Abstract
In recent years, RNA has reemerged as a versatile biological macromolecule capable of performing an astonishing number of biochemical activities. Initially described as the ubiquitous but transient carrier of genetic information in the Central Dogma, RNA has surprised scientists with its capacity to store genetic information, catalyze biochemical reactions, protect telomeres, guide proteins to their targets, help DNA replication and protein synthesis, scaffold ribonucleoprotein complexes, and transmit developmental and epigenetic information through mitotic and even meiotic cell divisions. The latest surprise came during the past decade with advances in deep sequencing technologies, which uncovered the pervasive world of noncoding RNAs (ncRNAs). Functional analysis of ncRNAs has revealed their wide-spread use in several biological pathways including the ones in the nucleus. We now know that nuclear ncRNAs of various sizes facilitate genome stability by inhibiting spurious recombination among repetitive DNA elements, repressing mobilization of transposable elements (TEs), templating or bridging DNA double-strand breaks (DSBs) during repair, and directing developmentally-regulated genome rearrangements in some ciliates. In this paper, we will survey the known mechanisms with which nuclear ncRNAs directly contribute to the maintenance of genome stability and outline the major advances in our understanding of the role of ncRNAs in the nucleus. These studies reveal an unexpected range of mechanisms by which ncRNAs contribute to genome stability and even potentially influence evolution by acting as templates for genome modification.
Keywords: noncoding RNA (ncRNA), genome stability, DNA repair, lncRNA, siRNA, piRNA, diRNA, DNA double-strand break (DSB), heterochromatin, transposon silencing
Introduction
All cells have evolved mechanisms to maintain the integrity of their genomes whose accurate duplication and transmission to progeny cells is critical for life. Exposure to genotoxic agents, metabolic byproducts and DNA replication errors can result in the formation of a variety of DNA lesions which must be repaired efficiently to maintain genome stability (Ciccia and Elledge, 2010). In addition to these, chromosome segregation errors or mobilization of transposable elements (TEs) cause aneupoloidies and genome-wide mutagenesis respectively, both of which threaten the stability and fidelity of eukaryotic genomes (Cordaux and Batzer, 2009). To manage these daunting challenges, cells have evolved a variety of molecular mechanisms to maintain genomic stability, some of which, as shown recently, utilize nuclear noncoding RNAs (ncRNAs).
Continued advances in next generation sequencing has uncovered a variety of new functional ncRNAs that have revolutionized our understanding of RNA biology. Transcription from previously-thought silent regions of the genome have demonstrated the pervasive nature of this process throughout all kingdoms of life, and the functional analysis of these ncRNAs have expanded the repertoire of known mechanisms by which these molecules contribute to biological processes (Morris and Mattick, 2014; Rinn and Chang, 2012; Sabin et al., 2013). These molecules have emerged as key cis- or trans-acting regulators of eukaryotic gene expression, governing diverse biological functions including metabolism, fertility, differentiation, oncogenesis and immunological responses, to name a few. These observations have sprouted a great deal of interest in ncRNAs not only as important regulators of biological activities, but also as potential therapeutic tools or pharmacological targets.
One of the most intriguing and unexpected recent discoveries about ncRNAs is their direct contribution to genome stability and DNA repair. In this Review, we describe the well-characterized and recently proposed mechanisms by which nuclear RNAs participate in genome-related functions beyond their capacity to encode proteins. Specifically, by using at least one example, we will describe their roles in the maintenance of genome stability, genome organization and rearrangement in ciliates and their emerging role in directly mediating double-strand break (DSB) repair in eukaryotes. These new findings underscore the biochemical versatility of RNA as an effector molecule in biological pathways, and yield unexpected insight into the mechanisms by which these molecules participate in a variety of biological pathways.
ncRNA classifications
Eukaryotic genomes produce a large number of ncRNAs, which with the exception of ncRNAs such as tRNAs, rRNAs, snoRNAs and snRNAs, are divided into two operational categories based on their size - long (>200bp) ncRNAs (lncRNA), and small (<200bp) ncRNAs (sRNA) (Table 1). The 200bp size is a convenient cutoff based on the biochemical fractionation properties of RNA, at which all known sRNAs are excluded from the lncRNA fraction (Kapranov et al., 2007).
Table 1.
Classes of ncRNAs implicated in repair and genome stability. All abbreviations are defined in the text.
| Classes of ncRNAs | Ago Effector subfamily |
Size (bp) |
Localization | Functions |
|---|---|---|---|---|
| lncRNA | N/A | >200 | Nucleus | TGS, heterochromatin formation, genome organization and DSB repair |
| miRNA | Ago | 19–24 | Cytoplasm | PTGS (mRNA degradation, translational repression) |
| siRNA | Ago | 21–24 | Nucleus | TGS, heterochromatin formation, chromosome segregation |
| piRNA | PIWI | 23–31 | Nucleus Cytoplasm |
Transposon silencing (TGS and PTGS) |
| diRNA | Ago | 21–24 | Nucleus | DSB repair |
lncRNAs are primary transcripts for which no protein product has been detected or predicted (but see (Banfai et al., 2012; Frith et al., 2006)). Nuclear lncRNAs often localize at their sites of synthesis and can act as platforms for the assembly of regulatory complexes in cis. sRNAs, on the other hand, are cleavage products of endogenous or exogenous primary transcripts and often target the recruitment of other proteins in trans. The most frequently studied groups of noncoding sRNAs are 20–30bp in length and are associated with Argonaute (Ago) proteins. These are further divided into three classes based on their mechanism of biogenesis. Because of the focus of this review, we will also include an additional class of sRNAs whose biogenesis is dependent on DSB formation (Table 1).
microRNAs (miRNAs) (19–24 nts) are cleavage products of endogenous hairpin ncRNAs by Drosha and Dicer proteins. miRNAs are loaded onto the Ago-bearing RNA-induced silencing complex (RISC) and direct RISC to complementary transcripts. RISC mediates posttranscriptional gene silencing (PTGS) by promoting degradation or inhibiting translation of the complementary transcripts. Because miRNAs are cytoplasmic and regulate DNA repair and genome stability indirectly (Wan et al., 2014), we will not discuss them further in this review.
Small interfering RNAs (siRNAs) (21–25 nts) are produced by Dicer-dependent cleavage of endogenous or exogenous double-stranded RNAs (dsRNAs). siRNAs specify the trans recruitment of Ago complexes to complementary genomic regions, which in some organisms contribute to genomic stability by repressing transcription and recombination at repetitive DNA elements (Discussed below in detail) (Castel and Martienssen, 2013; Malone and Hannon, 2009; Moazed, 2009).
PIWI-associated RNAs (piRNAs) (24–30 nts) are associated with the PIWI clade of Argonaute proteins. PIWI-piRNA complexes play a critical role in protecting genomes against instability by repressing transposon activity via transcriptional gene silencing (TGS) and/or PTGS mechanisms (Ross et al., 2014; Siomi et al., 2011).
Damage-induced sRNAs (diRNAs) are another class of recently discovered Ago-associated sRNAs (20–22 nts long) whose formation is DNA damage-dependent. Originally discovered in Neurospora (Lee et al., 2009), diRNAs appear to play a direct and/or indirect role in DSB repair in many organisms, including humans (Francia et al., 2012; Wei et al., 2012). This class of sRNAs has been given several different names, but for simplicity, we will refer to all damaged-induced sRNAs as ‘diRNAs’ in this review.
RNA – a versatile biological polymer with unique properties and mechanisms of action
Similar to DNA and proteins, RNA is a polymer built on a fixed backbone from which monomeric residues protrude. But unlike DNA, which is often found in its double-stranded (ds) form, RNA is predominately single-stranded (ss), availing its bases for hydrogen bonding with other molecules or intramolecularly to form complex structures. Three main features equip ssRNAs with their diverse range of molecular activities: (A) their base composition and order; (B) availability to basepair with complementary sequences; and (C) capacity to not only form three-dimensional structures and act as enzymes, but also, to create flexible scaffolds for the assembly of ribonucleoprotein particle (RNP) complexes (Zappulla and Cech, 2006).
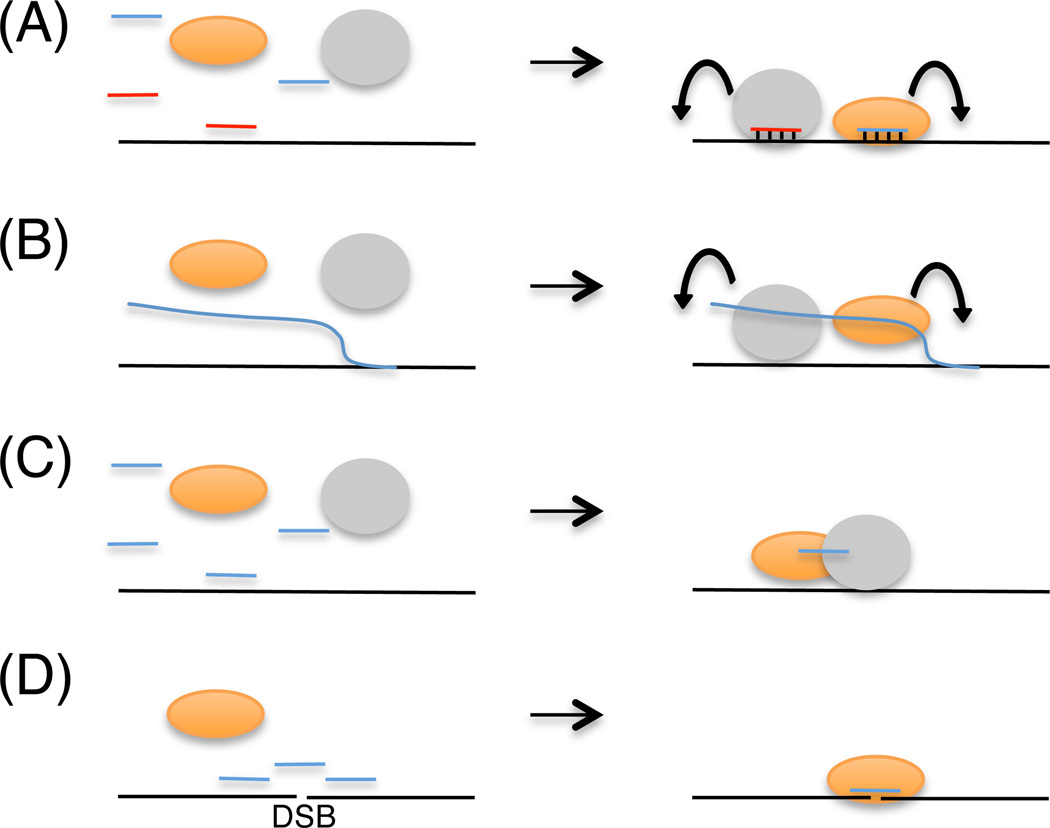
These features contribute to RNA’s unique biological activities via three well-established and a fourth emerging mechanisms (Figure 1). ncRNAs have been shown to (1) specify the trans recruitment of complexes by basepairing interactions with complementary sequences; (2) provide a platform for assembly of regulatory complexes in cis to the site of their synthesis; (3) act as a structural subunit of important biological complexes; and finally, (4) provide molecular bridges and/or act as genetic templates for genome reassembly and DSB repair. Initially described in ciliates and yeast, the fourth mechanism (Figure 1D) suggests that RNAs temporarily glue and/or act as transient archives of genetic information necessary for DSB repair or genome reassembly, respectively (Discussed in detail later).
Figure 1.
Mechanisms of ncRNA-mediated genome regulation. (A) Protein (orange and gray ovals)-sRNA (blue or red lines) interaction targets proteins to complementary sequences in trans. (B) Tethered ncRNAs form platforms for the assembly of regulatory protein complexes in cis. (C) ncRNAs facilitate protein-protein interactions by acting as a structural component of these complexes. (D) ncRNAs bridge or template the repair of double-strand breaks (DSBs).
Each of the above mechanisms provides an evolutionary benefit for the cell. For example in mechanism 1, it is important to point out that specificity can be encoded within a 20–30nt piece of RNA. This means that protein-sRNA interactions can be used to target the same enzyme to all complementary sequences (a strategy used for regulation of repetitive DNA elements in eukaryotes (Holoch and Moazed, 2015; Moazed, 2009), or, by varying the interacting sRNA, to different unique cytoplasmic or nuclear sequences (e.g. miRNA and siRNA pathways). Protein-sRNA interaction is regulated tightly at several steps, but most importantly by determining which transcripts become substrates for sRNA biogenesis. For example in the RNAi pathway, sRNAs biogenesis and amplifications are tightly controlled by the coordinated activities of several RNA processing and amplifying enzymes, which use unique features of transcripts for substrate recognition (Castel and Martienssen, 2013; Holoch and Moazed, 2015). These suggest that mechanism 1 provides an efficient adaptive solution to supplant (short term), or augment (long term), the more time-consuming process of co-evolving sequence-specific nucleic acid binding motifs in enzymes. Indeed, sRNAs have emerged as key components of stress and adaptive responses in organisms ranging from yeast to humans (Amaral et al., 2013), by acting as, among other things, specificity factors for the recruitment of regulatory complexes to different parts of the genome, thus permitting the transient or sustained regulation of genome functions globally.
Below, we will expand on how the aforementioned mechanisms equip ncRNAs to function in genome stability and repair pathways. (1) We begin by discussing three well-established examples in which nuclear ncRNAs regulate genome stability in organisms ranging from yeast to mammals. These provide a mechanistic snapshot of how nuclear ncRNAs of various lengths can establish amplification loops or platform to target genome stability functions to various regions of the genome via ncRNA-protein interactions. (2) Next, we discuss a series of elegant discoveries in ciliates in which ncRNAs not only orchestrate the developmentally-regulated reassembly of their fragmented genomes, but also can transmit somatic mutations to the next generation. (3) Finally, we summarize the emerging mechanisms through which small and long ncRNAs may assist in the repair of DSBs. Intriguingly, these data suggest that nuclear ncRNAs may act as bridges or templates for DSB repair by a mechanism that may involve the reverse transfer of genetic information from RNA to DNA. Together these examples reveal that the aforementioned mechanisms depicted in Figure 1 are recurrent themes in ncRNA-dependent pathways and underscore the biochemical versatility of RNAs in catalyzing biological processes.
(1) ncRNAs in genome stability
Heterochromatin and genome stability
A large fraction of the eukaryotic genome is composed of repetitive DNA elements, such as transposable elements (TEs), whose profusion have profoundly influenced eukaryotic evolution by affecting genome organization and stability (Cordaux and Batzer, 2009). To maintain genome stability by preventing transposition and homologous recombination (HR), the large tracks of repetitive DNA such as those found at telomeres (tel), centromeres (cen) and rDNA, are packaged into heterochromatin. Heterochromatin represses gene expression by TGS and PTGS mechanisms (Discussed below), but the mechanism involved in HR repression remains poorly understood. (Similar to transcription, HR repression appears to involve the exclusion or repression of HR activities within heterochromatic regions (Chiolo et al., 2011; Sinha et al., 2009; van Sluis and McStay, 2015). Heterochromatin also plays an important role in the proper incorporation of histone H3 variant CENP-A (Folco et al., 2008; Kagansky et al., 2009) at centromeres, a critical step in the assembly and attachment of kinetochores during cell division, and the proper cohesion of sister chromatids by recruiting cohesin to centromeres (Bernard et al., 2001; Gartenberg, 2009; Hahn et al., 2013; Nonaka et al., 2002). Cells defective for heterochromatin proteins show increased transposon activity, spurious recombination among repetitive DNA elements, chromosome segregation errors, and loss of telomere length regulation, all of which cause genome-wide instability and are associated with several human maladies including cancers. We will use the fission yeast (Schizosaccharomyces pombe) siRNA system, Drosophila piRNA system, and the telomeric RNA, TERRA, as examples of how nuclear ncRNAs help establish heterochromatin in eukaryotes (Figure 2).
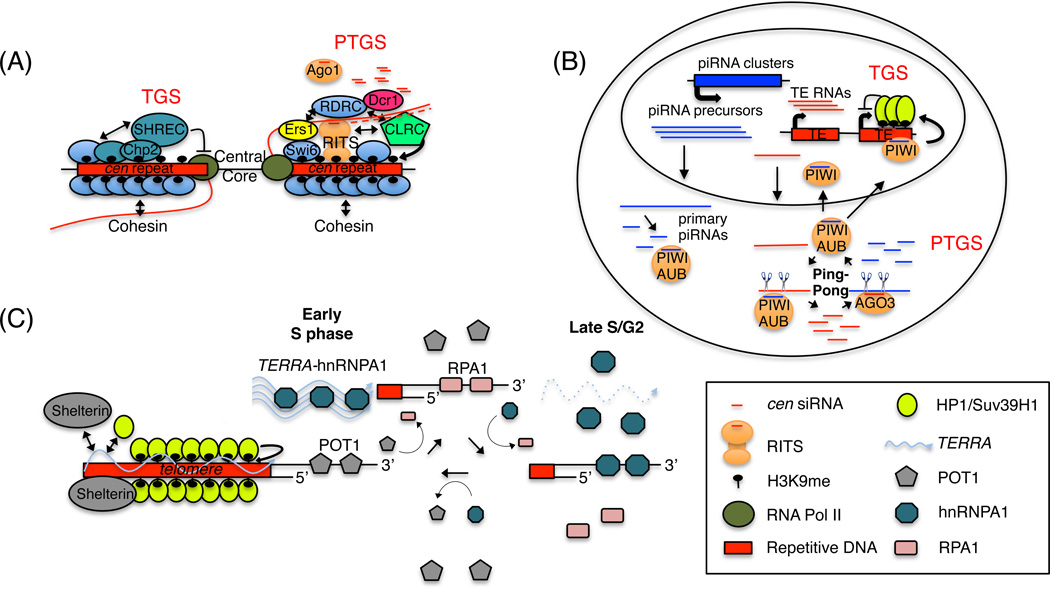
Figure 2.
ncRNA-mediated silencing and telomere regulation. (A) siRNA-dependent H3K9 methylation at the fission yeast pericentromeric regions. A network of interactions among RNAi complexes (RITS, RDRC, Dcr1), cen lncRNAs (red wavy line), and CLRC (green pentagon) creates siRNA and H3K9me amplification loops at centromeres. This leads nucleation and spreading of H3K9me and the recruitment of the heterochromatin protein 1 (HP1) proteins, Swi6 (blue oval) and Chp2 (turquoise). Chp2 in complex with SHREC represses RNA Pol II (green circle) transcription (TGS), and Swi6 facilitates RNAi-dependent PTGS via its Ers1-dependent interactions with RDRC. Swi6 also interacts cohesin and is required for its recruitment to cen repeats. In the absence of CLRC, RNAi complexes or Swi6, cohesin recruitment is lost and cells display chromosome segregation defects. (B) piRNA silencing in D. melanogaster maternal germ cells. piRNA clusters and transposons are shown in blue and red, respectively. Cytoplasmic degradation of TE transcripts by the Piwi family proteins (Aubergine (Aub), Piwi and Argonaute-3 (Ago3) is depicted. Iterative cycles of Piwi/Aub and Argonaute-3 (Ago3) cleavage events in the cytoplasm amplify the piRNA signal and degrade TE transcripts. Also, piRNA-Piwi complexes can be imported into the nucleus where they mediate TGS silencing via the recruitment of HP1/Su(var)3–9 proteins (yellow oval). (C) TERRA and telomere regulation. TERRA (blue wavy line) is a structural component of telomeres upon which Sheltrin (gray oval), HP1/Suv39H1 (lime green circle), and other silencing complexes assemble. TERRA also facilitates the RPA (pink rectangle)-to-POT1 (gray pentagon) transition via binding to hnRNAP1 (teal octagon). See text for details.
Centromeric silencing in fission yeast
The aforementioned functions of heterochromatin in genome stability are also true in fission yeast (Bernard et al., 2001; Ellermeier et al., 2010; Jia et al., 2004; Nonaka et al., 2002; Yamanaka et al., 2013), where centromeric heterochromatin formation requires the RNAi pathway (Volpe et al., 2002). In this system, centromeric lncRNAs (cen) tethered to their sites of synthesis act as platforms upon which complementary cen siRNAs target the assembly of silencing complexes: Nascent Transcript Model (Figure 2A and Figure 1A–C) (Buhler et al., 2006; Motamedi et al., 2004; Verdel et al., 2004). According to this model, lncRNA platforms establish two interdependent amplification loops - siRNA and histone H3 lysine 9 methylation (H3K9me) - both of which are essential for the formation, maintenance and propagation of epigenetically heritable chromatin states at centromeres. The molecular details of this pathway have been reviewed extensively elsewhere (Castel and Martienssen, 2013; Holoch and Moazed, 2015), so for this review, we limit our discussion to those features shared with other nuclear ncRNA pathways.
Heterochromatin formation in fission yeast requires H3K9 methylation (Rea et al., 2000), the hallmark of eukaryotic heterochromatin, to which several proteins bind (Bannister et al., 2001; Jacobs and Khorasanizadeh, 2002; Jacobs et al., 2001; Nakayama et al., 2001; Partridge et al., 2002). The sole H3K9 methyltransferase in S. pombe is Clr4 (Suv39H1/H2 homolog), whose efficient recruitment to centromeres requires the RNAi proteins (Volpe et al., 2002). In this system, dicer (Dcr1)-dependent cleavage of cen dsRNAs produces 22–24bp cen siRNAs which are loaded onto the Argonaute (Ago1)-bearing effector complex called RITS. RITS is a bivalent complex composed of Ago1, loaded with cen siRNAs, Chp1, an H3K9me-binding protein, and Tas3, a GW motif linker protein. Because RITS recruitment to centromeres requires siRNAs and H3K9me, current models propose that siRNA basepairing with nascent cen lncRNAs and Chp1 binding to H3K9me guide RITS to centromeres (Verdel et al., 2004). Once at centromeres, RITS binds to and recruits an RNA-dependent RNA polymerase (RdRP) complex called RDRC, which converts single-stranded cen transcripts to dsRNAs (Motamedi et al., 2004). These dsRNAs are processed by Dcr1, amplifying the cen siRNA pools (Colmenares et al., 2007; Sugiyama et al., 2005) and recruiting more RITS to centromeres. Also, RITS physically interacts with the Clr4 complex called CLRC (Bayne et al., 2010; Gerace et al., 2010; Zhang et al., 2008), through whose recruitment the H3K9me signal is amplified and spread to the surrounding regions (Noma et al., 2004). Artificial tethering of RITS to a euchromatic gene induces RNAi- and H3K9me-dependent TGS and PTGS silencing (Buhler et al., 2006), demonstrating that RITS acts as a recruitment hub for the establishment of H3K9me and siRNA amplification loops. Indeed much has been discovered about how several TGS and PTGS pathways converge at the fission yeast centromeres (Alper et al., 2013; Buhler et al., 2007; Buscaino et al., 2013; Egan et al., 2014; Fischer et al., 2009; Lee et al., 2013; Motamedi et al., 2008; Reyes-Turcu et al., 2011; Sugiyama et al., 2007; Zhang et al., 2011), but here, this example serves to illustrate how small and long nuclear ncRNAs are used to provide specificity for the recruitment of silencing factors, or a platform for the assembly of complexes, critical for safeguarding the genome against instability (Figure 1A–C and Figure 2A).
Transposable element (TE) silencing in flies
Activation of TEs can lead to insertional mutagenesis, genomic rearrangements and genome-wide instability in somatic and germ cells (Burns and Boeke, 2012). In organisms ranging from flies to mammals, germ cell TE silencing uses an Ago-mediated sRNAs (piRNA) pathway (Dumesic and Madhani, 2014; Siomi et al., 2011), which bears many similarities to the S. pombe pathway described above. The piRNA pathway is best understood in flies in which nuclear TGS and cytoplasmic PTGS pathways cooperate to silence TEs (Figure 2B). In Drosophila, piRNAs (22–30 nts), which associate with the PIWI clade of Argonaute proteins (Aub, Piwi and Ago3), orchestrate TE silencing in germ cells (Dumesic and Madhani, 2014; Siomi et al., 2011). This process is initiated by transcription from piRNA clusters: distinct genes encoding lncRNAs, which are antisense to TEs (Vagin et al., 2006). piRNA precursor lncRNAs are transported to and degraded in the cytoplasm, producing primary piRNAs, antisense to TE transcripts. These piRNAs bind to Aub and Piwi protein, which direct the cleavage of complementary TE RNAs in the cytoplasm (Figure 2B). This cleavage in turn produces sense piRNAs, which, once loaded onto Ago3, cleave piRNA precursor transcripts. Iterative cycles of Ago3 cleavage of piRNA precursors and Piwi/Aub cleavage of TE transcripts (so called Ping-Pong mechanism) amplifies the piRNA signal and leads to cytoplasmic degradation of TE transcripts (PTGS) (Figure 2B) (Brennecke et al., 2007; Gunawardane et al., 2007). These create a piRNA amplification loop which, even though cytoplasmic and using different enzymes, produces sRNAs as PTGS degradation products of TE transcripts, similar to the S. pombe siRNA amplification pathway.
The piRNA-Piwi complexes also are imported into the nucleus, triggering TGS silencing in the nucleus at the complementary sequences (TEs) by a mechanism similar to the one described in the fission yeast. In the nucleus, Piwi has been reported to physically interact with heterochromatin protein1a (HP1a), and is required for H3K9me and RNA Pol II repression at TEs (Brower-Toland et al., 2007; Le Thomas et al., 2013; Sienski et al., 2012; Wang and Elgin, 2011). Ectopic insertion of TE sequences in euchromatic regions or expression of ectopic piRNAs complementary to a reporter gene triggers H3K9me and TGS silencing at the target locus (Le Thomas et al., 2014; Sienski et al., 2012), suggesting a model in which piRNA-lncRNA basepairing interactions recruit PIWI and chromatin-based TGS silencing activities to TEs (Le Thomas et al., 2014; Mohn et al., 2014). (It is also possible that piRNAs or siRNAs interact with the complementary DNA sequences at the target loci, but a direct test of this has not been performed in either system.) Overall, the fly piRNA model extends the Nascent Transcript Model in S. pombe suggesting that cytoplasmic sRNA amplification loops (PTGS) can contribute to and cooperate with nuclear (TGS) silencing pathways to repress TE expression fully in germ cells and provide an efficient means to safeguard the genome against TE-induced genomic instability during gametogenesis.
Telomere function and genome stability
Telomeres are another chromosomal feature, which are packaged into heterochromatin and use ncRNAs to regulate their function and stability. Telomere formation prevents the recognition of chromosome ends as DSBs, thus repressing their inappropriate recombination and erosion. Loss of telomere function is linked to telomere shortening, genomic instability, cancer and premature aging. A nuclear telomeric lncRNA called TERRA (Telomeric Repeat-containing RNA which contains subtelomeric and telomeric sequences) is an integral component of the telomere structure (Figure 1B and Figure 2C), contributing to several telomeric functions (Cusanelli and Chartrand, 2015). Here we will use TERRA as an example to describe how properties of lncRNAs alone (without processing to sRNAs) create an interface between telomeric DNA and the regulatory proteins which maintain the integrity of these structures in eukaryotes.
Even though different in sequence, TERRA expression is a conserved feature of eukaryotic telomeres, and its transcription moves from the subtelomere to telomere direction (Azzalin et al., 2007; Schoeftner and Blasco, 2008). TERRA plays critical roles in regulating (1) telomeric heterochromatin formation (Arnoult et al., 2012; Deng et al., 2009), (2) proper capping of telomeres (Flynn et al., 2011) and (3) R-loop formation. TERRA is postulated to act as a platform for the assembly of several heterochromatin factors at telomeres (Figure 1B and Figure 2C). It physically interacts with TRF1 and TRF2, components of the hexamerous Shelterin complex (required for telomeric heterochromatin formation), heterochromatin protein 1 alpha (HP1α), H3K9me3 (Deng et al., 2009), SUV39H1 (Porro et al., 2014) and MORF4L2 (Scheibe et al., 2013), whose budding yeast homolog, EAF3, is required for formation of telomeric boundaries (Babiarz et al., 2006). A decrease in TERRA levels correlates with a decrease in H3K9me3 and other heterochromatic marks at telomeres (Deng et al., 2009), suggesting a model similar to lncRNA-dependent centromeric heterochromatin formation in S. pombe with TERRA acting as a scaffold for the recruitment of heterochromatin factors to telomeres (Figure 2C).
In addition to these proteins, TERRA is needed for proper capping of telomeres potentially by regulating the hnRNPA1- and cell cycle-dependent RPA to POT1 transition (Flynn et al., 2011). During DNA replication, the ssDNA binding protein RPA binds to the exposed ssDNA regions at telomeres. But, RPA must be removed and replaced by POT1 (a Shelterin component) at telomeres to maintain telomeric integrity by preventing the RPA- and ATR-dependent activation of the DNA damage response (DDR) pathway (Denchi and de Lange, 2007). RPA replacement by POT1 requires hnRNPA1 in a TERRA-regulated manner. In early S phase when TERRA levels are high, TERRA binds to and sequesters hnRNPA1, allowing for RPA binding to single-stranded telomeric DNA. But in late S and early G2 as TERRA levels decrease, hnRNPA1 is free and can displace RPA from telomeric ends, allowing for POT1 loading to telomeres (Flynn et al., 2011). These results suggest that TERRA acts as a sponge for hnRNPA1 protein pools, through which RPA and POT1 binding to telomeres may be regulated. Some support for this model exist (Flynn et al., 2015; Redon et al., 2013); however, several aspects of this model remain to be tested. Here this serves to illustrate that in addition to the mechanism shown in Figure 1, transcriptional regulation of lncRNA pools may act as sponges to regulate the availability of RNA-binding proteins in biological processes.
Work in yeast and mammalian cells has revealed that TERRA basepairs with its DNA template, forming RNA:DNA structures called R-loops (Arora et al., 2014; Balk et al., 2013; Pfeiffer et al., 2013; Yu et al., 2014). R-loops are recombinogenic and their stability in wild-type cells is tightly controlled by several pathways including the RNase H1 (RNH1) and RNase H2 (RNH201) proteins, which degrade the RNA component of R-loops (Aguilera and Garcia-Muse, 2012). In telomerase defective yeast and mammalian cells, telomeres shorten after each round of DNA replication, but the surviving cells enact an HR-dependent pathway to maintain telomeric length (Bryan et al., 1995; Lundblad and Blackburn, 1993). Interestingly loss of RNase H activity in these cells improves survival and delays senescence by increasing HR at telomeres (Arora et al., 2014; Balk et al., 2013). Even though the mechanism by which R-loops promote recombination remains elusive, their accumulation is linked to genomic instability, which, together with their role at telomeres, suggest that TERRA:DNA hybrids play a critical role in telomere stability. These results reveal that nuclear lncRNAs such as TERRA, in addition to providing a stable interface for the assembly of complexes (Figure 1B), their abundance in the form of RNA or RNA:DNA hybrids can be used as biological sensors for regulating functions important for genome stability.
(2) ncRNAs in genome organization
In recent years, one of the most surprising and intriguing discoveries about ncRNAs has come from studies of ciliates. In these organisms small and long nuclear ncRNAs mediate the reorganization and elimination of up to 95–98% of their genomes during sexual reproduction. Beyond insights into new mechanisms by which ncRNA regulate genome functions, ciliate studies have also revealed that ncRNAs can be agents of transgenerational epigenetic inheritance (TEI) (Discussed later). Because of these findings, we devote a significant part of our discussion to this work.
Genome reorganization in ciliates
One of the most unique features of ciliates is their nuclear dimorphism - the presence of two distinct nuclei per cell: the germline micronucleus (MIC) and the somatic macronucleus (MAC). MIC is compact (small), diploid, transcriptionally inert, contains repetitive TEs and archives all genetic information. The MIC genomes of some species (Oxytricha and Stylonychia) are scrambled - up to 20–30% of the genes are stored as fragmented pieces, scattered non-contiguously throughout the MIC genome (Figure 3A). On the other hand, MAC is highly transcribed, polyploid, devoid of TEs, consisting of amplified, neatly organized genes derived from the fragments found in the MIC genome (Chang et al., 2005). Following sexual reproduction, the progeny MAC genome is derived from the parental MIC genome by two developmentally-regulated processes: (1) repetitive DNA elements found in the MIC genome are removed, and (2) scattered gene fragments in the MIC nucleus are brought together, ordered and sewn forming contiguous genes in the MAC genome (Figure 3A–B) (Nowacki et al., 2011).
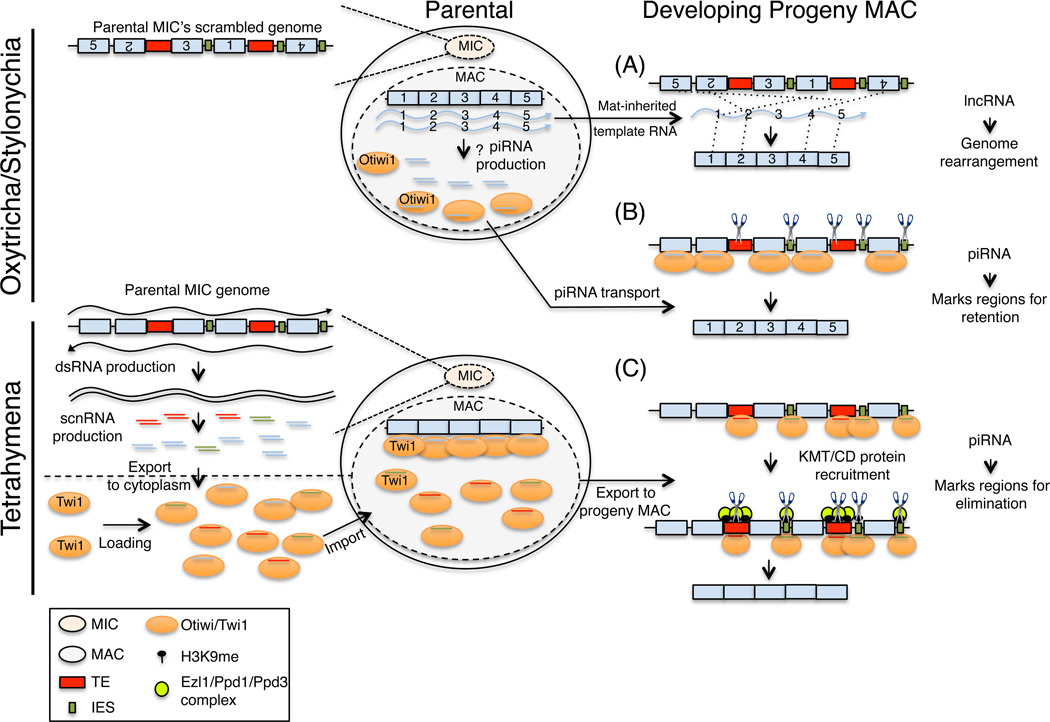
Figure 3.
ncRNA-mediated genome reorganization in ciliates. Genome rearrangement in Oxytricha involves the (A) unscrambling of the developing MAC genome. This requires the maternal inheritance of long noncoding template RNAs (blue wavy lines) from the parental MAC nucleus into the developing progeny MAC. Numbers depict segments of a gene which must be ordered to make a functional open reading frame. (B) DNA retention uses maternally inherited piRNA-Otiwi complexes (orange ovals with blue lines) to mark the regions of the genome which are retained in the developing progeny MAC nucleus. (C) DNA elimination in Tetrahymena requires the bidirectional transcription (wavy black lines) of the parental MIC genome. dsRNAs are degraded into scnRNAs (shown in red, blue and green lines) by Dcl1 and exported to the cytoplasm where they are loaded onto the PIWI protein Twi1 (orange oval). scnRNA-Twi1 complexes are imported into the parental MAC nucleus where they find the complementary genomic regions. All ‘self’ scnRNAs (blue) are eliminated, and the remaining scnRNAs (red and green), complementary to TEs (red rectangle) and IESs (green rectangle), are exported into the developing progeny MAC nucleus. scnRNA-Twi1 complexes basepair with complementary TEs and IESs in the MAC genome and recruit Ezl and Ppd1/3 proteins (lime green circle). These regions are packaged into heterochromatin and later eliminated from the MAC genome by an unknown mechanism.
Gene unscrambling and copy number regulation
MAC development in Oxytricha and possibly Stylonychia uses maternally-inherited lncRNAs (called template RNAs) to template genome unscrambling. These lncRNAs arise during conjugation in the parental MAC nucleus, transported to the developing progeny MAC where they direct the accurate unscrambling of genes (Figure 3A) (Nowacki et al., 2008). In Oxytricha, the artificial introduction of a template RNA (sense, antisense, or double-stranded) directs the unscrambling of the cognate gene in the MAC nucleus, and its RNAi knockdown prevents its unscrambling. These unscrambling events are transgenerationally inherited (as far as F3), suggesting the existence of a mechanism responsible for the inheritance of template parental lncRNAs into the developing progeny MAC (Nowacki et al., 2008). Interestingly, template RNAs also determine gene copy number in the developing MAC. Injection or RNAi knockdown of wild-type RNA templates during conjugation results in an increase or decrease in the DNA copy number of the corresponding gene in the progeny MAC, respectively (Heyse et al., 2010; Nowacki et al., 2010). Moreover, introduction of a synthetic RNA carrying mutations at the gene segment junctions in Oxytricha (Nowacki et al., 2008) (or modified telomeres in Stylonychia (Fuhrmann et al., 2016)), causes the corresponding alterations in the progeny MAC genome. Taken together, these observations demonstrate that maternal lncRNAs instruct gene unscrambling, dosage and sequence in the developing MAC genomes (Figure 3A), thus transmitting the genic, dosage and transcriptional program of parents to progeny. In principle, this is similar to the TEI pathways observed in C. elegans (Buckley et al., 2012; Burton et al., 2011; Gu et al., 2012) whereby maternally-inherited siRNAs bound by nuclear Ago proteins transmit the transcriptional memory of the parental animals to offspring. Even though different in size, the inheritance of these nuclear ncRNAs provides an important adaptive advantage – it tunes the transcriptional programs of progeny to those of its parents, improving its chances of survival through the early and often vulnerable time points in life. This is another unique molecular function of RNA, whose capacity to specify and establish transcriptional outputs is harnessed to relay important epigenetic adaptive responses to future generations.
Mechanistically, this process might use RNA:DNA hybrid intermediates to guide the cutting and pasting of the MAC genome. Precedents for RNA-templated genome modification also exit in yeast (Keskin et al., 2014; Storici et al., 2007) and potentially other eukaryotes (Onozawa et al., 2014; Shen et al., 2011), in which the repair of a DSB involves the formation of damage-induced lncRNAs, hypothesized to bridge or template repair via an RNA:DNA hybrid intermediate (see below).
DNA retention in Oxytricha
In addition to gene unscrambling and copy number regulation, small and long ncRNAs orchestrate a DNA elimination process in Oxytricha by which large portions of the progeny MIC genome (satellite repeats, transposons, and intergenic regions) are eliminated in the developing progeny MAC (Nowacki et al., 2011). During early conjugation, developmentally-regulated transcription of the parental MAC produces lncRNAs which are presumably degraded into 27 nt piRNAs (Bracht et al., 2013; Yerlici and Landweber, 2014). A PIWI homolog, Otiwi1, is loaded with these piRNAs (Fang et al., 2012; Zahler et al., 2012) and transported from the parental MAC to the progeny MIC, where piRNA interaction with complementary genomic sequences marks regions of the MIC genome protected from elimination during MAC genome development (Figure 3B). This piRNA-dependent pathway is similar to the one described in flies, in that piRNAs are utilized as sequence-specific guides for the regulation of the complementary sequences, but differ in that in Oxytricha, their association leads to the retention of these sequences instead of their transcriptional silencing. Many important questions remain unanswered in this fascinating field. For example, it would be interesting to determine how the lncRNA-dependent genome rearrangement (Figure 3A) and piRNA-dependent DNA retention mechanisms (Figure 3B) are coordinated in the developing Oxytricha MAC nucleus. It is plausible that the two pathways share common components through which the two can be coordinated temporally in the progeny MAC.
DNA elimination in Tetrahymena
In Tetrahymena thermophila, progeny MAC is derived by the programmed deletion of roughly 6000 unique internal eliminated sequences (IESs), which are intergenic, intronic or transposon-like repeats found in the MIC genome (Figure 3C) (Fillingham et al., 2004; Heinonen and Pearlman, 1994; Wuitschick et al., 2002). Similar to DNA retention mechanism outlined above, IES elimination is epigenetically regulated by the RNA sequences found in the parental MAC genome (Mochizuki and Gorovsky, 2004; Yao et al., 2003). During early conjugation, bidirectional transcription of the entire MIC genome produces double-stranded micronuclear non-coding RNAs (dsmic-ncRNAs) (Aronica et al., 2008; Chalker and Yao, 2001) which are processed by the meiosis-specific dicer-like enzyme called, Dcl1, into small RNAs (26–31nts) called scan RNAs (scnRNAs) (Malone et al., 2005; Mochizuki and Gorovsky, 2005). scnRNAs are exported to the cytoplasm where they complex with a PIWI clade Ago protein, Twi1p, and generate active Twi1p-scnRNA complexes by a process which requires the Twi1 slicer activity. The Twi1-scnRNA complexes are then transported to the parental MAC (Noto et al., 2010), undergo 2’-O-methylation by Hen1p, stabilizing scnRNAs (Kurth and Mochizuki, 2009), and scan the parental macronuclear ncRNAs (pmac-ncRNAs) transcriptome. Complementary scnRNAs are selectively degraded (Aronica et al., 2008), and the remaining scnRNAs (homologous to MIC-specific sequences (mostly IESs)) move to the developing MAC where they scan and later interact with new macronuclear ncRNAs (nmac-ncRNAs). This results in the recruitment of the histone methyltransferase Ezl1p and the chromodomain proteins Pdd1 and Pdd3 to these IESs, which cause the packaging of these regions into heterochromatin followed by their elimination from the MAC genome by an unknown mechanism (Coyne et al., 1999; Liu et al., 2004; Liu et al., 2007; Taverna et al., 2002). Pdd1 accumulation at heterochromatic IESs is dependent on scnRNAs and RNAi, demonstrating a direct role for scnRNAs in Pdd1 recruitment. Also, Pdd1 can interact with nuclear lncRNAs, potentially tethering these transcripts to their site of synthesis (Kataoka and Mochizuki, 2015). Interestingly, ectopic tethering of Pdd1 to a genomic locus that is normally retained leads to the elimination of this sequence in the progeny MAC (Taverna et al., 2002). This process also can occur in trans such as sRNAs derived from one TE can recognize, form heterochromatin and direct the elimination of other complementary TEs (Noto et al., 2015). Together these observations demonstrate a direct role for Ago-associated sRNAs in guiding chromatin-modifying proteins and the subsequent DNA elimination machinery to IESs in Tetrahymena (Figure 3C). How IESs are precisely marked and excised from within the gene-rich MIC genome has remained poorly understood, but interestingly in a recent paper, a model is proposed in which RNA:DNA hybrids may help demarcate the correct excision boundaries of a subset of IESs flanked by guanine-rich DNA (Carle et al., 2016).
The scnRNA-mediated IES elimination in Tetrahymena and the piRNA-mediated DNA retention in Oxytricha use PIWI-bound sRNAs to scan the developing MAC nucleus to mark eliminated or retained parts of the developing genome, respectively. The parental inheritance of these molecules transmits the genomic and transcriptional instructions of the previous generation to progenies. In addition to TEI, this pathway is thought to promote genomic stability by eliminating potentially harmful DNA elements (such as new transposon integration events) from the transcribed MAC nucleus. Collectively, this elegant body of work in ciliates has uncovered several unexpected roles for nuclear ncRNAs in genome regulation. Cells can (1) harness the ‘coding’ capacity of RNAs to instruct the reassembly of their genomes, (2) use sRNAs to demarcate parts of their genomes for elimination or retention, (3) set gene dosage, and (4) relay the adaptive memories of previous generations to progenies. Even though the conservation of these processes in other eukaryotes remains unknown, these discoveries add to the repertoire of mechanisms, which nuclear ncRNAs can use to participate in genome functions.
(3) ncRNAs and DSB repair
In this section, we will summarize and discuss the emerging connection between DSB repair and biogenesis of nuclear ncRNAs including the formation of damage-induced long and small ncRNAs, which, in some instances, appear to play a direct role in DSB repair.
sRNA-mediated DSB repair
The first evidence that small ncRNAs contribute to efficient repair of DSBs in eukaryotes was made in Neurospora crassa. Similar to S. pombe, N. crassa uses an RNAi-dependent mechanism called quelling to mediate the PTGS silencing of its repetitive genes (Chang et al., 2012). In this organism, DSBs induce the production of a class of Ago-associated sRNAs (20–22nts) called qiRNAs (Figure 4A, pathway 1) (Lee et al., 2009). (We will refer to this class of sRNAs as ‘diRNAs’.) Work in this organism has revealed that in response to DSBs, an HR-dependent mechanism targets RNAi factors to repetitive rDNA elements, where their coordinated activities generate diRNAs. diRNAs are Dicer (DCL-1 and DCL-2) cleavage products of double-stranded aberrant rDNA transcripts (aRNAs), produced by an RdRP, called QDE-1 (Lee et al., 2010; Salgado et al., 2006; Zhang et al., 2013b). Interestingly, recent data has shown that HR is also critical for the endogenous quelling (PTGS) pathway for repeat silencing (Zhang et al., 2013b), which together with the HR-dependency of diRNA production suggest that RNAi proteins use HR to identify repetitive regions for silencing. This is an intriguing possibility which, if conserved in other eukaryotes, would provide a new function for HR in sRNA biogenesis and function.
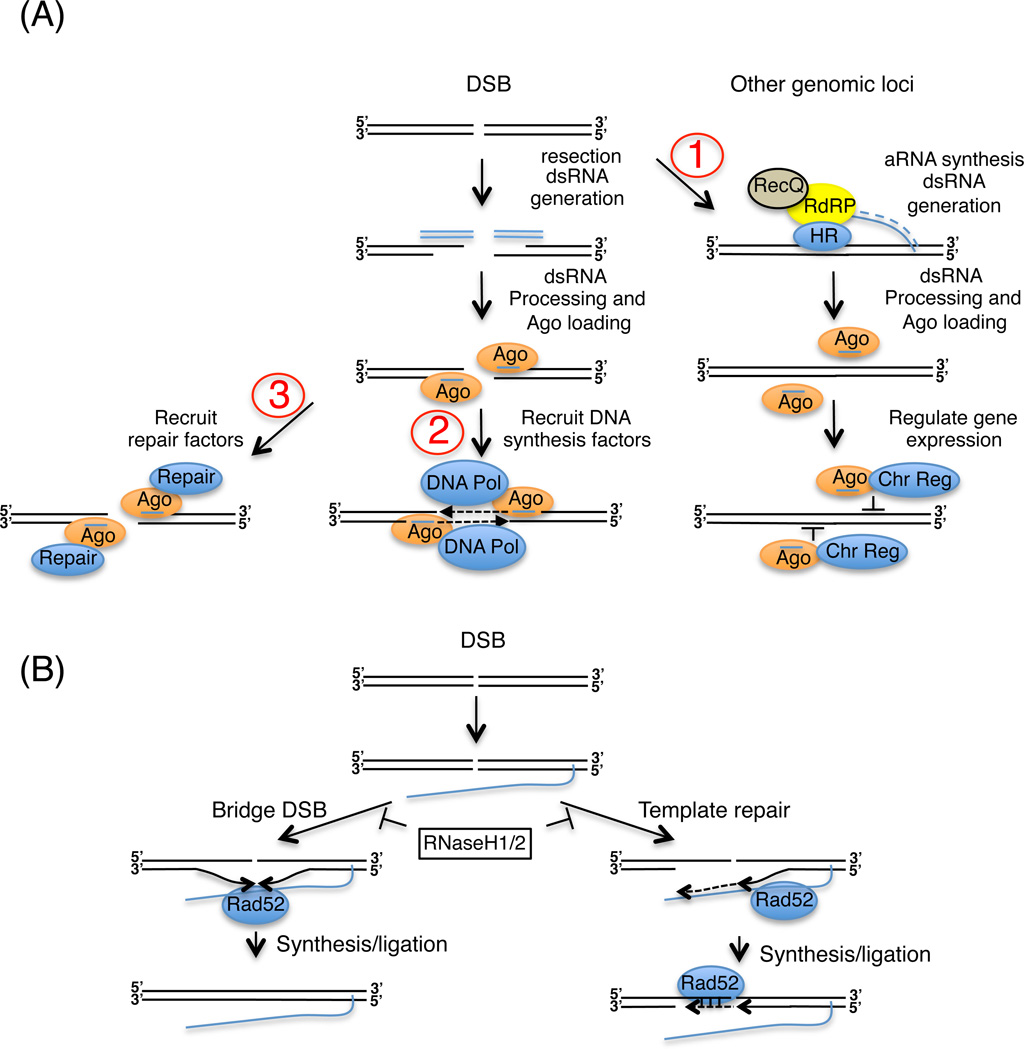
Figure 4.
ncRNAs regulation of DSB repair. (A) diRNA-mediated DSB repair. DSBs induce RNAi-dependent diRNA production, which once loaded onto Ago (orange oval) target Ago recruitment to DSBs (pathways 2 and 3) or other sites around the genome (pathway 1). In 1, diRNAs recruit Ago to sites (other than the DSB) around the genome (for example rDNA in N. crassa), potentially regulating their transcription in a manner that favors DSB repair. In 2, diRNAs recruit Ago and DNA polymerases (blue oval) to the DSB to catalyze a synthesis-dependent repair process. In 3, diRNA-programmed Ago complexes recruit repair proteins to the DSB. (B) lncRNA-mediated DSB repair. RNA molecules (blue line) complementary to the DSB may serve as a bridge (left) or template (right) for repairing DSBs. RNase H1/H,2 inhibit this repair mechanism by removing RNA:DNA hybrids. On the other hand, Rad52 (blue oval), which promotes RNA:DNA hybrid formation in vitro, is proposed to stimulate DSB repair by annealing the RNA to the DSB. RNA:DNA hybrids are thought to promote the precise re-ligation of a DSB (left) or reverse transcriptase-dependent synthesis and re-ligation (right) at the DSB. These depicted mechanisms may operate in non-dividing mammalian cells (Wei et al., 2015).
diRNA sequencing revealed that most map to the rDNA locus. DSB-induced rDNA silencing is conserved in eukaryotes; for example in mammals, DSBs cause an ATM-dependent translocation of NBS1-TCOF1 to nucleoli resulting in the global silencing of rRNA transcription (Kruhlak et al., 2007; Larsen and Stucki, 2016). Similarly in N. crassa, diRNAs silence rDNA expression in response to DSBs, however whether this inhibition is mediated through a PTGS (akin to how miRNAs contribute to DSB repair in metazoans (Wang and Taniguchi, 2013)) or TGS mechanism remains to be determined. More importantly, future work is required to test whether diRNAs complementary to the site of a DSB are produced. The latter question is especially pertinent because several recent papers have reported the occurrence of diRNAs complementary to DSBs (see below) in plants and mammalian cells (Francia et al., 2012; Gao et al., 2014; Wei et al., 2012).
diRNAs in mammals and plants
Studies in human cells, zebra fish larvae and plants suggest the occurrence of diRNAs beyond Neurospora (Francia et al., 2012; Gao et al., 2014; Wei et al., 2012). In these organisms, DSBs induce diRNA production, which in human and plant cells maps to the vicinity (0.5–2.5Kb) of DSBs. The requirement for DICER and the detection of sense and antisense diRNAs suggest the formation of dsRNA intermediates from the DSB; however how these putative dsRNAs are produced remains purely speculative at this time. But recently using an inducible, site-specific DSB system in a human cell line, it was shown that Ago2 interacts with the repair protein Rad51, whose accumulation at the DSB requires the RNA binding and catalytic activities of Ago2 (Gao et al., 2014). This suggests that diRNAs program Ago2-Rad51 complexes to the vicinity of the DSB via basepairing interactions with complementary sequences (Figure 4A, pathways 2 and 3).
Overall, the evidence in different model systems suggests that DSBs induce the synthesis of diRNAs mapping to the DSB or other parts of the genome (e.g. rDNA). The production of these diRNAs requires the activity of RNAi factors, DNA helicases, chromatin remodelers, ssDNA binding protein and, in Neurospora and plants, RNA-dependent RNA polymerases. diRNAs appear to target Ago complexes to complementary sequences to the DSBs or other parts of the genome (e.g. rDNA) through which the recruitment of repair (e.g. Rad51) or transcriptional regulatory complexes could occur (Figure 4A). The aforementioned direct and indirect mechanisms are not mutually exclusive and future studies are needed to determine how these nuclear sRNAs contribute to the repair of DSBs.
Alternatively, sRNAs may serve as bridges or templates for DSB repair. Work in E. coli, yeast and human cells indicate that artificial introduction of short RNA oligonucleotides homologous to the site of the DSB can direct their repair (Onozawa et al., 2014; Shen et al., 2011; Storici et al., 2007). It is important to point out that this pathway also operates in S. cerevisiae (which lacks an Ago homolog), suggesting that sRNAs arising from processing of primary transcripts can bridge or template DSB repair directly, instead of recruiting repair proteins to DSBs. The details of how these putative RNA:DNA complexes are formed or regulated or contribute to repair are unknown at this time (For a model see Figures 4B and 1D), but recent evidence from yeast provides interesting insights into how lncRNAs can contribute to DSB repair via bridging breaks, forming RNA:DNA hybrid intermediates and modifying the genome by a reverse-transcriptase-dependent mechanism, which, by the virtue of its capacity to modify the genome, is similar to the genome rearrangement pathway described in ciliates.
lncRNA-mediated DSB repair
Recent evidence from S. cerevisiae (Keskin et al., 2015; Keskin et al., 2014) suggests that in addition to sRNAs, long antisense transcripts originating from a DSB (cis) or artificially produced from a complementary genomic region (trans) can template DSB repair through a mechanism which requires the formation of RNA:DNA hybrid intermediates (Figure 4B). Using HIS− haploid cells, an elegant recombination assay was set up such that the only way that the repair of a DSB within a defective his3 reporter gene can produce a HIS+ cell is by using an antisense HIS3 transcript (carrying an artificial intron) as its template for repair. This antisense HIS3 allele was provided in trans or cis to the DSB. The budding yeast has its own reverse transcriptase activity (Boeke et al., 1986; Teng et al., 1996), and this activity is required for DSB repair in this assay, suggesting that cDNA intermediates derived from intact transcripts of actively transcribed genes can template repair, and potentially act as major sources of genome modifications at DSBs (Keskin et al., 2015). This result also suggests the formation of RNA:DNA hybrid intermediates during DSB repair. Consistent with this hypothesis, they showed that RNaseH1 (RNH1) and RNaseH2 (RNH201) proteins, which specifically degrade RNA:DNA hybrids inhibit this repair pathway – their removal improved repair efficiency dramatically in both the cis and trans systems. Even though repair in wild-type cells was dependent on reverse transcriptase activity, repair in rnh1 rnh201 cells could still proceed in the absence of reverse transcriptase. Unexpectedly, repair was more efficient if the antisense HIS3 transcript was produced in cis (rather than trans) to the DSB. Together these data suggest that stable RNA:DNA hybrids are recombinogenic, and in RNase H defective cells, RNA molecules complementary to a DSB can direct its HR-dependent repair without going through a cDNA intermediate. This suggests that long RNAs may act as a bridge or molecular glue to keep the ends of a DSB (complementary to the tethered transcript) near each other, thus facilitating HR repair. Also these data show that the proximity of the transcript to the DSB increases the efficiency of this repair mechanism. Furthermore, Rad52 protein, which was shown to promote RNA:DNA hybrid formation on DSB templates in vitro, is required for this type of DSB repair in vivo, whereas Rad51, which competes with Rad52 for ssDNA binding in vivo, is inhibitory (Figure 4B). Rad52 is also required for HR-dependent survival of telomerase defective cells in which telomeric RNA:DNA hybrids are thought to promote survival via HR among telomeres (Balk et al., 2013). Collectively these data suggest a model in which transcripts, tethered to the DSB by a stalled RNA polymerase (cis) or transcribed from a paralogous gene (trans), may provide a physical bridge or template for repair of DSBs (Figure 4B).
Two observations suggest that a similar RNA-dependent mechanism may contribute to mammalian DSB repair. First, it was shown that a fraction of the DSBs in mammalian cells are repaired by insertion of sequences derived from reverse transcribed RNAs originating from repetitive parts of the genome (Onozawa et al., 2015; Onozawa et al., 2014). Even though this repair pathway is mutagenic (inserting ectopic sequences into protein coding genes), these data suggest that long transcripts can be used as a template for repair across the DSB. Also recently, a non-mutagenic, HR-dependent and RNA-templated DSB repair in G0/G1 mammalian cells was described (Wei et al., 2015), which supports the occurrence of this RNA-templated repair in mammalian cells. Regardless of the mechanism, these observations suggest the existence of a conserved HR-dependent repair mechanism in which RNAs serve as templates or molecular bridges for DSB repair, providing yet another unexpected function for nuclear ncRNAs in cellular processes. In these instances, surprisingly, the flow of genetic information is reversed (RNA to DNA) such that any missing sequences at the DSB after resection is copied from the tethered RNA.
Conclusions and perspectives
The diverse mechanisms by which nuclear ncRNAs of different sizes are utilized in pathways that regulate genome stability highlight the versatility of RNA as a mediator of biological activity. RNA’s biochemical properties allow it to serve both as an enzymatic and genetic agent in nuclear functions. This is in contrast to proteins and DNA, which in their functional form lack this versatility, and are highly specialized to catalyze biochemical reactions or stably store genetic information, respectively. Despite their evolution, the primordial properties of RNA have continued to augment the biological activities of DNA and proteins, including the ones required for maintaining genome stability.
Noncoding sRNAs are best understood functionally in the context of their ability to basepair with complementary sequences, providing an efficient means for targeting enzymes to different sequences in the genome. lncRNAs, on the other hand, can act as scaffolds for the assembly of regulatory complexes often in cis to their site of synthesis. When used in concert, sRNA-lncRNA properties can generate amplification loops, ideally suited for establishing epigenetic states, including chromatin-based ones, and even transmitting these states transgenerationally. This makes sense evolutionary. In TEI, the inheritance of long or small ncRNAs imparts the transcriptional memory of the parental generation, through which parents pass their adaptive responses to their progeny, especially during the early, and often vulnerable times, of their offspring’s development. This helps progeny to quickly adapt by adjusting their transcriptional output based on their parents’ experiences.
In addition to this, long and/or small ncRNAs may bridge DNA breaks, or template their repair and modify the genome in a reverse transcriptase-dependent manner (Figure 4). These discoveries suggest that RNA to DNA information transfer may be a more general phenomenon than previously proposed (Crick, 1970). RNA-templated genome modifications are best characterized in ciliates, where parentally-inherited long and short ncRNAs cooperate to unscramble gene fragments, template their assembly, determine gene dosage, and exclude repetitive DNA elements, and in doing so, stabilize the maturing MAC genome (Figure 3). This mechanism differs from the chromatin-based TEI described previously, because in ciliates the inheritance of these parental nuclear ncRNAs determines the genetic makeup of the MAC genome. Based on these, it is intriguing to speculate that RNA-templated genome modifications, if induced as an adaptive response, could have served to accelerate DNA-based evolution via a reverse transcriptase-based mechanism (Mattick, 2009). Such a mechanism may even have a basis in human evolution. The recently discovered phenomenon of constitutional (germline) or cancer-associated chromothripsis provides strong evidence that a single event (such as the formation of micronuclei (Zhang et al., 2015)) could generate hundreds if not thousands of DNA breaks within the human genome. If the repair of such catastrophic events is mediated via an RNA-based mechanism, similar to the developmentally-regulated genome rearrangement mechanism studied in ciliates, one can speculate that such germline events could drive evolution via a punctuated equilibrium mechanism (Gould and Eldredge, 1977; Zhang et al., 2013a).
It is apparent that research in ncRNAs has wiped away our previous views in RNA biology, broadening our understanding about the mechanisms by which these molecules participate in biological activities (Morris and Mattick, 2014). Moreover, the recent discovery that RNA methylation has functional consequences extends our understanding of RNA’s biochemical capacity (Sergiev et al., 2015). Indeed if recent history is any indication, the functional analysis of ncRNAs should continue to produce several unexpected breakthroughs in the coming years.
Acknowledgments
We apologize to all colleagues whose work we were unable to cite due to size limitation. We thank Shobha Vasudevan, Nick Dyson, Lucia Aroncia and Capucine Van Rechem for discussion and critical reading of the manuscript. Research in the lab is supported by NCI Proton Beam Grant (C06 CA059267), a V Scholar Award (to MM) and Beatriu de Pinos postdoctoral fellowship to IAC. ITH is an NSF GRFP awardee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MM conceived and along with the rest of the authors wrote and edited this review.
References
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF. Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. The EMBO journal. 2013;32:2321–2335. doi: 10.1038/emboj.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12:254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nature structural & molecular biology. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes & development. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes & development. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nature structural & molecular biology. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Styles CA, Fink GR. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Molecular and cellular biology. 1986;6:3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht JR, Fang W, Goldman AD, Dolzhenko E, Stein EM, Landweber LF. Genomes on the edge: programmed genome instability in ciliates. Cell. 2013;152:406–416. doi: 10.1016/j.cell.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. The EMBO journal. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. The EMBO journal. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle CM, Zaher HS, Chalker DL. A Parallel G Quadruplex-Binding Protein Regulates the Boundaries of DNA Elimination Events of Tetrahymena thermophila. PLoS Genet. 2016;12:e1005842. doi: 10.1371/journal.pgen.1005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes & development. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Zhang Z, Liu Y. RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol. 2012;66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WJ, Bryson PD, Liang H, Shin MK, Landweber LF. The evolutionary origin of a complex scrambled gene. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15149–15154. doi: 10.1073/pnas.0507682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Molecular cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Nikiforov MA, Smothers JF, Allis CD, Yao MC. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Molecular cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015;6:143. doi: 10.3389/fgene.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Molecular cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Madhani HD. Recognizing the enemy within: licensing RNA-guided genome defense. Trends Biochem Sci. 2014;39:25–34. doi: 10.1016/j.tibs.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. Rna. 2014;20:867–881. doi: 10.1261/rna.044479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. RNAi and heterochromatin repress centromeric meiotic recombination. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham JS, Thing TA, Vythilingum N, Keuroghlian A, Bruno D, Golding GB, Pearlman RE. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot Cell. 2004;3:157–169. doi: 10.1128/EC.3.1.157-169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O'Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann G, Jonsson F, Weil PP, Postberg J, Lipps HJ. RNA-template dependent de novotelomere addition. RNA Biol. 2016 doi: 10.1080/15476286.2015.1134414. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wei W, Li MM, Wu YS, Ba Z, Jin KX, Liao YQ, Adhikari S, Chong Z, Zhang T, et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24:532–541. doi: 10.1038/cr.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. Heterochromatin and the cohesion of sister chromatids. Chromosome Res. 2009;17:229–238. doi: 10.1007/s10577-008-9012-z. [DOI] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Molecular cell. 2010;39:360–372. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S, Eldredge N. Punctuated Equilibria: The Tempo and Mode of Evolution Reconsidered. Paleobiology. 1977;3:115–151. [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Worz S, Sadic D, Schulte M, Mallm JP, Maiser A, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes & development. 2013;27:859–872. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen TY, Pearlman RE. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J Biol Chem. 1994;269:17428–17433. [PubMed] [Google Scholar]
- Heyse G, Jonsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. Embo J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Yamada T, Grewal SI. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell. 2004;119:469–480. doi: 10.1016/j.cell.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Mochizuki K. Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination. Developmental cell. 2015;35:775–788. doi: 10.1016/j.devcel.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Meers C, Storici F. RNA supports precise repair of its own DNA gene. RNA Biol. 2015:0. doi: 10.1080/15476286.2015.1116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2'-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. Rna. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Stucki M. Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2016;44:538–544. doi: 10.1093/nar/gkv1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes & development. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes & development. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Aalto AP, Yang Q, Chang SS, Huang G, Fisher D, Cha J, Poranen MM, Bamford DH, Liu Y. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–1074. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & development. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Molecular and cellular biology. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Has evolution learnt how to learn? EMBO Rep. 2009;10:665. doi: 10.1038/embor.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & development. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & development. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature cell biology. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K. Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination. Molecular cell. 2015;59:229–242. doi: 10.1016/j.molcel.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 2010;140:692–703. doi: 10.1016/j.cell.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF. RNA-Mediated Epigenetic Programming of Genome Rearrangements. Annu Rev Genomics Hum Genet. 2011;12:367–389. doi: 10.1146/annurev-genom-082410-101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozawa M, Goldberg L, Aplan PD. Landscape of insertion polymorphisms in the human genome. Genome Biol Evol. 2015;7:960–968. doi: 10.1093/gbe/evv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozawa M, Zhang Z, Kim YJ, Goldberg L, Varga T, Bergsagel PL, Kuehl WM, Aplan PD. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7729–7734. doi: 10.1073/pnas.1321889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. The EMBO journal. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41:9117–9128. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nature structural & molecular biology. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Delas MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Molecular cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe M, Arnoult N, Kappei D, Buchholz F, Decottignies A, Butter F, Mann M. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 2013;23:2149–2157. doi: 10.1101/gr.151878.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature cell biology. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Sergiev PV, Golovina AY, Osterman IA, Nesterchuk MV, Sergeeva OV, Chugunova AA, Evfratov SA, Andreianova ES, Pletnev PI, Laptev IG, et al. N6-methylated adenosine in RNA: From bacteria to humans. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Shen Y, Nandi P, Taylor MB, Stuckey S, Bhadsavle HP, Weiss B, Storici F. RNA-driven genetic changes in bacteria and in human cells. Mutation research. 2011;717:91–98. doi: 10.1016/j.mrfmmm.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Teng SC, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- van Sluis M, McStay B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes & development. 2015;29:1151–1163. doi: 10.1101/gad.260703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wan G, Liu Y, Han C, Zhang X, Lu X. Noncoding RNAs in DNA repair and genome integrity. Antioxid Redox Signal. 2014;20:655–677. doi: 10.1089/ars.2013.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Taniguchi T. MicroRNAs and DNA damage response: implications for cancer therapy. Cell Cycle. 2013;12:32–42. doi: 10.4161/cc.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Nakajima S, Bohm S, Bernstein KA, Shen Z, Tsang M, Levine AS, Lan L. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3495–E3504. doi: 10.1073/pnas.1507105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- Yerlici VT, Landweber LF. Programmed Genome Rearrangements in the Ciliate Oxytricha. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0025-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TY, Kao YW, Lin JJ. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3377–3382. doi: 10.1073/pnas.1307415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS One. 2012;7:e42371. doi: 10.1371/journal.pone.0042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes & development. 2013a;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nature structural & molecular biology. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chang SS, Xue Z, Zhang H, Li S, Liu Y. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes & development. 2013b;27:145–150. doi: 10.1101/gad.209494.112. [DOI] [PMC free article] [PubMed] [Google Scholar]