Abstract
The use of principal component regression, a multivariate calibration method, in the analysis of in vivo fast-scan cyclic voltammetry data allows for separation of overlapping signal contributions, permitting evaluation of the temporal dynamics of multiple neurotransmitters simultaneously. To accomplish this, the technique relies on information about current-concentration relationships across the scan-potential window gained from analysis of training sets. The ability of the constructed models to resolve analytes depends critically on the quality of these data. Recently, the use of standard training sets obtained under conditions other than those of the experimental data collection (e.g., with different electrodes, animals, or equipment) has been reported. This study evaluates the analyte resolution capabilities of models constructed using this approach from both a theoretical and experimental viewpoint. A detailed discussion of the theory of principal component regression is provided to inform this discussion. The findings demonstrate that the use of standard training sets leads to misassignment of the current-concentration relationships across the scan-potential window. This directly results in poor analyte resolution and, consequently, inaccurate quantitation, which may lead to erroneous conclusions being drawn from experimental data. Thus, it is strongly advocated that training sets be obtained under the experimental conditions to allow for accurate data analysis.
Keywords: Fast-scan cyclic voltammetry, principal component regression, calibration, training sets, multivariate data analysis, chemometrics, data analysis
Graphical Abstract
1. INTRODUCTION
In the analysis of fast-scan cyclic voltammetry (FSCV) data, the use of standard training sets (i.e., sets of cyclic voltammograms obtained from electrodes, recording sessions, and/or subjects other than those used for experimental data collection) for conditioning principal component regression (PCR) calibration models has increased in popularity.1–12 However, theoreticians have long warned about the dangers associated with this approach, emphasizing the need for either consistency between the conditions for obtaining model training and experimental data, or the use of transfer methods when such consistency cannot be obtained.13–16 Here, we aim to provide a clear theoretical explanation of PCR, with particular focus on using this discussion to evaluate the appropriateness of the use of standard training sets in analysis of FSCV data.
FSCV is an electroanalytical technique that allows for the real-time recording of the subsecond dynamics of electroactive neurotransmitters.17 Cyclic voltammograms (CVs) obtained through its use can be used for analyte concentration change identification and subsequent quantification, both of which are critical to proper calibration. In practice, however, CVs obtained in vivo often have contributions from multiple analytes that require resolution before positive identification, much less quantification, can be made. As a multivariate calibration technique, PCR is equipped to handle this issue, as information across the scan-potential window can be used to separate these overlapping signals. As such, considerable work has gone into the development of PCR as a robust data analysis technique for FSCV.18–21
In using PCR, proper training set construction for model conditioning is a critical consideration.22 The current-concentration relationships at each potential for a given analyte, which are essential information for accurate signal resolution, are directly estimated from the CVs in the training set. As these relationships are known to vary between experimental sessions, it has been advised that training set data be collected under conditions resembling those of the experimental data collection as closely as possible (e.g., in the same animal and anatomical region, with the same equipment, and within the recording session).23–25 However, procedural constraints have increased the use of standard training sets, which consist of data collected under circumstances significantly differing from those of experimental data collection. The questions then arise as to what degree these training sets can capture these experiment-specific relationships, and whether their use leads to a detrimental effect on the ability of derived models to resolve analyte contributions. To this end, recent work from our laboratory demonstrated undesirable practical consequences stemming from their use, including systematic misestimation of analyte concentration changes and failure of model validation procedures.22 However, the specific origin of these issues within the framework of PCR was not investigated.
The goal of this paper is multifold. First, we seek to increase the interpretability of PCR models by elucidating how principal component regression accomplishes the calibration goals of analyte resolution and quantification, particularly in the analysis of FSCV data. While there are many excellent resources for understanding the technique,23–25 this discussion aims to avoid the heavy reliance on the language of vector calculus or linear algebra often found in these guides, instead substituting visual explanations where possible. Understanding of these calculations can be used to help guide training set construction, enabling proper PCR use. Second, this discussion is used to build on our previous work, focusing here on identification of the source of the previously demonstrated shortcomings of models built with standard training sets.22 Particularly, it is shown that the use of standard training sets indeed leads to improper assignment of the current-concentration relationships that define multivariate analysis, leading to poor analyte resolution. Ultimately, these results serve to highlight the need and practical utility of training set construction within a given experimental session.
2. THEORY
2.1. Calibration
2.1.1. Calibration Basics
The goal of analytical calibration is to determine the relationship between an instrumental response and the corresponding value of the sample property to be estimated (e.g., concentration of an analyte) under a given set of conditions for future prediction of the latter.16 This is done in two steps: calibration method selection and model training. Method selection is driven by the expected characteristics of the data (e.g., dimensionality and presence of interference), assumptions made about the relationship between the instrumental response and sample property (e.g., linearity), and practical considerations (e.g., computational intensity and ease of use). Within that method’s framework, estimated model parameters are determined by training the calibration model using data for which both the response and sample property value are known (i.e., a training set). This relationship can then be used to convert experimental measurements into estimations of the unknown value of the sample property. The accuracy of this procedure is limited by the underlying assumptions of the calibration method, and the validity of the relationship found in the training data to the experimental data.
2.1.2. Univariate Linear Regression
The univariate linear regression calibration method uses a single instrumental response value (e.g., a current measurement) and assumes a linear relationship between it and the value of the sample property. In the analysis of FSCV data, the peak faradaic current, the most sensitive measurement, is typically used for prediction. Additionally, due to the large charging current that arises at the high scan rates employed in FSCV, background subtraction, where currents are reported relative to a “background” measurement, is performed to allow for reliable detection of the relatively small analytical signal, limiting quantitation to relative changes in analyte concentration.26 Here, this is specified by the use of the symbols Δi and Δ[analyte], indicating the changes in current and analyte concentration, respectively, relative to their absolute values at the time of the “background” measurement. The calibration equation of classical least-squares regression for background-subtracted FSCV dopamine quantitation can thus be written as
| (1) |
where ΔiDA,peak is the background-subtracted dopamine peak current, SenDA is the analytical sensitivity, Δ[DA] is the dopamine concentration change, and Δi0 is the background-subtracted peak current in the absence of a relative dopamine concentration change (ideally, or possibly constrained to be, zero). Model training consists of estimation of the parameter values (i.e., the sensitivity and blank measurement) that minimize the sum of the squares of the residuals. A more useful formulation is the inverse model, eq 2:
| (2) |
where CFDA is the calibration factor, Δ[DA]0 is the dopamine concentration change in the absence of a change in current (again, ideally, or constrained to, zero), and all other variables retaining their meaning from eq 1. Note that eq 2 can be converted into eq 1 (i.e., CFDA and Δ[DA]0 are equal to Senanalyte −1 and −Δi0/Senanalyte, respectively); however, the estimated model parameters differ when regression is done in the two formulations.27–29
2.1.3. Multivariate Analysis
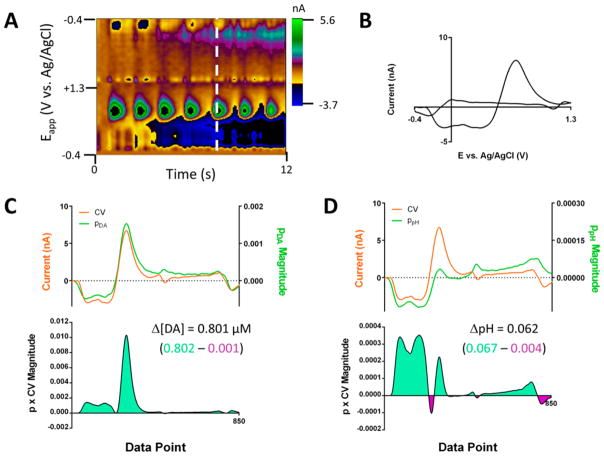
The simplicity and interpretability of the univariate linear regression model make it a powerful analytical tool. However, univariate models fail to make accurate predictions for signals containing multiple analyte contributions. For example, Supporting Information Figure 1 is a background-subtracted color plot of voltammograms obtained in a rodent brain with a carbon fiber microelectrode during an electrically stimulated dopamine release event, in which signals corresponding to dopamine and pH changes are present. A univariate model would interpret the basic pH shift as a decrease in DA. This error arises from the failure of the model to resolve the various analyte contributions prior to quantitation. Assuming linear superposition of analyte signals, a correction to this equation is
| (3) |
However, for a given peak current, an infinite number of dopamine and pH concentration pairs satisfy this equation, as there are more unknown than known values. As this discussion shows, for multicomponent calibration, both component identification and its quantitation must be accomplished, and this requires use of multiple measurements to succeed.
The multivariate model, using the entire scan-potential window to provide more known values, can be stated in the classical formulation as a system of N equations, where N is the number of measurements taken into account in the model (850 in this paper, sampling frequency × waveform duration):
| (4) |
The omission of an intercept term is intentional and standard in practice. Again, a more useful calibration equation is that of the inverse formulation:
| (5) |
where p is referred to as the calibration coefficient for the Nth data point (N = 1,2,…850). Again, eqs 4 and 5 can be interconverted but differ in the estimated model parameters obtained from their use in regression. The assumption of this model is, “The value of the current measurements at each potential depends only, and linearly, on the concentration changes of dopamine and pH.” If the data fail to meet this assumption, a prediction error will occur. Statistical techniques (i.e., residual analysis) allow for identification of data with significant additional signal contributions, which is referred to as the first-order advantage.30 However, if there is any measurement contribution not included in the model, increased error should be expected in model predictions.
2.2. Principal Component Regression
2.2.1. Principal Component Analysis
The question now arises of how to train the model (i.e., optimize the p parameters of eq 5). The ideal solution would spread the burden of prediction across the scan-potential window and not be overly specific to the training data. Furthermore, the solution should minimize noise in the calibration data as it degrades the quality of the relationships in the training data, decreasing confidence in the model predictions.
Principal component analysis (PCA) addresses these issues. In PCA, each training CV is restated as the linear combination of a collection of scaled vectors called principal components (PCs). The advantage of this reformulation is that some PCs correspond to relevant deterministic variance (i.e., that carrying information about the analytes), while others correspond to nondeterministic noise. Exclusion of the latter ideally allows for the production of noise-free training set CVs, allowing for parameter estimation with greater confidence. This estimation is done in the inverse formulation (inverse least-squares regression, or ILS) with the data stated in the terms of PCs, simplifying the calculations. Collectively, the use of PCA and ILS is referred to as principal component regression (PCR).
There are four major steps in the use of PCR:
Construction and selection of the relevant subset of principal components (PCA);
Scoring of standards on generated PCs (PCA);
Regression of PC scores against concentrations (ILS);
Application to experimental data.
2.2.2. Principal Component Construction
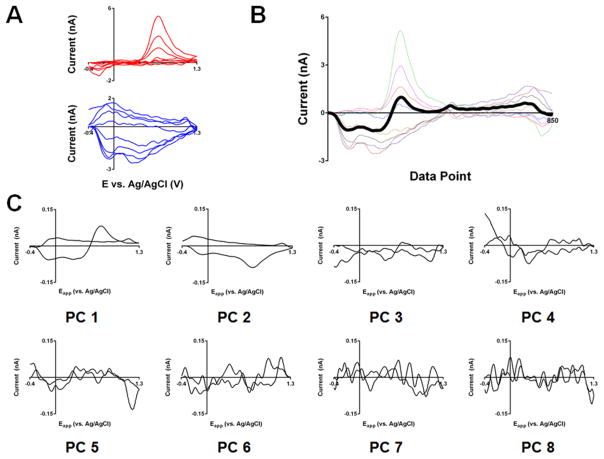
For generation of PCs, a training set must be constructed. We illustrate this with the training set shown in Figure 1A that contains both dopamine and pH background-subtracted CVs (n = 4/analyte). The PCR model is entirely defined by the relationships found in the training set. A good training set (ref 24) spans the concentration range of interest, contains mutually exclusive voltammograms of all expected analytes, and matches the characteristics of the experimental data (e.g., noise and peak positions).23,24 The limitations of in vivo training set construction make collection of single-analyte CVs preferable to allow for reliable estimation of the concentrations of the standards. This approach, however, mandates that the analyte voltammetric responses be independent of one another and the matrix, or model performance will suffer. This limits the use of models built in this way to conditions where these assumptions hold. Of note, as there is a known dependence of the dopamine voltammetric response on pH, use of PCR models built with pure DA and pH voltammogram standards should be limited to pH ranges around those used for training data collection.31 However, given the tight regulation of the neuronal environment, significant excursions of pH to extreme values are relatively rare.32
Figure 1.
Generation of principal components. (A) Dopamine (top) and pH (bottom) training set voltammograms shown in the traditional electrochemical format. (B) First principal component (dark black line) overlaid on the training set voltammograms in the “unwrapped” format. (C) Eight principal components generated by the singular value decomposition algorithm for the training set shown in (B).
The goal of PC construction is to calculate a set of vectors that can describe the entirety of the training set (i.e., entirely reconstruct the training standards through combinations of scaled PCs). These PCs retain the dimensions of CVs (i.e., have a single value corresponding to each applied potential), allowing visualization using traditional electrochemical representation. 20 (To minimize confusion, both the data and PCs are presented here “unwrapped” (Figure 1B)). Construction is done with a linear algebra technique termed single value decomposition, or SVD (see the Supporting Information for more details). In brief, the first PC is calculated to span the maximum variance possible (Figure 1B and as “PC 1” in 1C), without consideration of analyte identity or corresponding concentration change. The following PC then captures the most remaining variance and is constrained to being orthogonal (i.e., describe distinct information) to its successor. This process continues until all variance is described, resulting in a set of unique PCs equal to the number of the CVs in the training set (Figure 1C). Of note, without pretreatment (e.g., mean-centering and scaling), CVs with the largest currents have a disproportionately greater influence on the shape of the PCs.20,24 Thus, the quality (e.g., how representative it is of the average data collected, how free it is from interferents, and the noise levels in the CV) of these CVs is critical.
2.2.3. Scoring of Standards on PCs
After PC generation, the “scores” of each of the training standards on each of the PCs can be calculated as the sum of the pointwise product of the principal component and background-subtracted current magnitudes using the following formula:
| (6) |
where n is the data point, wn is the magnitude of the principal component loading vector at a given data point, and Δin is the background-subtracted current magnitude at that data point. This calculation is referred to as the projection of CV onto the PC, or, more generally, the dot product of the CV and PC vectors.
This calculation can be decomposed into its constituent steps and illustrated graphically (Figure 2A–D). First, the product of the current and PC magnitudes is calculated at each point, as shown in Figure 2C and D. These products are then summed across the scan-potential window to obtain the score value. As the positive and negative contributions balance one another in this latter calculation, the areas beneath the product of the CV and PC magnitudes at each point are shaded different colors (green, positive; pink, negative) to assist in conceptualization of the regions contributing most to the score value.
Figure 2.
Scoring process of principal component analysis. (A) CV of dopamine standard (orange) and the first principal component (green) shown in the “unwrapped” format. (B) As in (A) but with a pH standard. (C) Plot showing the value of product of the principal component and current amplitudes (wnΔin from eq 6) from (A) for each data point. The regions beneath positive and negative values of this calculation are shown in green and purple, respectively, to assist with conceptualization of the score calculation, which is calculated from summation across the data window (see text). (D) As in (C) but using the PC and CV amplitudes from (B). (E) Cook’s distance plot showing the scores for each CV on the first two principal components for the training set presented in Figure 1. Each axis corresponds to the adjacent principal component, and each training CV is shown as a point on the plot whose coordinates are defined by its scores on the corresponding principal component.
When comparing CVs that differ in shape, higher score magnitudes indicate a higher degree of similarity in shape between the PC and CV. For instance, as seen in Figure 2C, both the PC and dopamine CV magnitudes are positive and large, leading to an area with a large positive contribution to the final score. For similarly shaped CVs, higher scores simply imply higher intensities within the regions heavily weighted by the PC. The polarity of the score is a secondary consideration. For a given PC polarity, regions in which the PC and CV are alike or differ in sign tend to make the score more positive or negative, respectively. However, if all PC magnitudes were reversed in sign (i.e., the PC polarity was flipped), the scores would be identical in magnitude but differ only in sign. One merely expects that similar CVs (e.g., the training CVs for a given analyte) are alike in sign.
2.2.4. Selection of PC Subset for Model
After scoring, a set of N scores, where N is equal to the number of principal components, is produced for each training CV (i.e., the dimensionality of the data is reduced). Indeed, if all scores for a training CV on all unique PCs are taken, the original CV can be entirely reconstructed by the linear combination of each PC scaled by its corresponding score (Supporting Information Figure 2). However, we retain only the PCs that correspond to analytically relevant features of the training set voltammograms (i.e., primary PCs), while those composed primarily of noise are discarded (i.e., secondary PCs. PCs 3–8 in Figure 1). Thus, through use of only the primary PCs, the original CV can be reconstructed with the noise described by the secondary PCs excluded (2 PC model in Supporting Information Figure 2).
The selection of primary PCs typically relies on statistical analysis of the proportion of information described by each PC (i.e., variance-based methods). Of note, for FSCV data, a previous study from our laboratory compared two such selection methods, concluding that use of the Malinowski’s F-test was preferable for reliable identification and removal of PCs corresponding to noise.20 Such variance-based tests rely on the assumption that the analytically relevant portion of the signal is considerably larger than that attributable to noise. However, if the noise levels become too high, too many PCs, including those describing noise, are likely to be retained. Estimates place this transition for the Malinowski’s F-test between signal-to-noise ratios of 10–100.33
A useful tool to examine the model is the Cook’s distance plot (Figure 2E), which has coordinates corresponding to a CV’s score on a given PC (shown next to the corresponding axis). For simplicity, these are often two-dimensional plots where only coordinates corresponding to first two PCs (i.e., the two PCs that describe the most variance in the data) are shown. The utility of such plots is in the detection of outliers, which is discussed in detail elsewhere.21,34 Generally, one expects to observe linear groups of data corresponding to a given analyte. These linear groups tend to spread more along one coordinate, indicating that the corresponding PC captures more of the variance in that analyte than the other PC. Additionally, these groups should be sufficiently separated from one another in the plot; otherwise, it is likely that the analyte signals are highly similar (and not easily resolvable), or some training set spectra are not sufficiently “pure”.
2.2.5. Regression of PC Scores against Concentration
For a two-analyte, two-PC system, the calibration equations in terms of PCs are stated in the inverse formulation as
| (7) |
The values of m are estimated through least-squares regression, which is equivalent to estimating the p parameters in eq 5. This approach, however, limits the regression solutions to the subspace defined by the PCs and greatly simplifies the calculations. After this stage, the model, consisting of the retained PCs and the values of the m parameters, is completely defined, having only knowledge of the training set data.
2.2.6. Application to Experimental Data
For concentration prediction, the scores of the experimental CVs on the retained PCs are simply calculated and used with eq 7. These scores can also be used construct the model estimate of the CVs for residual analysis. The difference of the current of the original CV and this PC-reconstructed estimate forms the residual value at each potential. Statistical methods exist for the evaluation of the sum of the squares of the residual values for each voltammogram (Qt) to determine when there is sufficient residual current that the model-produced concentration estimate is significantly flawed. For FSCV data, the use of a Qα residual threshold is recommended.23,35 Qα is a value estimated from the secondary PCs (i.e., those excluded from the model and considered to be noise) that can be compared to Qt for each measurement, serving as a threshold of significance for the Qt values at a given confidence level. If Qt exceeds Qα, significant residual current remains unaccounted for in the PCR model, and the model may be an inaccurate description of the data.
It is now useful to return to the calibration equations stated in either the classical or the inverse forms (eqs 4 and 5, respectively), either of which can be used to evaluate the now-defined calibration model. In examining the classical formulation (eq 4), it can be seen that, for each analyte, the model provides a fixed value for the sensitivity at each potential. The set of all these values across the scan-potential window corresponding to analyte j is referred to as the kj vector. These vectors can be visually analyzed by plotting them either of our two visual conventions and thought of as the model estimates of the voltammetric response corresponding to a unit concentration change in analyte j.21 Inspection of plots of each of a PCR model’s kj vectors allows the user to verify that the model predicts, for each analyte, the expected shape of the cyclic voltammogram.
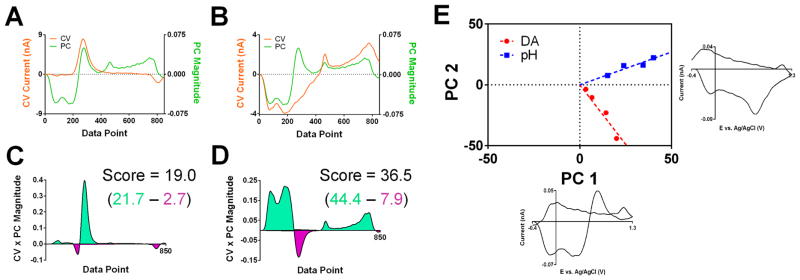
The regression model also provides coefficients, p values, explaining the contribution of the current at each potential to the calculation of each analyte concentration. These p values, which may be negative, are the coefficients in the inverse formulation (eq 5). The set of these coefficients across the scan-potential window corresponding to analyte j will be referred to as the pj vector. Both the origin of the pj vectors and its relationship to the kj vectors are given in the Supporting Information. With the knowledge of the pj vector, the analyte concentration prediction for an unknown CV (Figure 3B) proceeds in a manner analogous to the determination of scores. The pointwise product of the pj vector magnitude and the CV amplitude is calculated at each potential (Figure 3C and D). The concentration of the analyte is then determined from the sum of these values.
Figure 3.
Use of the p vectors in the calculation of analyte concentrations. (A) Color plot showing dopamine and pH shifts from a lever press-induced electrical stimulation for an animal performing intracranial self-stimulation. (B) Cyclic voltammogram, showing contributions from pH and dopamine, collected at the white vertical line shown in (A). (C) Plots of the dopamine p vector and the CV from (B) (upper) and the value of the product of their amplitudes (lower) for each data point. Analogous to the scoring process show in Figure 2, the concentration is calculated from summation across the data window, and the regions below positive and negative values of the p and CV product are shown in green and purple, respectively. (D) As in (C) but with the ppH matrix.
Analysis of the values of the pj vector and the pointwise product allows for the determination of the potentials weighted heavily for concentration calculation. Plots of the pj vectors, like those of the PCs, have defined shapes and features (e.g., peaks and zero crossings) that are derived from features in the CVs of multiple analytes in the training sets. For example, the plot of the pDA vector (Figure 3C, green) has an initial strong pH-like feature followed by a feature resembling the DA oxidation peak. In the analysis of a CV containing pH and DA contributions, which highly resembles the plot of the pDA vector, these two regions also make the major contributions to the dopamine concentration calculation. The dopamine oxidative wave, which directly provides information about the dopamine concentration, creates a large positive contribution, as expected. The pH-like region additionally generates a positive contribution to the dopamine concentration estimation, which is accounting for the current decrease in the dopamine oxidative region caused by the presence of pH contributions. A similar pattern is seen in the analysis of the calculation of the pH concentration with the ppH vector (Figure 3D). Within the early data region, where the signal contribution of pH is most pronounced, there is a large positive contribution to the concentration value. However, a positive contribution is also found in the region of the dopamine oxidation wave. This, in an analogous manner, is augmenting the pH concentration estimation to account for changes in the signal due to the presence of dopamine.
3. RESULTS AND DISCUSSION
3.1. Theoretical Failings of Standard Training Sets
Training sets that consist of CVs collected from different electrodes, recording sessions, and/or subjects (i.e., “standard” training sets) have been increasingly used in the analysis of in vivo FSCV data. This trend is particularly popular for the analysis of data collected with chronically implanted microelectrodes due to difficulties for training set construction at the site of recording. While this approach eases the procedural burden, it violates a fundamental assumption of calibration–that the relationships in the training set are the same as in the experimental data (see subsection 2.1.1). Correction for this error requires either knowledge of the differences between the experimental and calibration data, which is difficult to obtain, or the use of calibration transfer methods, which would require additional data collection and reintroduce the experimental complexity which is aimed to be avoided by the use of standard training sets.14 Indeed, recent work from our lab showed that the use of a training set collected in a different subject with a different electrode for PCR models resulted in significant, and unpredictable, misestimations of concentrations.22 Additionally, standard training sets (i.e., those with CVs collected from multiple subjects) resulted in high numbers of retained PCs (i.e., high rank) with kj vectors that no longer resembled the analyte voltammograms. Attempts to limit the rank of the models resulted in unreasonably high residual thresholds, removing the possibility of proper residual analysis and nullifying the first-order advantage. Here, this line of inquiry is furthered with focus on the origins of the shortcomings of standard training sets.
In the case of FSCV data, the primary differences in the data collected in different recording sessions can be seen in the voltammetric peak characteristics (e.g., peak width, position, and relative intensity), the electrode selectivity, and the noise levels.22 There are several sources for this variability. Regarding the electrochemical aspects of the data, there is an inherent level of variability between electrodes, particularly with disorganized surfaces such as those found on carbon fibers, while implanted reference electrodes differ in quality and are known to drift with time.36–38 Between subjects, differences in mass transport kinetics within the biological matrix or at the electrode surface may increase this variability further. These considerations affect voltammetric peak characteristics, sensitivity, and selectivity for electrochemical reactions.39 It is also standard practice to use extended anodic limits to electrochemically introduce oxide functionalities to the surface of the carbon fiber microelectrodes prior to measurements, which results in favorable conditions for catecholamine detection.40 However, the level of control over this process is limited, leading to varied electrochemical responses. This is particularly pronounced in the case of pH changes.41 Finally, noise levels are expected to differ between instruments. Thus, there are unavoidable sources of variation in the collection of FSCV data.
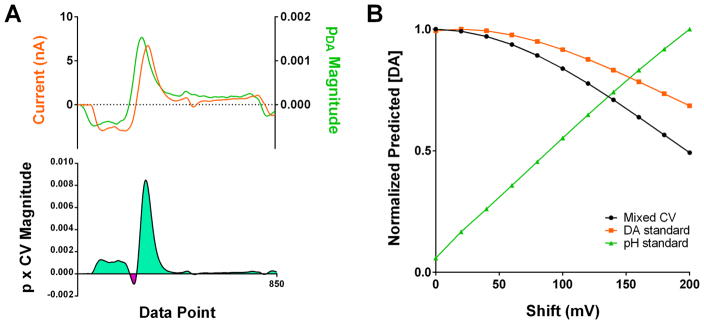
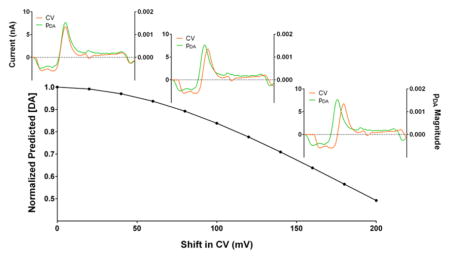
To illustrate the importance of this, we used the pDA vector to analyze the concentration prediction for the CV from Figure 3B with an artificial rightward shift introduced (Figure 4A), as might be introduced through reference electrode drift. In the case of DA and pH differentiation, the regions heavily weighted for the DA concentration feature sharp changes in the “p” values, including a zero crossing and polarity switch. A relatively minor shift leads to misalignment of these features, leading to significant changes in the predicted DA concentration. Such errors also arise in the analysis of DA and pH training standards (Figure 4B), leading to artificially low and high DA estimations, respectively. This illustrates that analyte resolution critically depends on correct assignment of the current-concentration relationships across the scan-potential window and, in particular, directly demonstrates the sensitivity of PCR to variations in the applied potential.
Figure 4.
Errors in the use of nonexperimental training sets. (A) Plots of the pDA matrix and the CV from Figure 3B with an artificial rightward 100 mV shift (upper), and the value of the product of their amplitudes (lower) for each data point. (B) Plot of the predicted DA concentration, normalized to the maximum predicted value, as a function of the artificial shift magnitude introduced for the mixed CV from Figure 3B (black) and a DA (orange) and pH (green) standard from Figure 1A.
3.2. Concatenation of Training Sets from Multiple Animals
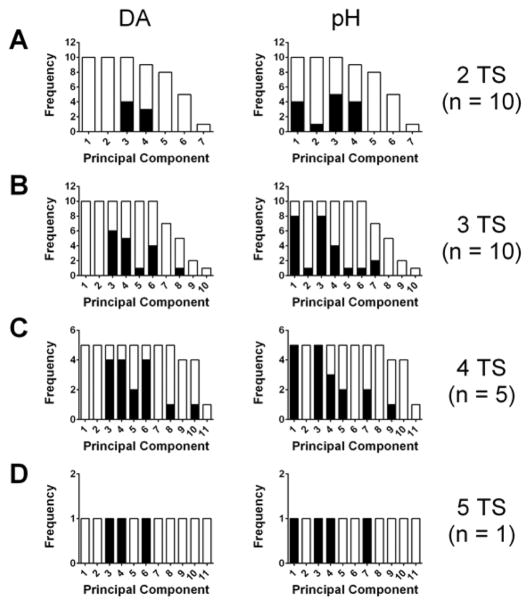
To demonstrate the incompatibility of data collected from different animals within a single model framework, training sets were constructed by the concatenation of entire training sets (TS, consisting of 10 CVs each) constructed in different animals (n = 5). One training set was selected as the primary TS (labeled A), while the number of training sets added was varied between one and four. (The data and labels used for these investigations are identical to those reported in ref 22.) The number of PCs retained (i.e., primary PCs by the Malinowski F-test) and the Qα thresholds were calculated for the PCR models. The results (Supporting Information Figure 3A,B) agree with those described in ref 22. The number of primary PCs tended to increase with the inclusion of more training set CVs, while the Qα thresholds tended to decrease, suggesting overfitting of the data. Limitation of the number of primary PCs to the first two (Supporting Information Figure 3) resulted in a drastic increase in the Qα threshold, nullifying the first-order advantage.
To carry this analysis further, the assumptions that go into the regression step of PCR (subsection 2.2.5) were tested. First, we examined whether there was a linear relationship, as measured by the R2 values for the zero-intercept linear regression model, between the scores on all retained PCs and the concentration values (eq 7). It was found that only the first two components tended to have a linear relationship with the concentrations for both analytes (Supporting Information Figure 3C). A Student’s t test of the significance of the estimated “m” parameters revealed that 80% of the third or higher primary PCs were not significantly different from zero, suggesting that the use of these PCs in regression is inappropriate.42
Next, we examined whether the proportionality constants (“m” values in eq 7) were shared between data collected from different animals. This was evaluated by an F-test comparing models in which the “m” parameters were either global (shared between data from different subjects) or subject-specific (see Supporting Information).43 This regression was done using scores calculated from PCs defined using all training set voltammograms, leading to the expectation of some degree of generality. However, it was found that the subject-specific constants were preferred in a large number of cases (Figure 5). When considering both the dopamine and pH parameters, preference for group-specific parameters was found in a considerable number of the third and higher primary PCs, highlighting the fact that these are likely capturing variance specific to certain subjects. Additionally, in the case of the pH parameters (right column of Figure 5), there were a large number of models in which group-specific parameters were preferred for the first two primary PCs. Concatenation of just one additional training set resulted in four out of the ten models showing preference for subject-specific parameters in the first PC, the one that describing the largest amount of variance. All but two of models with higher number of training sets exhibited this preference as well. This result can likely be traced to the wide variability of the characteristics of the pH voltammograms between electrodes, highlighting that, even with global definition of the PCs, the uniqueness of data from a single animal would lead to poor estimates as compared with a training set derived from the experimental animal.
Figure 5.
Results of F-test comparing models with subject-specific or global parameters for eq 7 for concatenated training sets. (A) Cumulative histogram showing the number of PCR models, built from concatenated training sets of two single-subject training sets, containing a given number of principal components or greater (frequency). The black portion of the bars corresponds to the number of principal components that were found to have subject-specific parameters preferred over global parameters (F-test, α = 0.05) for relating scores on that principal component and the dopamine (left) or pH (right) concentrations. (B) As in (A) but using concatenated training sets from three subjects. (C) As in (A) but using concatenated training sets from four subjects. (D) As in (A) but using concatenated training sets from five subjects.
3.3. Evaluation of “Library” Approach to Generalized Training Set Generation
To investigate the use of a library for the model generation, the construction of randomly generated CVs, derived from separate animals, was revisited. In this experiment, animal B’s experimental session was analyzed by a training set consisting of 10 CVs randomly selected from the other four animals (animals A, C, and D), resulting in 2.4 × 108 possible training sets. Of these, 19 704 were studied due to computational limitations, with the total analysis time being 10.1 days. These data were recorded during an intracranial self-stimulation trial and were binned into 75 5-s snippets.22 As an initial measure of the appropriateness of the constructed model in the description of the data, the total sum of all residual values calculated at each analyzed time point was chosen, a measure of the amount of signal captured by the PCR model. Additionally, to gain insight into the models constructed, the number of primary PCs and the Qα thresholds were again calculated.
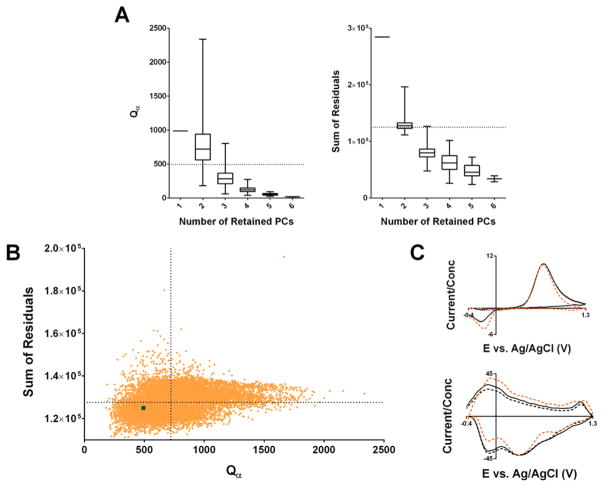
The summary of the generated models is shown in Figure 6A as a function of the number of primary PCs retained for the model. Just under half of the models (45.8%) had more than two primary components, with the remaining, with one exception, having two primary PCs. As expected, both the Qα threshold and the sum of the residuals decreased as a function of the number of the PCs, suggesting overfitting. This is supported by evaluation of the average kj vectors for the two analytes for a given number of PCs, which shows degradation in the quality of these model estimates with an increase in the number of components retained (Supporting Information Figure 4). The inappropriateness of such high rank models in the analysis of subject B’s data is further illustrated by the majority of Qα values and residual sums for 3+ PC models falling below those from Subject B’s model (i.e., that trained with data from the experimental session, shown as the dashed line in the figure).
Figure 6.
Summary of “library” approach to generate randomized standard training sets. (A) Box plots showing the distribution of Qα threshold values (left) and the residual sum for all snippets (right) shown as a function of the number of PCs retained in the constructed PCR model. The values for the training set constructed within the experimental subject (subject B) are shown as a horizontal dashed lines. (B) Scatterplot showing the residual sums plotted against Qα values for all 2 PC models. The dashed lines correspond to the medians of the distributions in each variable. The model constructed with subject B’s training set is shown as a green square in the lower left quadrant. (C) Average kj vectors constructed from all 2 PC models (solid black line) and 2 PC models in falling in the lower half of the distributions for both the residual sum and Qα values (lower left quadrant of (B), dashed black line), as well as subject B’s PC model (orange dashed line).
Given this, we limit the remaining discussion to the subset of models that retained two primary PCs (n = 10 679 training sets). Within the distribution defined by this subset, subject B’s PCR model had a Qα threshold (the green square in Figure 6B) falling in the lower 16th percentile. This suggests that most random models tended to overestimate this threshold, which would result in invalid data sets passing residual analysis undetected. Furthermore, the sum of residual values for training set B’s model falls in the lower 38th percentile, suggesting that the majority of these models do not capture as much information from the data as the proper training set.
Further requirements for model selection could be implemented. In this experiment, the subset of models considered can be refined to those having both a low total residual sum and Qα thresholds like subject B’s model (defined as those falling in the lower 50th percentile on both distributions–the lower left quadrant of Figure 6B). However, even this requirement fails to select models with current–concentration relationships like subject B’s PCR model. This can be seen by inspection of the plots of the kj vectors shown in Figure 6C, which directly summarize these relationships. The kj vectors for subject B’s PCR model (dashed orange line) differ noticeably in both absolute and relative peak amplitudes from the average kj vectors of this low total residual sum, low Qα threshold 2 PC subset (dashed black line). Instead, the latter closely resembles that obtained from all 2 PC models (solid black line).
Such a library approach also suffers from a number of theoretical drawbacks in its implementation. Here, even with knowledge of how the within-subject training set performed, criteria for selection of models that had similar performance were unable to be found. One could alternatively implement selection criteria on the CVs for model training. However, this would require the collection of information about “pure” analyte responses, minimizing the advantage of such an approach. Implementation would also require the generation of large library of spectra, as CVs could differ in a number of different characteristics (e.g., peak location, relative peak height, etc.) and multiple CVs are needed for each analyte. Finally, one of the principal advantages of using PCR is the ability to detect interferents through residual analysis. This process relies critically on estimation of the noise from the training set. Even if a combination of library analyte spectra matching the analytical characteristics of the data were found, these spectra will likely have unrepresentative noise within them, preventing proper residual analysis.
4. CONCLUSIONS
The strength of PCR in analysis of FSCV data comes from its ability to separate and quantitate analyte contributions, with the success of the latter critically dependent on the former. Resolution of analyte signals relies on knowledge of the current-concentration relationships across the scan-potential window (the k and p values of eqs 4 and 5), which are improperly assigned when standard training sets are used. This error results in incorrect signal attribution, as illustrated by the predicted dopamine concentration changes for the shifted CVs in Figure 4B. This problem is not unique to PCR, but rather all of multivariate linear analysis, for which eqs 4 and 5 hold. Avoidance of this issue requires the collection and use of training set voltammograms that contain accurate information about the experimental current-concentration relationships for all expected analytes, as information about each analyte is used for the resolution of one (which was illustrated directly by the analysis of the pj vectors in Figures 3C and D). The variability of these relationships between voltammetric data collected in different animals is such that unification within a single model framework results in easily detectable differences in model behavior (Figure 5), while their replication is not possible even with prior knowledge of derived model performance (Figure 6C). Thus, the only known reliable way to capture these relationships in a manner that allows for proper signal resolution is the collection of training set voltammograms under the experimental conditions. This can be most readily accomplished by including training set collection in the experimental protocol, minimizing the time between experimental and training data collection to avoid drift in experimental parameters (e.g., the state of the reference electrode), and collecting training data that satisfy the general recommendations given in ref 24 (and repeated in subsection 2.2.2). Other approaches for training set collection risk poor analyte resolution, precluding accurate assessment of neurochemical dynamics.
5. METHODS
Data collection is described in ref 22. Animal procedures were approved by the UNC-Chapel Hill Institutional Animal Care and Use Committee (IACUC). Data and statistical analyses were performed in GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA), LabView (National Instruments, Austin, TX), and MATLAB (Mathwork, Natick, MA) with an α value of 0.05.
Supplementary Material
Acknowledgments
Funding
This research was supported by grants from the NIH (DA010900 to R.M.W.).
ABBREVIATIONS
- FSCV
fast scan cyclic voltammetry
- PCR
principal component regression
- CV
cyclic voltammogram
- DA
dopamine
- PC
principal component
- PCA
principal component analysis
- ILS
inverse-least-squares regression
Footnotes
Author Contributions
J.A.J. carried out the analysis. N.T.R. conducted the in vivo data collection, discussed the analysis results, and edited the manuscript. J.A.J., N.T.R., and R.M.W. designed the project. J.A.J. and R.M.W. wrote the paper. All authors read and approved the final version of this manuscript.
The authors declare no competing financial interest.
- Representative color plot and voltammograms of electrically stimulated dopamine release, reconstruction of a cyclic voltammograms from principal components and scores, summary of parameters of interest for PCR models built from concatenated training sets and R2 values for linear regression between the models’ principal component scores and concentration, average kj vectors for PCR models built from randomly constructed training sets, a matrix algebra summary of principal component regression, and a description of the F-test used for statistical model comparison (PDF)
References
- 1.Parker JG, Zweifel LS, Clark JJ, Evans SB, Phillips PE, Palmiter RD. Absence of NMDA receptors in dopamine neurons attenuates dopamine release but not conditioned approach during Pavlovian conditioning. Proc Natl Acad Sci U S A. 2010;107:13491–13496. doi: 10.1073/pnas.1007827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flagel SB, Clark JJ, Robinson TE, Mayo L, Czuj A, Willuhn I, Akers CA, Clinton SM, Phillips PE, Akil H. A selective role for dopamine in stimulus-reward learning. Nature. 2011;469:53–57. doi: 10.1038/nature09588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasrallah NA, Clark JJ, Collins AL, Akers CA, Phillips PE, Bernstein IL. Risk preference following adolescent alcohol use is associated with corrupted encoding of costs but not rewards by mesolimbic dopamine. Proc Natl Acad Sci U S A. 2011;108:5466–5471. doi: 10.1073/pnas.1017732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willuhn I, Burgeno LM, Everitt BJ, Phillips PE. Hierarchical recruitment of phasic dopamine signaling in the striatum during the progression of cocaine use. Proc Natl Acad Sci U S A. 2012;109:20703–20708. doi: 10.1073/pnas.1213460109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howe MW, Tierney PL, Sandberg SG, Phillips PE, Graybiel AM. Prolonged dopamine signalling in striatum signals proximity and value of distant rewards. Nature. 2013;500:575–579. doi: 10.1038/nature12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanat MJ, Bonci A, Phillips PE. CRF acts in the midbrain to attenuate accumbens dopamine release to rewards but not their predictors. Nat Neurosci. 2013;16:383–385. doi: 10.1038/nn.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark JJ, Collins AL, Sanford CA, Phillips PE. Dopamine encoding of Pavlovian incentive stimuli diminishes with extended training. J Neurosci. 2013;33:3526–3532. doi: 10.1523/JNEUROSCI.5119-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart AS, Rutledge RB, Glimcher PW, Phillips PE. Phasic dopamine release in the rat nucleus accumbens symmetrically encodes a reward prediction error term. J Neurosci. 2014;34:698–704. doi: 10.1523/JNEUROSCI.2489-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willuhn I, Burgeno LM, Groblewski PA, Phillips PE. Excessive cocaine use results from decreased phasic dopamine signaling in the striatum. Nat Neurosci. 2014;17:704–709. doi: 10.1038/nn.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollon NG, Arnold MM, Gan JO, Walton ME, Phillips PE. Dopamine-associated cached values are not sufficient as the basis for action selection. Proc Natl Acad Sci U S A. 2014;111:18357–18362. doi: 10.1073/pnas.1419770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willuhn I, Tose A, Wanat MJ, Hart AS, Hollon NG, Phillips PE, Schwarting RK, Wohr M. Phasic dopamine release in the nucleus accumbens in response to pro-social 50 kHz ultrasonic vocalizations in rats. J Neurosci. 2014;34:10616–10623. doi: 10.1523/JNEUROSCI.1060-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goertz RB, Wanat MJ, Gomez JA, Brown ZJ, Phillips PE, Paladini CA. Cocaine increases dopaminergic neuron and motor activity via midbrain alpha1 adrenergic signaling. Neuropsychopharmacology. 2014;40:1151–1162. doi: 10.1038/npp.2014.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YD, Kowalski BR. Calibration Transfer and Measurement Stability of near-Infrared Spectrometers. Appl Spectrosc. 1992;46:764–771. [Google Scholar]
- 14.Feudale RN, Woody NA, Tan H, Myles AJ, Brown SD, Ferre J. Transfer of multivariate calibration models: a review. Chemom Intell Lab Syst. 2002;64:181–192. [Google Scholar]
- 15.Woody NA, Feudale RN, Myles AJ, Brown SD. Transfer of multivariate calibrations between four near-infrared spectrometers using orthogonal signal correction. Anal Chem. 2004;76:2595–2600. doi: 10.1021/ac035382g. [DOI] [PubMed] [Google Scholar]
- 16.Booksh KS. Encyclopedia of Analytical Chemistry. John Wiley & Sons, Ltd; New York: 2006. Chemometric Methods in Process Analysis. [DOI] [Google Scholar]
- 17.Bucher ES, Wightman RM. Electrochemical Analysis of Neurotransmitters. Annu Rev Anal Chem. 2015;8:239–261. doi: 10.1146/annurev-anchem-071114-040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heien MLAV, Johnson MA, Wightman RM. Resolving neurotransmitters detected by fast-scan cyclic voltammetry. Anal Chem. 2004;76:5697–5704. doi: 10.1021/ac0491509. [DOI] [PubMed] [Google Scholar]
- 19.Keithley RB, Heien ML, Wightman RM. Multivariate concentration determination using principal component regression with residual analysis. TrAC, Trends Anal Chem. 2009;28:1127–1136. doi: 10.1016/j.trac.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keithley RB, Carelli RM, Wightman RM. Rank estimation and the multivariate analysis of in vivo fast-scan cyclic voltammetric data. Anal Chem. 2010;82:5541–5551. doi: 10.1021/ac100413t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keithley RB, Wightman RM. Assessing principal component regression prediction of neurochemicals detected with fastscan cyclic voltammetry. ACS Chem Neurosci. 2011;2:514–525. doi: 10.1021/cn200035u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodeberg NT, Johnson JA, Cameron CM, Saddoris MP, Carelli RM, Wightman RM. Construction of Training Sets for Valid Calibration of in Vivo Cyclic Voltammetric Data by Principal Component Analysis. Anal Chem. 2015;87:11484–11491. doi: 10.1021/acs.analchem.5b03222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keithley RB, Heien ML, Wightman RM. Multivariate concentration determination using principal component regression with residual analysis. TrAC, Trends Anal Chem. 2009;28:1127–1136. doi: 10.1016/j.trac.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer R. Chemometric Techniques for Quantitative Analysis. Marcel-Dekker; New York: 1998. [Google Scholar]
- 25.Lavine BK, Workman J., Jr Chemometrics. Anal Chem. 2013;85:705–714. doi: 10.1021/ac303193j. [DOI] [PubMed] [Google Scholar]
- 26.Robinson DL, Venton BJ, Heien MLAV, Wightman RM. Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo. Clin Chem. 2003;49:1763–1773. doi: 10.1373/49.10.1763. [DOI] [PubMed] [Google Scholar]
- 27.Centner V, Massart DL, de Jong S. Inverse calibration predicts better than classical calibration. Fresenius’ J Anal Chem. 1998;361:2–9. [Google Scholar]
- 28.Krutchkoff R. Classical and Inverse Regression Methods of Calibration. Technometrics. 1967;9:425–439. [Google Scholar]
- 29.Krutchkoff R. Classical and Inverse Regression Methods of Calibration in Extrapolation. Technometrics. 1969;11:605–608. [Google Scholar]
- 30.Olivieri AC. Analytical advantages of multivariate data processing. One, two, three, infinity? Anal Chem. 2008;80:5713–5720. doi: 10.1021/ac800692c. [DOI] [PubMed] [Google Scholar]
- 31.Kawagoe KT, Garris PA, Wightman RM. Ph-Dependent Processes at Nafion(R)-Coated Carbon-Fiber Microelectrodes. J Electroanal Chem. 1993;359:193–207. [Google Scholar]
- 32.Chesler M. Regulation and modulation of pH in the brain. Physiol Rev. 2003;83:1183–1221. doi: 10.1152/physrev.00010.2003. [DOI] [PubMed] [Google Scholar]
- 33.Malinowski ER. Adaptation of the Vogt-Mizaikoff F-test to determine the number of principal factors responsible for a data matrix and comparison with other popular methods. J Chemom. 2004;18:387–392. [Google Scholar]
- 34.Cook RD. Detection of Influential Observation in Linear-Regression. Technometrics. 1977;19:15–18. [Google Scholar]
- 35.Jackson JE, Mudholkar GS. Control Procedures for Residuals Associated with Principal Component Analysis. Technometrics. 1979;21:341–349. [Google Scholar]
- 36.Chand S. Carbon fibers for composites. J Mater Sci. 2000;35:1303–1313. [Google Scholar]
- 37.Moussy F, Harrison DJ. Prevention of the Rapid Degradation of Subcutaneously Implanted Ag/Agcl Reference Electrodes Using Polymer-Coatings. Anal Chem. 1994;66:674–679. doi: 10.1021/ac00077a015. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XJ, Wang J, Ogorevc B, Spichiger UE. Glucose nanosensor based on Prussian-blue modified carbon-fiber cone nanoelectrode and an integrated reference electrode. Electroanalysis. 1999;11:945–949. [Google Scholar]
- 39.McCreery RL. Advanced carbon electrode materials for molecular electrochemistry. Chem Rev. 2008;108:2646–2687. doi: 10.1021/cr068076m. [DOI] [PubMed] [Google Scholar]
- 40.Heien MLAV, Phillips PEM, Stuber GD, Seipel AT, Wightman RM. Overoxidation of carbon-fiber microelectrodes enhances dopamine adsorption and increases sensitivity. Analyst. 2003;128:1413–1419. doi: 10.1039/b307024g. [DOI] [PubMed] [Google Scholar]
- 41.Takmakov P, Zachek MK, Keithley RB, Bucher ES, McCarty GS, Wightman RM. Characterization of Local pH Changes in Brain Using Fast-Scan Cyclic Voltammetry with Carbon Microelectrodes. Anal Chem. 2010;82:9892–9900. doi: 10.1021/ac102399n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krazanowski W. Statistical Principles and Techniques in Scientific and Social Investigations. Oxford University Press; Cary, NC: 2007. [Google Scholar]
- 43.Glatting G, Kletting P, Reske SN, Hohl K, Ring C. Choosing the optimal fit function: Comparison of the Akaike information criterion and the F-test. Med Phys. 2007;34:4285–4292. doi: 10.1118/1.2794176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.