Abstract
AMPK is a central energy sensor linking extracellular milieu fluctuations with the autophagic machinery. In the current study we uncover that Poly(ADP-ribosyl)ation (PARylation), a post-translational modification (PTM) of proteins, accounts for the spatial and temporal regulation of autophagy by modulating AMPK subcellular localisation and activation. More particularly, we show that the minority AMPK pool needs to be exported to the cytosol in a PARylation-dependent manner for optimal induction of autophagy, including ULK1 phosphorylation and mTORC1 inactivation. PARP-1 forms a molecular complex with AMPK in the nucleus in non-starved cells. In response to nutrient deprivation, PARP-1 catalysed PARylation, induced the dissociation of the PARP-1/AMPK complex and the export of free PARylated nuclear AMPK to the cytoplasm to activate autophagy. PARP inhibition, its silencing or the expression of PARylation-deficient AMPK mutants prevented not only the AMPK nuclear-cytosolic export but also affected the activation of the cytosolic AMPK pool and autophagosome formation. These results demonstrate that PARylation of AMPK is a key early signal to efficiently convey extracellular nutrient perturbations with downstream events needed for the cell to optimize autophagic commitment before autophagosome formation.
Macroautophagy (hereafter called autophagy) is an evolutionarily conserved pathway involving the formation of a double-membrane vesicle, the autophagosome, which engulfs cytoplasmic components and delivers them to the lysosome for degradation.1 Autophagy is also a major mechanism by which starved cells reallocate nutrients from non-vital pathways to more essential processes2 and its disruption is associated with multiple disease states, including neurodegenerative diseases, cancer, infection, and several myopathies.3, 4, 5
The intracellular mechanisms that spatially and temporally fine-tune the initiation of autophagy still remain poorly understood. Poly(ADP-ribose) polymerase-1 (PARP-1) catalyzes the conversion of NAD+ to polymers of Poly(ADP-ribose) (PAR) in a process called PARylation which plays different pleiotropic cellular roles ranging from DNA damage sensing to transcription, chromatin relaxation or cell death.6 We have recently shown that during starvation-induced autophagy PARP-1 activation is involved in amplifying autophagy by feeding-back ROS production/DNA damage/NAD+consumption axis.7 In the present study we uncover a new and unexpected role for PARylation in the early signalling of autophagy: PARP-1 activation leads to AMPK PARylation, dissociation of PARP-1-AMPK complex and the nuclear-to-cytosolic export of AMPK, an event needed to induce mTORC1 inactivation/ULK1 phosphorylation in the cytosol. Together, these findings identify a new regulatory mechanism in autophagy and expand the known functions of AMPK and PARP-1 to include spatial regulation of the early signals of autophagy in mammalian cells.
Results
PARylation regulates starvation-induced autophagy
To analyze the importance of PARylation in starvation-induced autophagy we used the breast cancer cell line MCF7 cells stably transfected with GFPLC3. PARP-1 is the best studied member of the PARP protein family accounting for around 90% of cellular PARylation activity following different stimuli.8, 9 Recent studies have demonstrated its involvement in the regulation of DNA damage- or starvation-induced autophagy.10, 11
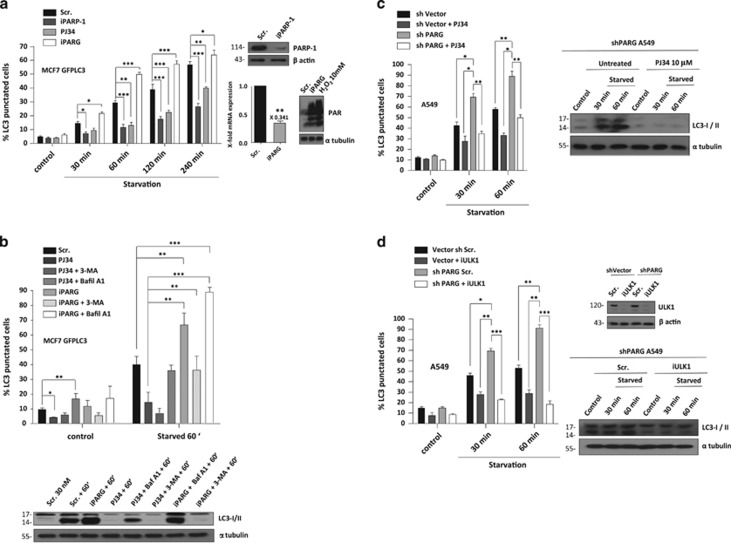
To correlate PAR production with starvation, we used the PARP inhibitors PJ34, DPQ and olaparib, iPARP-1 and iPARG (Poly(ADP-ribose)glycohydrolase) (Figure 1a, Supplementary Figure S1a, b and c; the efficiency of iPARP-1 and iPARG are shown in Figure 1a and Supplementary Figure S1c). Supplementary Figure S1a show that starvation induced PAR synthesis and in iPARP cells or after treatment with different PARP inhibitors autophagy was reduced (Figure 1a, Supplementary Figure S1b). In contrast in PARG-depleted cells, the accumulation of PAR accelerated autophagy after nutrient deprivation (Figure 1a). It has been reported that PAR accumulation may induce cell death (called PARthanatos),12 however this was not the case as PAR accumulation after nutrient deprivation did not compromise cell viability (Supplementary Figure S1d). Thus, the increased level of autophagy was not ascribed to a cellular attempt to detoxify the excess of PAR in autophagosomes, but there may be a mechanism of fine-tuning in the induction of PARylation-mediated autophagy. Fluorescent microscopic images showed an increased accumulation of autophagosomes in siPARG cells while inhibition of PARylation or PARP-1 knock-down abrogated starvation-induced autophagy (Supplementary Figure S1e). To analyze if PARylation-associated autophagy was an active process, we studied the membrane traffic associated to nutrient deprivation in presence of PARP inhibitors or PARG knock-down in combination with the autophagy inhibitors 3-MA, Chloroquine and Bafilomycin A1. Doses of autophagy inhibitors were established in MCF7 GFPLC3 during nutrient deprivation (Supplementary Figure S1f). By using the co-treatment of PARP inhibitors or siPARG in combination with 3-MA and Bafilomycin A1, we demonstrated that starvation-induced autophagy is an active process dependent of PAR levels and PARP activity (Figure 1b), as Bafilomycin A1 recovered the rate of autophagic cells and increased LC3-II translocation in starved cells treated with PJ34 or siPARG. To confirm the role of PAR in autophagosome formation, we used A549 cells stably-depleted for PARG13 which showed increased starvation-associated PARylation. Besides, PAR accumulation during nutrient deprivation did not affect to cell survival (data not shown) and induced a rapid translocation of LC3-II protein, compared with starved A549 shVector cells (Figure 1c and data not shown).
Figure 1.
Poly(ADP-ribose) regulates autophagy during nutrient deprivation. (a) Effect of PARG silencing, PARP silencing and PARP inhibition on starvation-induced autophagy. MCF7 GFPLC3 cells were transfected with PARP-1 siRNA (60 nM) or PARG siRNA (30 nM). 48 h later cells were starved with HANK Buffer. Other cells were pre-treated with 10 μM PJ34 for 1.5 h and maintained during the nutrient deprivation. * P<0.05, **P<0.01, ***P<0.001 versus the indicated group in MCF7 GFPLC3 cells by t-test. Western blot and qPCR for PARP-1 and PARG knock-down are shown. (b) Autophagy as active process dependent of PARylation levels. MCF7 GFPLC3 cells were transfected with PARG siRNA (30 nM). 48 h later, 3-MA (5 mM) and Bafilomycin A1 (0.2 μM) were used as autophagy inhibitors during the starvation treatment. 10 μM PJ34 for 1.5 h and maintained during the nutrient deprivation was used as PARylation inhibition control. * P<0.05, **P<0.01, ***P<0.001 versus the indicated group by t-test in MCF7 GFPLC3 cells. Lower panel shows endogenous LC3-II protein conversion during starvation, PARG siRNA or PJ34, alone or in co-treatment with 3-MA and Bafilomycin A1. α-tubulin was used as loading control. (c) Effect of PAR accumulation on starvation-induced autophagy. ShVector and shPARG A549 cells were starved with HANK buffer. The right panel shows the effect of PARPs inhibitor on the endogenous starvation-induced LC3-II conversion. α-tubulin was used as loading control. *P<0.05, **P<0.01 versus the indicated group in shVector and shPARG A549 cells by t-test. (d) Effect of ULK1 silencing on starvation-induced autophagy. ShVector and shPARG A549 cells were transfected with ULK1 siRNA (50 nM). 48 h later cells were starved with HANK buffer. The right panel shows the effect of ULK1 silencing on ULK1 protein levels and the endogenous starvation-induced LC3-II conversion in shPARG A549 cells. α-tubulin was used as loading control *P<0.05, **P<0.01, ***P<0.001 versus the indicated group in shVector and shPARG A549 cells by t-test. In all cases, at least 250 cells were counted in a Zeiss fluorescent microscope and data were represented as mean±S.E.M. of 3 independent experiments. In A, B and D figures scrambled or non-specific siRNA was used as negative control
A549 shPARG cells accumulated more autophagosomes compared with A549 shVector cells (Figures 1c and d and Supplementary Figure S2a). To further evaluate the impact of PAR, cells were treated with PJ34, which reduced the levels of starvation-induced autophagy in parental and shPARG cells (Figure 1c). Accordingly, LC3-II translocation in A549 shPARG cells disappeared after PARP inhibition (Figure 1c). Next, we evaluated the role of PAR synthesis in starvation-induced autophagy after silencing ULK1 which blocked autophagosome formation and LC3II translocation during starvation in both A549 cell lines (Figure 1d). These results suggest a dynamic interaction between PAR and the initiation of autophagy. In addition, silencing of other ATG-related protein as ATG7, confirmed the results obtained in starved A549 shPARG cells (Supplementary Figure S2b). All these data indicated that starvation-induced PARylation is directly and actively involved in autophagy formation.
Inhibition of PARylation down-regulates AMPKα activation and autophagy induction
The cellular metabolic status is sensed by two essential proteins, AMPK as an energetic sensor and mTORC1 complex as a nutritional sensor.14, 15, 16 Under energy rich levels, active mTORC1 acts as a negative regulator of autophagy, allowing proliferation and cell growth. On the contrary, AMPK is activated under energy depletion, and it is a positive regulator of autophagy, through in part inhibiting mTORC1.17
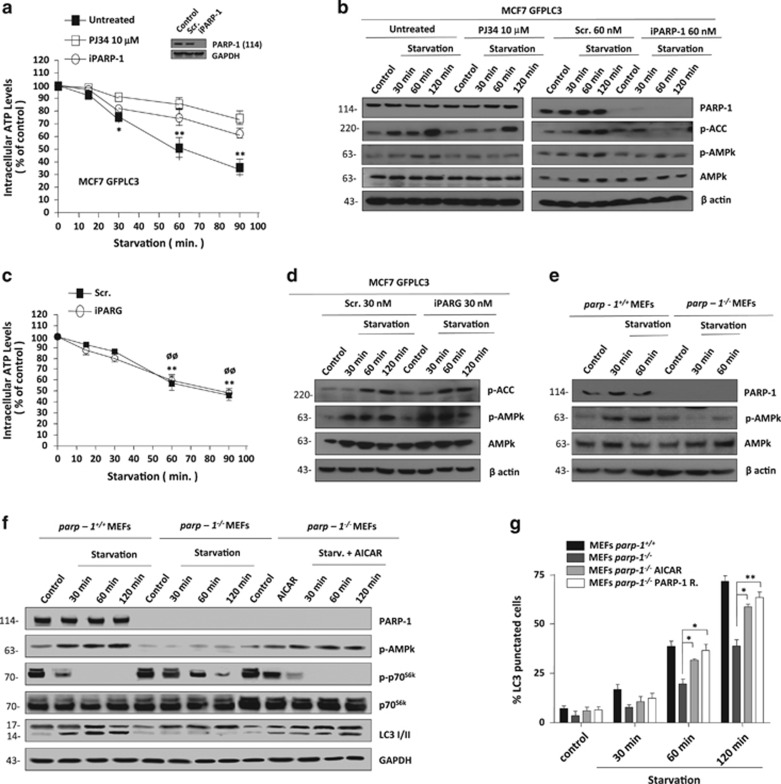
In view of the connection between PAR synthesis and the energy status of the cell we aimed to evaluate AMPK (hence forth AMPKα) during nutrient deprivation; we firstly observed a time-dependent decrease on ATP levels in MCF7 GFPLC3 cells following starvation (Supplementary Figure S3a, left panel) which correlated with an increase in AMPKα activation (Supplementary Figure S3a, right panel). Because PARP-1 is activated during nutrient deprivation (Supplementary Figure S1a), we hypothesized that the levels of PAR could regulate AMPKα activation. In line with this result, both PJ34 treatment or iPARP1 prevented starvation-induced ATP depletion (Figure 2a), implying that over-activation of PARP-1 during starvation was the main cause of energy drop in the starved cells. In addition, PJ34 treatment and iPARP-1 recovered ATP levels while AMPKα and autophagy were fully inactive (Figure 1a and Figure 2b). PARG silencing had no effect on ATP levels during starvation (Figures 2c and d). In starved MCF7 GFPLC3 cells, inhibition of autophagy in the initial steps with 3-MA, totally abrogated the activation of AMPKα (Supplementary Figure S3b); however there was no effect on AMPK activation after combination with Bafilomycin A1 suggesting that PARylation was affecting early autophagy events where ATP depletion and AMPK activation is taking place. To confirm the functional link between PARP-1 and AMPK, we used parp-1+/+ and parp-1−/− MEFs cells. parp-1−/− cells displayed a significant down-regulation in AMPKα activation during starvation (Figure 2e), and the percentage of autophagic cells decreased in the absence of PARP-1 (Supplementary Figure S3c and S3d). Moreover, in MEFs PARP1 knockout, Bafilomycin A1 recovered LC3 punctated pattern and LC3II translocation, thereby confirming that autophagy is an active process which is strongly delayed in absence of PARP-1 (Supplementary Figure S3c). To further confirm this result, we treated cells with the potent AMP mimetic AICAR, which reactivated AMPK (Figure 2f and Supplementary Figure S3d). Using AICAR in parp-1−/− MEF cells resulted on restoration of AMPK activity, mTORC1 inactivation and LC3-II lipidation, confirming the data obtained in starved parp-1−/− MEF cells co-treated with Bafilomycin A1 (Figure 2f and Supplementary Figure S3c). Starved parp-1−/− MEFs co-treated with AICAR, showed an important increase in the levels of autophagy similar to starved parp-1+/+ MEFs (Figure 2f). After restoring PARP-1 in parp-1−/− MEF cells we obtained similar levels of starvation-induced autophagy as in PARP-1 wild type cells (Figure 2g). All these data suggest that the presence and proper function of PARP-1 was needed for the adequate activation of AMPKα and for starvation-induced autophagy.
Figure 2.
Inhibition of PARylation down-regulates AMPk pathway. (a, b) Effect of PARylation inhibition on ATP levels and AMPk activation. Treatment with PJ34 and PARP-1 silencing (60 nM) strongly delays starvation-induced loss of ATP levels. Right panel shows the efficiency of PARP-1 silencing.*P<0.05, **P<0.01 versus the indicated group in MCF7 GFPLC3 cells by t-test (a). MCF7 GFPLC3 cells treated with PJ34 (left panel) or silenced for PARP-1 (right panel) during starvation showed down-regulation of AMPk activation (b). (c) Effect of PAR accumulation on ATP levels. Accumulation of PAR polymer hadn't effect on starvation-induced loss of ATP in starved MCF7 GFPLC3. The efficiency of PARG knock-down was previously shown in Figure 1a.**P<0.01 comparing between control and starved MCF7 GFPLC3 cells and øøP<0.01 comparing between control and starved PARG silenced MCF7 GFPLC3 cells. (d) Activation of AMPk in absence of PARG. The accumulation of PAR polymer in MCF7 GFPLC3 cells increased the activation of AMPK under nutrient deprivation. (e) AMPK activation is delayed in absence of PARP-1. Cell extracts of parp-1+/+and parp-1-/ -MEFs after different times of starvation were used to measure the levels of AMPk activation. (f) AICAR reactivated the AMPk/mTORC1/LC3 pathway in absence of PARP-1. Cell extracts from parp-1+/+, parp-1−/− and parp-1−/− MEFs pre-treated with AICAR 1 mM were used to measure the levels of AMPk activation (phospho-AMPK), mTORC1 activation (phospho-p70S6k) and autophagy induction (endogenous LC3-II translocation). (g) Starvation-induced autophagy is increased in parp-1−/−MEFS after AICAR treatment or PARP-1 reconstitution. *P<0.05, **P<0.01 versus the indicated group by t-test. In B, D, E and F figures total AMPk and total p70s6 kinase were used to normalize for the non-phosphorylated protein and β-actin or GAPDH were used as loading control. Similar results were obtained in three independent experiments
Activation of nuclear and cytosolic AMPK requires PARylation-dependent dissociation of the PARP-1/AMPK complex from PARP-1
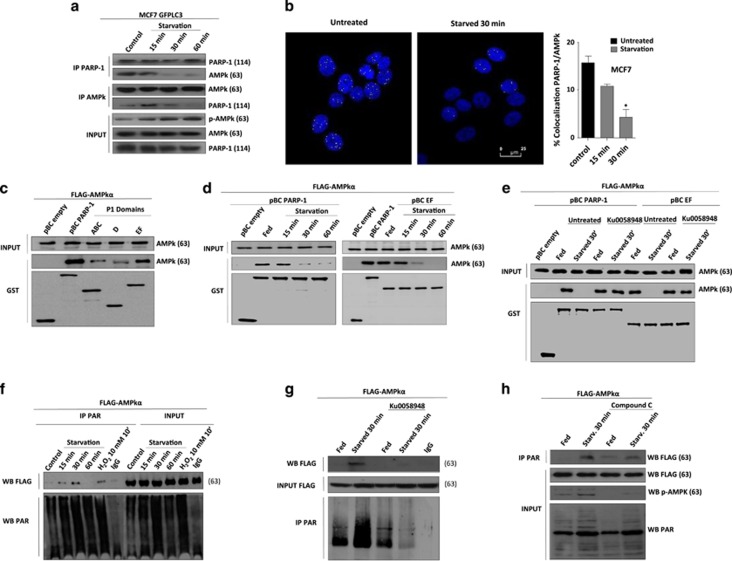
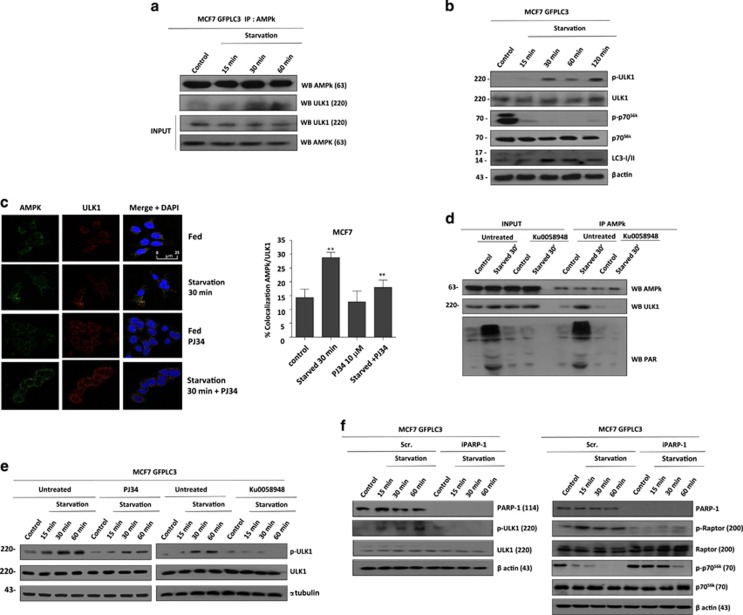
We then aimed to investigate whether PARP-1 interacts with AMPKα and if PARP-1 modifies its kinase activity by PARylation. To this end we performed co-IP assays at different times after starvation. Co-IP experiments revealed an interaction between PARP-1 and AMPKα in fed cells (Figure 3a). After starvation we observed a gradual and time-dependent dissociation of the PARP-1-AMPK complex (Figure 3a) with complete dissociation reached 30 min upon nutrient deprivation. This dissociation, in turn, allowed AMPKα activation (Figure 3a INPUT), concomitant with the inactivation of mTORC1 and autophagy induction shown above (Figure 2). To substantiate this physical interaction between PARP-1 and AMPKα we measured the percentage of co-localization in starved MCF7 GFPLC3 cells observing that diminished to 6% after 30 min of starvation (Figure 3b).
Figure 3.
Activation of AMPkα requires interaction with PARP-1 and its PARylation. (a) Endogenous co-Immunoprecipitation of AMPkα1 and PARP-1. MCF7 GFPLC3 cells were starved with HANK buffer for 15, 30 and 60 min. INPUT shows total levels of AMPk, PARP-1 and the activation of AMPk. (b) Confocal microscopy to detect colocalization between PARP-1 and AMPkα1. MCF7 GFPLC3 cells were starved with HANK buffer for 15 and 30 min. Colocalization percentages were obtained, using a LEICA LCS SP5 confocal microscope. The bar chart shows the percentage of cells in which colocalization foci were observed at control or upon treatment with HANK buffer.*P<0.05 comparing between control MCF7 GFPLC3 and starved MCF7 GFPLC3 cells by t-test. (c) Interaction by GST-pulldown assay of AMPkα1 protein with PARP-1 catalytic domain. COS1 cells were co-transfected with FLAG-AMPkα1 and pBC plasmids that over expressed different domains of PARP-1: pBC-ABC (Zn fingers), pBC-D (Automodification domain), pBC-EF (catalytic domain) and PARP-1 full protein. INPUT shows total levels of AMPkα. (d) Interaction between AMPkα1 and PARP-1 full protein or catalytic domain disappears during starvation. COS1 co-transfected with FLAG-AMPkα1 and pBC-empty, pBC-PARP-1 full and pBC-EF were starved for 15, 30 and 60 min with HANK buffer. (e) Disruption of AMPkα1/PARP-1 complex during starvation depends on PARylation levels. COS1 co-transfected with FLAG-AMPkα1 and pBC-empty, pBC-PARP-1 full and pBC-EF were pre-treated with KU0058948 100 nM 2 h before starved. (f) AMPkα1 is PARylated during starvation-induced autophagy. Immunoprecipitation of PAR polymer in HeLa shPARG transfected with FLAG-AMPkα and starved with HANK buffer for 15, 30 and 60 min. FLAG antibody shows levels of expression of FLAG-AMPkα1 in HeLa shPARG cells. INPUT shows starvation-induced PARylation. (g) KU0058948 blocked AMPkα1 PARylation during starvation. HeLashPARG transfected with FLAG-AMPkα1 were pretreated with 100 nM KU0058948 for 2 h and starved for 30 min with HANK buffer. PAR antibody reveals general starvation-induced PARylation and PAR-AMPkα modified protein. (h) AMPK inhibition using Compound C prevented AMPKα PARylation. HeLa shPARG were transfected with FLAG-AMPkα1. Cells were pretreated with 10 μM Compound C for 4 h and starved for 30 min with HANK Buffer. INPUT shows total AMPkα. Anti-PAR antibody reveals general starvation-induced PARylation and PAR-modified protein. In all cases, similar results were obtained in at least three independent assays
To get more insights about the interaction between PARP-1 and AMPKα, we mapped the domains of PARP-1 involved in this interaction. PARP-1 was dissected in three domains cloned with GST fusion protein18 as explained in Supplementary Figure S4a. These constructs were expressed in Cos1 cells and the co-IP of endogenous AMPKα was analyzed by GST-pulldown. Starvation-induced autophagy was confirmed to be efficient in Cos1 cells (Supplementary Figure S4b). The results indicated that AMPKα interacts efficiently with the catalytic domain of PARP-1 and to a lesser extent with the ABC and D domains (Figures 3c and d). Next, we starved COS1 cells co-transfected with both pcDNA3-FLAG-AMPKα and GST-fused PARP-1 to confirm that the AMPK/PARP-1 complex dissociated shortly after starvation (Figure 3d). Indeed, the interaction between AMPKα and PARP-1 moved-out confirming the endogenous co-IP in MCF7 GFPLC3 cells (Figure 3a). Finally we analyzed the role of PARP activity in PARP-1/AMPK complex stability and found that PJ34 prevented dissociation of the PARP-1/AMPK complex after starvation (Figure 3e). Together, these results suggest that PARylation of AMPKα takes place after PARP activation and is needed to disrupt the PARP-1/AMPK complex, allowing the activation of AMPKα and thereby, induction of autophagy (Figure 3e).
To demonstrate PARylation of AMPKα during starvation-induced autophagy, we immunoprecipitated PAR in shPARG HeLa cells. We found that AMPKα was PARylated at 15 and 30 min of starvation corresponding with the disruption of the PARP-1/AMPKα complex (Figure 3f). 15 min of nutrient deprivation induced PARylation of AMPKα, at 60 min PARylation of AMPKα completely vanished, but at 30 min of starvation we observed that the levels of PAR associated with AMPKα were critical to induce phosphorylation of AMPK, inactivation of mTORC1 and translocation of LC3-II (Figure 3f and Figure 2). PARP inhibition prevented AMPK PARylation during starvation and was co-incident with stabilization of PARP-1/AMPK complex (Figure 3g). To evaluate the possibility of a mutual interaction between AMPK and PARP-1 leading to PARP-1 phosphorylation, as has been described before,19 we treated the cells with the AMPK inhibitor Compound C which indeed prevented AMPK activation and decreased PARylation associated to AMPK (Figure 3h) suggesting that full AMPK activity was needed for PARP activation. Taken together, these data suggest that PARP-1 activation during starvation signals PARP-1/AMPK complex dissociation, to activate AMPKα in a PARylation-dependent manner.
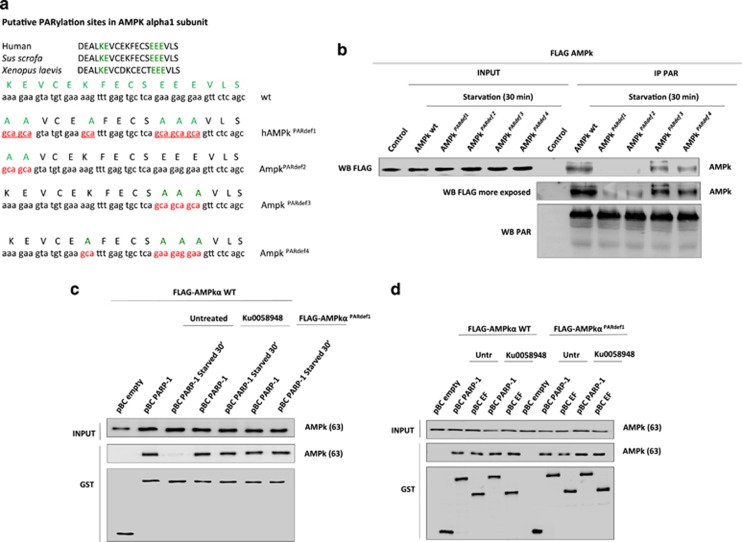
To strengthen the previous findings we generated different mutants of AMPK deficient for Poly(ADP-ribosyl)ation putative sites (Figure 4a). Expression of mutated proteins, followed by coIP with the anti-PAR antibody confirmed that AMPK mutants (AMPKPARdef) were indeed PARylation deficient in the following order: 1>2>4>3 (Figure 4b and Supplementary Fig. S5a). The most important residues were K291 and E292 whose change to A almost completely abolished AMPK PARylation (Figure 4b). We also verified that AMPKPARdef1 interacted with PARP-1 under basal (non starved) situation. By GST-pulldown assay in Cos1 cells we confirmed that the AMPKPARdef1 formed a stable complex with the full protein and the catalytic domain of PARP-1 under optimal nutrient conditions (Figures 4c and d). After PARP inhibition and in AMPKPARdef1 this interaction remained in starved cells (Figure 4c). The interaction between AMPKPARdef1 and PARP-1 took place in the domains EF of PARP-1 and after PARP inhibition the complex increased under nutrient-rich conditions (Figure 4d).
Figure 4.
Specific PARylation of AMPkα1 during starvation-induced autophagy. (a) Putative PARylation sites in AMPKα1 subunit. Comparing with the Sus scrofa and Xenopus laevis AMPKα1 subunit sequence, human AMPkα1 subunit was specifically mutated in the PARylated putative position E 315, 316 and 317, K 310 and K 305 and E 306, generating 4 different mutants (AMPKPARdef1, 2, 3, 4). Finally the mutants were tagged with FLAG oligopeptide and subcloned in pCDNA3 plasmid, obtaining two specific vector pCDNA3-FLAG-AMPkα wt and pCDNA3-FLAG-AMPkα mut. Each mutant represents the following changes: PARdef1: The residues K305,E306,K310,E315,E316,E317 were mutated in A: PARdef2: The residues K305,E306 were mutated in A: PARdef3: The residues E315,E316,E317 were mutated in A: PARdef4: The residues K310 was mutated in A. (b) AMPKPARdef blocked AMPkα PARylation during starvation. MCF7 cells were transfected with either FLAG-AMPkα wildtype or AMPKPARdef1,2,3,4 plasmids. INPUT shows total levels of AMPK. PAR antibody reveals starvation-induced PARylation of AMPKα is disrupted when PARylated putative sites are mutated (AMPKPARdef1,2,3,4). (c) PARP inhibition or mutation in putative PARylation sites of AMPkα1 prevents the disruption of PARP1/AMPkα1 complex during starvation. COS1 cells were pre-treated with 100 nM KU0058948 for 2 h, or co-transfected with FLAG-AMPkα1 wildtype/mutant protein and pBC-empty/pBC-PARP-1 full protein. Cells were starved for 30 min with HANK buffer. GST antibody shows the interaction PARP-1/AMPkα1 in basal and starved situations. (d) AMPkα1 mutated in PARylation sites interacts with PARP-1 in basal conditions. Interaction by GST-pulldown in COS1 cells co-transfected with FLAG-AMPkα1 wildtype and mutant isoforms and pBC PARP-1 or pBC EF plasmids that over expressed full protein and the catalytic domain of PARP-1. Transfected COS1 cells were pretreated with 100 nM KU0058948 for 2 h. In B, C, D figures similar results were obtained in three independent assays
All these data suggest that PARP-1 and AMPK form a stable complex under rich energy situation. The correct activation of PARP-1 and the modification by PAR of AMPKα are essential events in the disruption of the complex to induce total activation of AMPK under nutrients poor status.
PARylation and nuclear export of AMPKα
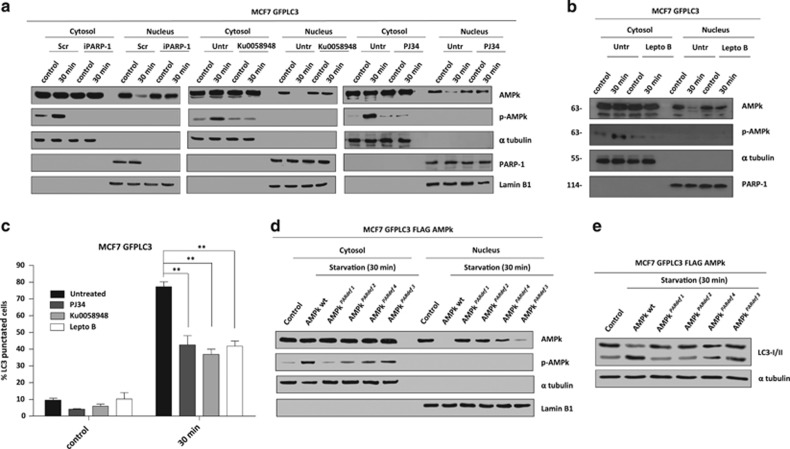
Autophagy is a predominantly cytosolic pathway; thus we hypothesized that AMPKα modified by PAR (PAR-AMPK) would be exported from the nucleus to the cytosol to regulate the initial steps of starvation-induced autophagy. To determine if PARylation of AMPKα induced cytosolic AMPK activation during autophagy, we measured the levels of total and activated AMPKα in nuclear and cytosolic fractions during starvation–induced autophagy. Sub-fractionation assays showed that nuclear AMPKα declined at 30 min of starvation. Nuclear export of AMPKα was co-incident in time with activation of cytosolic AMPKα (Figure 5a Left Panel). Silencing of PARP-1 prevented AMPKα vanishing from the nucleus and the cytosolic activation of AMPKα (Figure 5a Left Panel). To confirm this result we treated cells with PARP inhibitors which retained AMPKα in the nucleus and abolished the activation of AMPKα in cytosol after starvation (Figure 5a Central and Right Panels). The carrier protein Crm1 is involved in the nuclear export of a large number of proteins and is efficiently inhibited by Leptomycin B. To confirm the impact of the nuclear-cytosolic export of PAR-AMPKα in autophagy, we used Leptomycin B which led to nuclear accumulation of AMPKα. Moreover, the activation of AMPKα after 30 min of starvation declined after Leptomycin B treatment (Figure 5b). The levels of autophagic cells (and autophagosomes per cell) at 30 min of starvation decreased with Leptomycin B compared to untreated starved (Figure 5c). Finally we demonstrated that AMPK PARylation was necessary to induce its nuclear export during starvation using the AMPKPARdef mutants which were unable to undergo nuclear export, similar to the results obtained after PARP inhibition (Figure 5d). The presence of AMPKPARdef abrogated the activation of AMPK in cytosol (Figure 5d) and totally inhibited the induction of autophagy (Figure 5e).
Figure 5.
PARylation and nuclear export of AMPk. (a) Nuclear export of AMPk during starvation. The nuclear and cytosolic fractions were prepared from fed or starved MCF7-GFPLC3 cells, co-treated or not with PJ34 (10 μM) or KU0058948 (100 nM) or silenced for PARP-1. PJ34 and KU0058948 were used as pre-treatment during 2 h and maintained during starvation assay. (b) Leptomycin B prevents nuclear export of AMPkα1 during starvation. MCF7-GFPLC3 were pre-treated with Leptomycin B 20 ng/ml during 3 h and maintained during starvation with HANK buffer for 30 min. (c) Effect of Leptomycin B, PJ34 and KU0058948 on autophagy levels. MCF7 GFPLC3 cells were co-treated with 20 ng/ml Leptomycin B, 10 μM PJ34 or 100 nM KU0058948 and starved with HANK Buffer 30 min. Cells with the typical punctated pattern of starvation-induced LC3 was counted by fluorescence microscopy. Similar results were obtained in three independent experiments. At least 250 cells were counted in a Zeiss fluorescent microscope in 3 independent experiments. **P<0.01 versus the indicated group by t-test. (d) Nuclear export of AMPk during starvation is blocked by PARylation sites mutation in AMPkα1. MCF7 GFPLC3 cells were transfected with FLAG-AMPkα1 wildtype and AMPKPARdef mutants. (e) Mutation in PARylated sites of AMPkα1 blocks autophagy induction. MCF7 cells transfected with FLAG-AMPkα1 wildtype and mutants (PARdef 1-4), and starved for 30 min. Autophagy induction was assessed by specific endogenous LC3-II translocation. Similar results were obtained in three independent experiments. In A, B and D figures, total levels and activation of AMPk (Phospho-AMPk) were analyzed in both fractions. α-tubulin and PARP-1 and/or Lamin B1 were used as cytosolic and nuclear fractions respectively. Similar results were obtained in three independent experiments
Inhibition of PARylation prevents mTORC1 down-regulation and ULK1 activation during autophagy
Once activated by falling energy status, AMPK switches-on ATP-producing catabolic pathways such as autophagy. Under nutrient-rich conditions mTORC1 interacts with ULK1 complex inactivating the autophagosome formation. However, during starvation the energy depletion induces AMPK activation and mTORC1 inactivation.
We asked for the consequences of PAR-AMPKα nuclear export on ULK1 activation. First we analyzed the interaction AMPKα/ULK1 during starvation and the effect of PARP inhibitors. Co-IP and co-localization assays in MCF7 cells, showed that after 30 min of starvation AMPKα interacted with ULK1, confirming the sequence of events from activation/PARylation and nuclear export of AMPKα after starvation (Figures 6a and b). Starvation during 30 min was sufficient to provoke activation of ULK1, total inhibition of mTORC1 and endogenous LC3-II translocation (Figure 6b). Interestingly PARP inhibition disabled the interaction AMPKα/ULK1 during starvation, as the percentage of endogenous co-localization of both proteins decreased in starved cells treated with PARPi (Figure 6c). We have demonstrated that PAR-AMPK shuttled from nucleus to cytosol during starvation and interacted with ULK1 to promote its proper activation favouring the initial steps in the autophagosomes formation. Immunoprecipitation of AMPK in presence of the PARP inhibitor Ku0058948 demonstrated that PARylated-AMPK formed a stable complex with ULK1 protein (Figure 6d). But, was this modification needed to maintain AMPK/ULK1 complex? To address this question we measured the activation of ULK1 following starvation in the presence of PARP inhibitors and we found that PARPi prevented ULK1 phosphorylation (Figure 6e). Consistently, PARP1 knockdown in starved MCF7 GFPLC3 cells impeded ULK1 (as well as Raptor) phosphorylation, leading to the recovery of mTORC1 activity and the inhibition of autophagy (Figure 6f).
Figure 6.
PARP inhibitors and PARP-1 silencing modulate ULK1 activation. (a, b) Interaction AMPk/ULK1 during starvation. MCF7 GFPLC3 cells were starved with HANK Buffer for 15, 30 and 60 min. INPUT shows total levels of AMPk and ULK1 (a). Different times of starvation were used to measure the levels of ULK1 activation (phospho-ULK1), mTORC1 inactivation (phospho-p70S6k) and autophagy induction (endogenous LC3-II translocation) (b). (c, d) Effect of PARylation inhibition on AMPk/ULK1 interaction. MCF7 cells were starved with HANK Buffer for 30 min and co treated with PJ34 10 μM. Colocalization percentages were obtained quantifying the Immunofluorescence from the z-stacks on every imagein a LEICA LCS SP5 confocal microscope. The histogram shows the percentage of cells in which colocalization foci were observed. **P<0.01 versus the indicated group by t-test (c). Immunoprecipitation of AMPK in HeLa shPARG transfected with FLAG-AMPkα wildtype. Cells were starved with HANK Buffer for 30 min. 90 min pre-treatment with 100 nM KU0058948 was maintained during starvation. FLAG antibody shows levels of expression of FLAG-AMPkα1 and the specific PARylation of AMPkα1 in HeLa shPARG cells. ULK1 antibody shows AMPkα1/ULK1 interaction is prevented by PARP inhibition. INPUT shows starvation-induced PARylation (d). (e) Effect of PARP inhibitors on ULK1 activation. MCF7 GFPLC3 cells were starved with HANK Buffer for 15, 30 and 60 min. Total ULK1 kinase was used to normalize for the non-phosphorylated protein. (f) Silencing of PARP-1 down-regulated ULK1 activation (left panel) and maintained mTORC1 activation during starvation-induced autophagy (right panel). MCF7 GFPLC3 cells were transfected with PARP-1 siRNA (60 nM). scrambled or non-specific siRNA was used as negative control, using the same protocol as for siRNA transfection. 48 h later were starved with HANK Buffer for 15, 30 and 60 min. In A, C, E and F figures total ULK1, p70s6 and Raptor kinase were used to normalize for the non-phosphorylated protein. α-tubulin and β–actin were used as loading control. Similar results were obtained in three independent experiments
All these data suggest that PARylation is a crucial PTM on AMPK to induce an optimal activation and loading of ULK1 complex eventually leading to induce autophagosomes formation.
Discussion
Autophagy is regulated by a highly interconnected arrangement of proteins are involved in the control of key cellular processes. Autophagic responses in conditions of nutrient deprivation serve metabolic purposes and various intracellular entities can be degraded to provide the cell with metabolic substrates for survival. In response to nutrient deprivation AMPK is activated upon the accumulation of AMP at the expense of ATP. Multiple signal transduction pathways that ultimately accelerate or inhibit the autophagic flux do so by impacting on the activation state of AMPK and/or mTORC1.16, 20
AMPK functions as a fuel gauge and maintains energy homeostasis during cellular stress21 and is activated by changes in the cellular AMP:ATP and ADP:ATP ratios. AMPK functions to restore energy homeostasis by switching-off biosynthetic pathways consuming ATP while switching-on catabolic pathways that produce ATP. Upstream kinases phosphorylate the critical Thr172 activating site and include liver kinase B1 (LKB1), calcium–calmodulin-dependent kinase kinase 2 (CaMKK2) and transforming growth factor-β-activated protein kinase-1 (TAK1).22 PTM regulation of autophagy has been extensively reported and mechanisms including phosphorylation, glycosylation, ubiquitination, acetylation and lipidation have been shown to be critical in the ability of ATGs and other autophagy-related proteins to interact with regulators of this route.23
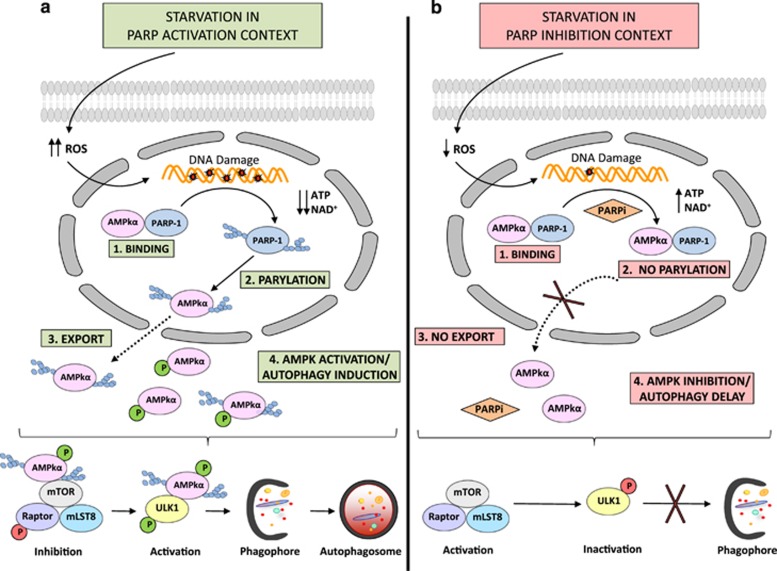
In the present study we demonstrate that the modification of AMPK by PARylation and the mutual interaction between PARP-1 and AMPK function as a multifaceted molecular switch to optimize the initiation of autophagy summarized in Figure 7: (1) non-PARylated nuclear AMPK is inactive when in complex with PARP-1 in non-starved cells; (2) following starvation PARP-1 is activated and AMPK is transiently PARylated and detached from PARP-1; (3) complex dissociation and AMPK PARylation trigger kinase activation which is needed to undergo AMPK nuclear export and activation of the cytosolic AMPK pool.
Figure 7.
PARylation regulates autophagy through AMPK activation:. PARP-1 forms a complex with AMPK in nucleus (1). During the starvation-induced autophagy ROS production induces DNA damage and overactivation of PARP-1. Auto PARylated PARP-1 is able to modify by PARylation AMPK in AMPKα1 subunit (2). The complex is disrupted and PAR-AMPK is exported to cytosol (3). Presence of PAR-AMPK and continuous absence of Aminoacids and ATP depletion, favours total activation of AMPK population, inhibition of mTORC1, interaction PAR-phospho-AMPK/ULK1 and autophagosomes formation (4). (a). Starvation-induced ROS production was abrogated during the treatment with PARP inhibitors. Following AMPKα1/PARP-1 interaction (1), AMPKα1 subunit is not PARylated and nuclear export of AMPK is inhibited (2 and 3). In spite of nutrient and energy depletion, AMPK is inhibited, mTORC1 is partially activated and interacts with ULK1 favouring it's inhibition (4). Finally the autophagosomes production will be delayed (b)
Interaction between PARP-1 and AMPK has been previously shown to be needed for DNA-damage dependent PARP-1 activation and PARP-1 has been reported to be a substrate of AMPK.19, 24 The molecular insights for this mutual interaction still remain to be fully elucidated. Our results suggest that AMPK activation is hindered by its interaction with PARP-1 that most likely will be in turn phosphorylated by AMPK following starvation. Indeed, both global and AMPK-associated PARylation are decreased in the presence of the AMPK inhibitor compound C (Figure 4e). DNA binding domain of PARP-1 has been reported to be phosphorylated by AMPK at Ser-177 leading to PARP-1 dissociation from Bcl-6 promoter.24 Given the highly elaborated nature of the autophagic pathway, the mutual regulation of AMPK and PARP-1 is likely to be complex and future studies are required to have a comprehensive understanding of this crosstalk in response to the wide range of autophagy-inducing signals.
One of the most remarkable finding in this study is the need of AMPK nuclear export to perform its cytosolic function during the induction of starvation-induced autophagy but how PARylated AMPK coming out of the nucleus ‘hits' cytosolic AMPK remains to be clarified. The nucleus has been implicated in the initiation of autophagy as a primary site of conditions that are potentially toxic for mammalian cells, as they cause DNA damage or perturb/prevent proper DNA replication.25 Nuclear sensors, including PARP-1, detect perturbations in nuclear and genetic homeostasis, to activate mechanisms for the repair of cellular damage and, if these fail, trigger different types of cell death for the benefit of organismal homeostasis. Increasing evidences indicate that repair of DNA lesions activate a non-selective autophagic response.26 The fine-tuning of key biological processes, other than DNA repair, through PARylation is still largely unknown. Our study provides new implications of PARylation as a dynamic process in autophagy beyond PARP-1 activation and its association with DNA repair and energy balance, as has been reported previously.7, 27 Our results place the nucleus and the interaction between PARP-1 and AMPK as an initial sensor of the metabolic alterations derived from perturbations in the nutritional extracellular status not necessarily related with the alterations in genomic integrity.
PARylation as a PTM process may also be involved in the regulation of more downstream autophagy events. Our results show that activation of ULK1/inactivation of mTOR might also be modulated by PARylation, although the precise substrates have not been defined. PARylation has also been recently implicated as regulatory signal for proteasomal-mediated protein degradation.28 In summary, our study supports a model where PARP activation and PAR levels during starvation-induced autophagy leads to global modulation of autophagic fluctuations and more specifically induce de-repression of AMPK activity (Figure 7). Until now, nuclear events have not been considered to be of critical importance for autophagy. Our data, however, uncover an essential connection between the induction of autophagy and AMPK PARylation which determines its export out of the nucleus to unleash the early cytosolic events of autophagy, including inhibition of mTORC1 and de-repression of ULK1-kinase (Figure 7b). Previous results have shown that in cytoplasts/enucleated cells the autophagic process is still feasible.29 Our findings do not contradict the previous conclusions about functionality of the autophagic process in enucleated cells, but add a new early regulatory axis influencing the outcome of autophagy. The discovery of tightly regulated PAR-modified proteins associated with the autophagic process offers a striking conceptual context to understand the short-term response to stimuli eliciting autophagy. Participation of nuclear AMPK in autophagy is needed to convey a rapid response of the alteration in the metabolic status, indicated by the level of PARylation as an alert indicator of changes in the energy balance. Cells deficient in PARP-1 are still able to undergo starvation-induced autophagy but the outcome of this delay may imply important consequences for the time-frame of drug treatment and the sensitization of tumor cell in their microenvironment to different combinatorial regimes including PARP inhibitors.
Materials and Methods
Cell cultures and treatments
MCF7 GFP-LC3 cells were cultured in RPMI 1640 GlutaMAX (GIBCO Invitrogen) with 6% inactive fetal bovine serum (FBSi, Gibco Invitrogen) and Pen/Strep (P4333, Sigma-Aldrich, St Louis, MO, USA). Immortalized mouse embryonic fibroblast (MEFs 3T3), derived from both wild-type and PARP-1 KO mice, cell lines were cultured in Dulbeco's Modified Eagle's Medium (DMEM) supplemented with 10% FBSi. ShVector and shPARG A549 cell lines were cultured in DMEM supplemented with 10% FBSi, L-Glutamine 2 mM, Pen/Strep and 5 μg/ml Puromycin and selection antibiotic. ShVector and shPARGHeLa cells were cultured in DMEM supplemented with 10% FBSi, Gentamicin 50 ng/ml and Hygromycin B 1:400 as antibiotic selection. All cell lines were maintained at 37 °C in a humidified 5% CO2 atmosphere. Cells were starved with Balanced HANK Buffer without Aminoacids (140 mMNaCl, 5 mM KCl, 1.3 mM MgCl2-6H2O, 2 mM CaCl2-2 H2O, HEPES 10 mM, 5 mM D-Glucose), at different times as stated in the figures.
Cells were treated with the PARP inhibitors PJ34 ([N-(6-Oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethylacetamide]), DPQ (3,4-Dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone]) (Alexis Biochemicals, San Diego, CA),Olaparib (KU0059436, Selleckchem) and KU0058948 (KU0058948, supplied from Francoise Dantzer's Lab). DPQ and Olaparib were dissolved in DMSO while KU0058948 and PJ34 were dissolved in miliQ H2O2 and stored at -20 ºC. For treatments, cell were pre-treated with 40 μM DPQ, 10 μM PJ34, 100 nM KU0058948 and 10 μM Olaparib for 1.5 h before starvation and maintained all the time of the experiments.
The autophagy inductors Rapamycin (553210 Calbiochem, Germany) and Concanamycin A (C9705 Sigma-Aldrich) were dissolved in DMSO at 1 mM (Rapa) and 100 nM (Con A) and stored at -20 ºC. Autophagy inhibitors 3-Methyladenine (M9281Sigma-Aldrich), Chloroquine (C6628 Sigma-Aldrich) and Bafilomycin A1 (B1793 Sigma-Aldrich) were dissolved in PBS1x (Chloroquine) RPMI 1640 (3-MA) and DMSO (Baf A1) and maintained at -20 ºC. For treatments, cell were pre-treated with 5 mM 3-MA, 10 μM CQ, 0.2 μM Baf A1, 100 nM Rapa and 4 nM Con A for 1.5 h before starvation and maintained all the time of the experiment.
The mimetic of AMP, AICAR (5-Aminoimidazole-4-carboxamide 1-β-D-ribofuranoside, Acadesine, N-(β-D-Ribofuranosyl)-5-aminoimidazole-4-carboxamide, A9978, Sigma-Aldrich) was dissolved in H2O: >7 mg/ml and stored at −20 ºC. Compound C (P5499 Sigma-Aldrich) AMPK inhibitor, was dissolved in DMSO at 4 mg/ml and maintained at -20 ºC. For treatments, cell were pre-treated with 1 mM AICAR for 1.5 h and 10 μM 4 h before starvation and maintained all the time of the experiment. Nuclear export inhibitor Leptomycin B (L6417 Sigma-Aldrich) was dissolved in methanol:water (7:3) and maintained at -20 ºC; was used in a final concentration 20 ng/ml during 3 h.
Western blot analysis
Cell lines were plated in 6-well plates with a density of 3 × 105 cells per plate and treated the next day with HANK buffer. After the nutrient starvation, cells were washed twice with PBS1x, resuspended in 200 μl of Lysis Buffer TR3 (10 mM Na2HPO4, 10% Glycerol, 20% SDS) and harvested. The samples were sonicated 10 s at 50% power in ‘constant' mode. Finally it was added a pre-heated Blue-β-Mercaptoethanol solution (1:1) 1:10 under the safety hood. All the samples were maintained at -20 ºC. Proteins were resolved on SDS-10%, SDS-5% polyacrylamide gels and AmershamTMECLTM Gels 4-12% (28-9898-06 GE Healthcare, Sweden) and transferred onto PVDF Membrane (Biorad, Berkeley, CA). The blots were blocked with 5% milk powder in PBS1x with 0.1% Tween-20 for 60 min, washed with PBS1x/Tween, and incubated overnight with the different antibodies anti-poly(ADP-ribose) (PAR) (20591E10, TREVIGEN, My), anti-PARP-1 (C2-10) (ALX-804-210, Alexis Biochemicals, LA), anti-LC3 (PD014, MBL, NY), anti- phospho-p70s6 kinase (Thr389) (#9205, Cell Signaling Technology Beverly, MA), anti-p70s6k (sc-8418, Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-AMPkα (Thr172) (#2535, Cell signalling Beverly, MA), anti-AMPkα (#2603, Cell Signaling, Beverly, MA), anti-phospho-ACC (Ser79) (#3661, Cell signalling Beverly, MA), anti-ATG7 (#2631, Cell signalling Beverly, MA), anti-Beclin1 (612112, BD Biosciences), anti-phospho-ULK1 (Ser317) (#6887, Cell signalling Beverly, MA), anti-ULK1 (#4776, Cell signalling Beverly, MA), anti-phospho-Raptor (Ser792) (#2083, Cell signalling Beverly, MA), anti-Raptor (N-19) (sc-27744, Santa Cruz Biotechnology, Santa Cruz, CA), LAMP1 (sc-20011 Santa Cruz Biotechnology, Santa Cruz, CA) α-tubulin (Sigma-Aldrich), β-Actin (Sigma-Aldrich) and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) were used as loading control. Bands were visualized by ECL-PLUS (Amersham Biosciences) and the pictures were taken with the imaging system Image Quant LAS 4000 (GE Healthcare)and medical X-ray films (AGFA).
Immunofluorescence
Immunostaining for poly (ADP-ribose) (PAR) was performed on cells grown on glass cover-slips (density of 3.5 × 104 cells per well and in 6-well plates) and fixed with 3% paraformaldehyde, 2% sucrose, PBS1x solution for 10 min RT. Fixed cells were permeabilized with 0.5% Triton-X100, PBS1x solution for 5 min RT and blocked with 2% BSA PBS1x for 1 h at 37 ºC. Poly (ADP-ribose) was detected using monoclonal antibody (PAR) (20591E10, TREVIGEN, My) 1:600 and cy3-conjugated goat anti-mouse immunoglobulin (C2181-1 ML, Sigma-Aldrich) 1: 800 in 2% BSA PBS1x solution. Nuclear counterstaining with DAPI (1:5000 in PBS1x) was performed after removal of excess secondary antibody. Cells were mounted on microscope slides with Vectashield mounting medium for fluorescence (H-1000, Vector, CA). Immunostaining was visualized with a Zeiss Fluorescence Microscope.
ATP determination
Cell lines were plated in 6-well plates with a density of 5 × 105 cells per plate. Intracellular ATP was measured using a luciferin/luciferase-based assay (Adenosine 5'-triphosphate (ATP) Bioluminescent Assay Kit, Sigma-Aldrich) following manufacturer's guidelines. A standard curve was generated rather than known concentrations of ATP and used to calculate sample ATP concentrations. Protein concentration was determined using Bradford protein assay reagents (Bio-Rad). The content of ATP was normalized for protein content and expressed as percentage of control.
Autophagy assays
GFP-LC3-expressing cells have been used to demonstrate induction of autophagy. We used MCF7 cells expressing stably GFPLC3 and the transitory GFP-LC3 expression vector, kindly supplied by Dr. T. Yoshimori, (National Institute for Basic Biology, Okazaki, Japan), in parp-1+/+ and parp-1−/− MEFs cells transiently transfected with this vector with jetPEITM (Polyplus Transfection, Illkirch, France), according to the manufacturer's protocol. The assay was performed on cells grown on glass cover-slips (4 × 104 cells per well) in 6-well plates and after the different treatment with HANK buffer, these cells were washed 3 times with sterile PBS1x and fixed with Paraformaldehyde Solution (4%, wt/vol in PBS1x with 2% Sucrose) for 10 min at room temperature. To determine LC3 localization GFP-LC3-transfected cells were observed under a Zeiss Fluorescence Microscope and to determine the endogenous LC3-II translocation in MCF7 GFPLC3, parp-1+/+ and parp-1−/−MEFs cells, we performed a western blot of LC3-I and its proteolytic derivative LC3-II (18 and 16 KDa, respectively), using a monoclonal antibody against LC3, from MBL (PD014, MBL, NY). Rapamycin and Concanamycin A were used as autophagy inductors, positive controls.
RNA interference
These assays were performed on cells grown on glass cover-slips (6 × 104 cells per well) in 6-well plates for fluorescence microscopy and 9 × 104 cells in 6-well plates for western blot assays in RPMI 1640 GlutaMAX (GIBCO Invitrogen) with 6% FBSi without antibiotic. Cells were transfected with the indicated siRNAs at 50 nM (iBclin1 and iATG7), 60 nM (iPARP-1 and iULK1), 30 nM (iPARG and iLAMP1) using LipofectamineTMreagent (Invitrogen) or JetPrime reagent (Polyplus Transfection, Illkirch, France), according to the manufacturer's guide. SiRNA corresponding to the cDNA sequences were: ATG7, ULK1 from Sigma, Human PARP-1 from Ambion Applied Biosystems and Beclin1, PARG and LAMP1 from Santa Cruz Biotechnology. After 48 h after of transfection, cells were treated as described and observed under a Zeiss Fluorescence Microscope or the proteins were extracted.
Cell viability assay
MCF7 GFPLC3 cells were seeded in 24-well plates with a density of 1.2 × 104 cells per well, with different treatments with HANK buffer. The levels of cell viability were determined using MTT Cell Proliferation Kit I (1-65-007, Roche, Mh, Germany). MTT assay (3-(4, 5-Dimethylthiazol-2-yl)-2,5-diphenyl Tetrazolium Bromide,) was performed following manufacturer's instructions.
Co-Immunoprecipitation
MCF7 GFPLC3 cells were plated in p100 plates with a density of 1 × 106 cells per plate. After starvation, cells were washed 2 times with PBS1x Pefabloc 0.5 mM, resuspended in 200 μl of Lysis Buffer EBC (50 mMTris-HCl pH8, 120 mMNaCl, 0.5% NP-40, 1 mM PMSF, Protease Inhibitor Complete Mini Roche), on ice 30 min, vortex 3 times. Centrifuge 13.000 rpm 20 min 4 °C and obtained the INPUT, the rest of supernatant was incubated with 35 μl of Protein Sepharose A (PAS) 30 min 4 °C rocking. 13.000 rpm 5 min 4 °C to obtain the Pre-Immune (PI) supernatant using anti-Cyt C (sc-7159, Santa Cruz Biotechnology, Santa Cruz, CA) and pellet in 50 μl EBC Buffer with PAS, rocking 1-2 h 4 ºC. Co-IP of AMPk and PARP-1 or AMPk and ULK1 endogenous proteins performed using AMPk, PARP-1 and ULK1 (1:100) antibodies previously described. Co-IP was incubated at 4 °C o/n rocking and centrifuged 13.000 rpm 5 min 4 ºC. Pellets were washed 5 times with 500 μl NET-N buffer per sample (20 mMTris-HCl pH8, 100 MNaCl, 0.5% NP-40, 1 mM EDTA, 1 mM PMSF, Protease Inhibitor Complete Mini Roche) 13.000 rpm 4 °C 5 min. Finally the pellet was resuspended in Loading Buffer 2x and maintained at -20 ºC.
AMPk mutation in PARylation sites
Amino acids which can be potentially PARylated in AMPK were searched in silico and selected according to previously described PARylation consensus sequences and in particular the PAR acceptor aminoacids found for the transcription factor CTCF that were conserved in AMPKα.30 Mutations of such individual amino acids (K to A or E to A as specified in Figure 4a) generated 4 mutants (AMPKPARdef1, 2, 3, 4) and Wild Type constructs were tagged with FLAG oligopeptide and sub-cloned in pCDNA3 expression vector (pCDNA3-AMPKα1-FLAG). Total structure from AMPk alpha 1 protein was screened comparing with different putative PARylated motives in different proteins. Putative PARylation site in AMPK alpha1 subunit (DEALKEVCEKFECSEEEVLS) was identified comparing with CTCF transcription factor, previously described as Poly ADP-Ribose acceptor TEEG-KDVDVSVYDFEEEQQ.31 AMPkα deficient in PARylation1 mutant (AMPKPARdef1) was generated using de novo synthesis and sub cloning in a XhoI site in pCDNA3 expression vector (pCDNA3-AMPKalpha1-FLAG).
AMPkα deficient in PARylation mutants 2, 3 and 4 were carried out using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA). A pcDNA3-flag plasmid carrying the wild type AMPK cds was used as template. Orientation, integrity and fidelity of all the cloned fragments were checked by Sanger sequencing.Plasmids were transfected with JetPEI co-transfection according to the manufacturer's guide.
PARylation assay
ShPARG HeLa cells were plated in p100 plates with a density of 1 × 106 cells per plate and transfected with JetPEI reagent (Polyplus Transfection, Illkirch, France) with the plasmid pcDNA3-FLAG-AMPkα containing the wildtype and mutant forms of AMPk alpha 1 subunit, according to the manufacturer's guide. Twenty-four hours later, cells were starved at different times with HANK Buffer. After starvation, cells were washed 2 times with PBS1x Pefabloc 0.5 mM, resuspended in 500 μl of Lysis Buffer EBC (50 mMTris-HCl pH8, 120 mMNaCl, 0.5% NP-40, 1 mM PMSF, 1 mM Na3VO4, 10 mMNaF, Protease Inhibitor Complete Mini Roche), on ice 20 min. Centrifuge 13.000 rpm 20 min 4 °C and obtained the INPUT, the rest of supernatant was incubated with 35 μl of Protein Sepharose A (PAS) 1 hour 4 °C rocking. IP of endogenous PAR polymer performed using PAR (1:50) polyclonal antibody (4336-APC-050, TREVIGEN, My). IP was incubated at 4 °C o/n rocking and centrifuged 13.000 rpm 5 min 4 ºC. Pellets were washed 4 times with 500 μl of EBC 250-120 mMNaCl. Finally the pellet was resuspended in Loading Buffer 2x and maintained at -20 ºC.
GST-Pulldown
COS1 cells were seeded (1 × 106) on p100 plates and transfected with JetPEITM co-transfection (Polyplus Transfection, Illkirch, France) with the plasmids expressing FLAG-AMPkαwildtype and mutant, GST-PARP-1 full fusion proteins and ABC, D and EF PARP-1 domains and GST fusion protein as transfection control; according to the manufacturer's guide. Forty-eight hours later, cells were washed with PBS1x Pefabloc and lysed in B buffer (20 nMTris-HCl (pH 7.5), 400 mMNaCl, 20% glycerol, 5 mM DTT, 0.5 mMPefabloc,NP40 0.5%, protease inhibitors (Complete Mini; Roche, Mannheim, Germany)). Lysates were cleared by centrifugation (10.000 rpm 15 min 4 ºC) and incubated for 2 h with glutathione-Sepharose 4B (Amersham). Beads were washed 3 times with DB buffer (200 mMTris-HCl (pH 7.5), 150 to 500 nMNaCl, 0.1% NP-40, and protease inhibitors). All samples were resuspended in charge buffer, boiled for 5 min and analysed western blot. Blots were subsequently incubated with anti-GST antibody (A190-121A, Bethyl Lab, Mt, Texas) and anti-FLAG (F3175 Clone M2, Sigma-Aldrich).
Confocal microscopy co-localization
MCF7 cells were grown on cover-slips (density 4.5 × 103) and starved at different times. Cells were fixed with 3% paraformaldehyde, 2% sucrose, PBS1x solution for 10 min RT. Fixed cells were permeabilized with 0.5% Triton-X100, PBS1x solution for 5 min RT and blocked with 2% BSA PBS1x for 1 hour at 37 ºC. Incubation with primary antibodies anti-AMPkand anti-PARP-1 were performed in 2% BSA PBS1x for 30 min at 37 ºC, and second antibodies (FITC labelled anti-mouse, Cy3 labelled anti-rabbit IgG, both from SIGMA) in 2% BSA PBS1x for 20 min at 37 ºC. After extensive washing with PBS1x, coverslips were mounted with Vectashield mounting medium. Images were captured on a Leica LCS SP5 confocal microscope. Colocalization percentages were quantifying the immunofluorescence obtained from the z-stacks on every image. The bar chart shows the percentage of cells in which colocalization foci were observed at control or upon treatment with HANK buffer. *P<0.05.
Subfractionation cytosol nucleus
MCF7 GFPLC3 cells were seeded in p100 plates (density 1 × 106) and starved at different times. To obtain the nuclear and cytosolic fraction, cells were washed 2 times with PBS1x and lysed with 300 μl of Buffer A (0.5 M Hepes-KOH pH7.8, 1 M MgCl2, 1 M KCl, 1M DTT). Cells in eppendorfs were gently pipetting up and down 10 times and were incubated in ice 10 min. All samples were mixed with 10% TritonX-100 (vortex 30 s) and were precipitated 11.000 rpm 4 °C during 1 min. Take off the supernatant (Cytosolic Extract CE) and the pellet (Nuclear Extract NE). The NE fraction was washed with 100 μl of Buffer A 11.000 rpm 4 °C 1 min. NE extracts were mixed with 1/5 vol of Buffer C (0.5 M Hepes pH7.8, Glycerol, 5 M NaCl, 1 M MgCl2, 0.25 M EDTA, 1 M DTT) and CE extracts with 1/10 vol of Buffer B (0.5 M Hepes pH7.8, 1 M KCl, 1 M MgCl2). Finally all the samples were precipitated 15.000 g 4 °C 15 min. All the supernatants were mixed with loading buffer and maintained at -20 ºC.
Acknowledgments
We would like to acknowledge Laura López and Salvador Guerrero for technical assistance. JMRV has been in part funded by ASTF 32-2014 (Short-Term Fellowships EMBO Program 2014) and Ramón Areces Foundation (Postdoctoral Grants Program 2015); JMM has been funded by a predoctoral fellowship by the programme FPU, Spanish Ministry of Education Culture and Sport. This work was supported by Junta de Andalucía, project of Excellence from Junta de Andalucía P07-CTS-0239, P10-CTS-0662, to FJO, Spanish Ministry of Economy and Competitiveness SAF2012-40011-C02-01, SAF2015-70520- R to FJO, BIO-778) and SAF2015-64383-P to ALR, and RTICC RD12/0036/0026 to ALR and FJO. GI, FD and VS were supported by LABEX ANR-10-LABX-0034-Medalis.
Glossary
- 3-MA
3-Methyl Adenine
- AICAR
5-Aminoimidazole-4-carboxamide 1-β-D-ribofuranoside
- ACC
Acetyl-CoA carboxylase
- ATG7
Autophagy-Related Gen 7
- AMP
Adenosine Monophosphate
- AMPk
AMP-Activated Kinase
- ATGs
Autophagy Related Genes
- ATP
Adenosin 5'-Triphosphate
- Crm1
Nuclear Export Receptor 1
- CTCF
Transcriptional repressor CTCF or 11-zinc finger protein
- DMSO
Dimethyl sulfoxide
- DPQ
(3,4-Dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone])
- HANK
Hank's Balanced Salt Solution
- HCQ
Hidroxy-Chloroquine
- LAMP1/2
Lysosomal-Associated Membrane Protein ½
- LC3
Light Chain of the Microtubule-Associated Protein 1
- LKB1
Liver Kinase B1
- MEFs
Murine Embrionary Fibroblasts
- mTOR
Mammalian Target of Rapamycin
- MTT
3-(4, 5-Dimethylthiazol-2-yl)-2,5-diphenyl Tetrazolium Bromide
- NAD+
Nicotinamide adenine dinucleotide
- p70S6k
Ribosomal protein S6 kinase beta-1 (S6K1)
- PAR
Poly ADP-Ribose
- PARG
Poly (ADP-Ribose) Glycohydrolase
- PARP-1
Poly (ADP-Ribose) Polymerase 1
- PBS
Phosphate-buffered saline
- PI3K
Type III Phosphatidylinositol 3-kinases
- PJ34
([N-(6-Oxo-5,6-dihydro-phenanthridin-2-yl)-N,N- dimethylacetamide])
- PTM
Post-Translational Modification
- ROS
Reactive species of oxygen
- ULK1
Unc-51-Like Kinase 1 or ATG1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev 2011; 25: 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews 2012; 11: 709–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 2012; 36: 933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F et al. Autophagy in malignant transformation and cancer progression. The EMBO journal 34: 856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7: 517–528. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Vargas JM, Ruiz-Magaña MJ, Ruiz-Ruiz C, Majuelos-Melguizo J, Peralta-Leal A, Rodríguez MI et al. ROS-induced DNA damage and PARP-1 are required for optimal induction of starvation-induced autophagy. Cell Research 2012; 22: 1181–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Molecular cell biology 2012; 13: 411–424. [DOI] [PubMed] [Google Scholar]
- Kalisch T, Ame JC, Dantzer F, Schreiber V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends in biochemical sciences 2012; 37: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Gamez JA, Rodríguez-Vargas JM, Quiles-Pérez R, Aguilar-Quesada R, Martín-Oliva D, de Murcia G et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 2009; 5: 61–74. [DOI] [PubMed] [Google Scholar]
- Chang PC, Wang TY, Chang YT, Chu CY, Lee CL, Hsu HW et al. Autophagy pathway is required for IL-6 induced neuroendocrine differentiation and chemoresistance of prostate cancer LNCaP cells. PloS one 2014; 9: e88556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci 2009; 14: 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi K, Bai P, Kovács I, Szabó E, Mocsár G, Kakuk A et al. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 2009; 23: 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clohessy JG, Reschke M, Pandolfi PP. Found in translation of mTOR signaling. Cell Res. 2012; 22: 1315–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology 2011; 13: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta physiologica 2009; 196: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Molecular and cellular biology 1998; 18: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JW, Jijon HB, Madsen KL. AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun 2006; 342: 336–341. [DOI] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nature reviews 2014; 15: 155–162. [DOI] [PubMed] [Google Scholar]
- Carling D, Viollet B. Beyond energy homeostasis: the expanding role of AMP-activated protein kinase in regulating metabolism. Cell Metab 2015; 21: 799–804. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 2015; 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015; 11: 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongol B, Marin T, Peng IC, Woo B, Martin M, King S et al. AMPKalpha2 exerts its anti-inflammatory effects through PARP-1 and Bcl-6. Proc Natl Acad Sci U S A 2013; 110: 3161–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012; 12: 587–598. [DOI] [PubMed] [Google Scholar]
- Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-Specific Initiation of Autophagy. Mol Cell 2015; 59: 522–539. [DOI] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 2014; 157: 882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell 2013; 153: 614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M et al. Regulation of autophagy by cytoplasmic p53. Nature cell biology 2008; 10: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar D, Chernukhin I, Klenova E. Generation of poly(ADP-ribosyl)ation deficient mutants of the transcription factor, CTCF. Methods in molecular biology 2011; 780: 293–312. [DOI] [PubMed] [Google Scholar]
- Farrar D, Rai S, Chernukhin I, Jagodic M, Ito Y, Yammine S et al. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol 2010; 30: 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.