Abstract
De-ubiquitylating enzymes (DUBs) reverse protein ubiquitylation and thereby control essential cellular functions. Screening for a DUB that counteracts caspase ubiquitylation to regulate cell survival, we identified the Drosophila ovarian tumour-type DUB DUBA (CG6091). DUBA physically interacts with the initiator caspase death regulator Nedd2-like caspase (Dronc) and de-ubiquitylates it, thereby contributing to efficient inhibitor of apoptosis-antagonist-induced apoptosis in the fly eye. Searching also for non-apoptotic functions of DUBA, we found that Duba-null mutants are male sterile and display defects in spermatid individualisation, a process that depends on non-apoptotic caspase activity. Spermatids of DUBA-deficient flies showed reduced caspase activity and lack critical structures of the individualisation process. Biochemical characterisation revealed an obligate activation step of DUBA by phosphorylation. With genetic rescue experiments we demonstrate that DUBA phosphorylation and catalytic activity are crucial in vivo for DUBA function in spermatogenesis. Our results demonstrate for the first time the importance of de-ubiquitylation for fly spermatogenesis.
Posttranslational modification of proteins with mono- or poly-ubiquitin is a versatile way to influence protein fate and cellular pathways. Ubiquitin gets conjugated to lysine (K) residues of target proteins by a cascade of an activating enzyme (E1), a conjugating enzyme (E2) and a ligase (E3).
Poly-ubiquitin chains can be formed through conjugation of ubiquitin to one of the seven lysines in its own sequence (K6, K11, K27, K29, K33, K48 and K63) and to the α-amino group of the first methionine (M1) of ubiquitin.1 Different linkage types of poly-ubiquitin chains have distinct consequences for the target protein. K48-linked chains are considered the main signal for proteasomal degradation of proteins,1, 2 whereas K63-linked poly-ubiquitin chains are often involved in non-degradative signalling processes1, 3, 4 or can target proteins for lysosomal degradation.5, 6, 7 Other chain types can also trigger proteasomal degradation but are less well understood.2, 8 De-ubiquitylating enzymes (DUBs) counteract protein ubiquitylation. The human genome encodes for 91 DUBs.9 Model organisms such as Drosophila melanogaster with genomes of lower complexity and a lower number of identified DUBs10 are a valuable tool to discover new functions of these enzymes.
Caspases are the enzymes that execute apoptosis by cleavage of various cellular substrates. Tight regulation of caspases is crucial to prevent cell death under non-apoptotic conditions and one way to achieve this is ubiquitylation.11, 12 In Drosophila, caspase activity is restrained by the action of the E3 ligase Drosophila inhibitor of apoptosis-1 (DIAP1),13, 14 a member of the inhibitor of apoptosis (IAP) protein family, which binds and ubiquitylates initiator and effector caspases.15, 16, 17, 18 Ubiquitylation of the initiator caspase death regulator Nedd2-like caspase (Dronc), homologue of caspase-9, inactivates Dronc by non-degradative mechanisms.17, 18, 19 Moreover, DIAP1 may also regulate Dronc levels by ubiquitylation under certain conditions, to prevent its activation in the activating platform apoptosome.20 For the Drosophila effector caspase death-related ICE-like caspase (DrICE; homologue of caspase-3/-7), DIAP1-mediated ubiquitylation has been shown to impair caspase activity directly in a non-degradative manner.16
It has become evident that caspases are also involved in non-apoptotic processes, such as migration, immunity, learning and memory, and differentiation.21, 22, 23 A well-studied example is the individualisation of spermatids in Drosophila, where activity of caspases is required to remove connections between elongated spermatids and to clear out cytoplasmic contents, in order to generate mature sperm.24, 25, 26 This process does not only require active effector caspases but also other parts of the apoptotic machinery such as Dronc and the apoptosome component DARK (death-associated APAF1-related killer).24, 25, 27 Interestingly, a ubiquitin E3 ligase complex of the Cullin family is required for caspase activation during the individualisation process.28
Searching for DUBs that regulate caspase activity, we have identified the D. melanogaster gene CG6091 in an in vivo RNAi screen. It is the orthologue of the human de-ubiquitylating enzyme DUBA/OTUD5 and will be named Duba in accordance with the human gene. DUBA carries a protease domain of the ovarian tumour (OTU) family.
Found as modulator of IAP-antagonist-induced apoptosis, we assessed DUBA's role as caspase regulator. DUBA co-immunoprecipitated with the initiator caspase Dronc and de-ubiquitylated it. Non-apoptotic caspase activity is required for spermatid individualisation in Drosophila and male sterile Duba-null allele flies showed reduced caspase activity and defects in the individualisation process. DUBA function in spermatogenesis relied on its de-ubiquitylating activity. Interestingly, DUBA critically required phosphorylation of a conserved serine residue for catalytic activity.
Our data support a model, in which DUBA is activated through phosphorylation, to specifically de-ubiquitylate tissue-specific substrates such as caspases and their regulators, to mediate essential processes during spermatogenesis.
Results
DUBA is required for GMR-Rpr- and GMR-Hid-induced apoptosis
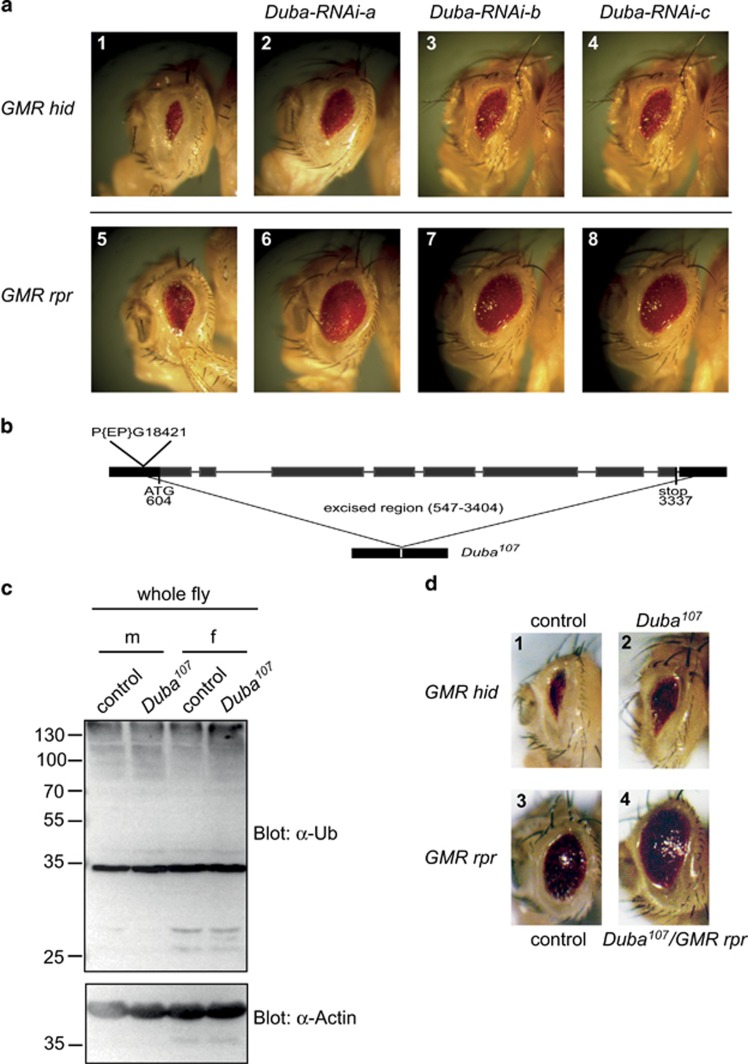
GMR (glass multimer reporter)-driven expression of the pro-apoptotic IAP-antagonists Reaper (Rpr) and Head involution defective (Hid) in the developing Drosophila eye represents a well-established model for apoptotic cell death and is an ideal tool for the identification of new regulators of caspase activity. To identify DUBs that are involved in the regulation of caspases, we have performed an in vivo screen for modifiers of the Rpr- and Hid-induced small eye phenotype.29
Using an RNAi collection for Drosophila DUBs, we have identified Duba (CG6091) as modulator of Rpr- and Hid-induced cell death in the eye. Knockdown of Duba in the developing eye using three different RNAi fly lines suppressed photoreceptor cell death and led to an increase in eye size compared with control Rpr- or Hid-expressing flies (Figure 1a). To genetically validate the RNAi data we generated a Duba-null mutant, Duba107, by imprecise P-element excision, where the entire coding region of CG6091 was removed (Figure 1b). Duba107 flies are homozygous viable and develop normally. Developmental cell death seemed to occur normally in Duba107 flies and we could not observe defects, for example, in the removal of interommatidial cells in the developing eye30 or concerning the architecture of arista31 (Supplementary Figures 1A–D). A mild accumulation of ubiquitylated proteins was observed in whole fly lysates from Duba107 males and females compared with controls (Figure 1c), which suggests that DUBA in fact de-ubiquitylates one or several proteins in vivo. Of several tissues probed by immunostaining for ubiquitin, we noted a slight increase in ubiquitylated proteins in eye discs and ovaries from Duba107 flies but not in larval or adult brains (Supplementary Figure 2).
Figure 1.
Loss of DUBA suppresses Rpr- and Hid-induced cell death in the eye and leads to accumulation of poly-ubiquitylated proteins. (a) GMR-hid- or GMR-rpr-expressing flies were crossed with Duba-RNAi fly lines. Genotypes: (1) GMR-hid, GMR-Gal4/+ ey-Gal4/TM3; (2) GMR-hid, GMR-Gal4/+ ey-Gal4/Duba-RNAi-a; (3) GMR-hid, GMR-Gal4/Duba-RNAi-b; ey-Gal4/+ (4) GMR-hid, GMR-Gal4/Duba-RNAi-c; ey-Gal4/+ (5) ey-Gal4/+ GMR-rpr, GMR-Gal4/TM3; (6) ey-Gal4/+ GMR-rpr, GMR-Gal4/Duba-RNAi-a; (7) Duba-RNAi-b/+ GMR-rpr, GMR-Gal4/+ (8) Duba-RNAi-c/+ GMR-rpr, GMR-Gal4/+. (b) Generation of a Duba-null allele (Duba107). Shown is the longest transcript (CG6091-RE). The complete coding region was removed by imprecise P-element excision of P{EP}CG6091[G18421]. (c) Loss of Duba leads to increased levels of poly-ubiquitylated proteins. Male (m) or female (f) Duba107 or control flies (precise excision) were lysed in SDS-loading buffer and analysed by western blotting. (d) GMR-hid- or GMR-rpr-expressing flies were crossed with Duba107 flies. Genotypes: (1) GMR-hid, GMR-Gal4/cyo; ey-Gal4/TM6b; (2) GMR-hid, GMR-Gal4/+ Duba107; (3) GMR-rpr, GMR-Gal4/TM6b; (4) GMR-rpr, GMR-Gal4/Duba107
Although normal eye development is not affected by loss of Duba (Supplementary Figures 1E and F), GMR-Rpr- and GMR-Hid-induced apoptosis in the eye is suppressed in Duba mutants (Figure 1d). This indicates that DUBA is required for efficient induction of apoptosis in this system.
DUBA interacts with DIAP1 and the initiator caspase Dronc
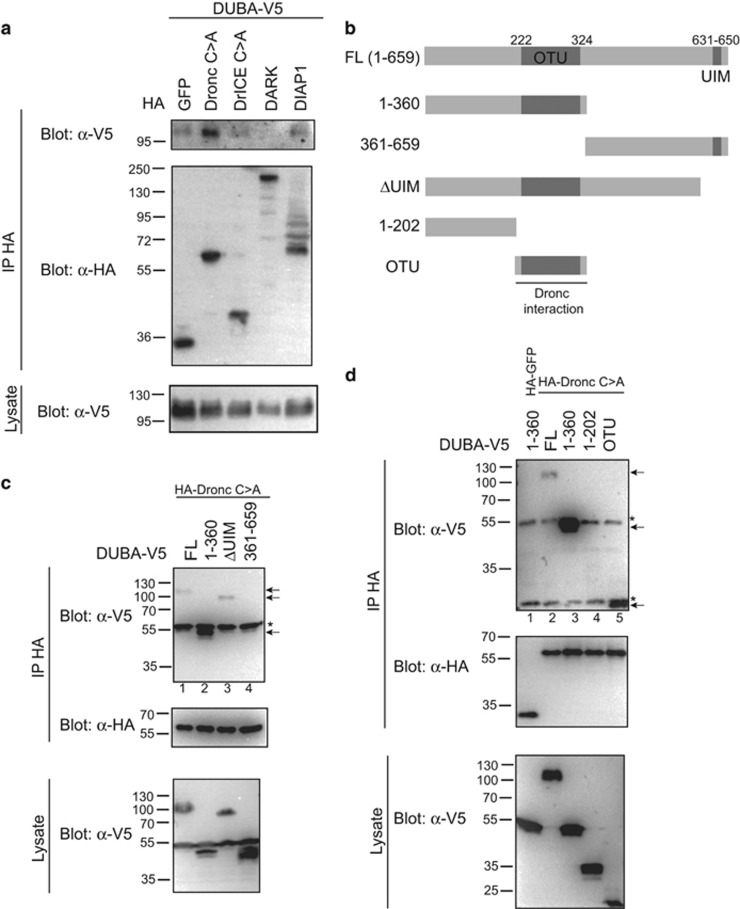
IAP-antagonist-induced cell death depends largely on inactivation of the anti-apoptotic DIAP1 protein and subsequent activation of the caspases Dronc and DrICE. Dronc activation occurs in the multimeric protein complex apoptosome, requiring the adaptor protein DARK.32 We sought to test whether DUBA affects Rpr- and Hid-induced apoptosis by biochemical interaction with any of these proteins. We expressed V5-tagged DUBA in Drosophila S2 cells together with HA-tagged green fluorescent protein as control, HA-Dronc, HA-DrICE, HA-DARK or HA-DIAP1. Anti-HA-immunoprecipitation (IP) revealed co-IP of DUBA with Dronc and also weakly with DIAP1. In contrast, an interaction with DrICE or DARK could not be detected (Figure 2a). DUBA carries an OTU domain and a C-terminal ubiquitin-interacting motif (UIM) (Figure 2b). We generated several truncated constructs of DUBA to identify the region interacting with Dronc. Co-IP experiments indicated a strong interaction between Dronc and the DUBA N-terminal region containing the OTU domain (Figure 2c, lane 2). A fragment encompassing the C-terminal half of DUBA did not interact (Figure 2c, lane 4). Further analysis indicated a weak Dronc interaction with the isolated OTU domain but not with the region before (Figure 2d, compare lane 4 and 5). Deletion of the UIM did not affect binding to Dronc (Figure 2c, lane 3). These results suggest the OTU domain as the main interaction site between DUBA and Dronc.
Figure 2.
DUBA interacts with DIAP1 and the initiator caspase Dronc. (a) S2 cells were transfected with expression vectors for DUBA (V5-tagged) and HA-GFP, HA-Dronc C>A (caspase-inactive), HA-DrICE C>A (caspase-inactive), HA-DARK or HA-DIAP1 as indicated, and lysed for anti-HA IP. IP eluates and lysates were analysed by immunoblotting with α-HA or α-V5 antibodies. Co-IP of DUBA was detected with HA-Dronc and HA-DIAP1 (weak). (b) Schematic overview of DUBA domain structure (OTU, ovarian tumour domain; UIM, ubiquitin-interacting motif) and deletion constructs used in c). (c and d) Co-IP to determine the interaction domain of DUBA with Dronc. The experiment was performed as in a, with the indicated V5-tagged fragments of DUBA. Positive Co-IP of DUBA fragments is indicated by arrows. Interaction is mediated through the N-terminal half of DUBA (c, lane 2) and within this part by the OTU domain (d, lane 5). Cross-reacting IgG bands are marked with *
DUBA requires phosphorylation at Serine 183 for catalytic activity
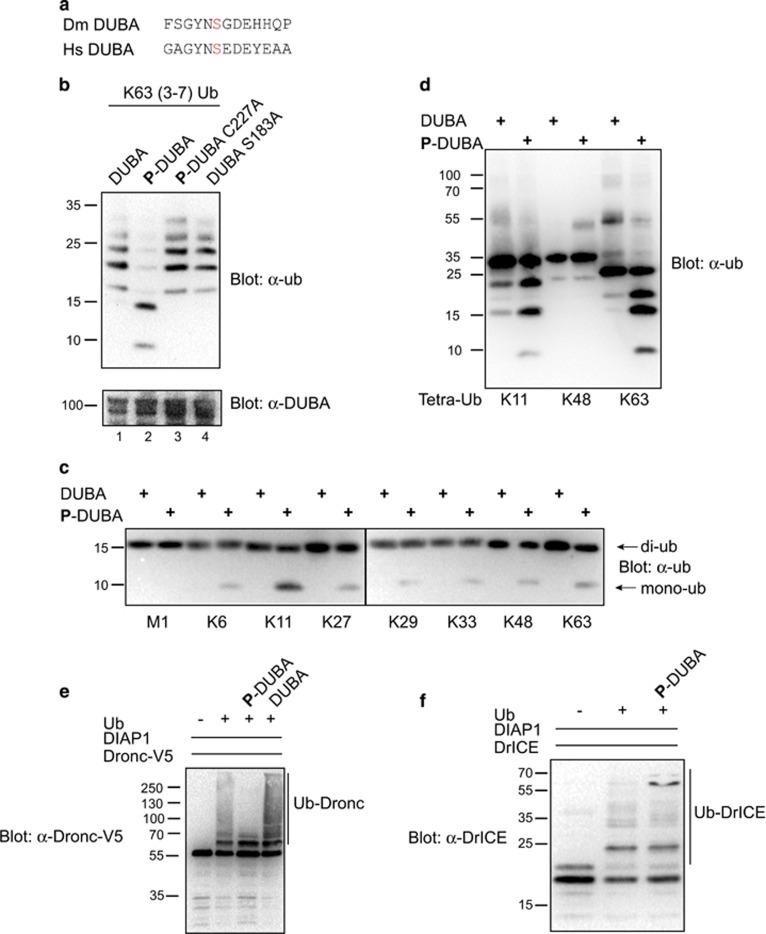
DUBA is the orthologue of the mammalian protein DUBA/OTUD5, which requires a phosphorylation-dependent activation step.33 In vitro, serine 177 of DUBA/OTUD5 is phosphorylated by casein kinase 2 (CK2) and the phosphate assists in putting the ubiquitin substrate in the right orientation for cleavage. Likewise, Drosophila DUBA contains a phosphorylation motif for CK2 around serine 183 (Figure 3a and Huang et al.33). Recombinant DUBA purified from Escherichia coli was not able to cleave ubiquitin chains. In contrast, in vitro phosphorylation with Drosophila CK2 generated a catalytically active enzyme, which was able to process K63-linked ubiquitin chains (Figure 3b). Mutation of serine 183 in the phosphorylation motif or cysteine 227 in the predicted catalytic triad of the OTU domain abolished cleavage activity. Together, this indicates the requirement for activation of DUBA by phosphorylation and identifies serine 183 as the only phosphorylation site for activation in vitro. In addition, DUB activity depends on the catalytic cysteine C227.
Figure 3.
Biochemical analysis of DUB activity in vitro. (a) Phosphorylation motif of D. melanogaster (Dm) DUBA, phosphosite S183 shown in red and human (Hs) DUBA, phosphosite S177 in red. (b) Bacterially expressed DUBA was incubated with CK2 for phosphorylation (lane 2/3/4) and tested for cleavage of K63-linked ubiquitin chains. DUB activity can only be detected following in vitro phosphorylation (lane 2). Mutation of C227 (catalytic cysteine in OTU domain) and S183 (phosphorylation site) to alanine abolishes activity. (c) Determination of chain-type preference of DUBA. Recombinant phosphorylated (P-DUBA) or non-phosphorylated DUBA was incubated with di-ubiquitin molecules of the indicated linkage type. Ubiquitin cleavage occurred only with phosphorylated DUBA. K11- and K63-di-ubiquitin was cleaved most efficiently (production of mono-ub and reduction of di-ub band). (d) Chain type preference of DUBA on K11-, K48- and K63-tetra-ubiquitin. The experiment was performed as in (c). (e) De-ubiquitylation of Dronc. Recombinant Dronc (caspase-inactive C>A) was ubiquitylated in vitro by DIAP1. Addition of active P-DUBA removed ubiquitin chains from Dronc, while non-phosphorylated DUBA did not change the ubiquitylation pattern. (f) DUBA does not de-ubiquitylate DrICE. Recombinant active DrICE was ubiquitylated by DIAP1. Addition of P-DUBA did not lead to removal of ubiquitin chains. Shown is the p20 of DrICE
DUBs of the OTU family can be selective for cleavage of one or several ubiquitin linkage types.34 To further analyse DUBA's catalytic activity and specificity, we tested cleavage of di-ubiquitin molecules of all known linkage types. Phosphorylated DUBA was able to process K11 di-ubiquitin and K63-linked di-ubiquitin (Figure 3c). For other chain types, only background activity was detected. This was further confirmed by cleavage of tetra-ubiquitin chains, where K11 and K63 chains were processed but K48-tetra-ubiquitin were not (Figure 3d). In general, DUBA seems to process longer chains more efficiently than di-ubiquitin under the tested conditions.
Our results indicate a biochemical interaction of DUBA with Dronc (Figure 2). Dronc and also the effector caspase DrICE are negatively regulated by ubiquitylation.15, 16, 18, 19 As our data suggest that DUBA is required for IAP-antagonist-induced cell death, ubiquitylated caspases represent a probable target of DUBA. To test this, Dronc and DrICE were ubiquitylated by DIAP1 in vitro. The ubiquitylation reaction was stopped and DUBA or active P-DUBA was added. P-DUBA, but not non-phosphorylated DUBA, efficiently removed ubiquitin from Dronc (Figure 3e). In contrast, P-DUBA was not able to de-ubiquitylate DrICE under these conditions (Figure 3f).
Loss of DUBA impairs spermatogenesis
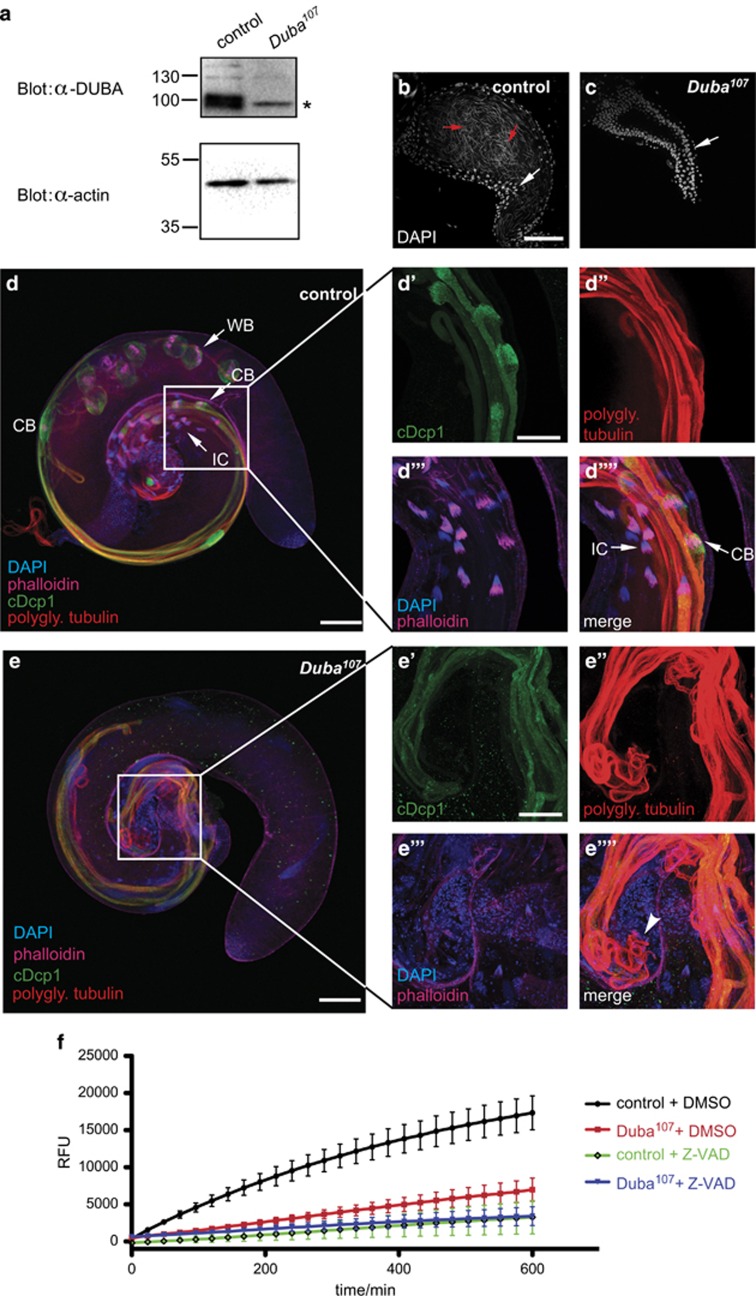
As DUBA might also have a role in regulation of caspase activity in a non-apoptotic context such as spermatid individualisation, it was highly interesting to discover that homozygous male Duba107 flies were sterile. Control flies in which only the P-element was removed but no Duba coding region, as well as female Duba107 flies, were fertile. The fact that Duba expression is highest in testes (flyatlas.org and modENCODE.org) supports an important role of Duba for male fertility. Lysates of testes of Duba107 flies were analysed and the absence of DUBA protein was confirmed by western blotting (Figure 4a).
Figure 4.
Testes from Duba-null flies display morphological defects and reduced caspase activity. (a) Testes lysates from 1- to 2-day-old homozygous Duba107flies or controls were immunoblotted to confirm a complete lack of DUBA protein. * indicates a nonspecific signal of the α-DUBA antiserum. Actin levels are shown as loading control. (b and c) Seminal vesicles of Duba107 testes do not contain mature sperm. Seminal vesicles of control (b) and Duba107 (c) testes, stained with DAPI, are shown. White arrows indicate round-shaped nuclei from cells of the sheath of the seminal vesicles; red arrow shows single needle-shaped nuclei of individualised spermatids that are only present in control testes. Scale bar: 50 μm. Repeated three times. (d and e) Confocal projections of testes of 1-day-old control (d) and Duba107 (e) flies reveal structural differences in individualising and elongated spermatids. CB, cystic bulge; IC, individualisation complex; WB, waste bag. Testes were stained with DAPI (blue), phalloidin (magenta) and antibodies against active caspase (cDcp1, green) and polyglycylated tubulin (AXO49, red). Enlarged in (d'–d''''): individualisation complex and cystic bulge. Enlarged in (e'–e''''): Elongated spermatids of Duba107 testes lack IC and CB and show degrading cysts (e'''', white arrowhead). Scale bar: 100 μm in (d and e), 50 μm in (d'–d'''') and (e'–e''''). Repeated four times, four to six testes per condition each time. (f) Caspase activity in lysates from Duba107 testes was measured by DEVDase assay in vitro and compared with control lysate or lysate treated with the caspase inhibitor z-VAD-fmk. Means of three independent experiments and S.E.M. are shown. RFU, relative fluorescence units
Examining Duba107 testes, we noticed that seminal vesicles do not contain mature, needle-like sperm, in contrast to properly developed sperm in controls (Figures 4b and c). Therefore, spermatogenesis in Duba mutants seems to be impaired.
Caspases have a crucial, non-apoptotic role during a late remodelling step in Drosophila spermatogenesis, the individualisation process.24, 25 As DUBA was found in a screen for caspase regulators and biochemically interacted with the initiator caspase Dronc, a potential function of DUBA might be the control of caspases during spermatid individualisation.
Immunostaining with an antibody for the active form of the effector caspase death caspase-1 (DCP-1), cleaved DCP-1 (cDCP-1) marks the individualising spermatids in testes from control flies (Figure 4d). In contrast, DUBA-deficient testes show only weak staining for active caspase (Figure 4e). Furthermore, spermatids seem to be thicker and disorganised (e' and e'' in comparison with d' and d'').
During individualisation, cytoplasmic connections and organelles are removed from the initially interconnected elongated spermatids. Testes of control flies show typical structures of the individualisation process, such as the actin-based individualisation complex, cystic bulges (CBs) and waste bags (WB), containing excess cytoplasmic contents (Figures 4d–d''''). In contrast, ICs, CBs and WBs are absent in testes of Duba107 flies (Figures 4e–e''''). Degrading, arrested cysts, rather than mature sperm, were detected at the base of the testes from Duba107 flies (Figure 4e'''').
A marker for initiation of the individualisation process is the polyglycylation of axonemal tubulin,35 detected with the AXO49 antibody.36 This process occurs independently of caspase activation.28 DUBA-deficient testes showed AXO49 staining, indicating that spermatids have developed until the onset of individualisation. However, in Duba107 testes elongated spermatid cysts appear thicker and more disordered than in the control sample (Figure 4d'' versus Figure 4e''), possibly because of additional roles of DUBA earlier in sperm development.
To quantitatively assess the reduction of caspase activity, we measured activity in testes lysates in vitro by cleavage of the fluorogenic caspase substrate DEVD-AMC. DEVDase activity in lysates from Duba107 testes was markedly reduced by up to 70% compared with controls (Figure 4f). Treatment with the caspase inhibitor z-VAD-fmk further reduced DEVDase activity in Duba107 lysates, which is in line with residual caspase activity that was also seen by immunostaining (Figure 4e).
With the aim to increase caspase activity and potentially rescue the Duba phenotype, we expressed Dronc and DrICE, and their catalytically inactive mutants (C>A) in Duba107 and in a wild-type background. Interestingly, although expression of Dronc was lethal, DroncC>A, DrICE and DrICEC>A did not have any effects on fly viability and spermatogenesis (Supplementary Figures 3c, e and g). In contrast, when DroncC>A, DrICE and DrICEC>A were expressed in the absence of Duba, spermatogenesis seemed to be severely disrupted at early stages, as mature spermatids were completely missing (Supplementary Figures 3d, f and h). This was also the case for catalytically dead caspases, which did not display dominant-negative effects in wild-type testes. Therefore, the observed effects cannot be explained by aberrantly high caspase activity. We conclude that DUBA is required to buffer toxicity effects induced by high caspase levels in the testes, potentially due to its ability to biochemically interact with Dronc.
Catalytic activity of DUBA is required for spermatogenesis
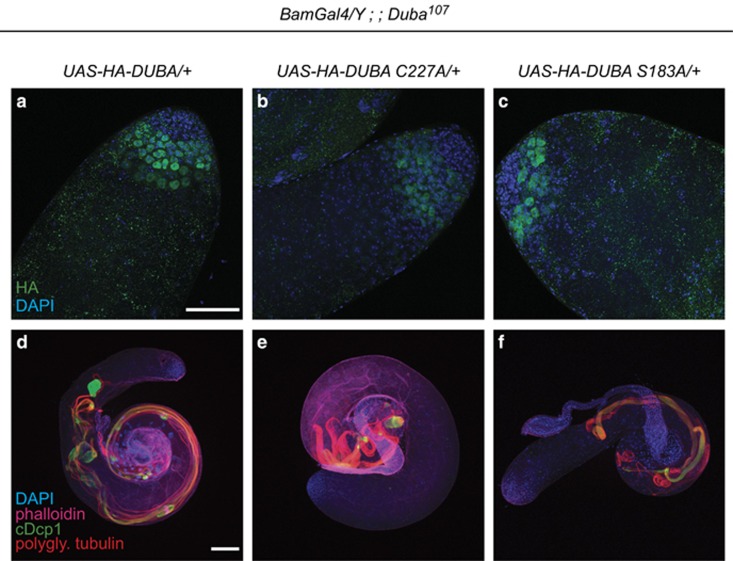
To confirm that defects in spermatogenesis are indeed caused by loss of Duba and to test whether catalytic activity is required in this context, we performed genetic rescue experiments in the Duba107 background. Transgenic fly lines for HA-tagged Duba or point mutants HA-Duba C227A (catalytic cysteine) and HA-Duba S183A (phosphorylation-deficient) were created, with all transgenes inserted at the same landing site. Two different driver lines for the UAS-Gal4 system were used: Bam-Gal4 that expresses early in spermatogenesis37 and Hsp83-Gal4 that drives expression throughout spermatogenesis.24, 38 DUBA expression re-stored fertility in Duba107 flies. In contrast, both DUBA mutants failed to re-establish fertility. Bam-Gal4-driven expression of the transgenes in advanced spermatogonial cysts and spermatocytes was confirmed by immunofluorescence (Figures 5a–c). Immunostaining of Duba107 testes, expressing HA-DUBA, HA-DUBA C227A or HA-DUBA S183A, revealed strong differences in cDCP-1 staining and morphology. Although expression of wild-type HA-DUBA fully restored spermatid development, formation of individualisation complexes and WBs, as well as overall structure, C227A- and S183A-DUBA, failed to do so (Figures 5d–f, compare with Figure 4d). In contrast, spermatid structures seemed even more disrupted than in Duba107 testes without transgene expression, suggesting a potential dominant-negative effect. These results are in line with rescue of fertility by the wild type but not the point mutants of the DUBA transgene. Together, we demonstrate that DUBA's catalytic activity as de-ubiquitylating enzyme is required for its role in sperm development and that phosphorylation of serine 183 is necessary for this activity in vivo. Although CK2 phosphorylates and activates DUBA in vitro, CK2 knockdown did not affect fertility, and Rpr and Hid eye phenotypes (data not shown), raising the possibility for a different kinase regulating DUBA in vivo.
Figure 5.
Rescue of male sterile phenotype by ectopic expression of wild-type DUBA in Duba107 testes but not by inactive mutants. (a–c) Confocal projections of testes stained with DAPI (blue) and against HA-tag of transgenes (green), genotypes: (a) Bam-Gal4/Y; UAS-HA-Duba/+ Duba107, (b) Bam-Gal4/Y; UAS-HA-DubaC227A/+ Duba107 and (c) Bam-Gal4/Y; UAS-HA-DubaS183A/+ Duba107. (d–f) Confocal projections of testes stained with DAPI (blue) and phalloidin (magenta), and antibodies against active caspase (cDcp-1, green) and polyglycylated tubulin (AXO49, red): (d) Bam-Gal4/Y; UAS-HA-Duba/+ Duba107, (e) Bam-Gal4/Y; UAS-HA-DubaC227A/+ Duba107 and (f) Bam-Gal4/Y; UAS-HA-DubaS183A/+ Duba107. Scale bars: 50 μm (a–c) and 100 μm (d–f). The experiment was repeated two times. Three to six testes were investigated each time per condition
Loss of DUBA leads to accumulation of ubiquitylated proteins in vivo
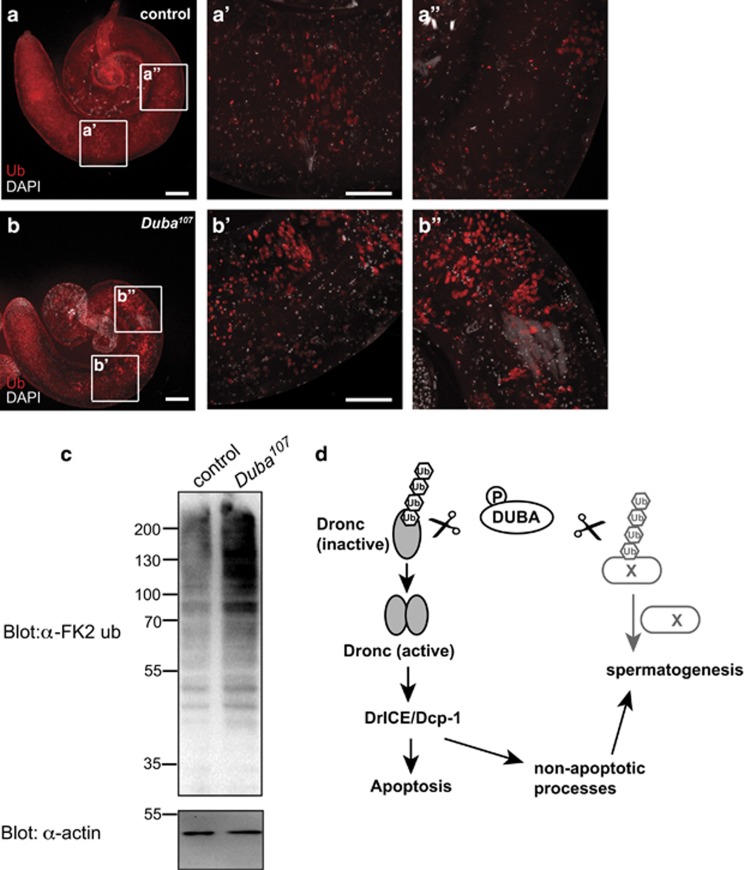
As DUBA's catalytic activity is essential for spermatogenesis, we tested whether loss of DUBA leads to accumulation of ubiquitylated proteins in testes. Immunostaining of DUBA-deficient testes with FK2 anti-ubiquitin antibody (recognising preferentially ubiquitin conjugated to substrate proteins) revealed strong accumulation of ubiquitylated proteins, whereas testes of control flies showed a much weaker and more diffuse pattern (Figures 6a and b).
Figure 6.
Ubiquitylated proteins accumulate in Duba107 testes. (a and b) Confocal projections of (a) control and (b) Duba107 testes of 1-day-old flies stained with DAPI (grey) and for ubiquitylated proteins (FK2, red). Enlarged sections in (a'–a''' and b'–b'''). The experiment was repeated two times. Five testes were investigated each time per condition. Scale bars: 100 μm (a and b), 50 μm (a'–a''' and b'–b'''). (c) Western blotting to detect levels of ubiquitylated proteins (α-FK2) in lysates of wild-type and Duba107 testes of 1-day-old flies; α-actin staining as loading control. (d) Model. DUBA is activated by phosphorylation and de-ubiquitylates Dronc to activate it for subsequent activation of effector caspases, to regulate apoptotic and non-apoptotic processes. ‘Protein X' denotes potential further substrate protein(s) of DUBA during spermatogenesis
Testes lysates were also analysed by western blotting and this confirmed a stronger poly-ubiquitin signal in lysates from Duba107 testes than in control samples (Figure 6c).
Our data suggest that DUBA – by its ability to interact and de-ubiquitylate Dronc – contributes to activation of non-apoptotic caspase activity during spermatogenesis. In addition, DUBA might also act through de-ubiquitylation of further protein(s), to promote sperm development in Drosophila (Figure 6d).
Discussion
Caspases are best known as executioners of cell death but are indispensable also in non-apoptotic processes. Caspases can be regulated by ubiquitylation and this has been well studied for their function in Drosophila cell death pathways.12, 39
One example is the initiator caspase Dronc, which is subject to DIAP1-mediated ubiquitylation and inactivation – in most cases without being targeted for proteasomal degradation.17, 18, 19 In addition, the Drosophila effector caspase DrICE is ubiquitylated by DIAP1 and this leads to non-degradative inactivation.16 Under apoptotic conditions, the cell will have to counteract ubiquitylation for efficient caspase activation. We postulated that a DUB might be required for this step. A systematic RNAi screen identified DUBA as a candidate DUB for the regulation of caspases. RNAi-mediated downregulation and genetic loss of Duba impaired IAP-antagonist-induced apoptosis, suggesting that DUBA might contribute to the activation of caspases or other pro-apoptotic factors. Our experiments showed that DUBA biochemically interacts with Dronc and de-ubiquitylates it. These results provide a possible mechanism by which DUBA is able to influence IAP-antagonist-induced cell death in the eye. In contrast, the consequence of the biochemical interaction between DIAP1 and DUBA remains to be clarified. Counter-regulation of DIAP1 auto-ubiquitylation by DUBA would lead to DIAP1 stabilisation and loss of DUBA would most probably enhance apoptosis, in contrast to the phenotypes observed here. Interestingly, two DUBs were identified previously that blocked apoptosis in flies presumably by stabilisation of DIAP1, namely DUBAI and scrawny/emperor's thumb/USP36,40, 41 thereby acting as negative regulators of apoptosis in contrast to the suggested pro-apoptotic role of DUBA.
Surprisingly, Duba107-null mutant flies did not show obvious defects in developmental apoptosis. However, screens that targeted the apoptotic cascade have previously also identified regulators of non-apoptotic caspase activity.42, 43, 44
The requirement of non-apoptotic caspase activity for sperm individualisation in Drosophila has been well characterised24, 25, 28, 45 and the individualisation process is also regulated through ubiquitylation.28 Therefore, we postulated that sterility of Duba107 males could be explained by defects in spermatid individualisation. Interference with caspase activity by expression of DIAP1, the caspase inhibitor p35 or alleles impairing Dronc and Dark function was previously shown to affect formation of CBs and WBs.24, 25, 27 Because of the observed biochemical interaction between DUBA and Dronc, it is interesting to note that active Dronc is present in individualising spermatids, and that inactivation of Dronc leads to partial individualisation failure.25, 27 A role for ubiquitylation of Dronc in this context has not been investigated so far and is technically challenging to address in vivo.
A Cullin-3-based E3 ligase complex is required for caspase activation during sperm individualisation by regulating levels or localisation of the IAP protein BRUCE.28, 45 However, a role for DUBA in BRUCE de-ubiquitylation can not be concluded from our data, as BRUCE ubiquitylation and DUBA activity both seem to be required for caspase activation and individualisation of spermatids. Although our findings suggest that DUBA might regulate spermatogenesis due to its interaction with and de-ubiquitylation of Dronc, it is technically difficult to provide a full proof for this concept in vivo. Our attempt to elevate caspase activity in testes of Duba mutants to rescue the observed defects had the opposite effect and led to a complete disruption of spermatogenesis specifically in Duba mutant flies. Although this genetic interaction between Duba and caspases implies a role for DUBA in caspase regulatory processes, we cannot exclude that DUBA might also act through other proteins to promote spermatid maturation.
Ubiquitylated proteins accumulate in Drosophila testes in the absence of DUBA. A general requirement for the Drosophila ubiquitin gene Ubi-p63E and the importance of proper ubiquitin homeostasis in spermatogenesis was shown recently.46 Deletion of the Ubi-p63E locus leads to strongly reduced ubiquitin levels specifically in testes and an arrest of spermatocytes in meiosis. Several ubiquitylation events in Drosophila testes have been characterised, for example, for histone removal during chromatin remodelling47 or during late stages of spermatogenesis.28, 45 DUBs counteracting ubiquitylation in these or other processes during fly spermatogenesis have not been identified so far.
In the future, an analysis of a potential role of mammalian DUBA in male fertility will be highly interesting. So far, mammalian DUBA has been described only for its role in immune settings.48, 49
Our analysis revealed that DUBA cleaves preferentially K11- and K63-linked ubiquitin. Human DUBA/OTUD5 has been shown to cleave K48, K63 and K11 di-ubiquitin in vitro,33 whereas another study observed only K48 and K63 cleavage.34 Whereas K48 and K63 ubiquitylation has been widely studied, much less is known about K11-linked ubiquitylation. Several studies came to conflicting results regarding the abundance of K11-linked chains in cells,2, 50, 51 but it seems clear that K11-linked ubiquitin chains can target proteins for proteasomal degradation.2, 52 In addition, as part of a heterotypic, ‘mixed' ubiquitin chain, K11 linkages can also contribute to endocytosis53 or promote NF-κB signalling in a non-degradative way.54 The function of K11- and K63-linked ubiquitin in spermatogenesis has not been studied so far. In flies, no K11-specific DUB has been described to date. No homologues of Cezanne (OTUD7B) and Cezanne2 (OTUD7A), K11-selective mammalian DUBs,34, 55 seem to exist in Drosophila; thus, our findings that DUBA cleaves K11-linked ubiquitin are highly interesting.
Taken together, our data provide the first characterisation of the Drosophila DUB DUBA. Our findings prove an obligate requirement for DUBA activation by serine phosphorylation in vivo and propose a novel role for K11/K63-linked ubiquitin chains in sperm development. We suggest a function for de-ubiquitylation of the initiator caspase Dronc for efficient induction of IAP-antagonist-induced cell death in the eye, as well as for regulation of non-apoptotic caspase activity during sperm development.
Materials and Methods
Fly stocks
GMR-rpr, GMR-Gal4 and GMR-hid, GMR-Gal4 and Ey-Gal4 lines,56, 57 and UAS-FL-DroncC>A (Meier et al.58) and UAS-DrICE-V5 lines were kindly provided by P Meier (ICR, London). Bam-GAL4 flies were described;37 Hsp83-Gal4 was also described.24 RNAi lines for CG6091 were obtained from VDRC: Duba-IR-a=27 558, Duba-IR-b=27 559 and Duba-IR-c =109 912.
Duba107 flies, lacking the entire coding sequence of CG6091/Duba, were generated by imprecise P-element excision from yw;P{EP}CG6091[G18421] (#26936, Bloomington Stock Center, IN, USA), see Figure 1. As control, flies with precise P-element excision from the same stock were used. Transgenic flies for HA-Duba and HA-Duba C227A, HA-Duba S183A were created by PhiC31-integrase-mediated insertion to 51C (BestGene Inc., Chino Hills, CA, USA) from pUASTattB HA-Duba plasmids. All genetic crosses were performed at 25 °C.
DNA constructs
Expression constructs for DUBA (longest isoform, transcript variant F) were cloned in pMT vectors (Invitrogen/Thermo Scientific GmbH, Schwerte, Germany) with V5-tag or in pGEX-6P1 for expression in bacteria. pMT HA-DIAP1, HA-Dronc (C>A), HA-DrICE (C>A) and HA-DARK were kindly provided by T Tenev (ICR, London, UK). Constructs were verified by DNA sequencing. Point mutants of DUBA were created by standard site-directed mutagenesis.
Antibodies
Primary antibodies used for western blottings were as follows: α-actin C-11 (St. Cruz Biotechnology, Dallas, TX, USA), α-DUBA (directed against aa 1-166, guinea pig, Eurogentec, Seraing, Belgium), α-DrICE, kindly provided by P. Meier and colleagues,59 α-mono-polyubiquitin conj. FK2 (Enzo Life Sciences, Lörrach, Germany), α-Ubiquitin (07-375) (Merck Millipore, Darmstadt, Germany), α-HA 3f10 (Sigma-Aldrich, Munich, Germany) and α-V5 tag (Bio-Rad Serotec, Puchheim, Germany). Secondary HRP-coupled antibodies were from St. Cruz. Primary antibodies used for fluorescent staining were as follows: α-HA-probe (Y11) (St. Cruz), α-cleaved Drosophila Dcp-1 (Asp216) (Cell Signaling Technology, Danvers, MA, USA), α-mono-polyubiquitin conj. FK2 (Enzo Life Sciences), all 1 : 100 and α-polyglycylated tubulin (AXO49), 1 : 1000 (Merck Millipore). Fluorescent secondary antibodies used were α-mouse 488, α-rabbit 488 (both DyLight, Thermo Scientific), and α-mouse 633 and α-rabbit 568 (all Alexa, Thermo Scientific), all 1 : 200.
Cell culture, transfections, co-IP and immunoblot analysis
Schneider 2 (S2) cells60 were cultured in Schneider's Drosophila Medium (Gibco, Thermo Scientific) complemented with 10% FBS (Biochrom, Merck Millipore) and 1% Penicillin/Streptomycin (Invitrogen) at 25 °C.
Transfections with expression vectors were done using Effectene Transfection Reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. For Co-IPs, three wells of a six-well plate were transfected. Expression was induced by addition of 350 μM CuSO4 overnight. Cells were lysed in lysis buffer (125 mM Tris pH 7.5, 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM EDTA, Complete Protease Inhibitor Cocktail (Sigma-Aldrich) and 1 mM DTT). Cell lysates were incubated with α-HA-coupled agarose beads (Sigma-Aldrich) for 2 h at 4 °C. Beads were washed and bound proteins eluted with 0.1 M Glycine pH 2.5.
For immunoblot analysis, samples were run on standard SDS-polyacrylamide gels or 4–12% BisTris gradient gels (NuPAGE, Thermo Scientific) and proteins transferred to PVDF membrane. For detection of poly-ubiquitylated proteins from testes lysates with FK2 antibody, membranes were treated with 6 M GuHCl, 20 mM Tris pH 7.5, 1 mM PMSF and 5 mM β-ME for 30 min at 4 °C, before blocking and antibody incubation.
Recombinant proteins
cDNAs for DIAP1, Dronc-V5, DrICE-V5-His, Drosophila CK2 and DUBA (wt and C227A and S183A mutants) were cloned into pGEX vectors (GE Healthcare, Freiburg, Germany) for protein expression in E. coli (BL-21). Expression was induced with 0.1 mM IPTG (18 °C, o/n) and proteins were purified by GST affinity purification. The GST tag was cleaved off with PreScission protease (GE Healthcare). DrICE was purified using Ni-NTA agarose (Qiagen).
In vitro ubiquitylation/de-ubiquitylation assays
For in vitro phosphorylation of recombinant DUBA, 20 μg of the protein were incubated with 10 mM MgCl2, 3 mM ATP and 0.1 μg recombinant CK2 for 3 h at 30 °C. The reaction was stopped by addition of 20 mM EDTA. A control reaction was carried out without ATP. Before de-ubiquitylation experiments, 10 mM DTT were added.
For the in vitro ubiquitin cleavage assay, 2 μg of DUBA or P-DUBA were used. Two hundred and fifty nanograms of K63 chains (3–7) or 100 ng of the respective di-ubiquitin (all Boston Biochem, Cambridge, MA, USA) were added to the reaction in cleavage buffer (50 mM Tris pH7.5, 1 mM EDTA, 1 mM DTT). After incubation at RT for 3 h, the reaction was stopped by addition of SDS sample buffer and heating of samples at 65 °C for 5 min.
Ubiquitylation of Dronc and DrICE was performed in a 30 μl reaction volume as described.16 The reaction was stopped by addition of 20 mM EDTA. For de-ubiquitylation, 2 μg DUBA or 2 μg P-DUBA were added to 4 μl of the ubiquitylation reaction in cleavage buffer (as above) and incubated overnight at RT.
Immunostaining of Drosophila testes
Whole testes of 1-day-old flies, anaesthetised on ice, were dissected in testes buffer24 and fixed in 4% formaldehyde for 30 min. Testes were washed three times in PBS for 5 min. Permeabilisation was done in PBT (PBS+0.3% Triton X-100) for 30 min. Testes were washed three times in PBT for 5 min. The tissue was blocked in 1% BSA in PBS for 45 min. All steps were performed at RT. Testes were incubated with the primary antibody in PBS for 2 nights at 4 °C. After washing three times for 1 h in PBT, incubation with the secondary antibody (and 3 U phalloidin (Thermo Scientific) and 1 μg/ml DAPI (Sigma-Aldrich), diluted in PBS, was performed overnight at 4 °C. After washing trice in PBT for 1 h each, testes were mounted on microscope slides in VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA, USA).
Confocal microscopy
Confocal imaging was performed with a Zeiss LSM780 microscope with Zen2010 software (Carl Zeiss, Jena, Germany). Images were taken with × 10 and × 40 magnifications and recorded as z-stacks in 2 μm intervals. Images were processed using ImageJ (NIH, Bethesda, MD, USA) and PhotoshopCS5 (Adobe, Dublin, Ireland).
Preparation of lysates from Drosophila and Drosophila testes
Whole fly lysates were prepared by homogenisation of 10 flies in 100 μl 2 × SDS loading buffer. Ten microlitres were loaded for WB analysis. Testes of 1–2-day-old flies were dissected as above. Forty testes were lysed in 100 μl lysis buffer (125 mM Tris pH 7.5, 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM EDTA, Complete Protease Inhibitor Cocktail (Sigma-Aldrich), 1 mM DTT and 5 mM N-ethylmaleimide) and were homogenised for 30 s. Lysis was performed for 30 min on ice, followed by centrifugation (14 000 r.p.m. at 4 °C for 10 min). Twenty-five micrograms of protein was used for western blotting analysis.
DEVDase assay from testes lysate
Per genotype, 180 testes (1–3-day-old flies) were dissected in testes buffer.24 Testes were collected in testes buffer with 1 mM PMSF on ice. Testes were disrupted in 70 μl lysis buffer (125 mM Tris, 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF) with pestles for 30 s and additional 20 manual strokes. The pestle was rinsed with 20 μl lysis buffer to reduce loss of sample. Testes were lysed as above. Lysates of each genotype were split and incubated either with DMSO or 20 μM z-VAD-fmk (Enzo Life Sciences) for 25 min at room temperature. Between 57 and 75 μg total protein/sample were used for an assay in a total volume of 200 μl. DEVDase activity was monitored by cleavage of 20 μM Ac-DEVD-AMC (Enzo Life Sciences) in 10 mM Tris pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 5% glycerol, 1 mM DTT, 1 mM PMSF. Fluorescence was measured at 380 nm excitation/460 nm emission in 12-min intervals for 10 h with a microplate reader (Infinite F200 Pro, Tecan Group, Männedorf, Switzerland).
Fertility test
Fertility of flies was tested as described.24
Acknowledgments
We thank Pascal Meier (ICR, London) and members of his laboratory for reagents and outstanding support. Many thanks to Daniele Bano, Gaia Tavosanis and Lukas Schwintzer for critical reading of the manuscript. We are also grateful to Bloomington (NIH P40OD018537) and VDRC stock centres for providing fly lines. The study was supported by Deutsche Forschungsgemeinschaft (DFG) (BR 3442/2-1).
Glossary
- CB
cystic bulge
- CK2
casein kinase 2
- Dark
death-associated APAF1-related killer
- DCP-1
death caspase-1
- DIAP1
Drosophila inhibitor of apoptosis-1
- DrICE
death-related ICE-like caspase
- Dronc
death regulator Nedd2-like caspase
- DUB
de-ubiquitylating enzyme
- GMR
glass multimer reporter
- Hid
head involution defective
- IAP
inhibitor of apoptosis
- OTU
ovarian tumour
- Rpr
reaper
- UIM
ubiquitin-interacting motif
- WB
waste bag
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Salvesen
Supplementary Material
References
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem 2012; 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009; 137: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K. Diverse roles of the ubiquitin system in NF-kappaB activation. Biochim Biophys Acta 2014; 1843: 129–136. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 2013; 49: 795–807. [DOI] [PubMed] [Google Scholar]
- Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio JP et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J 2006; 25: 1635–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T, Wooten MW. TrkA receptor endolysosomal degradation is both ubiquitin and proteasome dependent. Traffic 2008; 9: 1146–1156. [DOI] [PubMed] [Google Scholar]
- Lauwers E, Jacob C, Andre B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 2009; 185: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieser E, Cordier SM, Walczak H. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci 2013; 38: 94–102. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiol Rev 2013; 93: 1289–1315. [DOI] [PubMed] [Google Scholar]
- Tsou WL, Sheedlo MJ, Morrow ME, Blount JR, McGregor KM, Das C et al. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS ONE 2012; 7: e43112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Steller H. Regulation of cell death by the ubiquitin-proteasome system. Curr Opin Cell Biol 2009; 21: 878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol 2009; 19: 130–140. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 2000; 19: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 1999; 98: 453–463. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA et al. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol 2003; 10: 892–898. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell 2008; 32: 540–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ 2007; 14: 861–871. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 2002; 4: 445–450. [DOI] [PubMed] [Google Scholar]
- Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P et al. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet 2011; 7: e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol 2008; 10: 1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y, Arama E. Can't live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis 2009; 14: 980–995. [DOI] [PubMed] [Google Scholar]
- Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell 2009; 16: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol 2007; 17: 135–144. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 2003; 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Huh JR, Vernooy SY, Yu H, Yan N, Shi Y, Guo M et al. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol 2004; 2: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 2006; 133: 3305–3315. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J 2006; 25: 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol 2007; 5: e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J et al. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell 2010; 40: 810–822. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol 1989; 136: 346–362. [DOI] [PubMed] [Google Scholar]
- Cullen K, McCall K. Role of programmed cell death in patterning the Drosophila antennal arista. Dev Biol 2004; 275: 82–92. [DOI] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ 2008; 15: 1132–1138. [DOI] [PubMed] [Google Scholar]
- Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol 2012; 19: 171–175. [DOI] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013; 154: 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bre MH, Redeker V, Vinh J, Rossier J, Levilliers N. Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in paramecium. Mol Biol Cell 1998; 9: 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bre MH, Redeker V, Quibell M, Darmanaden-Delorme J, Bressac C, Cosson J et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci 1996; 109(Pt 4): 727–738. [DOI] [PubMed] [Google Scholar]
- Yacobi-Sharon K, Namdar Y, Arama E. Alternative germ cell death pathway in Drosophila involves HtrA2/Omi, lysosomes, and a caspase-9 counterpart. Dev Cell 2013; 25: 29–42. [DOI] [PubMed] [Google Scholar]
- Yue L, Karr TL, Nathan DF, Swift H, Srinivasan S, Lindquist S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics 1999; 151: 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A. The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ 2010; 17: 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaya JP, Ranmuthu M, Copeland J, Boyarskiy S, Blair AP, Hay B et al. The deubiquitinase emperor's thumb is a regulator of apoptosis in Drosophila. Dev Biol 2009; 329: 25–35. [DOI] [PubMed] [Google Scholar]
- Yang CS, Sinenko SA, Thomenius MJ, Robeson AC, Freel CD, Horn SR et al. The deubiquitinating enzyme DUBAI stabilizes DIAP1 to suppress Drosophila apoptosis. Cell Death Differ 2014; 21: 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SK, Chen P, Link N, Galindo KA, Pogue K, Abrams JM. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature 2009; 460: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Brot A, Chen P, Vaishnav M, Yuan S, Akey CW, Abrams JM. Tango7 directs cellular remodeling by the Drosophila apoptosome. Genes Dev 2013; 27: 1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M et al. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 2006; 126: 583–596. [DOI] [PubMed] [Google Scholar]
- Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev Cell 2010; 19: 160–173. [DOI] [PubMed] [Google Scholar]
- Lu C, Kim J, Fuller MT. The polyubiquitin gene Ubi-p63E is essential for male meiotic cell cycle progression and germ cell differentiation in Drosophila. Development 2013; 140: 3522–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007; 120(Pt 9): 1689–1700. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O'Rourke KM et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 2007; 318: 1628–1632. [DOI] [PubMed] [Google Scholar]
- Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 2015; 518: 417–421. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem 2011; 286: 10457–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics 2011; 10: M111 009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell 2010; 39: 477–484. [DOI] [PubMed] [Google Scholar]
- Boname JM, Lehner PJ. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses 2011; 3: 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 2010; 29: 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 2010; 17: 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell 2008; 14: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D et al. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ 2006; 13: 1663–1674. [DOI] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J 2000; 19: 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 2005; 7: 70–77. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 1972; 27: 353–365. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.