Abstract
High affinity aptamer-based biomarker discovery has the advantage of simultaneously discovering an aptamer affinity reagent and its target biomarker protein. Here, we demonstrate a morphology-based tissue aptamer selection method that enables us to use tissue sections from individual patients and identify high-affinity aptamers and their associated target proteins in a systematic and accurate way. We created a combinatorial DNA aptamer library that has been modified with thiophosphate substitutions of the phosphate ester backbone at selected 5′dA positions for enhanced nuclease resistance and targeting. Based on morphological assessment, we used image-directed laser microdissection (LMD) to dissect regions of interest bound with the thioaptamer (TA) library and further identified target proteins for the selected TAs. We have successfully identified and characterized the lead candidate TA, V5, as a vimentin-specific sequence that has shown specific binding to tumor vasculature of human ovarian tissue and human microvascular endothelial cells. This new Morph-X-Select method allows us to select high-affinity aptamers and their associated target proteins in a specific and accurate way, and could be used for personalized biomarker discovery to improve medical decision-making and to facilitate the development of targeted therapies to achieve more favorable outcomes.
Keywords: morphology-based tissue aptamer selection, thioaptamer, laser microdissection, targeted protein, ovarian cancer
Biomarker-based diagnosis, prognosis, and targeted therapy are essential parts of personalized medicine. Identifying molecular profiles of individual patients and developing biomarkers specific to those patients will improve clinical decision making and benefit targeted therapies. Our Morph-X-Select technology, a morphology-based tissue aptamer selection method, enables us to identify high-affinity aptamer reagents and their associated protein biomarkers from individual patients. It opens up a new avenue for identification of high binding-affinity aptamer reagents and their target biomarker proteins for personalized targeted cancer therapy.
Although therapeutic and imaging antibodies have achieved remarkable clinical success (1–3), current antibody biomarker imaging agents are limited by an antibody's immunogenicity, stability, reusability, and ability to be modified with imaging labels or drug warheads. An attractive alternative to antibody-based techniques is the use of assays incorporating aptamers (functional nucleic acid ligands). Aptamers are structurally distinct RNA and DNA molecules that can bind with high affinity (picomolar to low nanomolar) to protein targets. Aptamers are robust, easily labeled, prepared in vitro, and generally not immunogenic. They can bind specifically to their targets and have a high potential for diagnostic, imaging, and therapeutic applications (4,5). The aptamer binding affinity-based strategy has been demonstrated by simultaneously identifying CypB as a serum biomarker and generating a new reagent to recognize it in body fluids (6,7). Although limited progress has been made using non-modified aptamers, new generations of modified aptamers are being rapidly developed for detecting and manipulating molecular changes, biomarker discovery, and targeted therapies. Research on phosphate- and sugar-modified aptamers led to the FDA-approved drug Macugen, an inhibitor of vascular endothelial growth factor (VEGF) for the treatment of age-related macular degeneration (AMD) (8).
Over the past several years, we have shown that a combinatorial DNA aptamer library modified with a thiophosphate substitution of the phosphate ester backbone (thioaptamer, TA) on the 5′-side of many of the dA positions increased the binding affinity to targeted cells (9–11). The monothiophosphate substitutions, which also enhance nuclease resistance, can be introduced into the aptamer library by including thio-substituted dNTP-α–S (commercially available) into the enzymatic Taq amplification step during PCR amplification. This allows for selection of both backbone and sequence. We have successfully applied the modified TA library to solution- and bead-based selections in our laboratory (9–14). Using purified primary human ovarian cancer endothelial cells from patient tumors, we successfully selected high-affinity TAs binding to tumor endothelial cells and identified annexin A2 as one of the potential target proteins (15).
To overcome the limitations of cell-based systematic evolution of ligands by exponential enrichment (Cell-SELEX) (16), which can only use cell lines or isolated cells as targets, we present a morphology-based tissue aptamer selection method (Morph-X-Select) that enables us, for the first time, to use targeted tissue sections from individual patients and identify high binding-affinity aptamer sequences and their associated target proteins in a systematic and accurate way. We combined our modified TA library with Morph-X-Select to simultaneously select TAs specifically binding to ovarian tumor vasculature or tumor cells, but not to the tumor stromal cells. Unlike traditional aptamer tissue selection using whole tissue sections (17,18), we used an image directed laser microdissection (LMD) technique to dissect only regions of interest (ROIs) bound with TAs based on morphological assessment of the tissue, identified the high-affinity TA sequences by next-generation sequencing (NGS), and further identified the targeted proteins by mass spectrometry (MS). Using the Morph-X-Select approach, we are able to select tissue-specific TAs in a rapid and cost-effective way from large TA libraries. Our strategy offers a novel way to select aptamers and their target proteins from ROIs for an individual patient.
Materials and methods
Reagents
Oligonucleotide primers were synthesized by Midland Certified Reagents (Midland, TX). Streptavidin-coated magnetic particles were purchased from Pure Biotech (Middlesex, NJ). Taq polymerase and the chirally pure Sp isomer of dATP-α–S were obtained from Axxora LLC (San Diego, CA). Anti-human CD31 and CD44 antibodies were purchased from eBioscience, Inc. (San Diego, CA). Anti-human vimentin polyclonal antibody (Cat. #AF2105) and normal goat IgG (Cat. #AB-108-C) were both purchased from R&D Systems (Minneapolis, MN).
Human tissue samples and cell lines Human epithelial ovarian cancer tissue was collected at the time of standard care surgical intervention at the University of Texas M.D. Anderson Cancer Center (MDACC). All tumor samples in the study were phenotyped by the Department of Pathology and Laboratory Medicine at MDACC. This study has been approved by the MDACC Institutional Review Board. We specially focused on high-grade serous ovarian cancer (HGSC), which is the most common and deadly epithelial ovarian cancer (19). Fresh residual ovarian tumor tissues were embedded with optimal cutting temperature (OCT) (Thermo Fisher Scientific, Waltham, MA) compound. All tumor samples for this study were collected prior to initiation of any therapy. Five ovarian tumor tissue samples and five normal ovarian tissue samples were used for this study, although only one pair of tumor-normal tissue was used for Morph-X-Select. Other tissues were used for validation of TA binding and target protein expression. Ovarian cancer IGROV cells and OVCAR3 cells (ATCC, Manassas, VA) were maintained in RPMI-1640 medium supplemented with 15% fetal bovine serum (FBS) and 0.1% gentamicin sulfate (tissue culture reagents purchased from Life Technologies, Grand Island, NY). Human microvascular endothelial cells (HMVECs) were a kind gift from Rong Shao (University of Massachusetts, Amherst, MA). HMVECs were grown in M131 medium with microvascular growth supplement (MVGS) (Life Technologies). All experiments were performed at 70%–80% cell confluence with 5% CO2 at 37°C.
DNA thioaptamer library
The random single-stranded DNA (ssDNA) library with a 30-nucleotide random region flanked by PCR primers was chemically synthesized using standard phosphoramidite chemistry, and the oligonucleotide library was purified by HPLC under reverse-phase conditions. DNA TA combinatorial library synthesis was described previously (9). The library was PCR-amplified with dATP-α–S in place of dATP and with normal dNTPs for the remaining bases; sense or anti-sense primers were labeled with 5′-biotin. The ssDNA was isolated by treating the PCR products with streptavidin-coated magnetic beads and alkaline denaturation. The ssDNA TA library was taken up in binding buffer (PBS with 5 mM MgCl2) and heated at 95°C for 5 min and then cooled slowly at room temperature to form the proper secondary structure. The library was purified by 10,000 MWCO centrifugal filter columns (Millipore, Billerica, MA), and size was confirmed by gel electrophoresis (15% polyacrylamide, Bio-Rad, Hercules, CA).
Morph-X-Select thioaptamer tissue selection
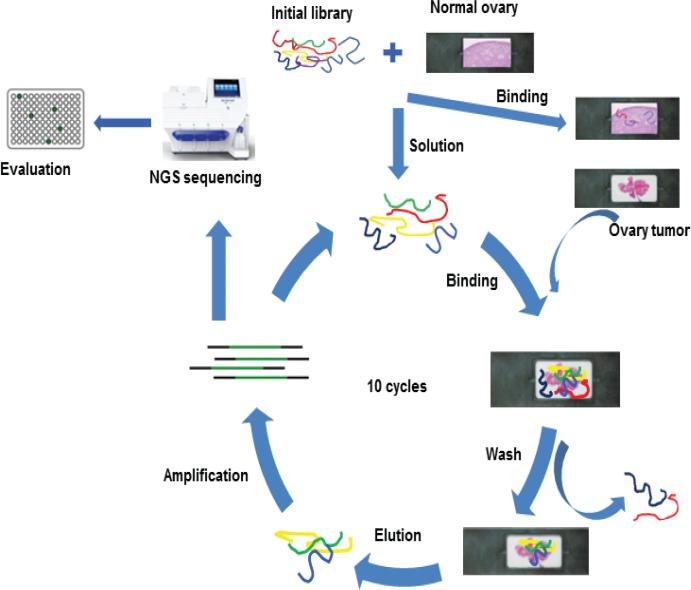
One pair of HGSC tumor tissue and normal ovarian tissue was used for Morph-X-Select. Fresh HGSC tumor or normal ovarian tissue was embedded in OCT compound prior to cryostat sectioning. Sections were cut to 5–10 microns in thickness, mounted on window slides suitable for LMD (JUNGWOO F&B Corp, Kyonggi-Do, South Korea), and then stained with hematoxylin and eosin (H&E). Based on cell morphology and pathological diagnosis, ROIs were defined directly on a touch screen display and subsequently cut out by a laser beam and dropped into a collection cap. TAs bound to LMD-harvested tissues were released by heating at 95°C for 5 min and then amplified by PCR with a dNTP mix containing dATP-α–S and a biotin-labeled primer. The ssDNA was separated from the PCR products with streptavidin magnetic beads and was used in the next selection cycle (Figure 1). Ten Morph-X-Select cycles were performed using tissue from the same patient. The selection stringency was increased by gradually decreasing the incubation time for tumor sections and by increasing the number of washes after TA library binding to tissue sections.
Figure 1.
Scheme of the tissue Morph-X-Select method.
Sequence analysis
After the 10th round of Morph-X-Select, we PCR-amplified the TA pools and used 100 ng of the gel-purified products for fragment library construction (Ion Plus Fragment Library Kit, Cat. #4471252; Life Technologies). We then sequenced the library using the Ion Personal Genome Machine (PGM) System (Life Technologies). The sequence data were analyzed using Aptaligner (20), which builds Markov models for each library and uses noise and length filters to reduce computation time. Based on the optimal alignment and statistical analysis, TA sequences with a high frequency of occurrence and high sequence homology were selected. The top 250 sequences were grouped using bottom-up hierarchical clustering by Aptaligner and the Clustal W program (www.clustal.org/clustal2/). Their secondary structures were predicted using the Mfold program (http://unafold.rna.albany.edu/?q=mfold).
Validation of specific binding of selected thioaptamers to human endothelial cells or tumor cells
After sequence analysis, a TA panel was selected, chemically synthesized with a 5′-Cy3 dye, and tested for specific binding to tumor endothelial cells or ovarian tumor cells. Sections of frozen human ovarian tumor were first incubated with Universal Blocker blocking buffer in TBS (Cat. #36000; Thermo Fisher Scientific) and then incubated with 50 nM TA for 30 min at room temperature with subsequent washing, fixing, and nuclei counterstaining. The relative extent of TA binding to the tissue was assessed by fluorescence microscopy analysis using a Nikon Eclipse TE2000-E inverted microscope (Nikon Instruments Inc., Melville, NY) on the tumor vasculature or tumor cells. CD31 and CD44 antibodies were used as positive controls for specific binding of TAs to endothelial cells and tumor cells. Hoechst 33342 (Thermo Fisher Scientific) was used to counterstain nuclei. Cells were incubated with TAs first, then washed, fixed, and the nuclei were counterstained. Several TAs that showed high binding affinity for human ovarian tumor vasculature or tumor cells were selected for further studies.
Affinity pull-downs and mass spectrometry
Based on a previous aptamer-associated biomarker study (21) and the use of chromatin immunoprecipitation (ChIP) for identifying protein–DNA interactions (22), we developed an MS-based pull-down method to identify TA-associated target proteins. The TA-associated target proteins were pulled down by streptavidin magnetic beads after cross-linking biotinylated TAs with HMVECs or IGROV cells and then analyzed by MS. Cells at 90%–95% confluence were used for all TA pull-down experiments. After being washed with cold PBS, cells were incubated with 200 nM biotinylated TAs, or the TA library as a control, in Dulbecco's PBS (Life Technologies) and gently agitated for 2 h at 4°C. After incubation, bound TAs and targeted proteins in cells were cross-linked with 1% formaldehyde for 10 min at ambient temperature and then quenched with glycine. Cells were harvested, washed, lysed with lysis buffer (Pierce Biotechnology, Waltham, MA), and treated with protease inhibitors (Sigma-Aldrich Corp., St. Louis, MO). The lysates were freeze-thawed for 30 min on ice and cleared by centrifugation at 10,000 × g for 2 min at 4°C. To pull down the labeled proteins, equal amounts of cell lysate were incubated with pre-washed streptavidin magnetic beads for 1 h at ambient temperature under continuous rotation. On-bead protein digestion was performed to isolate fragments of the targeted proteins for MS analysis (LTQ-Orbitrap-XL; Thermo Fisher Scientific). Each sample was analyzed in triplicate. The raw data files were processed to generate a Mascot Generic Format file with Mascot Distiller and searched against the SwissProt_2012_01 (Human) database using the Mascot search engine v2.3.02 (www.matrixscience.com/search_form_select.html) run on an in-house server. Identified proteins were ranked according to their scores. After comparing the protein lists from different selected TAs with a non-specific TA library control, a potential target protein was identified, and expression of the identified target protein was further validated by antibody detection and TA non-competition binding assays with antibodies against the target protein.
Aptamer-antibody non-competition binding
Aptamer-antibody non-competition binding was performed with a anti-human vimentin polyclonal antibody and Cy3-V5 (the selected TA with high binding affinity to tumor vessels) at different concentrations. IGROV cells (60%–70% confluent) were incubated with anti-human vimentin polyclonal antibody at 10 μg/mL, normal goat IgG, or serum-free RPMI medium at 37°C overnight. After washing to remove excess antibody, cells were incubated with Cy3-V5 at different concentrations (250 nM and 500 nM in PBS) for 1 h at 37°C with subsequent washing, fixing, and nuclei counterstaining with Hoechst 33342. The binding intensity of V5 to the cells was assessed by fluorescence microscopy (Nikon TE2000-E).
Results and discussion
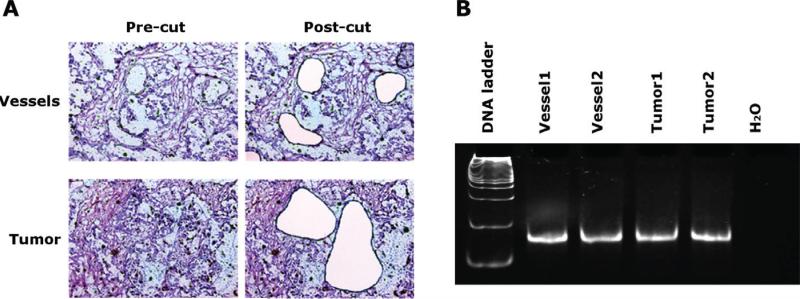
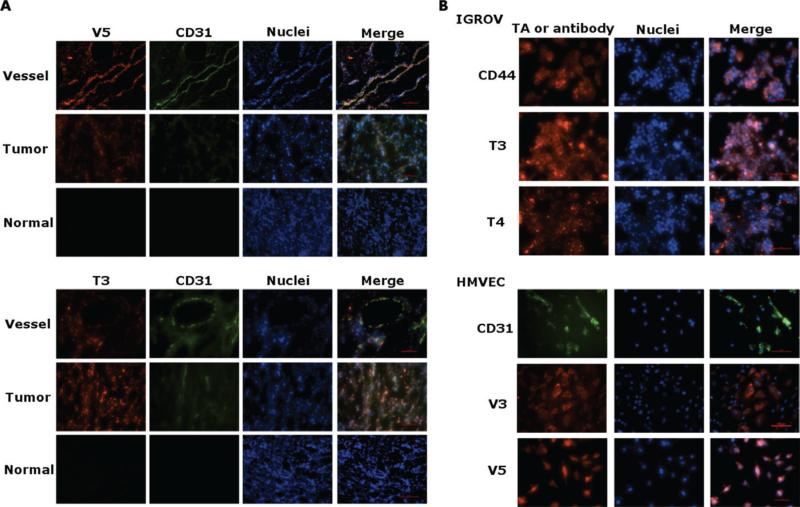
Our innovative morphology-based aptamer selection method, Morph-X-Select, enables us to use highly targeted heterogeneous tissue sections from individual patients and identify high binding affinity aptamer sequences as affinity reagents, along with their associated tumor biomarkers. Morph-X-Select has the advantage of precise dissection of targeted tumor cells compared to the traditional tissue SELEX method, which uses scraped tissue with highly heterogeneous cell populations. Morph-X-Select reduces the occurrence of false-positive aptamers and simultaneously selects high binding affinity aptamers for multiple ROIs. We screened our TA library for membrane-bound proteins in tissue sections that have intact structure. Based on cell morphology and pathological diagnosis, tumor vessels and tumor cells were defined on a touch screen, cut out by a laser beam, and collected (Figure 2A). TAs bound to LMD-dissected tumor cells and tumor vessels were eluted, PCR amplified, and enriched by 10 rounds of selection with tissue from the same patient. Figure 2B shows that the TA pool was recovered from the LMD-dissected tumor cells and tumor vessels. To identify high binding affinity sequences from LMD tissue, corresponding PCR products were submitted for NGS using the Ion PGM System. This method allows the discrimination of sequences that arise from experimental biases (such as the conventional SELEX-approach of cloning, colony picking, and Sanger sequencing of a small number of colonies) rather than true high-affinity target binding. Ten sequences showing the highest frequency of occurrence within each pool (tumor vasculature or tumor cells) were selected as TA candidates, synthesized, and tested for binding affinity to ovarian cancer tissue. Sequences identified from LMD-dissected tumor vasculature (V3) or tumor cells (T3) showed enhanced binding to only tumor vessels or to tumor cells, respectively (Figure 3A). To confirm specific binding of selected TAs to ovarian tumor tissue, a random TA sequence (R4) was used as a negative control for all binding assays (data not shown). Those selected and confirmed sequences with high binding affinity toward patient ovarian tumor tissue and cell lines were used for identifying the biomarker proteins associated with TA binding (Table 1).
Figure 2. Morph-X-Select procedure.
(A) Laser microdissection (LMD) of blood vessels (pre- and post-cut, upper row) and tumor cells (pre- and post-cut, lower row) from human ovarian cancer. Scale bar = 40 μm. (B) PCR amplification of eluted thioaptamers from LMD-harvested blood vessels and tumor cells.
Figure 3. Specific binding of selected thioaptamers (TAs) to human ovarian cancer tissue and cell lines.
(A) TAs identified by laser microdissection (LMD) and next-generation sequencing (NGS) analysis showed specific binding to ovarian tumor vessels (V5) and tumor cells (T3) but not to normal ovarian tissue. Scale bar = 50 μm or 100 μm. (B) High binding affinity of selected TAs has been confirmed with human microvascular endothelial cells (HMVECs) (V3, V5) or IGROV cells (T3, T4). Cells were fixed after TA–cell incubation. Red: Cy3-labeled TA or CD44 antibody. Green: CD31 antibody–labeled vessels. Blue: nuclei. Scale bar = 100 μm.
Table 1.
Laser microdissection (LMD)– and next-generation sequencing (NGS)– selected thioaptamer (TA) sequences.
| Group | Name | Frequency of occurrence | Sequences |
|---|---|---|---|
| Tumor Vessels | V3 | 9158 | 5′-CGCTCGGATCGATAAGCTTCG-ATCATCGATCACCTCTTCAAGAACATCCCT- GTCACGGATCCTCTAGAGCACTG-3′ |
| Tumor Vessels | V5 | 3684 | 5′-CGCTCGGATCGATAAGCTTCG-CATAGACCCAGCTGGTCCGGAAAATAAGAT-GTCACGGATCCTCTAGAGCACTG-3′ |
| Tumor Cells | T3 | 3374 | 5′-CGCTCGGATCGATAAGCTTCG-AGCCTGAGTTTTTGCCGCGATCTCGACCTCC- GTCACGGATCCTCTAGAGCACTG-3′ |
| Tumor Cells | T4 | 2189 | 5′-CGCTCGGATCGATAAGCTTCG-TCTTCAAGAACATCCCTGAAGTAACTTCCT-GTCACGGATCCTCTAGAGCACTG-3′ |
| Random Control | R4 | 5′-CCCACTTATCGTCCCTTAATGAGTTTACTCGCACACCGGACAGCCGTCGGA-TGGCTGGATCCGTAGCGGTCCGG-3′ |
Adenines that have monothioated phosphates on their 5′ sides are shown in red. The 5′- and 3′-primer regions of selected TAs are separated from the central random regions of the aptamers by dashes.
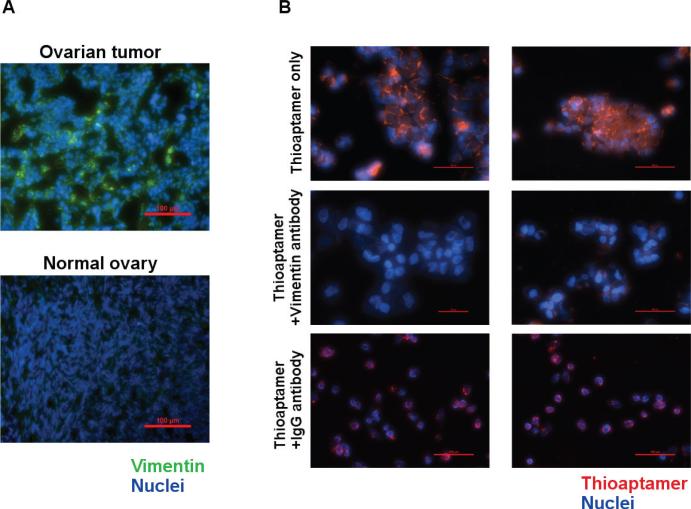
Limited by the amount of protein that can be isolated from LMD-dissected tumor vasculature or tumor cells, we used cell lines (HMVEC and IGROV) that have been validated with high binding affinity to selected TAs (Figure 3B) for target protein identification. An MS-based proteomics approach (21,22) has been used to identify target proteins of selected TAs binding with HMVECs. After comparing the protein lists of selected TAs with that of a non-specific TA library, vimentin was identified as a top potential target protein in three independent MS pull-down experiments using TA V5. As the result of a Mascot database search (www.matrixscience.com/search_form_select.html), vimentin was found to be the most probable target protein, with the highest overall abundance score of the TA V5 pull-down samples (Supplementary Table S1) compared with the low abundance score of the TA library control sample (Supplementary Table S2), with an abundance score ratio of 30:1. To validate vimentin as a target protein of TA V5, vimentin expression in human ovarian cancer tissues and cell lines (IGROV and HMVECs) was evaluated using an anti-vimentin antibody immunofluorescence assay. Five HGSC tumor tissue samples and five normal ovarian tissue samples were also examined for vimentin expression. High levels of vimentin expression were detected in human ovarian tumor tissues and vasculature, in contrast to low levels of vimentin expression in normal ovary tissues in all tissue samples we collected (Figure 4A), as well as overexpression of vimentin on IGROV cells and HMVECs (data not shown). An aptamer–antibody non-competition binding experiment was performed to confirm vimentin as the likely binding target of V5. Significantly reduced fluorescence intensities of Cy3-V5 binding to IGROV cells were observed at different concentrations of V5 after the cells were pre-incubated with anti-vimentin polyclonal antibody, but not with cells pre-incubated with goat IgG control antibody (Figure 4B). This demonstrated that V5 and the anti-vimentin antibody likely share binding sites on IGROV cells.
Figure 4. Overexpression of vimentin in human ovarian cancer tissue and tumor cell lines.
(A) Overexpression of vimentin was detected by anti-vimentin antibody on human ovarian tumor tissue (top panel) in contrast to low levels of vimentin expression on normal ovary tissues (bottom panel). (B) An aptamer–antibody non-competition binding assay was performed with anti-vimentin polyclonal antibody and cy3-V5 at different concentrations (250 nM and 500 nM respectively). Significantly reduced fluorescence intensity of V5 binding to IGROV cells was detected after incubation with the anti-vimentin antibody, but not in cells pre-incubated with goat IgG control antibody. Scale bar = 100 μm.
Although we identified many target proteins by the MS pull-down experiment using TA V5, we first focused on vimentin for validation since it had the highest protein abundance score. Vimentin is one of the most widely expressed mammalian intermediate filament proteins It can be attached to the nucleus, endoplasmic reticulum or mitochondria (23) and is frequently used as a marker of differentiation (24,25). Studies have demonstrated that vimentin is overexpressed in various human epithelial cancers (26–33). High-level expression of vimentin is associated with tumor invasion (34) and poor prognosis (35). Several studies have demonstrated that vimentin has potential as a molecular target for anticancer therapeutics (36-38). Using different approaches, Zamay et al. has also demonstrated that a selected DNA-aptamer combined with the carrier reagent arabinogalactan could target intracellular vimentin and inhibit adenocarcinoma growth in vivo (39).
Here, we demonstrated a TA selection procedure that allows direct incorporation of more nuclease-resistant thiophosphates into the aptamer sequence. By combining LMD technology with aptamer tissue SELEX, we have overcome the obstacles of tissue heterogeneity and improved the accuracy of aptamer-based tissue biomarker discovery. Compared to other whole-tissue aptamer selection methods, Morph-X-Select has the advantages of accurate ROI identification in heterogeneous tissue sections, especially for minor cell populations; customization for individual patients; and selection of high binding affinity aptamers from different ROIs simultaneously. In combination with NGS and MS analysis, true high binding affinity sequences and their associated target proteins could be identified without experimental biases.
Supplementary Material
METHOD SUMMARY.
We developed a morphology-based tissue aptamer selection method, Morph-X-Select, that combines a thiophosphoate-modified DNA aptamer library with image-directed laser microdissection (LMD) to dissect regions of interest bound with the thioaptamer (TA) library, allowing identification of the targeted proteins.
Acknowledgments
We gratefully acknowledge CompanionDx Lab for their support of next-generation sequencing experiments and service centers supported by our local CTSA (UL1 TR000371). This work was supported in part by grants from the National Cancer Institute [U54 CA151668, P50 CA083639]; the Department of Defense [W81XWH-09-01-0212 and W81XWH-10-2-0125], and the Welch Foundation [AU-1296]. This paper is subject to the NIH Public Access Policy.
Footnotes
Author contributions
H.W. conceived and designed the experiments, conducted thioaptamer tissue selection, binding validation, pull-down experiments, and prepared the manuscript. X.L. conducted cell binding assays and aptamer pull-down experiments. D.E.V. conducted thioaptamer synthesis, next-generation sequencing analysis, and manuscript editing. G.L.R.L. conducted thioaptamer synthesis. M.-A.E.-R conducted next-generation sequencing data analysis. L.L. conducted mass spectrometry analysis. A.M.N and A.K.S. provided tissue samples. K.P.R. and D.G.G. conceived the project and provided guidance.
Supplementary material for this article is available at www.BioTechniques.com/article/114473.
Competing interests
D.G.G. and the University of Texas Health Science Center at Houston have research-related financial interests in AptaMed Inc. and AM Biotechnologies LLC (Houston, TX).
References
- 1.Hughes B. Antibody–drug conjugates for cancer: poised to deliver? Nat. Rev. Drug Discov. 2010;9:665–667. doi: 10.1038/nrd3270. [DOI] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Shih T, Lindley C. Bevacizumab: an angiogenesis inhibitor for the treatment of solid malignancies. Clin. Ther. 2006;28:1779–1802. doi: 10.1016/j.clinthera.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Nutiu R, Li Y. In Vitro Selection of Structure-Switching Signaling Aptamers. Angew. Chem. Int. Ed. Engl. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DS, Szostak JW. In Vitro Selection of Functional Nucleic Acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 6.Ray P, Rialon-Guevara KL, Veras E, Sullenger BA, White RR. Comparing human pancreatic cell secretomes by in vitro aptamer selection identifies cyclophilin B as a candidate pancreatic cancer biomarker. J. Clin. Invest. 2012;122:1734–1741. doi: 10.1172/JCI62385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaCava J, Molloy KR, Taylor MS, Domanski M, Chait BT, Rout MP. Affinity proteomics to study endogenous protein complexes: pointers, pitfalls, preferences and perspectives. Biotechniques. 2015;58:103–119. doi: 10.2144/000114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gragoudas ES, Adamis AP, Cunningham ET, Jr., Feinsod M, Guyer DR, VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 9.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Wang H, Beasley DW, Volk DE, Zhao X, Luxon BA, Lomas LO, Herzog NK, et al. Selection of Thioaptamers for Diagnostics and Therapeutics. Ann. N. Y. Acad. Sci. 2006;1082:116–119. doi: 10.1196/annals.1348.065. [DOI] [PubMed] [Google Scholar]
- 11.Thiviyanathan V, Somasunderam A, Gorenstein DG. Combinatorial selection and delivery of thioaptamers. Biochem. Soc. Trans. 2007;35:50–52. doi: 10.1042/BST0350050. [DOI] [PubMed] [Google Scholar]
- 12.Fennewald SM, Scott EP, Zhang LH, Aronson JF, Gorenstein DG, Luxon BA, Shope RE, Beasley DW, et al. Thioaptamer Decoy Targeting AP-1 Proteins Influences Cytokine Expression and the Outcome of Arenavirus Infections. J. Gen. Virol. 2007;88:981–990. doi: 10.1099/vir.0.82499-0. [DOI] [PubMed] [Google Scholar]
- 13.Mann AP, Somasunderam A, Nieves-Alicea R, Li X, Hu A, Sood AK, Ferrari M, Gorenstein DG, et al. Identification of thioap-tamer ligand against E-selectin: potential application for inflamed vasculature targeting. PLoS One. 2010;5:e13050. doi: 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann AP, Bhavane RC, Somasunderam A, Liz Montalvo-Ortiz B, Ghaghada KB, Volk DE, Nieves-Alicea R, Suh KS, et al. Thioap-tamer Conjugated Liposomes for Tumor Vasculature Targeting. Oncotarget. 2011;2:298–304. doi: 10.18632/oncotarget.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangala LS, Wang H, Jiang D, Wu SY, Somasunderam A, Volk DE, Lokesh GLR, Li X, et al. Improving vascular maturation using noncoding RNAs increases antitumor effect of chemotherapy. JCI Insight. 2016;1:e87754. doi: 10.1172/jci.insight.87754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Li S, Liu F, Zhou L, Shao N, Zhao X. SELEX aptamer used as a probe to detect circulating tumor cells in peripheral blood of pancreatic cancer patients. PLoS One. 2015;10:e012192. doi: 10.1371/journal.pone.0121920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurman RJ. Origin and molecular pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 2013;10:x16–21. doi: 10.1093/annonc/mdt463. [DOI] [PubMed] [Google Scholar]
- 20.Lu E, Elizondo-Riojas MA, Chang JT, Volk DE. Aptaligner: automated software for aligning pseudorandom DNA X-aptamers from next-generation sequencing data. Biochemistry. 2014;53:3523–3525. doi: 10.1021/bi500443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-facilitated biomarker discovery (AptaBiD). J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 22.Carey MF, Peterson CL, Smale ST. Chromatin Immunoprecipitation (ChiP). Cold Spring Harb. Protoc. 2009;2009:pdb.prot527. doi: 10.1101/pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 23.Challa AA, Stefanovic B. A novel role of vimentin filaments: binding and stabilization of collagen mRNAs. Mol. Cell. Biol. 2011;31:3773–3789. doi: 10.1128/MCB.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay ED. Extracellular matrix, cell skeletons, and embryonic development. Am. J. Med. Genet. 1989;34:14–29. doi: 10.1002/ajmg.1320340107. [DOI] [PubMed] [Google Scholar]
- 25.Williams AA, Higgins JP, Zhao H, Ljunberg B, Brooks JD. CD 9 and vimentin distinguish clear cell from chromophobe renal cell carcinoma. BMC Clin. Pathol. 2009;9:9. doi: 10.1186/1472-6890-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korsching E, Packeisen J, Liedtke C, Hungermann D, Wulfing P, Van Diest PJ, Brandt B, Boecker W, et al. The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential. J. Pathol. 2005;206:451–457. doi: 10.1002/path.1797. [DOI] [PubMed] [Google Scholar]
- 27.Vora HH, Patel NA, Rajvik KN, Mehta SV, Brahmbhatt BV, Shah MJ, Shukla SN, Shah PM. Cytokeratin and vimentin expression in breast cancer. Int. J. Biol. Markers. 2009;24:38–46. doi: 10.1177/172460080902400106. [DOI] [PubMed] [Google Scholar]
- 28.Fuyuhiro Y, Yashiro M, Noda S, Kashiwagi S, Matsuoka J, Doi Y, Kato Y, Kubo N, et al. Clinical significance of vimentin-positive gastric cancer cells. Anticancer Res. 2010;30:5239–5243. [PubMed] [Google Scholar]
- 29.Zhao Y, Yan Q, Long X, Chen X, Wang Y. Vimentin affects the mobility and invasiveness of prostate cancer cells. Cell Biochem. Funct. 2008;26:571–577. doi: 10.1002/cbf.1478. [DOI] [PubMed] [Google Scholar]
- 30.Sethi S, Macoska J, Chen W, Sarkar FH. Molecular signature of epithelial-mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am. J. Transl. Res. 2010;3:90–99. [PMC free article] [PubMed] [Google Scholar]
- 31.Rho JH, Roehrl MH, Wang JY. Glycoproteomic analysis of human lung adenocarcinomas using glycoarrays and tandem mass spectrometry: differential expression and glycosylation patterns of vimentin and fetuin A isoforms. Protein J. 2009;28:148–160. doi: 10.1007/s10930-009-9177-0. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Zhang B, Sun B, Wang X, Ban X, Sun T, Liu Z, Zhao X. A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J. Exp. Clin. Cancer Res. 2010;29:109. doi: 10.1186/1756-9966-29-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, et al. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23:298–302. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- 34.Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, Foidart JM. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 2003;63:2658–2664. [PubMed] [Google Scholar]
- 35.Jin H, Morohashi S, Sato F, Kudo Y, Akasaka H, Tsutsumi S, Ogasawara H, Miyamoto K, et al. Vimentin expression of esophageal squamous cell carcinoma and its aggressive potential for lymph node metastasis. Biomed. Res. 2010;31:105–112. doi: 10.2220/biomedres.31.105. [DOI] [PubMed] [Google Scholar]
- 36.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahat G, Zhu QS, Huang KL, Wang S, Bolshakov S, Liu J, Torres K, Langley RR, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS One. 2010;5:e10105. doi: 10.1371/journal.pone.0010105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001;8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- 39.Zamay TN, Kolovskaya OS, Glazyrin YE, Zamay GS, Kuznetsova SA, Spivak EA, Wehbe M, Savitskaya AG, et al. DNA-aptamer targeting vimentin for tumor therapy in vivo. Nucleic Acid Ther. 2014;24:160–170. doi: 10.1089/nat.2013.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.