non-small lung carcinoma cell) line was obtained from ATCC in 2012; keratin positivity by immunoperoxidase staining was used to monitor cell phenotype. All cell lines were passaged for <6 months after resuscitation. The cells were grown in DMEM or RPMI (as appropriate for each) supplemented with 10% FBS plus 1X Antibiotic:Antimycotic Solution (Sigma-Aldrich; cat#A5955), and passaged following ATCC instructions. MMS, cisplatin (CDDP), etoposide, doxorubicin and paclitaxel were from Sigma-Aldrich, and 4-hydroperoxycyclophosphamide (4-HC) was from US Biologicals. Unless otherwise specified, chemotherapeutics doses used were as follows: MMS (40 µg/mL; i.e., 363 µM), 4-HC (50 µM), etoposide (20 µM), CDDP (50 µM), paclitaxel (0.2 µM) and doxorubicin (1 µM), which are in the range of IC40-IC50 for MDA-MB231, MCF-7 and U2OS cells as obtained from 72 h Cell-titer Glo assays (normalized as treated/control ratio of luminescence signal). When used, the GCLC/GCLM inhibitor buthionine sulfoximine (BSO, 1 mM) and the antioxidants N-Acetyl-Cysteine (NAC, 7.5 mM), glutathione ethyl-ester (GSH-E, 10 mM) and Trolox (200 µM) were pre-incubated for 6–8 h prior to treatment with alkylating agents and were maintained during the period of alkylating agent treatment.
RNA sequencing
MDA-MB231 and U2OS cells treated for 8 h with MMS (40 µg/mL), etoposide (20 µM), paclitaxel (0.2 µM) and doxorubicin (1 µM) were profiled by RNA-sequencing using Illumina HiSeq 2000 system (Illumina, San Diego, CA). Afterwards, the RNA was harvested using the RNeasy protocol (Qiagen), and its purity was determined using Agilent 2100 BioAnalyzer. Samples of 1–2 µg of total RNA were used for sequencing library preparation according to Illumina TruSeq Total RNA Sample Preparation Guide (Illumina Cat. #: RS-122-2201). Each library was bar-coded and then pooled for cluster generation and sequencing with 100bp single-end (SE) sequencing protocol. Short read sequences from RNAseq were first aligned to UCSC hg19 genome build using TopHat2 algorithm and then quantified for gene expression by HTSeq to obtain raw read counts per gene and then converted to RPKM (Read Per Kilobase of gene length per Million reads of the library) according to gene length and total mapped read count per sample. Log2-transformed RPKM measurements were used as gene expression level, and entities with low-read counts RPKM<2 in both MMS/chemotherapeutic and control samples (as determined by examining read counts in non-exonic regions) were removed from analysis. The RNA sequencing data have been deposited in Gene Expression Omnibus and are accessible through GEO Series accession number GSE84863.
Pathway Enrichment Analysis (PEA) and Pathway Signature Expression
Ingenuity PEA: MMS-induced gene expressions more than 2-fold (treated versus untreated control) were selected as differentially expressed genes and analyzed using the Ingenuity Pathway Analyzer (IPA) set at default parameters (Ingenuity Systems, Qiagen). Pathway enrichment significance was calculated using the right-tailed Fisher Exact Test (cut-off: p<0.01).
Pathway Signature Expression: from a list of canonical genes targets of NRF2 (SLC7A11, SRXN1, GCLM, TXNRD1, NQO1, FTH1, AKR1C1, GCLC, EPHX1, MAFG, PRDX1, GSR, GSTP1, TALDO1, FTL and HMOX1) and markers of ER stress pathway activation (ATF4, PPP1R15A, XBP1, TRIB3, DDIT3, HSPA5, ERN1, ERO1LB/ERO1B, HERPUD1, ASNS, SARS, SELS, SERP1, WARS, ATF3/JDP2, HSPA13, HSPA1A, HSPA1B, HSPA9, HSPB8, DNAJA4, DNAJB9, DNAJC3) curated from PubMed, Qiagen and Ingenuity databases, we calculated fold-change (FC) expression values for each gene in drug-treated (i.e., doxorubicin, etoposide, MMS and paclitaxel) versus untreated samples of RNA-sequenced MDA-MB231 and U2OS cells. These individual FC values were log2 transformed and pooled together with equal weights and plotted as a single group, therefore providing an estimate of the relative expression of ER stress and NRF2 pathways. Kruskal-Wallis followed by Dunn’s test was used to assess statistical significance at a p<0.05.
Cell growth and caspase activity assay
Cell growth and caspase-3 activity were assessed using Cell-Titer Glo Luminescent Cell Viability Assay and Caspase-Glo 3/7 (Promega), respectively, following manufacturer’s instructions. The cells were seeded in 96-well plates and assayed immediately at the end of treatments. Cell-Titer Glo data were expressed as Relative Luminescence (%) as compared to luminescence signal of experimental controls.
DCF and DHE assays for ROS production
Intracellular ROS production was detected using the cell-permeant 2',7'-dichlorodihydrofluorescein diacetate (DCFH-DA; Sigma) as described (24). Briefly, the cells seeded in black-edge/clear bottom 96-well plates were incubated with 20 µM DCFH-DA in DMEM containing 0.5 % FBS to allow probe incorporation. Subsequently, the treatments were added and the kinetics of DCF fluorescence was monitored (Ex/Em = 485/532 nm). Hydrogen peroxide (100 µM) was used as a positive control. Superoxide production was monitored using Dihydroethidium (DHE), a cell-permeable compound that interacts with superoxide anion to form oxyethidium and ethidium bromide, which in turn interact with nucleic acids to emit a red fluorescence. Cells were pre-incubated with 20 µM DHE for 1 h, treated with alkylating agents, and the fluorescence was monitored at Ex/Em 518/605 nm. Paraquat (10 µM) was used as a positive control for superoxide production. Delta fluorescence over a 12 h period was calculated and expressed as % fluorescence compared to control samples.
Glutathione assays
Intracellular glutathione levels (GSH+GSSG) were measured in accordance with Glutathione Fluorimetric Assay Kit protocol (BioVision Incorporated, CA). Total glutathione content was normalized by protein levels as determined by Bradford’s method. Data are expressed as fold compared to controls.
Preparation of cytoplasmic, nuclear, mitochondrial and microsomal/ER fractions from cell cultures
For analysis of protein-SH groups, cytoplasmic, nuclear and mitochondrial fractions were isolated from MDA-MB231 (2×107 cells) as previously described (24). Microsomal/ER fraction isolation followed the Endoplasmic Reticulum Isolation Kit protocol (Sigma-Aldrich, cat# ER0100). Briefly, 2×107 cells were trypsinized, centrifuged and resuspended in 1X hypotonic buffer (10 mM HEPES, pH 7.8, 1 mM EGTA and 25 mM KCl). After 20 min incubation on ice, the cells were centrifuged at 600 × g for 5 min, and the supernatant was discarded. Cell pellets were incubated with 1× Isotonic Extraction Buffer (10 mM HEPES, pH 7.8, 250 mM sucrose, 1 mM EGTA, and 25 mM KCl) and passed 10-times through a 27 gauge needle. The homogenates were centrifuged at 1,000 × g (10 min, 4 °C), and the resulting supernatant was re-centrifuged at 12,000 × g (15 min, 4 °C). This supernatant post-mitochondrial fraction was centrifuged for an additional 60 min at 100,000 × g in an ultracentrifuge at 4 °C. The resulting pellet (microsomal/ER fraction) was used for protein-SH measurements. Purity of cell fractions was determined by immunoblot against TATA-binding protein TBP (nucleus), Calnexin (ER/microssomal) and SOD2 (mitochondria).
Fluorescence labeling of protein thiol groups (Dibromobimane assays)
Depletion of protein thiols groups (protein-SH) following alkylating agent exposure was measured by dibromobimane assays with minor modifications (25, 26). Dibromobimane reacts with reduced thiols to generate a highly fluorescent adduct. MDA-MB231 cells plated in 6-well plates were treated with alkylating agents, harvested in 600 µL PBS, and then 200 µL perchloric acid (6 N) was immediately added to precipitate the proteins. The samples were incubated for 5 min on ice and centrifuged at 14,000 × g for 10 min. The supernatants were used to measure glutathione, and the pelleted proteins were solubilized with 100 µL of 0.1M NaOH and subsequently neutralized with 0.5 M Tris-HCl. A stock solution of 4 mM dibromobimane in DMSO was added to a final concentration 40 µM in PBS, and incubated for 40 min at 37° C. Dibromobimane bound protein-SH groups were measured in a Molecular Devices M5 reader set at Ex/Em 393/477 nm. Fluorescence was normalized by total protein (Bradford method) and expressed as a % of control samples.
Western blot
Protein lysates were prepared using RIPA buffer containing 1 mM PMSF, 1 mM sodium orthovanadate, 1 mM NaF, and 30 µL/mL aprotinin. The proteins (20–30 µg) were resolved in SDS-PAGE, electro-transferred onto nitrocellulose membranes (Hybond-ECL, GE Healthcare) and blocked with 5% BSA. Primary antibodies included NRF2 (D1Z9C), BiP/GRP78 (C50B12), IRE1α (14C10), CHOP (L63F7), Calnexin (C5C9) and PERK (D11A8) from Cell Signaling Technologies; p-IRE1α (Ser724; NB100-2323) from Novus Biologicals; GCLC (Ab41463), beta-actin (Ab8227), TBP (ab818) and ATF6 (Ab37149) from Abcam; p-PERK (Thr 981, sc-32577), NQO1 (C-19), and ATF3 (C19) from Santa Cruz. After secondary antibody incubation (1:3000 in TBS-T, 2 h), the proteins were detected using Lumiglo substrate (Cell Signaling Technology, CA) and X-ray films. Immunoblot images are representative of three independent experiments.
Small interference RNA (siRNA)
The siRNA duplexes (20 to 40 nM final concentrations) targeting human NRF2 (siRNA#1: sc-37030A and siRNA#2: sc-37030B), IRE1α (sc-40705), PERK (sc-36213), KEAP1 (sc-43878) and scrambled siRNA-A (sc-37007) were from Santa Cruz Biotechnology; ATF6 siRNAs (ID22926 trilencer-27) were from Origene. Reverse transfections were performed using the Lipofectamine RNAiMAX Reagent (Invitrogen) following manufacturer’s instructions. siRNAs were incubated for 24 h prior to treatments, and protein knockdown efficiency was assessed by immunoblotting.
Reporter gene assays
Cignal Antioxidant Response Reporter Assay kit (Qiagen) was used to measure Antioxidant Response Elements (ARE) activation by active NRF2. Cignal ERSE Reporter (luc) Kit (CCS-2032L; Qiagen) was used to measure ER stress pathway activation via ATF6, PERK-ATF4 and IRE1α-XBP1 axes. The cells were transfected with a mixture of ARE- or ERSE-driven firefly luciferase and constitutive Renilla-luciferase constructs (40:1 ratio, 100 ng reporter mixture) using Lipofectamine 3000 in 96-well plates. When used, siRNAs (30 nM) or cDNA constructs (1:1 Reporter gene cDNA mixture, total 100 ng DNA mixture) were co-transfected with ARE/ERSE-luciferase constructs. Twenty four hours post-transfection, the cells were treated for additional 8 h with alkylating agents, washed out and kept for an additional 16 h in drug-free culture medium prior to assessment using the Dual-Luciferase® Reporter Assay (Promega). Data were expressed as fold-induction of firefly luciferase/renilla luciferase ratio in treatments versus control cells.
Plasmid Construct Overexpression
The pcDNA3-EGFP-C4-NRF2 (plasmid: 21549, from Dr. Xiong lab (27); hereafter noted as NRF2-Myc) and pcDNA3.1(+)-GRP78/BiP (plasmid:32701, from Dr. Austin lab; hereafter noted as GRP78) were purchased from Addgene. The pCDNA3 and pCDNA3-EGFP constructs were used as empty vector and transfection efficiency control, respectively. The cells were transfected with Lipofectamine-3000 (Invitrogen) at a ~90% confluence for 24–36 h, trypsinized, and re-plated for 24 h before treatments. Protein overexpression was validated by immunoblot.
Bioinformatics
Patient risk/survival
To assess the relationship between NRF2 target genes expression and patients risk/survival in different types of cancer we used the SurvExpress tool (28). The SurvExpress database comprises microarray gene expression and matched clinical data of several solid and hematologic cancers. For an input gene list of NRF2 targets, the software calculates the prognostic index (PI), also known as the risk score, for each sample. The PI is known as the linear component of the Cox model PI = β1×1+ β2×2+…+βp×p (28), where xi is the quantile normalized gene expression value and the βi can be obtained from the Cox fitting. PI values were used to split samples and generate 3 risk groups (low-, medium- and high risk); Overall Survival was used as clinical endpoint. Cancer datasets were preferentially those with high number of samples (n>100) and coverage of >75% of genes/probes present in the input list. The input gene list comprised classical NRF2 targets (same list as above described in “Pathway Signature Expression” subsection). Quantile normalized values of probe expression were used and, for multiple probe genes, the maximum variance probe was selected. Log-rank test of differences between risk groups, hazard-ratio (HR) estimate, and gene expression fold-changes in high-, medium- and low-risk groups were calculated in order to identify the NRF2 targets with differential expression in the high-risk group. These fold-change values were used to estimate a NRF2 Pathway Signature Expression (above described) in high-risk versus low-risk patients in each cancer dataset evaluated. Cancer types with higher expression of NRF2 signatures/targets in the high-risk group were assumed as potential candidates for NRF2 pathway involvement in the disease outcome.
TCGA and E-MTAB2706 datasets analysis
TCGA provisional cohorts of RNA sequenced (RNAseq V2 RSEM) lung adenocarcinomas (n=230), lung squamous cell carcinoma (n=178) and head and neck squamous cell carcinoma (n=279) with matched information of KEAP1 and NFE2L2 mutational and copy-number alteration (CNA) status were selected. RNA-sequencing gene expression data of NRF2 targets as well as KEAP1, NFE2L2 gene mutational and CNA status for each patient were derived directly from cBioportal for Cancer Genomics (http://cbioportal.org/; (29, 30)). For a panel of lung cancer cell lines, the E-MTAB-2706 RNA sequencing dataset was downloaded from ArrayExpress, and KEAP1 and NFE2L2 mutational status in cell lines was obtained from the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic). Gene expression values were log2 transformed, and Z-score of the mRNA expression of each gene was calculated as compared to its expression distribution across the entire sample panel. For NRF2 pathway signature expression in TCGA datasets, average Z-scores of each NRF2 target (same gene list as described in “Pathway Signature Expression” subsection) in wild-type and mutant KEAP1 groups were plotted with equal weights, and pathway signature expressions were analyzed by Kruskal-Wallis/Dunn’s test (for multiple groups) or Mann-Whitney test (for 2-group comparison) at a p<0.05. For heatmap representation, the samples were grouped based on their KEAP1 and NFE2L2/NRF2 mutational status and hierarchical clustering was performed using Euclidean Distance and Ward’s linkage as dissimilarity measures and a clustering method, respectively.
Mutation Enrichment Analysis (MEA)
We used the “Enrichment” tool available with the cBioportal for Cancer Genomics (http://cbioportal.org/) to detect mutations associated with upregulated expression of NRF2 targets in the lung and head and neck cancer datasets described above. From an input list comprising the NRF2 targets (detailed above), the tool ranks the gene mutations (compared against all the mutations identified for the respective TCGA dataset) that co-occur or display mutual exclusivity with NRF2 target genes expression alterations. Z-score = ±1 was used as a cut-off for differential gene expression and P-values were derived from Fisher-Exact Test and corrected by Benjamini-Hochberg. In addition, to identify which cancers types display the highest frequency of NFE2L2 and KEAP1 genetic alterations, the cBioportal tool parameters were selected as follows: i) “All (126)” datasets; ii) “Select data type priority: Mutation and CNA”; iii) “Gene set: KEAP1, NFE2L2”. Thus, all TCGA cancer cohorts with DNA sequencing information were evaluated, and a ranked list of cancer datasets showing NFE2L2/KEAP1 alterations is provided.
Results
NRF2, GSH biosynthesis and UPR/ER stress transcriptional responses in cells treated with alkylating agents
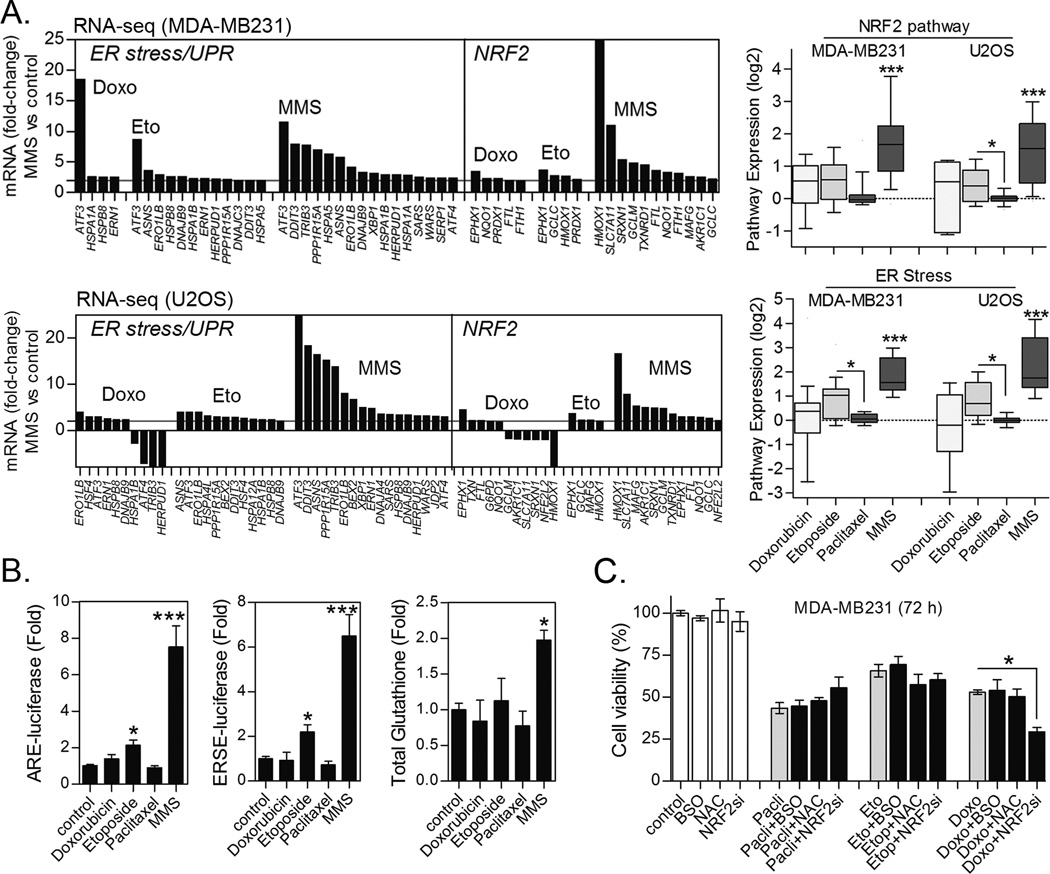
Initially, we performed RNA sequencing of MDA-MB231 and U2OS cell lines treated for 8 h with 40 µg/mL MMS. Pathway enrichment analysis (PEA) of MMS-induced genes indicated a high enrichment in targets of the NRF2 transcriptional program, GSH biosynthesis enzymes (GCLC/GCLM) and members of the unfolded proteins response (UPR)/ER stress transduction machinery as shown in figure 1A. In contrast, classic DNA damage responses showed a lesser level of enrichment (Figs. 1A and S1). We therefore decided to focus on delineating the survival roles of NRF2 and ER stress pathways in response to alkylating agents.
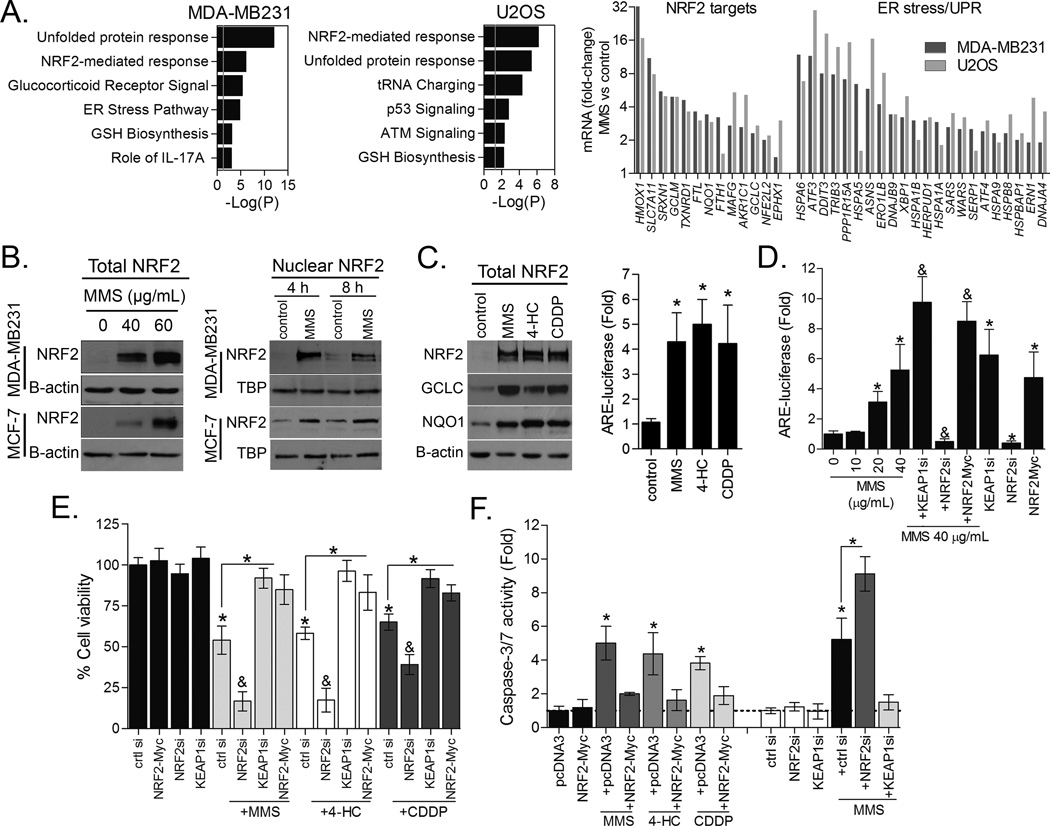
Figure 1. Transcriptional responses and NRF2 activation by alkylating agents.
(A) Pathway Enrichment Analysis (PEA) significance (−log (P-value)) of MMS-induced gene expressions in MDA-MB231 and U2OS cells as evaluated by RNA sequencing. Selected MMS-induced gene expression changes in NRF2 and ER stress markers are also shown. (B) Representative immunoblots showing the effect of MMS on total and nuclear NRF2 in MDA-MB231 and MCF-7 cells. (C) Effect of different alkylating agents on NRF2, GCLC and NQO1 proteins immunocontent and ARE-luciferase reporter activity in MDA-MB231 cells. (D) ARE-luciferase reporter assays showing the dose-dependent effect of MMS, the effect of NRF2 and KEAP1 depletion by siRNA, and NRF2-Myc overexpression upon NRF2 activity in MDA-MB231 cells. (E–F) Impact of NRF2 and KEAP1 knockdown by siRNA and NRF2-myc overexpression on (E) CellTiter-Glo cell growth assay and (F) caspase-3/7 activation in MDA-MB231 cells exposed to MMS, CDDP and 4-HC for 48 h. Unless otherwise specified, cells were treated for 8 h with alkylating agents at ~IC40–50 as described in Materials and Methods. Legends: si (siRNA). Data are represented as the average ± SD of a representative experiment performed in triplicate and repeated thrice. *different from untreated/controls or at indicated comparisons; &different from untreated and from alkylating agent-treated (p<0.05, ANOVA-Tukey).
We first confirmed that MMS promoted total protein accumulation and nuclear translocation of NRF2 (Fig. 1B), agreeing with the classical mechanism involving the inhibition of KEAP1-mediated NRF2 degradation. NRF2 activation was also detectable in cells treated with the clinically relevant alkylating agents 4-HC and cisplatin/CDDP (Fig. 1C), indicating a drug class effect. NRF2 protein accumulation and translocation correlated with increased ARE-luciferase reporter gene induction by these alkylating agents as well as protein increase of two classical NRF2 targets, GCLC and NQO1 (Fig. 1C). These results corroborate the induction of GCLC and NQO1 transcripts in MMS-treated cells (see Fig. 1A RNA sequencing). By modulating NRF2 activity through NRF2-myc overexpression, NRF2 knockdown, or knockdown of its negative regulator KEAP1 (Fig. 1D; Fig. S2 for NRF2 overexpression validation), we were able to determine that NRF2 activation is an important anti-apoptosis defense mechanism of cancer cells exposed to alkylating drugs. NRF2 silencing potentiated, whereas NRF2-myc overexpression or KEAP1 knockdown suppressed MMS, CDDP and 4-HC toxicity in cell growth assays (Fig. 1E) and blocked alkylating agents-induced caspase-3 activation (Fig. 1F).
NRF2-augmented GSH biosynthesis dictates cell survival to alkylating agents, but not via antioxidant mechanisms
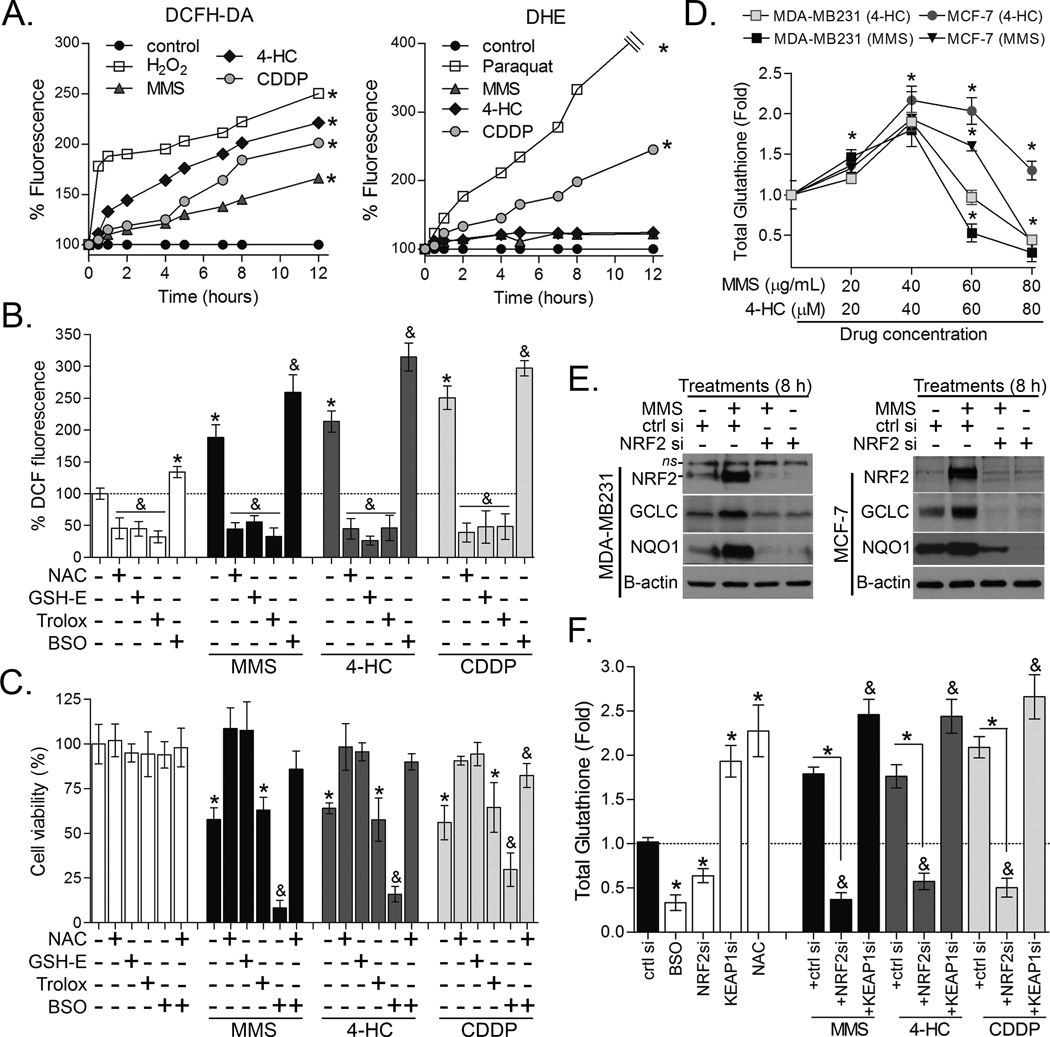
NRF2 is widely described as a transcription factor involved in antioxidant and phase-II detoxification responses (11, 12), two molecularly distinct mechanisms. In our model, all alkylating drugs induced a rapid increase in general ROS production as measured by DCF assay, whereas only CDDP increased superoxide anion production as inferred from DHE assays (Fig. 2A). To test whether ROS contributed to the cell death induced by alkylating agents, we quenched ROS by pre-treating cells with GSH precursors (NAC or glutathione-ethyl-ester (GSH-E)), or a thiol-unrelated antioxidant, Trolox (a vitamin E analogue). While all three antioxidants decreased basal and drug-induced ROS (Fig. 2B) only NAC and GSH-E, but not Trolox, protected against MMS, 4-HC and CDDP toxicity (Fig. 2C). This is very interesting because it shows that simply reducing ROS (as observed with Trolox) is not sufficient to retain cell viability; yet the GSH pool is key for survival. To confirm this data, inhibition of GSH production by BSO (validated in Fig. S2), an inhibitor of the GSH synthesis regulatory enzyme, GCLC/GCLM, caused robust cytotoxicity when combined with alkylating agents; an effect prevented by NAC (Fig. 2C). Interestingly, total glutathione content increased in cells exposed to lower levels of alkylating drugs whereas higher concentrations depleted glutathione (Fig. 2D). We previously reported that glutathione pools are depleted in MMS-treated D. melanogaster and primary mouse fibroblasts due to non-enzymatic or/and GST-mediated conjugation of MMS reactive methyl group to GSH thus producing methyl-glutathione (22). Corroborating that observation, it has previously been noted that formation of glutathione-cisplatin conjugates also take part of cisplatin detoxification in osteosarcoma cells (31). We also determined that NRF2 is a key regulator of glutathione synthesis in response to alkylating drugs exposure since NRF2 knockdown decreased both GCLC protein induction (Fig. 2E) and glutathione levels in the presence of MMS, CDDP and 4-HC (Fig. 2F). Conversely, induction of NRF2 via KEAP1 knockdown enhanced glutathione pools (Fig. 2F).
Figure 2. NRF2-GSH pathway is key for alkylating agent survival.
(A) Representative kinetics of DCF and DHE assays showing the effect of alkylating agents on cellular ROS production in MDA-MB231 cells. (B) DCF and (C) Cell-Titer Glo assays showing the effect of antioxidants (NAC, GSH-E and Trolox) and BSO on alkylating agents-induced ROS (12h treatment; DCF assay) and cell survival (48 h exposure), respectively. (D) Dose effect of alkylating agents on total glutathione levels in MDA-MB231 and MCF-7 cell lines after 12 h treatment. X-axis denotes 4-HC and MMS at µM and µg/mL units of concentration, respectively. (E) Effect of NRF2 knockdown on MMS-induced GCLC and NQO1 immunocontents in MDA-MB231 and MCF-7 cells (8 h treatment). (F) Effect of NRF2 and KEAP1 depletion by siRNA on glutathione content in MMS, CDDP or 4-HC treated MDA-MB231 cells (12 h treatment). Unless otherwise specified, the cells were treated with alkylating agents at ~IC40–50 as described in Materials and methods. Legend: si (siRNA); ctrl si (scrambled control siRNA). Data are represented as the average ± SD of a representative experiment performed in quadruplicate and repeated thrice. *different from untreated/controls or at indicated comparisons; &different from untreated and from alkylating agent-treated (p<0.05, ANOVA-Tukey).
In addition to glutathione, we also tested the involvement of MRP and MDR-mediated drug efflux pumps and thioredoxin/thioredoxin reductase (TXN/TR) systems on cell survival to alkylating drugs. MK571 (MRP1 and MRP2 inhibitor) and verapamil (MRP1 and MDR1 inhibitor) pre-treatments were used to block efflux protein activity, but no significant potentiation of alkylating agents toxicity was observed in MDA-MB231 and U2OS cells (Fig. S2). Noteworthy, though these cell lines express MRP1, mRNA levels of MDR1, MDR2 and MRP2 are very low, and these genes did not respond to MMS (Fig. S2). On the other hand, subtoxic concentrations of the TR inhibitor auranofin potentiated toxicity of MMS and 4-HC by ~20–25% and of CDDP by 30–35% in both cell lines (Fig S2). With the TXN/TR system, TR is a key responsible for maintaining the proper redox balance at the protein level, reducing TXN to allow TXN-mediated reduction of protein substrates (32). Collectively, these and our previous work (22) point to a pivotal role for glutathione-mediated drug detoxification in cell survival. As drug efflux appears not to be the major mechanism determining alkylation sensitivity in this model, nor ROS, yet there is a reliance upon TXN/TR, our results indicate an involvement of protein-SH homeostasis in cell survival.
Alkylating agents induce damage to ER proteins leading to ER stress-mediated cell death via PERK
PEA analysis showed a highly enriched ER stress signature in both MMS-treated MDA-MB231 and U2OS cells (Fig. 1A). We first validated the MMS-induced activation of the three ER membrane-bound sensors; i.e. IRE1α and PERK phosphorylation and ATF6 cleavage (Fig. 3A), and upregulation of ER stress downstream effectors GRP78/BIP, CHOP and ATF3 proteins (Fig. 3A); a drug class effect was also observed with 4-HC and CDDP (Fig. 3B). Alkylating agents-induced BiP/GRP78 and CHOP levels were decreased by knockdown of ATF6 and/or PERK, indicating activation of canonical ER stress machinery (Fig. 3C).
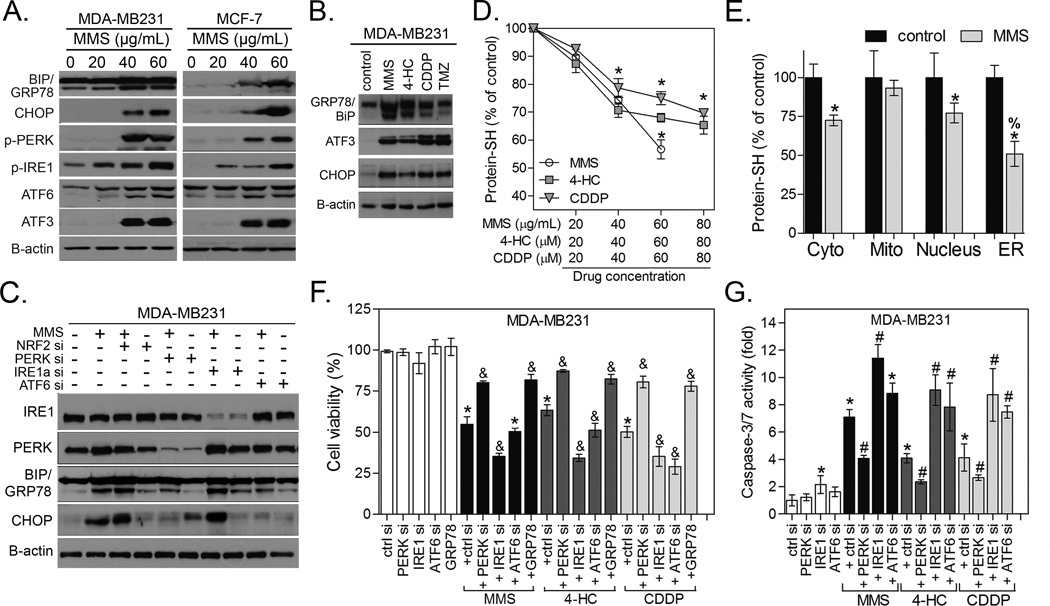
Figure 3. Alkylating agents induce protein-SH depletion and ER stress-dependent cell death.
(A) Immunoblots showing the dose-effect of MMS and (B) other alkylating agents on ER stress markers in MDA-MB231 and MCF-7 cells. (C) Representative immunoblots showing the impact of PERK, IRE1α and ATF6 depletion by siRNA upon CHOP and GRP78/BiP immunocontents in MDA-MB231 cells. (D) Dose-effect of alkylating agents on protein thiol (protein-SH) levels in MDA-MB231 cells. (E) Protein-SH levels in subcellular fractions from MMS-treated cells. Unless otherwise specified, the cells were treated for 12 h with alkylating agents at ~IC40–50 as described in Materials and Methods. (F–G) Representative experiment showing the impact of PERK, IRE1α and ATF6 depletion by siRNA or GRP78-pcDNA3 overexpression on (F) CellTiter-Glo cell growth assay and (G) caspase-3/7 activity in MDA-MB231 cells exposure to alkylating agents for 48 h. Legends: si (siRNA); ctrl si (scrambled siRNA), control (untreated cells), GRP78 (GRP78-pcDNA3 overexpression). Unaltered results with pcDNA3 empty vector and scrambled siRNA are omitted. Data are represented as the average ± SD of a representative experiment performed in triplicate and repeated thrice (for D,F,G) or twice (for E panel). *different from untreated; #&different from untreated and from alkylating agent-treated cells; %different from all other groups (p<0.05, ANOVA-Tukey).
We then went on to test the impact of alkylating agents on protein-SH as a possible trigger of ER stress, and whether NRF2-GSH status could modulate this proteotoxicity. In fact, alkylating agents decreased the amount of reduced protein-SH in whole cell extracts (Fig. 3D). Cellular fractionation revealed that protein-SH depletion by alkylating agents was more pronounced in ER proteins, followed by cytoplasm/nucleus, while it was not detected in mitochondrial fractions (Fig. 3E; Fig. S2 for cell fractionation controls). ER stress may exert a dual role in controlling cell fates, which depend on the balance of ER sensors activation and unfolded protein cargo (17, 33, 34). In our model, depletion of IRE1α or ATF6 potentiated, while PERK silencing (the classical pro-apoptotic arm in the ER (33, 34)) partially reversed, cell growth inhibition (Fig. 3F) and caspase-3 activation (Fig. 3G) by MMS, 4-HC and CDDP. Conversely, BiP/GRP78 chaperone overexpression (validated in Fig. S2B), which is known to improve ER protein folding capability (34), attenuated MMS, CDDP and 4-HC toxicity (Fig. 3F).
NRF2-mediated GSH synthesis controls the magnitude of ER stress via inhibition of protein thiol damage
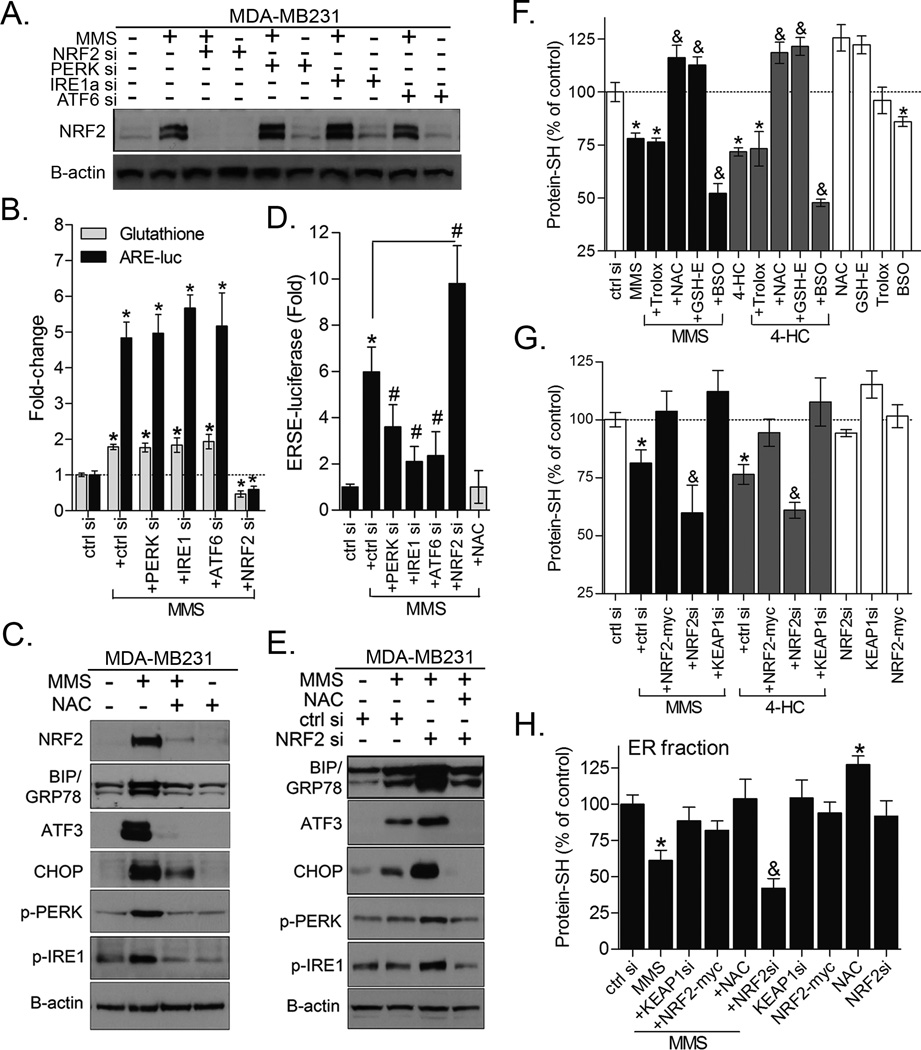
While both NRF2 and ER stress responses are activated by alkylating agents, NRF2 confers a cell survival response whereas the PERK axis of ER stress pathway promotes apoptosis. Based on these observations, we decided to evaluate a possible interdependence of NRF2 and ER stress. Firstly, even though PERK has been reported to promote phosphorylation-mediated NRF2 dissociation from KEAP1 (33), neither PERK and IRE1α nor ATF6 were necessary for MMS-induced NRF2 protein accumulation (Fig. 4A), ARE-luciferase activity or GSH production (Fig. 4B) in MDA-MB231 cells. This indicates that NRF2 is not downstream of ER stress, at least not in this context. On the other hand, NAC pre-treatment blocked the MMS-induced upregulation of ER stress markers (Fig. 4C) and ERSE-luciferase reporter activity (Fig. 4D); an effect not reversed by Trolox (data not shown). In keeping with this, NRF2 depletion by siRNA, in combination with MMS, potentiated ERSE-luciferase activation (Fig. 4D) and increased the magnitude of ER stress (i.e., greater CHOP, GRP78, ATF3 protein and increased PERK and IRE1α phosphorylation) compared to MMS alone; an effect rescued by NAC pre-treatment (Fig. 4E). With regard to alkylating agent decreased protein-SH levels, depleting intracellular GSH with BSO led to an enhanced impact, while NAC or GSH-E, but not Trolox, protected protein-SH (Fig. 4F). Corroborating these results, NRF2 knockdown emulated BSO, accentuating the impact of exposure to alkylating agents on protein-SH, while KEAP1 silencing and NRF2-myc overexpression were protective in both whole-cell (Fig. 4G) as well as ER protein extracts (Fig. 4H). It is key to note that our methodology does not directly measure alkylation of protein-SH residues by the alkylating agents used here. However, when whole-cell lysates from alkylating agent-treated cells were incubated with dithiothreitol (DTT, 10 mM), which converts disulfide bonds into free protein-SH groups, we still observed DTT-resistant protein-SH depletion by alkylating agents (Fig. S2). This effect was analogous to cell extracts treated with iodoacetamide, a classical alkylator that reacts with protein-SH groups to form stable S-carboxyamidomethyl-cysteine adducts (Fig. S2).
Figure 4. NRF2 and GSH control the magnitude of protein damage and ER stress response.
(A) NRF2 immunoblots and (B) assays for ARE-luciferase activity and total glutathione showing the impact of depleting different ER stress sensors by siRNA on MMS-induced NRF2 activation and GSH synthesis in MDA-MB231 cells. (C) Western blot analysis showing that NAC pre-treatment inhibits ER stress proteins expression in MMS-treated MDA-MB231 cells. (D) ERSE-luciferase reporter gene assay to demonstrate that NRF2 depletion exacerbates, while NAC blocks, MMS-induced ER stress activation in MDA-MB231 cells (8 h treatment). (E) Representative immunoblots showing the impact of NRF2 siRNA and the rescuing effect of NAC on ER stress markers in MDA-MB231 cells treated with MMS for 24 h. (F–G) Dibromobimane assays showing the effect of: (F) Antioxidants and BSO; (G) NRF2 and KEAP1 knockdown or NRF2-myc overexpression in the basal and alkylating agents-induced protein-SH depletion in MDA-MB231 cells. (H) Effect of NAC, KEAP1 or NRF2 knockdown, and NRF2-myc overexpression in the protein-SH levels of ER fractions of MMS-treated MDA-MB231 cells. Unless otherwise specified, the cells were treated for 12 h with alkylating agents at ~IC40–50 as described in Materials and Methods. Data are represented as the average ± SD of a representative experiment performed in triplicate and repeated thrice (for B,D,F,G) or twice (for H panel). *different from untreated; #&different from untreated and from alkylating agent-treated at equivalent conditions (p<0.05, ANOVA-Tukey).
NRF2-GSH and ER stress pathways are enriched as alkylating agent specific responses
While determining the role of NRF2-GSH and ER stress in the fate of cells exposed to alkylating agents, we considered whether these mechanisms could be a general cell toxicity response also relevant to other clinically used chemotherapeutics. We therefore performed RNA sequencing of MDA-MB231 and U2OS cells treated with etoposide, paclitaxel or doxorubicin and compared to MMS data (see Methods for details). Even though these non-alkylating drugs, especially etoposide, induced expression of some NRF2 and ER stress markers, the number of entities and magnitude of their upregulation was much lower than with MMS treatment (Fig. 5A). Consequently, changes in NRF2 and ER stress Pathway Signature expressions were not observed with paclitaxel and doxorubicin while etoposide only evoked a weak response (Fig. 5A). In keeping with RNA sequencing, only etoposide induced ARE- and ERSE-luciferase activities (Fig. 5B) but none of the non-alkylating compounds produced observable alterations in glutathione levels at either 8 h (Fig. 5B) or 24 h treatments (data not shown). Further, modulating the GSH levels by inhibition (BSO) or augmentation (NAC supplementation) had no impact in the cytotoxicity caused by these agents (Fig. 5C). Despite not inducing any obvious NRF2 activation, we noted that NRF2 depletion increase doxorubicin toxicity to MDA-MB231 cells, indicating that a NRF2 target other than the GSH synthesis enzymes may play a role in doxorubicin survival. Taken together, these data indicate that NRF2-GSH/ER stress response system is not likely a major determinant of cell survival to these non-alkylating drugs in our model.
Figure 5. Activation of NRF2 and ER stress pathways in response to alkylating agents as compared to other chemotherapeutics.
(A) The most significant mRNA fold-changes in ER stress and NRF2 pathway markers as determined by RNA sequencing of MDA-MB231 and U2OS cells treated for 8 h with chemotherapeutics and MMS (see Methods). NRF2 and ER stress pathway expression index for each tested drug is also shown. (B) Comparative effect of chemotherapies on ERSE- and ARE-luciferase reporter gene activities and total glutathione content in MDA-MB231 cells treated for 8 h. (C) Impact of NAC and BSO pre-treatments and NRF2 knockdown on the cytotoxicity of non-alkylating drugs in MDA-MB231 (72 h treatment). Figs. “B and C” data are represented as the average ± SD of a representative experiment performed in triplicate and repeated at least thrice. *different from untreated or at indicated comparisons; ***different from all other groups (p<0.05, ANOVA/Tukey for “B–C”; Kruskal-Wallis/Dunn’s for “A”).
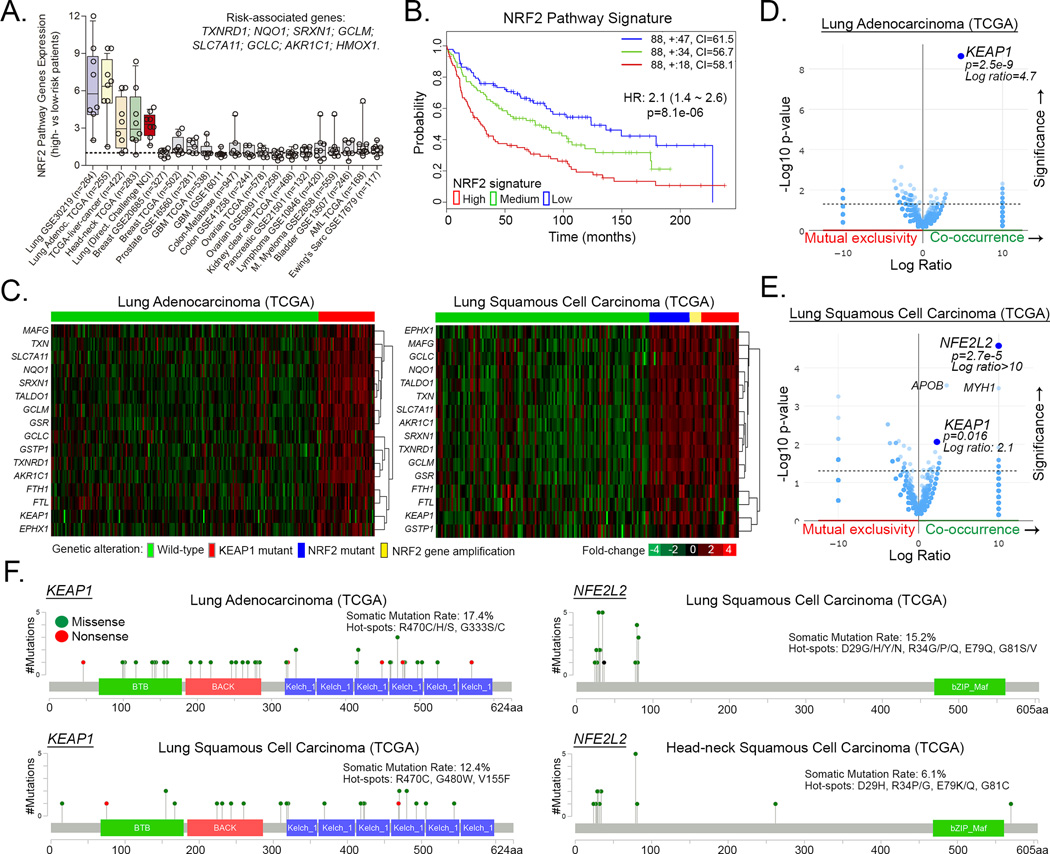
NRF2 pathway activating mutations drive poor survival in lung and head-neck carcinomas
Having elucidated the importance of NRF2-dependent attenuation of ER stress for alkylating agent cytotoxicity, we wanted to identify for which cancers this mechanism is most relevant. By using the SurvExpress tool, we found that upregulation of NRF2 targets/signature is mainly associated with high-risk/poor survival in lung adenocarcinoma, squamous cell carcinoma, liver cancer and head-neck carcinomas (Fig. 6A–B; and Fig. S3A–B); glioblastomas (GBM), colon, ovarian, pancreatic and other cancers showed no clear association (Fig. 6A). Comparative analysis of all TCGA cancer datasets indicated that lung adenocarcinoma, squamous cell carcinoma, and head-neck carcinoma, showed the highest frequency of genetic alterations in KEAP1 (mutation and/or deep-deletions) and/or NFE2L2/NRF2 (mutations and/or amplifications) (Fig. S3C; agreeing with (35, 36)). Noteworthy, tumors carrying mutant KEAP1 (17.4% and 12.4% of lung adeno- and squamous carcinomas, respectively) and/or NFE2L2/NRF2 (15.2% lung adenocarcinomas) or NFE2L2/NRF2 amplifications (5.4% lung squamous carcinoma) showed significant upregulation of various classical gene targets of NRF2 (for example, GCLC, GCLM, NQO1, TXNRD1, SRXN1, EPHX1, AKR1C1 and SLC7A11) and, consequently, displayed an upregulation of the NRF2 pathway signature (Fig. 6C, detailed in Fig. S4A). Mutation Enrichment Analysis (MEA) indicated that mutant KEAP1 is most associated with upregulation of NRF2 targets in lung adenocarcinomas (Fig. 6D), whereas both NFE2L2 and KEAP1 mutations are related to NRF2 pathway signature enrichment in lung squamous cells (Fig. 6E); and only NFE2L2 mutation in head and neck carcinomas (Fig. S4). The majority of KEAP1 gene alterations are missense mutations that occur in the Kelch (hotspots: R470*, G333*, G480*) or BTB domains of KEAP1 (Fig. 6F; and Fig. S5 for details), thereby impeding KEAP1 protein interaction with NRF2 (as reviewed by (37)). NFE2L2 mutations are mostly missense and occur within the first 100 amino acids (D29*, R34*, E79 and G81*) that contains the Neh2 domain which includes the two degrons that are specifically bound by KEAP1 and thus likely impede KEAP1-mediated NRF2 degradation (37).
Figure 6. NRF2 pathway activating mutations and prognosis in different cancers.
(A) NRF2 Pathway Gene Expressions Signature in high- vs low-risk/longer survival patients of different cancer datasets available with the SurvExpress database. (B) Kaplan-Meier curves showing the impact of NRF2 pathway expression signature upregulation (red), medium (green) or downregulation (blue) in lung cancer patients’ survival as determined by the SurvExpress tool. See Figure S3 for individual gene expressions across risk groups. (C) Heatmap representation of NRF2 target genes expressions in KEAP1 and NRF2/NFE2L2 altered versus wild-type tumor subsets of lung cancer. (D–E) Mutation Enrichment Analysis (MEA) showing that KEAP1 and NFE2L2 mutations are the genetic alterations most significantly associated with increased expression of the NRF2 targets in (D) lung adenocarcinomas and (E) lung squamous cell carcinomas, respectively. (F) Graphical representation of the localization, frequency and mutation hotspots with KEAP1 and NFE2L2 genes in lung and head-neck carcinomas; data from TCGA cBioportal.
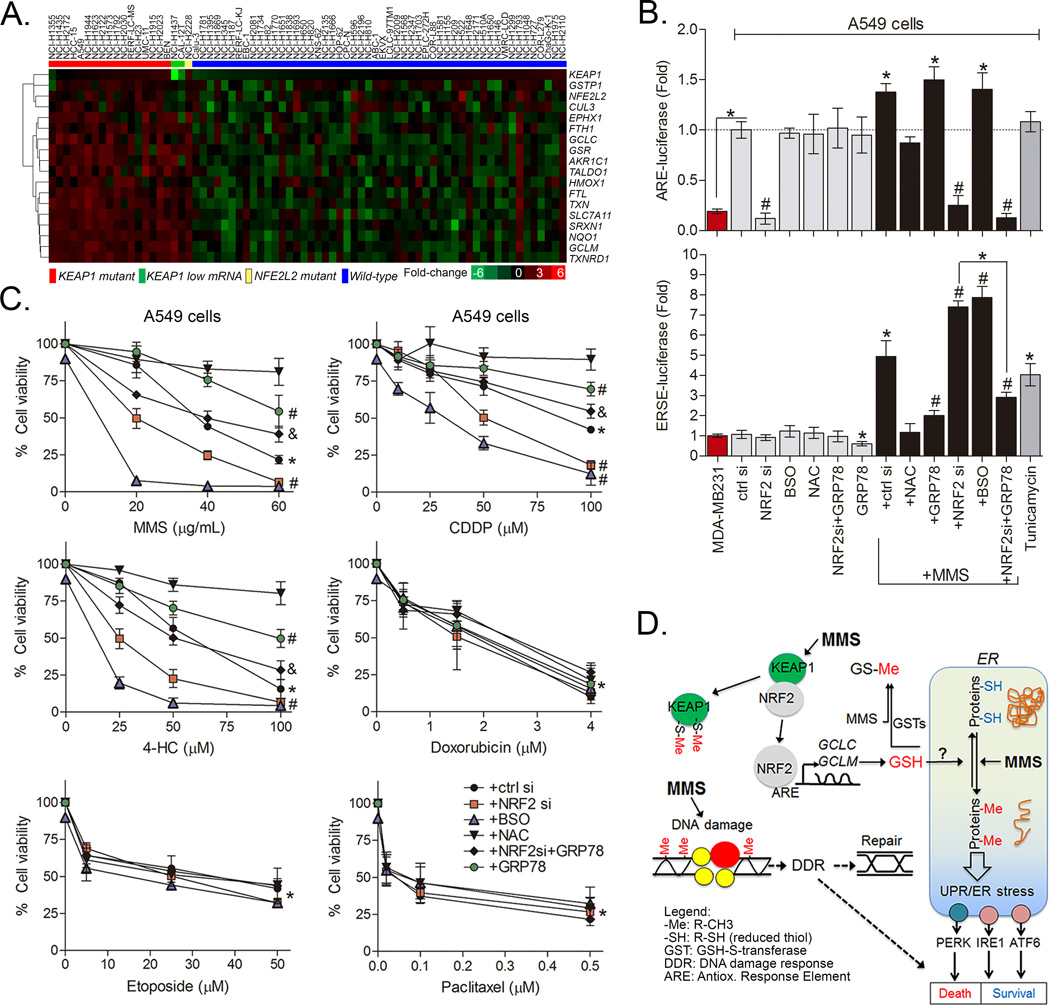
Constitutive NRF2 inhibits ER stress induction by alkylating agents in KEAP1 mutant lung cancer cells
In addition to lung tumor datasets, the NRF2 signature was examined by gene expression profiling of a panel of 77 lung cancer cell lines (E-MTAB-2706 RNA sequencing dataset) with known KEAP1/NFE2L2 mutational status (derived from Cosmic). The result confirmed an increase in NRF2 pathway signature in the KEAP1/NFE2L2 mutant cells (16 KEAP1 mutant; and 1 NFE2L2 mutant, the NCI-H2228 cell line) (Figs. 7A and S4); no change in ER stress markers was associated with KEAP1/NFE2L2 mutations (data not shown). Separate from mutation status in the Cosmic database, HCC15 was shown to carry a KEAP1 mutation (38). We also observed two other cell lines with the same activated NRF2 pathway signature (CAL-12T and NCI-H1437; indicated with green bar in Fig. 7A) that express low levels of KEAP1, which was previously confirmed for NCI-H1437 (35). The apparently KEAP1/NFE2L2 wild-type cell line NCI-H1793 displayed NRF2 gene target upregulation (Fig. 7A), but we have not been able to identify the basis of this phenotype. From our analysis we noted that the lung cancer cell line A549 carries a G333C KEAP1 hotspot mutation and could be used as a model to study the impact of NRF2 activating mutations on cell survival to different drug classes. As a consequence of KEAP1 mutation, basal ARE-luciferase is increased by ~5-fold in A549 compared to MDA-MB231 (Fig. 7B). Corroborating our aforementioned data, high NRF2 and glutathione/NAC protected while NRF2 knockdown and BSO sensitized A549 cells to MMS, 4-HC and CDDP but not doxorubicin, paclitaxel and etoposide (Fig. 7C). NAC, BSO and other controls alone are shown in Figure S4. In addition, MMS induced ERSE-luciferase reporter activation while NRF2 knockdown or GSH depletion with BSO potentiated this MMS effect upon ERSE reporter gene (Fig. 7B). Interestingly, a concomitant overexpression of GRP78 attenuated the impact of NRF2 knockdown on ERSE reporter gene activation by MMS (Fig. 7B). Together, these results indicate that NRF2 inhibition sensitizes KEAP1 mutant cells by potentiating ER stress induction following alkylating drugs exposure.
Figure 7. NRF2-GSH pathway activation inhibits ER stress and promotes survival to alkylating agents in the KEAP1 mutant lung cancer cell line A549.
(A) Heatmap representation of NRF2 target genes expressions in a panel of lung cancer cells (E-MTAB-2706 RNA-sequencing dataset) grouped based on their reported KEAP1 mutational status (Red, known KEAP1 mutation; Green, low KEAP1 expression; Yellow, NFE2L2 mutation; Blue, no known KEAP1/NFE2L2 mutation). See Figure S4 for individual gene expressions and NRF2 pathway signature. (B) Reporter gene assays showing the impact of NRF2 siRNA, GRP78 overexpression, BSO and NAC on ARE- and ERSE-luciferase activities in A549 cells; Tunicamycin (0.5 µM) was used as a positive control for ER stress induction. (C) Results of Cell-Titer Glo assays show the effect of NRF2 knockdown by siRNA, GRP78 overexpression, BSO and NAC pre-incubations (see methods) upon the dose-effect of chemotherapeutics in A549 cells (48 h treatment). Data are represented as the average ± SD of a representative experiment performed in triplicate and repeated twice. *different from untreated controls; #different from chemotherapy alone and from untreated cells. &different from chemotherapy+NRF2 siRNA (ANOVA/Tukey; p<0.05). (D) Schematic chart showing the proposed mechanism of NRF2-dependent control of protein-SH homeostasis and ER stress in cells exposed to alkylating agents.
Discussion
Despite decades of clinical experience with cytotoxic chemotherapy, the balance between tumor versus normal cell toxicity is still challenging and critical for successful treatment. Therefore, improving our understanding of drug specific mechanisms may lead to stratification of patients into those with tumors with genetic contexts that render them responsive to a particular chemotherapy versus those that are likely to be refractory. This knowledge may also provide insight into the adverse impact of these agents to healthy tissues.
Here we examined the gene expression responses of cancer cells to alkylating drugs followed by protein and cell function validations to reveal two pivotal processes of drug response: i) NRF2-dependent GSH synthesis and ii) activation of ER stress signaling. These are the same processes that we recently identified in D. melanogaster cells and primary mouse fibroblasts (22), indicating their conservation across species. While it is not novel that NRF2 is activated in response to alkylating agents (11, 12), ER stress induction by chemotherapies has not been extensively studied. We demonstrate a novel crosstalk between NRF2 and ER stress, where alkylating agent-induced NRF2 influences the magnitude of ER stress and PERK-induced apoptosis through maintenance of GSH pools.
GSH is a ubiquitous molecule that fulfills detoxification processes including xenobiotic conjugation by GSTs, MRP-mediated drug efflux and hydrogen peroxide detoxification via glutathione peroxidases among others (19, 21, 39). GSH is essential for conjugation with MMS as we previously determined in fly and mouse cells, and GST enzymes (GstE5 and GstE3 fly orthologues) knockdown strongly sensitized Kc167 cells to MMS (22). GSH conjugation also has been reported as a mechanism of CDDP detoxification (31). In addition, our cell models and CDDP studies (as in (39, 40)) have not observed a clear contribution of MRP1 (and MDR1) to cell survival in the context of alkylating drugs. On the other hand, MRP2 (which is low expressed in MDA-MB231 and U2OS cells) increased CDDP-glutathione conjugates efflux and decreased kidney injury in mice (41). Regarding the antioxidant role of GSH, alkylating agents are well-known inducers of ROS production (6, 42, 43), although our data and data from others (43) are contending that the contribution of ROS to toxicity is minor; likely a secondary phenotype consequent to GSH depletion. For instance, thiol antioxidants (GSH-E and dithiothreitol) completely protected COV434 granulosa cell lines exposed to 4-HC while ascorbic acid (thiol unrelated) provided only minor benefit (43). On the other hand, thiol unrelated antioxidants such as ascorbic acid and vitamin E have been described as preventing cyclophosphamide-induced ovary (44) and hepatic (45) toxicity in rats, indicating context dependent differences. Thus, it should be noted that long-term and tissue specific in vivo effects of alkylating agents upon oxidative stress may have more relevance and should not be discounted.
It is widely accepted that GSH/GSSG ratio in the ER is more oxidizing (i.e. concentration of GSSG is relatively higher) than in other cell compartments, although how glutathione specifically affects protein disulphide bond formation remains unsolved (19, 21). The oxidizing environment of the ER allows the formation of native disulphide bonds in a process mostly coordinated by protein disulphide isomerase (PDI). GSH/GSSG ratios within the ER are optimum to keep active site thiols of PDI in an oxidized state thereby allowing the enzyme to accept electrons from protein-SH residues (20), and the re-oxidation of PDI occurs through disulphide exchange with ER oxidoreductin-1 (ERO1) (46). Recently, it has been suggested that GSH (reduced form) might have a role in ensuring the redox homeostasis ER oxidoreductases so that they can catalyze reduction or isomerization reactions (19, 21). In our model, alkylating agent-induced ER stress could be counteracted by NAC while, if NRF2 is depleted or GCLC/GCLM enzyme is inhibited, ER stress is pronounced. It is also important to note that solely depleting GSH (with BSO or NRF2 knockdown) was not sufficient to affect cell survival (up to 72 h) or to cause ER stress; the presence of alkylating agent to trigger damage at the same time was required. MMS has been described as being able to directly alkylate proteins (47, 48) as well as GSH (49). In addition, alkylation of ER proteins such as glucose-regulated protein 94 (GRP94), calreticulin, valosin containing protein (VCP) and HSP90 has also been reported in CDDP-treated Cochlear and kidney cells (50). Even though we did not demonstrate that our treatment exposures directly alkylated proteins, our data indicates that the NRF2-mediated GSH synthesis response is key for alkylation detoxification; without this mechanism alkylating agents seem to react with proteins and impair ER homeostasis (Fig. 7D; Working model).
Moreover, if the NRF2/GSH-mediated inhibition of ER stress is a pleiotropic mechanism, one could expect that cancers harboring constitutive activation of NRF2 pathway are prone to be less sensitive to alkylating agent-induced ER stress and could display a lack of clinical efficacy of alkylating agents. From various screened cancer types, we identified subsets of lung and head-neck carcinomas harboring KEAP1/NFE2L2 mutations, which showed upregulated expression of NRF2 targets and high-risk/poor survival. In the KEAP1 mutant A549 cell line, we demonstrated that constitutive NRF2 activation inhibits alkylating agents-induced ER stress while GRP78 chaperone overexpression exerts a protective effect. In keeping with our findings, CDDP has also been shown to cause ER stress in different models (51–54) and GRP78 was found to reverse CDDP toxicity in melanoma (54) and ovarian cancer cells (55). Moreover, while modern alkylating agents such as temozolomide (TMZ) could be expected to bypass the NRF2-GSH requirement, it has been shown that BSO potentiates and NAC attenuates TMZ toxicity in gliomas cells (56). TMZ also induces NRF2 (57) and GRP78 upregulation in gliomas (58), supporting the concept of NRF2-GSH/ER stress as a general response to alkylating drugs.
Throughout our investigations, we found a specific requirement for NRF2-GSH pathway in alkylating agent survival, but not for paclitaxel, etoposide and doxorubicin. Corroborating our data, Wang et al showed that the classic alkylating agents chlorambucil, BCNU and the cyclophosphamide metabolite acrolein induced luciferase activity in MCF-7 AREc32 cells; an effect augmented by BSO (59). Paclitaxel, methotrexate and doxorubicin had no effect whereas etoposide acted as a weak inducer of ARE-luciferase in a GSH-independent mechanism (59). Even though it seems that non-alkylating drugs are unlikely to directly activate NRF2, other studies have demonstrated that elevated levels of NRF2 can confer resistance to these compounds in other contexts. For instance, NRF2 depletion sensitized gallbladder cancer cells to 5-FU (60), in contrast to the lack of effect of 5-FU treatment in A549 (61). NRF2-shRNA clones of A549 cells showed sensitivity to etoposide and carboplatin (62). Interestingly, NRF2 siRNA was effective in enhancing CDDP toxicity in A549 cells (high NRF2) but not in LC-AI and NCI-H292 (low NRF2) (61). CDDP and paclitaxel toxicities were enhanced by NRF2 siRNA in SPEC-2 (high NRF2) but not Ishikawa (low NRF2) endometrial cancer cells (63). These contrasts could be attributed to differences in NRF2 knockdown efficiency and timing (which may lead to varying levels of depletion of NRF2 targets), stable shRNA clones versus transient siRNA protocols as well as cell line specific phenotypes. Despite these varied results, the specific commitment of the GCLC/GCLM-GSH arm of the NRF2 pathway seems specific for surviving alkylating agents.
In summary, the data presented herein delineate a novel alkylating agents-related NRF2-dependent control of thiol damage to ER proteins, which dictates the magnitude of ER stress activation and PERK-mediated apoptosis. This NRF2 control is dependent upon maintenance/synthesis of GSH pools, which is a pivotal metabolite for both drug detoxification and protein-SH homeostasis. This effect, at least in the contexts used here, are independent of NRF2 antioxidant functions or ROS (Fig. 7D). Consequently, while NRF2 activating mutations could confer resistance and alkylating therapy failure, identifying an altered NRF2-GSH/ER stress status offers the opportunity to select alternative chemotherapeutics that may retain efficacy.
Supplementary Material
Acknowledgments
We thank support and comments from members of the Bishop Lab.
Financial Support: Funding was by the NIH (K22ES012264, 1R15ES019128 and 1R01CA152063), Voelcker Fund Young Investigator Award, GCCRI Ambassador’s Circle Research Support Award and San Antonio Area Foundation grant to A.J.R. Bishop; DOD-CDMRP Breast Cancer Research Program Postdoctoral Fellowship (W81XWH-14-1-0026) to A. Zanotto-Filho and MCTI/CNPq Universal (485758/13-0) to A. Zanotto-Filho and J.C.F. Moreira; NCI-T32 training grant (5T32CA148724) to S.S. Tonapi; Greehey Fellowship, a Translational Science Training Across Disciplines Scholarship from the UT System Graduate Programs Initiative, a CPRIT fellowship (RP101491) and a NCI-T32 training grant (5T32CA148724) to A. Gorthi; and a NIH-P30 Cancer Center Support Grant (CA054174) to Cancer Therapy & Research Center at UTHSCSA.
Footnotes
Conflict of Interest Statement: The authors declare that no conflict of interest exists.
References
- 1.Hernandez-Aya LF, Gonzalez-Angulo AM. Adjuvant systemic therapies in breast cancer. Surg Clin North Am. 2013;93:473–491. doi: 10.1016/j.suc.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Jones RB, Matthes S, Dufton C, Bearman SI, Stemmer SM, Meyers S, et al. Pharmacokinetic/pharmacodynamic interactions of intensive cyclophosphamide, cisplatin, and BCNU in patients with breast cancer. Breast Cancer Res Treat. 1993;26(Suppl):S11–S17. doi: 10.1007/BF00668355. [DOI] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 4.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6:2585–2597. [PubMed] [Google Scholar]
- 5.Chan BA, Coward JI. Chemotherapy advances in small-cell lung cancer. J Thorac Dis. 2013;5:S565–S578. doi: 10.3978/j.issn.2072-1439.2013.07.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CJ, Lee CC, Shih YL, Lin TY, Wang SH, Lin YF, et al. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med. 2012;52:377–391. doi: 10.1016/j.freeradbiomed.2011.10.487. [DOI] [PubMed] [Google Scholar]
- 7.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, et al. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Malet-Martino M, Gilard V, Martino R. The analysis of cyclophosphamide and its metabolites. Curr Pharm Des. 1999;5:561–586. [PubMed] [Google Scholar]
- 9.Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- 10.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Paonessa JD, Zhang Y. Mechanism of chemical activation of Nrf2. PLoS One. 2012;7:e35122. doi: 10.1371/journal.pone.0035122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore MJ. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet. 1991;20:194–208. doi: 10.2165/00003088-199120030-00002. [DOI] [PubMed] [Google Scholar]
- 14.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 19.Birk J, Meyer M, Aller I, Hansen HG, Odermatt A, Dick TP, et al. Endoplasmic reticulum: reduced and oxidized glutathione revisited. J Cell Sci. 2013;126:1604–1617. doi: 10.1242/jcs.117218. [DOI] [PubMed] [Google Scholar]
- 20.Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanotto-Filho A, Dashnamoorthy R, Loranc E, de Souza LH, Moreira JC, Suresh U, et al. Combined Gene Expression and RNAi Screening to Identify Alkylation Damage Survival Pathways from Fly to Human. PLoS One. 2016;11:e0153970. doi: 10.1371/journal.pone.0153970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolova T, Ensminger M, Lobrich M, Kaina B. Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair (Amst) 2010;9:1050–1063. doi: 10.1016/j.dnarep.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Zanotto-Filho A, Delgado-Canedo A, Schroder R, Becker M, Klamt F, Moreira JC. The pharmacological NFkappaB inhibitors BAY117082 and MG132 induce cell arrest and apoptosis in leukemia cells through ROS-mitochondria pathway activation. Cancer Lett. 2010;288:192–203. doi: 10.1016/j.canlet.2009.06.038. [DOI] [PubMed] [Google Scholar]
- 25.Cox DP, Cardozo-Pelaez F. High Throughput Method for Assessment of Cellular Reduced Glutathione in Mammalian Cells. J Environ Prot Sci. 2007;1:23–28. [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JS, Raines RT. Dibromobimane as a fluorescent crosslinking reagent. Anal Biochem. 1995;225:174–176. doi: 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]
- 27.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komiya S, Gebhardt MC, Mangham DC, Inoue A. Role of glutathione in cisplatin resistance in osteosarcoma cell lines. J Orthop Res. 1998;16:15–22. doi: 10.1002/jor.1100160104. [DOI] [PubMed] [Google Scholar]
- 32.Koharyova M, Kollarova M. Thioredoxin system - a novel therapeutic target. Gen Physiol Biophys. 2015;34:221–233. doi: 10.4149/gpb_2015006. [DOI] [PubMed] [Google Scholar]
- 33.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 36.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abazeed ME, Adams DJ, Hurov KE, Tamayo P, Creighton CJ, Sonkin D, et al. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73:6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp SY, Smith V, Hobbs S, Kelland LR. Lack of a role for MRP1 in platinum drug resistance in human ovarian cancer cell lines. Br J Cancer. 1998;78:175–180. doi: 10.1038/bjc.1998.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen DW, Pouliot LM, Hall MD, Gottesman MM. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev. 2012;64:706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen X, Buckley B, McCandlish E, Goedken MJ, Syed S, Pelis R, et al. Transgenic expression of the human MRP2 transporter reduces cisplatin accumulation and nephrotoxicity in Mrp2-null mice. Am J Pathol. 2014;184:1299–1308. doi: 10.1016/j.ajpath.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syu JP, Chi JT, Kung HN. Nrf2 is the key to chemotherapy resistance in MCF7 breast cancer cells under hypoxia. Oncotarget. 2016 doi: 10.18632/oncotarget.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai-Turton M, Luong BT, Tan Y, Luderer U. Cyclophosphamide-induced apoptosis in COV434 human granulosa cells involves oxidative stress and glutathione depletion. Toxicol Sci. 2007;98:216–230. doi: 10.1093/toxsci/kfm087. [DOI] [PubMed] [Google Scholar]
- 44.Gurgen SG, Erdogan D, Elmas C, Kaplanoglu GT, Ozer C. Chemoprotective effect of ascorbic acid, alpha-tocopherol, and selenium on cyclophosphamide-induced toxicity in the rat ovarium. Nutrition. 2013;29:777–784. doi: 10.1016/j.nut.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Cuce G, Cetinkaya S, Koc T, Esen HH, Limandal C, Balci T, et al. Chemoprotective effect of vitamin E in cyclophosphamide-induced hepatotoxicity in rats. Chem Biol Interact. 2015;232:7–11. doi: 10.1016/j.cbi.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 46.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F, Bartels MJ, Pottenger LH, Gollapudi BB. Differential adduction of proteins vs. deoxynucleosides by methyl methanesulfonate and 1-methyl-1-nitrosourea in vitro. Rapid Commun Mass Spectrom. 2005;19:438–448. doi: 10.1002/rcm.1806. [DOI] [PubMed] [Google Scholar]
- 48.Paik WK, DiMaria P, Kim S, Magee PN, Lotlikar PD. Alkylation of protein by methyl methanesulfonate and 1-methyl-1-nitrosourea in vitro. Cancer Lett. 1984;23:9–17. doi: 10.1016/0304-3835(84)90056-9. [DOI] [PubMed] [Google Scholar]
- 49.Trezl L, Park KS, Kim S, Paik WK. Studies on in vitro S-methylation of naturally occurring thiol compounds with N-methyl-N-nitrosourea and methyl methanesulfonate. Environ Res. 1987;43:417–426. doi: 10.1016/s0013-9351(87)80042-7. [DOI] [PubMed] [Google Scholar]
- 50.Karasawa T, Sibrian-Vazquez M, Strongin RM, Steyger PS. Identification of cisplatin-binding proteins using agarose conjugates of platinum compounds. PLoS One. 2013;8:e66220. doi: 10.1371/journal.pone.0066220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi S, Tan P, Yan B, Gao R, Zhao J, Wang J, et al. ER stress and autophagy are involved in the apoptosis induced by cisplatin in human lung cancer cells. Oncol Rep. 2016;35:2606–2614. doi: 10.3892/or.2016.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Wang C, Su J, Xie Q, Ma L, Zeng L, et al. Tolerance to endoplasmic reticulum stress mediates cisplatin resistance in human ovarian cancer cells by maintaining endoplasmic reticulum and mitochondrial homeostasis. Oncol Rep. 2015;34:3051–3060. doi: 10.3892/or.2015.4283. [DOI] [PubMed] [Google Scholar]
- 53.Rabik CA, Fishel ML, Holleran JL, Kasza K, Kelley MR, Egorin MJ, et al. Enhancement of cisplatin [cis-diammine dichloroplatinum (II)] cytotoxicity by O6-benzylguanine involves endoplasmic reticulum stress. J Pharmacol Exp Ther. 2008;327:442–452. doi: 10.1124/jpet.108.141291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang CC, Mao ZG, Avery-Kiejda KA, Wade M, Hersey P, Zhang XD. Glucose-regulated protein 78 antagonizes cisplatin and adriamycin in human melanoma cells. Carcinogenesis. 2009;30:197–204. doi: 10.1093/carcin/bgn220. [DOI] [PubMed] [Google Scholar]
- 55.Li W, Wang W, Dong H, Li Y, Li L, Han L, et al. Cisplatin-induced senescence in ovarian cancer cells is mediated by GRP78. Oncol Rep. 2014;31:2525–2534. doi: 10.3892/or.2014.3147. [DOI] [PubMed] [Google Scholar]
- 56.Rocha CR, Garcia CC, Vieira DB, Quinet A, de Andrade-Lima LC, Munford V, et al. Glutathione depletion sensitizes cisplatin- and temozolomide-resistant glioma cells in vitro and in vivo. Cell Death Dis. 2014;5:e1505. doi: 10.1038/cddis.2014.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma L, Liu J, Zhang X, Qi J, Yu W, Gu Y. p38 MAPK-dependent Nrf2 induction enhances the resistance of glioma cells against TMZ. Med Oncol. 2015;32:69. doi: 10.1007/s12032-015-0517-y. [DOI] [PubMed] [Google Scholar]
- 58.Zanotto-Filho A, Braganhol E, Klafke K, Figueiro F, Terra SR, Paludo FJ, et al. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett. 2015;358:220–231. doi: 10.1016/j.canlet.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 59.Wang XJ, Hayes JD, Wolf CR. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 60.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. 68 e1–68 e4. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 61.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 62.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.