Much of our understanding of the biology of MYCN amplification in neuroblastoma is based on the exploration of transcriptional activity of MYCN. Powers and colleagues, in a paper in Nature, now show that MYCN RNA also has an important role in neuroblastoma in addition to MYCN protein production. Their findings place the Let-7 microRNA family front and centre in neuroblastoma biology.
Amplification of MYCN has been recognized as a key oncogenic driver of high-risk neuroblastoma for nearly 30 years, and the detection of MYCN amplification is an essential part of the standard of care for any child presenting with neuroblastoma.1 With good reason, the study of the biology of MYCN amplification has focused on MYCN transcriptional functions and the molecular complexes in which it functions, notably as a heterodimer bound to MYC-associated factor X (MAX). However, the paper from Powers et al.,2 recently published in Nature, suggests that other mechanisms are also important in MYCN-dependent oncogenesis. Most strikingly, they show that MYCN mRNA has an independent function promoting neuroblastoma, by sponging up and inhibiting the activity of members of a tumour suppressor microRNA family, let-7. These findings provide a completely new insight into the ways MYCN drives neuroblastoma (Figure 1). In addition, they provide data to suggest that the key role that inhibiting members of the let-7 microRNA family has in the development of neuroblastoma can explain a number of the other recurrent genomic features of neuroblastoma beyond MYCN amplification.
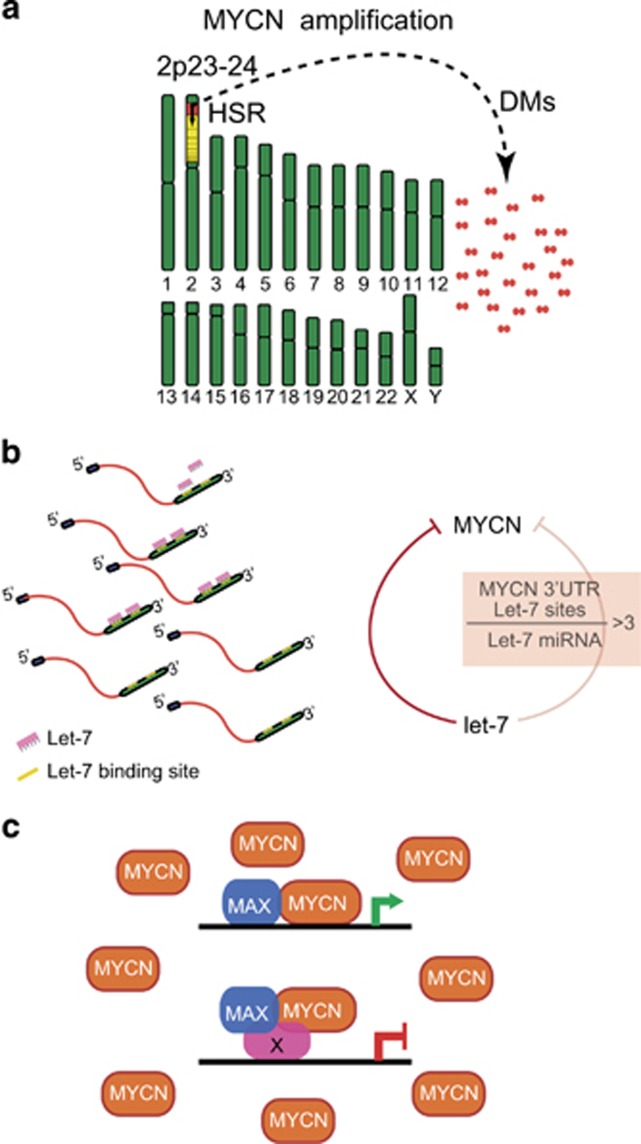
Figure 1.
(a) MYCN amplification in neuroblastoma arises either as a result of the formation of extrachromosomal double-minute chromosomes (DM) or as an intrachromosomal amplification identifiable as a homogenously staining region (HSR) in chr 2p. MYCN amplification is present when there is a fivefold or greater increase in MYCN copy number over control regions on chr 2. In most instances, the MYCN copy number is very much greater than this minimum value, and there is a direct relationship between higher MYCN copy number and poorer prognosis. (b) Elevated MYCN copy number results in abundant MYCN RNA, which, when it exceeds the amount of let-7 miRNA, can break the negative feedback exerted on MYCN by let-7. Now, MYCN RNA binds let-7 and both are targeted for destruction, but there is no impact on MYCN protein expression (c) because there is sufficient MYCN RNA for translation. MYCN, in complex with MAX, drives transcription and neuroblastoma. In complex with other factors, MYCN may also function as a transcriptional repressor of some genes. The net effect is high-risk neuroblastoma
As all roads lead to Rome, the oncogenic mechanisms in poor-prognosis neuroblastoma described in this paper converge on the repression of let-7 functions, of which the most important is the down-regulation of MYCN itself. It has been shown previously in neuroblastoma that let-7 expression is repressed by the LIN28B protein, which can bind the pri-miR let-7 (among other microRNAs) and prevent their processing into mature, active forms.3 As a starting point, Powers et al.2 have shown genetically that LIN28B is not required for repression of let-7 function in MYCN-amplified neuroblastoma cell lines. Instead, the abundant MYCN RNA is sufficient to soak up let-7 and still leave enough mRNA to be translated into elevated MYCN protein. This is a complex and unexpected oncogenic mechanism for a well-characterized oncogene, but the data are compelling. It also provides an explanation of at least one curious feature of MYCN biology in neuroblastoma. MYCN gene amplification has always been a more powerful marker of poor prognosis than was elevated MYCN protein expression itself, a fact that is at odds with the proposition that the most relevant function of a protein-coding gene is that of producing the protein. Furthermore, the selective pressure in favour of the often extraordinarily high copy number of MYCN genes in neuroblastoma has remained obscure. Now, Powers et al.2 have probably solved the puzzle. The amplification of MYCN copy number produces enough mRNA to effectively titrate down the amount of let-7. This breaks the negative feedback between let-7 and MYCN. MYCN protein produced in neuroblastoma cells is then derived from the MYCN mRNA not engaged with let-7 and that may significantly vary among tumours.
These findings place the let-7 microRNA family as central players in the complex biology of neuroblastoma. Indeed, circumstantial evidence presented in this paper suggests that other poor-prognosis molecular subtypes of neuroblastoma may also be unified by a loss of let-7 expression, in this case by the deletion of let-7 genes. Deletion of 11q23 in neuroblastoma is a recognized molecular subtype of the disease. It almost never co-occurs with MYCN amplification, but like MYCN-amplified disease, is an aggressive form of the tumour with 5-year survival rates significantly lower than tumours presenting at equivalent stages, but without 11q23 abnormalities.4 Deletion of chromosome 3p is also recurrent in neuroblastoma and may accompany 11q23 deletions.4 The minimal overlapping regions of both these chromosomal aberrations results in copy-number loss of let-7 family members. The proposition is that, although there is no MYCN gene copy-number amplification in 11q23 or 3p-deleted neuroblastoma, the net effect of the let-7 gene deletions is increased MYCN expression, and thus a similarly poor prognosis. Determining that MYCN expression levels really do correlate with these let-7 gene deletions in large expression data sets of human neuroblastoma, and that restoration of let-7 expression in tumours with 11q23 and 3p deletions alters MYCN expression and changes tumour behaviour in xenograft models, will go a long way to clinching the case. Thus, the evidence emerging from the Daley lab is that it is the loss of members of the let-7 family that is a common mechanism in poor-prognosis neuroblastoma,2 whether this arises by inhibition of miR transcription by LIN28B, by gene deletion or by the sponging up of let-7 pre-miRs by the MYCN 3′UTR.
One still cannot minimize the importance of MYCN transcriptional activity in neuroblastoma, and to be fair, Powers et al.2 do not dispute this. Transgenic mouse models of MYCN expression, that have been so important in advancing our understanding of this tumour, do not include the 3′UTR of MYCN, but still have a near 100% penetrance of a tumour that phenocopies human neuroblastoma very accurately.5 Further, if MYCN amplification occurs to principally mop up let-7 miRNA, one may wonder why it is only MYCN that is amplified in neuroblastoma and not any other gene that carries multiple let-7 sites in their 3′UTR region? Clearly, MYCN translation is not dispensable for the development of neuroblastoma, but MYCN transcription is doing much more than has previously met the eye.
Several other interesting implications and questions arise from this study. Are the silencing of let-7 and the activation of MYCN transcriptional activity truly epistatic events, or do other important consequences of the loss of let-7 contribute to neuroblastoma biology? Interrogation of let-7-regulated gene networks and targets, particularly, in the neuroblastoma cell of origin may reveal these and unexpected therapeutic options. Another intriguing line of investigation will be establishing whether the let-7 sponge function of MYCN RNA is more generalizable. Does this mechanism also operate in other MYCN-amplified tumours, such as some rhabdomyosarcomas, medulloblastomas and retinoblastomas? Further, although the data from Powers et al.2 clearly indicate that the two mechanisms, MYCN transcription and MYCN sponging, both contribute to the development of high-risk neuroblastoma, it would be fascinating to know if the 3′UTR of MYCN has an independent function to promote or accelerate the onset of neuroblastoma in transgenic animal models. This would provide powerful evidence that this mechanism is critical to the development of the disease.
A final point of interest for those studying MYC is that until now the biological activities of MYCN and c-MYC have been indistinguishable.6 However, the let-7 sites in MYCN 3′UTR are not conserved in c-MYC. It may be that the let-7 sponge function is the first truly distinctive difference between MYCN and c-MYC, and this is only now evident because Powers et al.2 had the insight to consider that the MYCN RNA had a biological function beyond the translation of MYCN protein.
Advanced stage neuroblastoma remains difficult to treat tumour with a dismal prognosis. There is no question that better, more targeted treatments and better patient outcomes will come from a deeper understanding of the underlying biology. The paper from Powers et al.2 is a significant step down this path.
The authors declare no conflict of interest.
References
- Brodeur GM. Nat Rev Cancer 2003; 3: 203–216. [DOI] [PubMed]
- Powers JT et al. Nature 2016; 535: 246–251. [DOI] [PMC free article] [PubMed]
- Madison BB et al. Genes Dev 2013; 27: 2233–2245. [DOI] [PMC free article] [PubMed]
- Spitz R et al. Clin Cancer Res 2003; 9: 52–58. [PubMed]
- Weiss WA et al. EMBO J 1997; 16: 2985–2995. [DOI] [PMC free article] [PubMed]
- Malynn BA et al. Genes Dev 2000; 14: 1390–1399. [PMC free article] [PubMed]