Abstract
Regardless of its etiology, once septic shock is established, survival rates drop by 7.6% for every hour antibiotic therapy is delayed. The early identification of the cause of infection and prognostic stratification of patients with sepsis are therefore important clinical priorities. Biomarkers are potentially valuable clinical tools in this context, but to date, no single biomarker has been shown to perform adequately. Hence, in an effort to discover novel diagnostic and prognostic markers in sepsis, new genomic approaches have been employed. As a result, a number of small regulatory molecules called microRNAs (miRNAs) have been identified as key regulators of the inflammatory response. Although deregulated miRNA expression is increasingly well described, the pathophysiological roles of these molecules in sepsis have yet to be fully defined. Moreover, non-human miRNAs, including two Kaposi Sarcoma herpesvirus-encoded miRNAs, are implicated in sepsis and may drive enhanced secretion of pro-inflammatory and anti-inflammatory cytokines exacerbating sepsis. A better understanding of the mechanism of action of both cellular and viral miRNAs, and their interactions with immune and inflammatory cascades, may therefore identify novel therapeutic targets in sepsis and make biomarker-guided therapy a realistic prospect.
Facts
Early diagnosis is a constant challenge in sepsis, as late diagnosis results in delayed therapy and increased mortality.
miRNAs work also as regulators of the immune response, with potentially important translational implications in sepsis.
Specific cellular and viral miRNAs expression is strongly associated with poor prognosis in sepsis.
DNA virus-encoded miRNAs (e.g., KSHV-miR-K-10b, KSHV-miR-K12-12*) are involved in sepsis by interacting with Toll-like receptor 8 (TLR8) as agonists establishing a positive feedback that may promote the sepsis-induced cytokine storm leading to increased inflammation and subsequent deadly immunosuppression.
Open questions
What are the underlying mechanisms of the TLR binding-mediated agonistic activity of cellular and viral miRNAs in triggering or enhancing sepsis progression?
Are there any therapeutic advantages to target cellular or viral miRNAs as part of new strategies for the treatment of sepsis?
Introduction
Sepsis is defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection.1 It is clinically diagnosed by the presence of infection with signs/and symptoms of the systemic inflammatory response syndrome (SIRS). Sepsis is a major cause of death with a total of 18 million people worldwide that develop sepsis every year.1, 2 In developed countries, the incidence of sepsis is 2% of all hospitalized patients and 6–30% of patients in intensive care units (ICU),3 while the incidence in developing countries is higher.4 In the US alone, sepsis is responsible for more than 750 000 deaths in the ICUs,5, 6 and linked to an estimated annual cost of over $20 billion and increasing.7, 8 In fact, recent reports have shown that overall mortality from sepsis exceeds that of many common cancers and approaches parity with heart diseases.7 Despite all these frightening data, the therapeutic options did not address any pathogenic mechanism except the bacterial or fungal infection.
The complex etiology of sepsis can be attributed to both community-acquired and health-care-associated infections in most of the cases with Gram-positive bacteria (e.g., Staphylococcus aureus, Streptococcus pneumoniae), Gram-negative bacteria (e.g., Escherichia coli, Klebsiella species, Pseudomonas aeruginosa) or fungi.9, 10 Pneumonia, intra-abdominal and urinary tract infections are considered to be some of the most common causes of sepsis.5, 6, 11, 12 Regardless of the etiology of sepsis, the clinical picture is dominated by a very severe reaction of the immune system with activation of pro-inflammatory cascades and compensatory anti-inflammatory response that phenotypically defines a final common pathway which leads to organ failure and death. Characteristic features of the hyper-inflammatory phase include increased production of pro-inflammatory cytokines, such as IL-1, IL-6, IL-8 and tumor necrosis factor α (TNF-α). By contrast, the compensatory anti-inflammatory response syndrome that characterizes the immunoparalysis phase, involves the production of IL-4, IL-10, IL-13, transforming growth factor-β (TGF-β), granulocyte-colony-stimulating factor and granulocyte–macrophage colony-stimulating factor (GM-CSF), soluble TNF-α receptors and IL-1 receptor antagonists.13, 14 Blood lymphocyte dysfunction during sepsis has long been recognized with significant lymphopenia and decreased lymphocyte CD4+, CD8+ T cells, natural killer (NK) cells and B cells.15
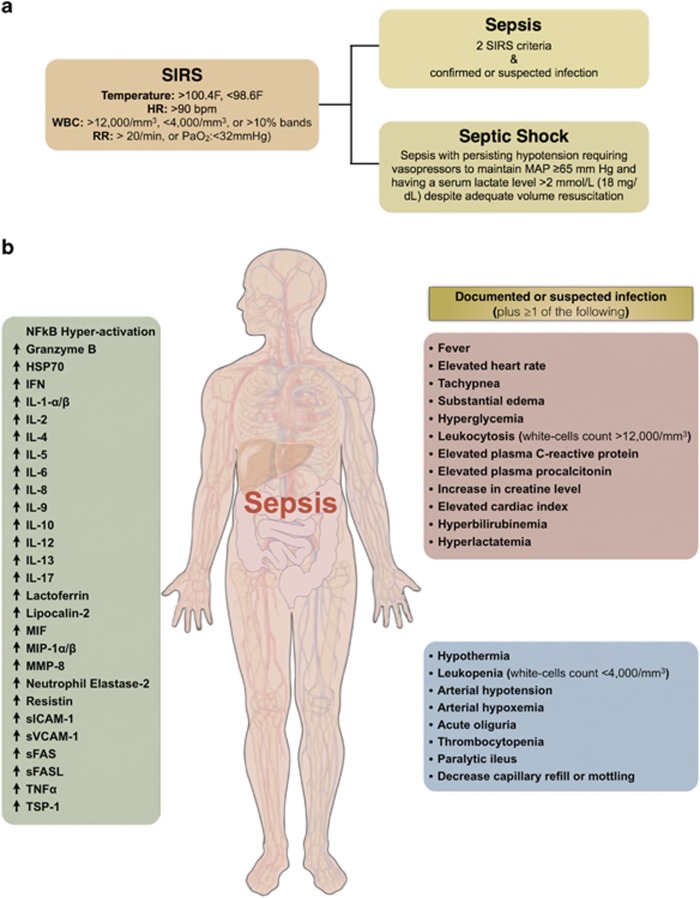
Owing to the fact that sepsis presents highly variable clinical manifestations (Figure 1), it is often difficult to provide an accurate early diagnosis.6, 16, 17 Late diagnosis and delayed therapy increase mortality considerably. Septic shock decreases the survival rate by 7.6% for every hour of delay of the appropriate antibiotic therapy.18 Inadequacy in the current understanding of sepsis pathophysiology remains evident by the lack of success in clinical trials for new therapeutic agents. Although there are many published research findings about biomarkers designed to identify sepsis at the earliest stage and about possible treatment strategies, many of them have failed to gain acceptance or be effective in clinical practice.19 Furthermore, since Eli Lilly voluntarily withdrew drotrecogin alfa (Xigris) from the market due to failure to show a survival benefit, there is currently no FDA approved drug to mitigate the damaging effects of dysregulated inflammation associated with sepsis.9 Therefore, timely intervention and accurate identification of sepsis are important priorities for both the biomedical research and biopharmaceutical industries.18
Figure 1.
Clinical manifestation of sepsis in patients. (a) Diagnostic criteria of systemic inflammatory response syndrome (SIRS), sepsis and septic shock. (b) The detailed diagnostic criteria for sepsis include the documentation or suspicion of possible infection with at least one or more other clinical manifestation as presented in the red and blue boxes. Sepsis also has been correlated with NF-κB signaling hyper-activation, secretion of pro-inflammatory (e.g., Eotaxin, IFNγ, IL-1α, IL-1β, IL-2, IL-5, IL-6, IL-8, IL-9, IL-12, IL-17, MIF, MIP-1α/β, TNFα) and anti-inflammatory (e.g., IL-4, IL-10, IL-13) cytokines, and secretion of other inflammatory biomarkers (Granzyme B, HSP70, Lactoferrin, Lipocalin-2, MMP-8, Neutrophil Elastase-2, Resistin, sFAS, sFASL, sICAM-1, sVCAM-1, TSP-1) as presented in the green box. Red arrow represents direct or indirect stimulation/upregulation, while dark blue line with flat end represents direct or indirect inhibition/downregulation. HSP70, heat-shock 70 kDa protein; IFNγ, interferon gamma; IL-1α, interleukin-1 alpha; IL-1β, interleukin-1 beta; IL-2, interleukin 2; IL-4, interleukin 4; IL-5, interleukin 5; IL-6, interleukin 6; IL-8, interleukin 8; IL-9, interleukin 9; IL-10, interleukin 10; IL-12, interleukin 12; IL-13, interleukin 13; IL-17, interleukin 17; MIF, macrophage migration inhibitory factor; MIP-1α/β, macrophage inflammatory protein 1 alpha and macrophage inflammatory protein 1 beta; MMP-8, matrix metalloproteinase 8 (also known as neutrophil collagenase); sFAS, soluble Fas (also known as soluble apoptosis antigen 1); sFASL, soluble Fas ligand; sICAM-1, soluble cell adhesion molecules 1; sVCAM-1, soluble vascular adhesion molecules 1; TNFα, tumor necrosis factor alpha; TSP-1, thrombospondin 1
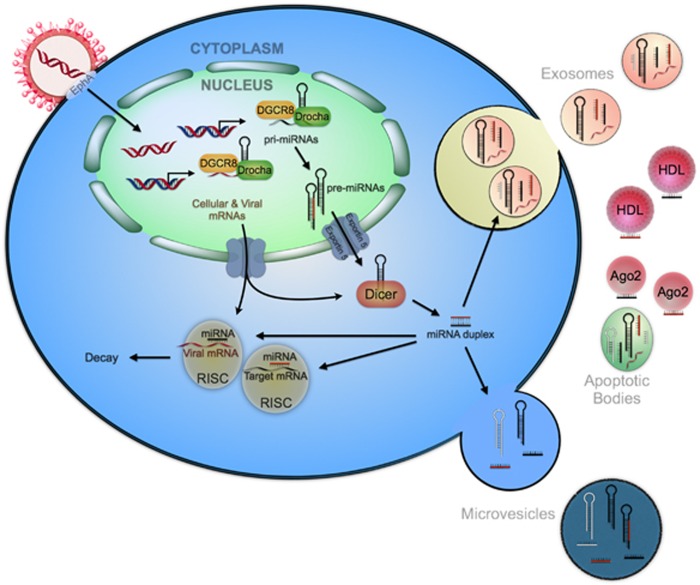
In a recent development, new genomic approaches involving DNA and RNA profiling have also been employed to identify novel classes of molecules such as microRNAs (miRNAs) as regulators of the immune response, with potentially important translational implications in sepsis.20 miRNAs are small (18–24 nucleotides) non-coding RNAs (ncRNAs), which function is to inhibit protein synthesis by degrading or inhibiting translation of messenger RNA (mRNA).21 Interaction between the miRNA and the mRNA (miRNA:mRNA) occurs in the 3′-untranslated region (3′-UTR) of the target mRNA. However, miRNAs can also bind to other mRNA domains. As a result of this interaction, both translational repression and mRNA cleavage can occur, and consequently protein expression is suppressed. Each miRNA can regulate multiple gene targets, while multiple miRNAs may target the same protein coding mRNA. Interestingly, miRNAs can also switch between repression and activation of the translation of targeted mRNAs 21, 22, 23 (Figure 2).
Figure 2.
Cellular and viral microRNAs biogenesis. The viral genome is integrated into the human genome to guarantee the preservation of genetic information codifying for viral genes and miRNAs. Cellular and viral long primary miRNA transcripts (pri-miRNAs) are transcribed by RNA polymerase II. Subsequently, the pri-miRNAs are processed by RNase III-type enzyme Drosha to yield a hairpin precursors (pre-miRNA, 70 nucleotides). After the pre-miRNA hairpins are exported to the cytoplasm mediated by exportin 5, the RNase III protein Dicer further processes them into unstable miRNA duplex structures (19–25 nucleotides).75, 76 Mature miRNAs are either incorporated into a multiple-protein nuclease complex (RNA-induced silencing complex, RISC) to regulate its target mRNAs, or released out of the cells in exosomes, microvesicles or apoptotic bodies, or bond to some high-density lipoprotein (HDL) and Argonaute protein 2 (Ago2). Viral miRNAs are processed in the same fashion as human cellular microRNAs modulating viral and/or host gene expression. Human cells lack the ability to distinguish between viral and human miRNAs sequences
In this comprehensive review we will discuss the most important studies that have identified miRNAs to be differentially expressed in sepsis, in cellular and animal models, and septic patients.24 We will present possible functional roles of miRNAs in the pathogenesis of sepsis and highlight promising avenues of potential clinical translation. Moreover, we will also summarize the most relevant clinical studies published by now on adult septic patients that correlated cellular and viral miRNAs expression with sepsis (Table 1), and summarize the mechanisms of actions for the immune miRNAs identified and proposed as sepsis biomarkers (Table 2). One of the greatest advantages when designing future diagnostic and therapeutic strategies in sepsis using miRNAs is their ability to target multiple components of the immune response pathways that results in an additive, stronger response. The current finding of the unexpected expression of viral miRNAs in septic patients is intriguing and provides arguments for further investigations of their implication in clinically relevant immunosuppression state in sepsis.25 If the viral loads are markedly elevated, then it is possible that the viral miRNAs may contribute to the pathology of sepsis. One probable mechanism by which viral miRNAs might be involved in sepsis is by functional mimicry mechanisms with cellular miRNAs produced by the human genome, sharing the regulation of same signaling pathways and regulating the same spectrum of mRNAs – target mimicry.
Table 1. Circulating miRNAs identified as a biomarker in different blood specimens.
| Number of patients |
miRNA expression |
References | |
|---|---|---|---|
| Increased in sepsis | Decreased in sepsis | ||
| 17 septic patients 32 healthy individuals | miR-486, miR-182 | miR-150, miR-342-5p | 20 |
| 50 septic patients 30 SIRS patients 32 healthy individuals | miR-223 and miR-146a (septic vs. SIRS) | 77 | |
| Patients with sepsis: 78 surviving 64 non-surviving | miR-297 (non-surviving) | miR-574-5p (non-surviving) | 48 |
| Patients with sepsis: 117 surviving 97 non-surviving | miR-15a, miR-122, miR-193, miR-483-5p (non-surviving) | miR-16, miR-223 (non-surviving) | 63 |
| 17 ICU non-septic and 36 septic patients | miR-181b (septic) | 78, 79 | |
| 166 septic patients 32 SIRS patients 24 healthy individuals | miR-15a (septic<SIRS) miR-116 (septic & SIRS) | 80 | |
| 43 septic patients 123 septic shock 24 healthy individuals | miR-15b (septic) miR-223 (septic & septic shock) miR-483-5p (septic) | miR-122 (septic & septic shock) miR-193b* (septic & septic shock) miR-499-5p (septic shock) | 81 |
| 5 septic patients 3 healthy individuals | miR-466I | 49 | |
| 22 septic patients 22 SIRS patients 17 healthy individuals | miR-122 (septic & septic shock) miR-4772 family (septic & SIRS) | miR-150, miR-342-3p, miR-3173-5p, miR-191-iso (septic & SIRS) | 64 |
| 138 septic patients 85 non-septic patients 76 healthy individuals | miR-150 (septic or non-septic ICU patients with death) | 47 | |
| 14 septic patients 14 SIRS patients | miR-146a (septic) | 62 | |
| 138 septic patients 85 non-septic patients 76 healthy individuals | miR-133a (septic) | 50 | |
| 99 septic patients 84 surgical patients 53 healthy individuals | miR-16-5p, miR-93-5p, miR-182-5p, miR-486-5p, KSHV-miR-K12-12-5p | miR-23a-3p, miR-26a-5p, miR-26b-5p, miR-146a-5p, miR-342-3p, miR-150-5p, KSHV-miR-K12-10b | 25 |
| 232 septic patients 24 healthy individuals | miR-122, miR-193b, miR-483-5p, miR-574-5p, | 82 | |
| 123 septic patients | miR-122 (coagulation disorder) | 83 | |
| 40 septic children 20 non-septic children 15 healthy individuals | miR-146a, miR-223 | 84 | |
| 138 septic patients 85 non-septic patients 76 healthy individuals | miR-122 | 85 | |
| 29 septic patients 40 septic shock 24 healthy individuals | miR-150 (septic shock) | 86 | |
| 22 septic patients 20 healthy individuals | let-7a, miR-150 | 87 | |
| 138 septic patients 85 non-septic patients 76 healthy controls | miR-15a, miR1622 | 88 | |
| 70 septic patients 30 SIRS patients | miR-25 (septic) | 89 | |
| 103 septic patients 95 SIRS patients 40 healthy controls | miR-143 (high correlation to SOFA scores ≥7 and APACHE II scores ≥10) | 65 | |
Abbreviations: SIRS, systemic inflammation response syndrome; KSHV, Kaposi sarcoma herpesvirus.
Table 2. The mechanisms of action for the immune miRNAs identified as sepsis biomarkers.
| miRNA | Sepsis involvement & mechanism of action | References |
|---|---|---|
| Let-7i | Negatively regulates TLR4 | 38 |
| miR-9 | Negatively regulate NF-κB (p50 subunit) | 90 |
| miR-15a/b, miR-16 | miR-15a and miR-16 promote NF-κB signaling by negatively regulating IKK-α. miR-15a potentially inhibits VEGFA, VEGFC and MYLK, key genes in increasing vascular permeability. It also target IRAK2 and NFKB1 in the NF-κB pathway, inhibiting these genes and thus inhibiting NF-κB. miR-15b induces p53phosphorylation, apoptosis, DNA repair and cell-cycle arrest | 77, 91, 92 |
| miR-21 | Regulates expression of PDCD4, increases IL-10 production and serves as a agonist of human endosomal TRL8 (murine TRL7). miR-21-3p is upregulated in cardiac tissue under LPS-induced sepsis | 66, 68, 93 |
| miR-23a-5p | Upregulated in sepsis patients; involved in acute respiratory distress syndrome, a byproduct of sepsis | 67 |
| miR-23b | Regulates NF-κB, TNFα, IL-6, ICAM-1, E-Selectin and VCAM-1 | 94 |
| miR-25 | Increases cell proliferation by targeting and inhibiting expression of CDKN1C protein; promotes cell survival by targeting pro-apoptotic proteins such as BIM, BAX and Caspase3 | 95, 96 |
| miR-29a | Agonist of human endosomal TRL8 (murine TRL7) | 93 |
| miR-33 | Decreases TNF-alpha and IL1beta expression | 49 |
| miR-93-5p | Targets STAT3, PTEN | 97, 98 |
| miR-122 | Suppresses interferon-stimulated response element (ISRE), which increases cellular response to interferon (IFN); increases expression of SOCS3 (suppressor of cytokine signaling 3) via promoter methylation | 99 |
| miR-133a | Increased in sepsis; targets EGFR and IGF1R | 50, 100, 101 |
| miR-143 | Increased in sepsis with high correlation to SOFA and APACHE II scores; targets TNF-alpha and IL-13 receptor | 65, 102 |
| miR-146 family (146a and 146b) | Negatively regulates IRAK1 and TRAF6; miR-146 upregulates IL-6 secretion by binding to the 3′-UTR of IL-6 mRNA; miR-146b negatively regulates TLR4 and MYD88 | 28, 33 |
| miR-149, miR-203 | Negatively regulates MYD88 | 39, 40 |
| miR-150 | Decreased in sepsis. Negatively regulates colony-stimulating factor 1 receptor (CSF1R) | 20, 47, 53 |
| miR-155 | Enhances LPS-induced translation of TNF-alpha | 27 |
| miR-181b | Targets importin-α3 inhibiting canonical NF-κB signaling in endothelial cells | 78, 79 |
| miR-193b | Inhibits TGF-beta2 signaling pathway | 103 |
| miR-195 | A member of the miR-15 family increased in sepsis. Increases multi-organ injury and worsens the survival in sepsis; negatively regulates survival factors (e.g., BCL-2, SIRT1 and PIM1) and prevents apoptosis | 104 |
| miR-199a | Negatively regulates IKK-beta promoting NF-κB signaling activation | 42 |
| miR-200 family (-200a, -200b, -200c) | Regulates TLR signaling and NF-κB; miR-200b and miR-200c negatively regulates MYD88 in differentiated monocyte THP-1 cell line | 30 |
| miR-221, miR-579, miR-125b | Targets pro-inflammatory cytokine TNF-alpha | 105 |
| miR-223 | Increased in sepsis. Negatively regulates IKK-alpha promoting NF-κB signaling activation | 63, 77, 106 |
| miR-483-5p | Targets TGF-B, Notch3 and MAPK3. Notch pathway in upregulated in septic shock. | 107, 108, 109 |
| miR-486 | Targets CD40 and TMED1 | 110 |
| miR-499-5p | Decreased in septic shock. | 81 |
| miR-574-5p | Upregulated by TLR9, which is associated with increased mortality in sepsis; increases cell-cycle progression by targeting checkpoint suppressor 1 (Ches1). | 111, 112 |
| miR-758-3p | Negatively regulate TLR7 | 113 |
| miR-4772-5p-iso | Increased in sepsis. | 64 |
| KSHV-miR-K12-10b, KSHV-miR-K12-12* | Direct agonists of TLR8 stimulating the production of IL-6 and IL-10 | 25 |
Cellular miRNAs mediating immune pathways in sepsis
In this subsection, we will first review the molecular and animal research studies that discuss the miRNAs with modified expression in experimentally induced sepsis (e.g., endotoxin tolerance studies), and their potential roles regulating inflammation. The immune response in endotoxin tolerance experiments is considered to be similar to the immune response in septic patients.26, 27 This can bring further insights on how miRNAs are mechanistically involved in regulating the expression of important targets within the immune signaling pathways. Innate immune cells like monocytes/macrophages, after being exposed to small amounts of endotoxin, become refractory to subsequent endotoxin challenge (‘endotoxin tolerance'), which resembles the ‘immunosuppression' state in sepsis.
The earliest study on expression profiling of miRNAs in human monocytes identified several miRNAs (e.g., miR-146a/b, miR-132 and miR-155) that respond to endotoxin exposure.28 These findings show that immune cells respond to inflammatory insult and that involves miRNA deregulation. Dysregulated miRNAs act on specific immune-regulatory genes, and play key roles in regulating Toll-like receptor (TLR) signaling pathway, which was shown to be upregulated in critically ill patients,29 and the NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells), which is involved in regulating the transcription of many of the immunomodulatory mediators involved in the development of sepsis-induced organ failure.30, 31
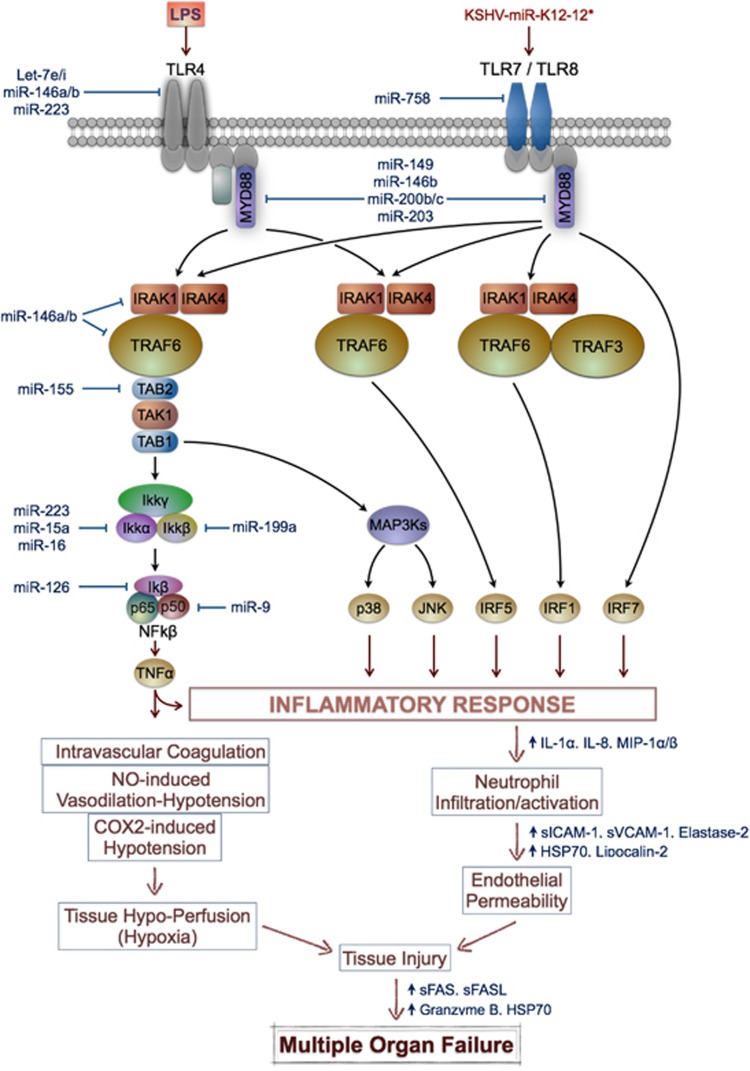
We will further summarize some of the known key miRNAs playing important roles in the TLR signaling and NF-κB inflammatory response (Figure 3). miRNAs such as miR-146a, miR-146b and miR-155 play an important role in regulating the immune response to bacteria by modulating components of NF-κB signaling.28, 32, 33 MiR-146 regulates TRAF6 and IRAK1, suggesting that miR-146 participates in a negative feedback loop to control NF-κB signaling in monocytes.34 Lipopolysaccharide (LPS) also increases the expression of miR-155, further targeting the SRC homology-2 domain-containing inositol 5-phosphatase 1 (SHIP-1), which is itself a negative regulator of NF-κB signaling.35 These findings have been validated in endotoxin tolerance studies, in which pretreated cells with bacterial LPS develop LPS tolerance, and miR-146 in particular was shown to be involved in TLR4/MYD88/IRAK1/TRAF6 signaling.28, 33
Figure 3.
Involvement of cellular miRNAs in the signaling pathway of the immune response in sepsis. Cellular immune miRNAs target important components of the NF-κB signaling pathway at different levels regulating the inflammatory response in the pathogenesis of sepsis. Lower part of the figure illustrates the pathophysiological events in sepsis that lead to tissue injury and subsequent multiple organs failure
TLRs are components of the innate immune system, expressed on macrophages, dendritic cells and various non-professional antigen-presenting cells, and recognize pathogen-associated molecular patterns (PAMPs), expressed by microbial pathogens, or danger-associated molecular patterns (DAMPs) that are released from necrotic or dying cells.36 TLR4 and other cellular surface receptors on mononuclear cells and neutrophils are dynamically regulated by LPS from Gram-negative bacteria across the different stages of sepsis and once engaged, these molecules stimulate signaling pathways resulting in the activation of downstream gene transcription factors, of which NF-κB is central.27 Moreover, the intensity of TLR stimulation was shown to have a prognostic value in the most severe patients, because activation of NF-κB has been reported to be higher in non-survivors than in survivors from septic shock.37
It was reported that let-7i miRNA regulates TLR4 expression in an in vitro model of human biliary cryptosporidiosis in H69 cells contributing to epithelial immune response against C. parvum infection.38 This highlights the importance of miRNA-mediated post-transcriptional pathways in host-cell regulatory responses to microbial infection in general. Other regulators of TLR4 include the miR-200 family members such as miR-200a, miR-200b and miR-200c.30 The myeloid differentiation primary response 88 (MYD88) is a cytosolic adapter protein that plays a central role in the innate and adaptive immune response. MiR-200b and miR-200c can interrupt TLR signaling by directly targeting the 3′UTR of MYD88, which is an essential signal transducer in the IL1R/TLR pathway.24 Interestingly, miR-149 can negatively regulate MYD88 expression upon stimulation with Mycobacterium bovis,39 whereas overexpression of miR-203 can significantly reduce protein levels rather than mRNA levels of MYD88 in RAW264.7 cells.40
Another important component of the NF-κB mediated inflammatory response is the IKK complex, which consists of IKK-α, IKK-β and IKK-γ (NEMO) subunits. MiR-15a, miR-16 and miR-223 were found to target the IKK-α gene, and decreased expression of these miRNAs led to an increase in IKK-α levels in human monocytes in vitro when stimulated with GM-CSF.41 Using target prediction algorithms and subsequent experimental validation, miR-199a and miR-126 were found to share sequence complementarity with the IKK-β and IκBα subunits, respectively.42
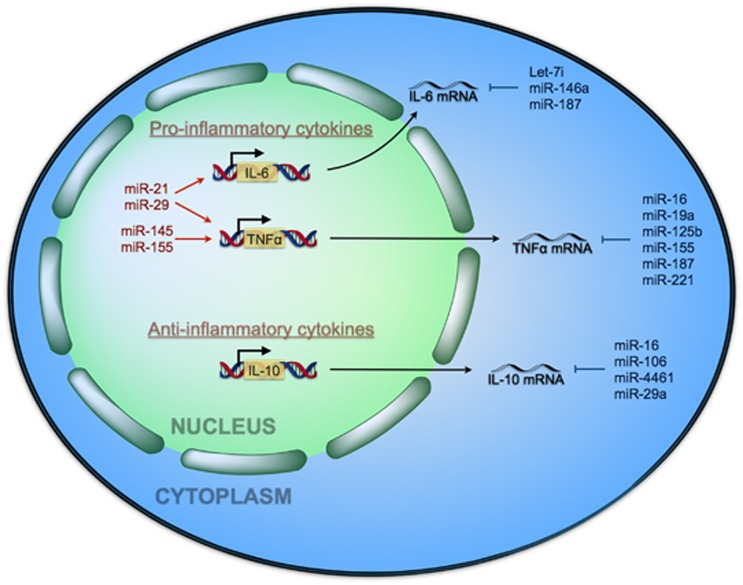
miR-221 was found to target TNF-α promoting its mRNA degradation, while miR-125 and miR-579 were found to target TNF-α by reducing its protein levels in LPS-tolerant THP-1 cells.26 Evidence of direct interaction of miR-125b to the TNFα 3′-UTR was supported by 3′-UTR-TNFα reporter gene experiments in HEK-293 cells.27 Moreover, ectopic expression of miR-146a/b decreases IL-6 secretion in primary human fibroblast,43 supporting the role of miRNAs regulating cytokine production contributing to the immune response and consequent pathogenesis in sepsis. In response to pro-inflammatory storm, the secretion of IL-10 protects cell from damage through JAK2-STAT3, while miR-29a can inhibit IL-10-induced cytokine release. Several miRNAs has been reported to be integrated in the pathophysiology of sepsis by regulating the expression of NF-κB downstream genes involved in the pro-inflammatory signaling (e.g., IL-6, by let-7i or miR-146a, and TNFα by miR-16, miR-125 or miR-155) and in the anti-inflammatory signaling (e.g., IL-10 by miR-16 or miR-29a) playing an important role in sepsis (see Figure 4). Although several mRNAs encoding for components of the TLR and NF-κB signaling pathways were found to be targeted by these miRNAs, the clinical implications of the abnormal miRNAs expression in sepsis has to be further investigated.
Figure 4.
Involvement of cellular miRNAs modulating pro-inflammatory cytokines of the immune response involved in sepsis. Several miRNAs have been reported to be integrated in the pathophysiology of sepsis by targeting (promoting or inhibiting) the expression of NF-κB downstream genes involved in the pro-inflammatory signaling (e.g., IL-6, TNF-α) and in the anti-inflammatory signaling (e.g., IL-10) playing an important role in sepsis
miRNAs as chemically stable biomarkers in septic patients: the lessons learned from the clinical studies
The study of the circulating microRNAome (defined as the full spectrum of expressed miRNAs) in SIRS and sepsis is consistent with simultaneously increased expression of genes involved in the systemic inflammatory, innate immune and compensatory anti-inflammatory responses.29 This gene expression pattern is easy to detect in the peripheral blood cells/plasma/serum of septic patients. Although, a non-coding RNA signature for diagnosing sepsis was proposed, there is yet no consensus regarding whether the use of a single miRNAs (validated in several clinical studies) or a whole panel of miRNAs will be of better use in clinical practice. Currently, there are not many ongoing clinical trials to investigate the potential therapeutic role of miRNAs. It is not yet clear if all the miRNA biomarkers proposed for sepsis perform as well or superior to other biomarkers (e.g., lactate, procalcitonin) that are currently used. Therefore, it is important to evaluate the dimension of these basic research findings and to identify how they can be integrated into current clinical practice.
In comparison with other biomarkers, miRNAs are stable in the circulation. Their association with different RNA-binding proteins and lipoprotein complexes, and their inclusion in microparticles make them resistant to RNAse, severe changes in temperature, repetitive freezing and thawing, and changes in pH.44 The presence of circulating miRNAs in body fluids is the result of a number of mechanisms: the passive release of miRNAs from cellular fragments/debris after cell death from apoptosis or necrosis,45 the active shedding of cell-derived microvesicles, that include exosomes,46 and active secretion by cells as protein-bound conjugates.44 In sepsis, it is not yet clear what cells release the miRNAs in blood samples of septic patients and by which mechanisms miRNAs made their way into the circulation. However, because of their relative small molecular size and lack of post-transcriptional processing, it was hypothesized that miRNAs might be a reliable class of prognostic and targeted biomarkers in sepsis.
A number of studies profiled the miRNAs expression in septic patients and found to be differentially expressed in sepsis versus healthy controls, and in non-survivors versus survivors. miR-146a, miR-150, miR-223, miR-574-5p and other miRNAs expressed in different type of samples (blood cells/plasma/serum) were shown to have a potential role as biomarkers in sepsis as detailed in Table 1. As with most of the biomarkers, it is expected that miRNAs also have some prognostic values; however, this is not an absolute rule. The first study to examine the clinical utility of circulating miRNAs in septic patients showed that miR-150 expression in leukocytes from healthy control patients was significantly different compared with septic patients, and that levels of miR-150 in serum were correlated with the severity of sepsis as determined by the SOFA (Sequential Organ Failure Assessment) score, an ICU scoring system used to determine the extent of a person's organ function or rate of failure20 (Table 1). In a further larger study on serum from critically ill patients (138 with sepsis and 85 without sepsis) and 76 healthy individuals, high expression of miR-150 did correlate with enhanced survival whereas low expression was associated with increased risk of organ dysfunction and mortality.47 When compared with other biomarkers proposed in sepsis, there were no correlations between miR-150 serum levels and markers of inflammation and bacterial infection, such as C-reactive protein (CRP) or procalcitonin. However, a clear correlation of miR-150 serum levels with serum concentrations of lactate and other classical indicators of hepatic and renal organ failure was identified. In a study published by Wang et al., microarray screens from 12 surviving and 12 non-surviving sepsis patients showed that only miR-297 and miR-574-5p were significantly differentially expressed between the two groups studied. In fact, after combining miRNA expression in sepsis with SOFA scores, only miR-574-5p was shown to have prognostic utility.48 Another miRNA with a potential prognostic role is miR-466I, whose expression in leukocytes but not serum, was higher in sepsis non-survivors than those who survived.49 Upregulation of miR-133a was correlated with the severity of sepsis or bacterial infection markers (e.g., CRP, procalcitonin) and SOFA scores.50 Using an alternative approach, an miRNA regulatory network using information from various clinical studies, and both experimental and in silico data was built.51 The authors identified several miRNAs with a diagnostic value in sepsis (e.g., miR-15a, miR-16, miR-122, miR-146a, miR-150, miR-223, miR-499-5p) or with some prognostic utility (e.g., miR-193b, miR-483-5p and miR-574-5p).51
One possible approach in assessing survival rates in sepsis might be better reflected by analyzing the miRNAs that integrate both pro-inflammatory and anti-inflammatory signals in critical illness, rather than by measuring single proteins alone. However, the exact functional mechanisms that allow cellular and viral miRNAs to determine the prognosis of septic patients are presently unclear and remain a matter of debate.
Viral miRNAs have biomarker potential in sepsis
In an extensive study conducted by Walton et al.,52 it was shown that 42% of the septic patients have reactivation of two or more viruses. A number of miRNAs appear to regulate the cross-talk between host tissue and viral pathogens. Viruses such as Kaposi sarcoma herpesvirus (KSHV) and Epstein-Barr virus (EBV) encode for miRNAs that regulate host gene expression promoting their virulence and carcinogenesis.53 KSHV, human herpesvirus 8 (HHV-8), is central to the pathogenesis of Kaposi sarcoma, primary effusion lymphoma and multicentric Castleman disease,54 all of which predominantly affect patients with human immunodeficiency virus/acquired immune deficiency syndrome.55 KSHV-derived miRNAs were shown to function as regulators of this process by maintaining viral latency and inhibiting viral lytic replication. Sepsis and surgical trauma induces tissue damage that may release host-derived single-stranded RNAs (cellular or viral miRNAs), which may then act as a molecular ‘switch', triggering the transition from one phase to another. In our recent study, the profiling of genome-wide miRNA expression in leukocytes from septic patients and non-septic controls identified differences in plasma levels of two KSHV miRNAs, miR-K-10b and miR-K12-12*.25 Owing to the fact that surgical trauma can also trigger SIRS, when plasma levels of KSHV miRNAs were measured pre- and postoperatively in two non-septic surgical cohorts, it was showed that surgical trauma increases plasma miR-K12-10b expression. These increases in expression levels of viral miRNAs in surgical patients suggest that surgical trauma may have triggered KSHV reactivation. By now, it is not clear if the increased expression of viral miRNAs is the trigger or the result of immunosuppression in sepsis, but we can hypothesize that reactivation of latent KSHV infection may be a positive feedback mechanism that contributes to the inappropriate inflammatory response associated with fatal sepsis.25
Some viral miRNAs share perfect ‘seed' homology with cellular miRNAs – functional mimicry in sepsis
Viral reactivation has been found to occur during the immunosuppression phase of sepsis.52 This supports the concept that sepsis leads to a functional immunosuppression, which requires more intensive monitoring than is currently done in most of the cases. Reactivated viruses may also contribute to the pathogenesis of sepsis and require active treatment. Viral miRNAs are implicated in sepsis pathogenesis, as it has emerged that HHV-8 or KSHV derived miRNAs contribute to the septic response in humans. Viral miRNAs found to be expressed in septic patients are proved to serve as ligands for TLR8 triggering pro-inflammatory response, in addition to functioning through the canonical mechanism via post-transcriptional repression of target mRNAs. This is supported by RNA immunoprecipitation experiment with Flag-tagged TLR8 that showed increased binding of KSHV-miR-K12-10b and KSHV-miR-K12-12* to TLR8 compared with other miRNAs.25
KSHV is also strongly associated with both endothelial and B-cell neoplasms. Interestingly, KSHV-miR-K12-11, a viral miRNA constitutively expressed in cell lines derived from KSHV-associated tumors, shares perfect seed sequence homology with the cellular miR-155.56 Since miR-155 is overexpressed in a number of human tumors, it has been suggested that miR-K12-11 can mimic miR-155 functions and may contribute to cellular transformation in KSHV-associated malignant diseases. Several other KSHV miRNAs were shown to share ‘seed' homology with cellular miRNAs, suggesting that they might function as viral analogs of these miRNAs.57
Viral miRNAs that share the same ‘seed homology' are potentially able to regulate the same targets as the cellular miRNA counterparts. More recently, the concept of ‘molecular mimicry' has also been explored in the context of viral derived malignancies. EBV induces the expression of miR-146a by the activation of NF-κB in infected cells, resulting in the downregulation of IRAK1 and the inhibition of genes involved in the interferon response. While EBV modulates the expression of NF-κB-induced cytokines by acting on cellular miRNAs (miR-146a and miR-155), KSHV encodes its own miRNAs that regulate the same cellular targets.55 Understanding the interplay between cellular and viral miRNAs targeting the same components in the same immune pathway (‘pathway mimicry') might bring new information about the implication of reactivated viruses on sepsis' outcome (Figure 3). Therefore, modulating the expression of miRNAs or mRNAs within the immune signaling pathway in this way may lead to a stronger additive immune response and may be relevant in the design of new therapies.
Cellular and viral miRNAs are direct agonists of TLRs in sepsis
In the context of viral infections, TLR 3, 7, 8, 9 are established as the predominant receptors for virus recognition and have been shown to play a role in KSHV reactivation.58 It has also been shown that secreted miRNAs regulate gene expression by canonical binding to receptors of the TLR family in immune cells, triggering a TLR-mediated inflammatory response.59 Fabbri et al.59, 60 showed that miR-21 and miR-29a trigger a TLR-mediated pro-metastatic inflammatory response by binding as ligands to TLR family leading to tumor growth and metastasis, regulating tumor microenvironment. More recently, it was shown that neuroblastoma cells release miR-21 in exosomes and transfer this miRNA to surrounding tumor-associated macrophages (TAMs) expressing TLR8. MiR-21 binding to TLR8 triggers NK-kB activation in TAMs and their secretion of miR-155 in exosomes that is transferred back to neuroblastoma cells, where miR-155 increases drug resistance by targeting TERF1, an inhibitor of telomerase activity.61 In a recently published work, it was shown that two KSHV miRNAs, miR-K12-10b and miR-K12-12*, are direct agonists of TLR8.25 Furthermore, both KSHV miRNAs were increased on first postoperative day, but returned to baseline on after one week.25 When evaluated the functional effect of these two KSHV miRNAs on cytokine production there was sustained increased secretion of IL-6 and IL-10 over time when compared with sham-transfected controls. Furthermore, these miRNAs appeared to cooperate in the induction of cytokine release upon exposure to LPS. Cellular and KSHV miRNAs were found to be differentially expressed in sepsis and in early postsurgical patients. Increased miR-K-10b and miR-K12-12* are functionally involved in sepsis as direct agonists of TLR8, leading to cytokine deregulation and participating in sepsis' pathogenesis.25 Further extensive experimental and clinical studies are required to validate their diagnostic and therapeutic purposes. Furthermore, additional work is needed to prove the direct involvement of viral KSHV miRNAs targeting TLR in sepsis mortality.
miRNAs as diagnostic and prognostic biomarkers for SIRS and sepsis
miRNAs have been used to identify a putative ‘diagnostic signature' that can distinguish between SIRS and sepsis. SIRS and sepsis are both potentially fatal conditions that may culminate in a whole-body inflammatory state leading to multiple organ dysfunction syndrome and death.45 While SIRS is frequently associated with non-infective pathology such as pancreatitis, trauma, drug and immunologic reactions, sepsis is triggered by microbial pathogens. Crucially, at the time of diagnosis, a primary infectious cause of an SIRS response may not be evident, but the differentiation of sepsis from non-infective SIRS is a priority as antimicrobial agents have to be administered in a timely and effective manner.
In a recent study, low serum expression of miR-146a and miR-223 distinguished patients with sepsis from those with SIRS (ROC curve analysis: AUC=0.858 and 0.804, respectively).62 Furthermore, a study of 214 sepsis patients found that a combination of four miRNA markers in serum (miR-15a, miR-16, miR-193* and miR-483-5p) alongside clinical prognostication scores predicted 28 days survival rate with a sensitivity of 88.5% and a specificity of 90.4%.63 Two novel miRNAs, namely miR-342-3p and miR-3173-5p, were also decreased significantly in septic patients when compared to SIRS.64
Among several miRNAs that seem to be upregulated in sepsis patients, miR-4772 family is significantly upregulated in sepsis as opposed to healthy control subjects and SIRS patients. It has also been noted that miR-143 levels are elevated in patients with sepsis as opposed to SIRS or healthy control subjects, with high correlation to SOFA (≥7) and APACHE II (≥10) scores.65 One study noted that miR-21-3p was significantly upregulated in plasma of septic patients and in heart tissue of mice with LPS-induced sepsis.66 Another study found that miR-23a-5p is upregulated in septic patients and in LPS-induced acute respiratory distress syndrome, which is a complication of sepsis that leads to death.67 These miRNAs could be a prognostic marker as well as a biomarker for sepsis.
To date, no miRNAs or combinations of miRNAs have been identified with clear diagnostic and prognostic utility. Some candidates including miR-150 and miR-146a, however, have been identified in several studies making them highly promising (there were at least two individual studies on the same type of biological sample, but on different number of patients that reported the same levels of expression). Unfortunately, most of the studies published by now, as summarized in Table 1, are designed to compare septic patients with healthy controls and not with SIRS patients, which would make miRNAs more trustable biomarkers in detecting early sepsis. Further work is required before introducing miRNAs as sepsis biomarkers in clinical practice.
Rationale for using miRNA therapeutics in sepsis
Therapeutic strategies, which employ miRNA mimics or antisense-RNA inhibitors, may represent a valid option for the treatment of certain patients with sepsis in the near future.68 However, the selection of target genes, the optimal design of therapeutic molecules and drug delivery systems remain major challenges.69, 70, 71, 72 Several companies are developing new approaches to target miRNAs, notably with antagomirs and locked nucleic acid miRNA inhibitors, with the aim of suppressing target mRNA expression.68 However, the only clinical trials currently being undertaken are designed to identify the diagnostic and predictive value of circulating microRNAs during sepsis (ClinicalTrials.gov Identifier: NCT00862290, NCT01207531, NCT01459822) and not the efficacy of RNA inhibition based therapy. There are some studies that point to possible miRNA targets for treatment. In one experiment it was identified that miR-195 is upregulated in the liver and lungs of mice with LPS-induced sepsis.73 Silencing this gene reduced apoptosis and increased sepsis survival rates in the mice. Thus, inhibiting upregulated miRNAs could be an avenue of treatment, depending on the miRNA molecule and its role in the disease. Preclinical and clinical data support the idea that targeting genes involved in the innate immune response might restrict inappropriate inflammation and prevent uncontrolled sepsis. Putative candidates to regulate these targets include miR-146, which targets TRAF6 and IRAK1, in the TLR signaling pathway, or miR-155, which targets SHIP-1 in the phosphoinositide 3 kinase pathway.74
New concept: antiviral miRNAs therapy in sepsis
Cellular and viral miRNAs, as it was shown, are implicated in sepsis by regulating the inflammatory and anti-inflammatory cytokine storm. Thus, the design of new therapies that will target these miRNAs might represent a new avenue in sepsis treatment. Furthermore, we propose that the next step in early treatment of sepsis will be to use candidate miRNAs (both viral and cellular transcripts) with clear pathogenic function as new therapeutic targets, by infusing the mimics or antagonists (antagomirs) in established and clinically relevant models of sepsis. For sepsis patients, timely diagnosis and early treatment are important factors when it comes to improving the prognosis. While the functions and the clinical value of the cellular miRNAs are still under investigation, the viral miRNAs identified as sepsis biomarkers are new in the field and their roles are not yet completely understood. Therefore, targeting genes regulated by these miRNAs may represent potential new therapeutic avenues in sepsis patients.
Summary discussion
The increasing research findings supporting the important role of miRNAs in the immune system strongly suggest their contribution to host immunity in response to pathogens (e.g., viruses, bacteria) and to inflammatory disease pathogenesis. Several miRNAs have been shown to be abnormally expressed in septic patients and some of them were correlated with a decreased survival, supporting their possible use as targetable biomarkers for early diagnosis and prognosis of sepsis. However, an outstanding question to answer is which is the underlying mechanistic role of viral miRNAs in the immune regulation during sepsis. One of the new scenarios that emerged is that during sepsis or immunosuppression triggered by surgical trauma, a latent subclinical virus is reactivated contributing to sepsis' mortality and morbidity rates by interfering with normal immune response pathway. However, larger patient cohorts and other laboratory tests for detecting viral infection, such as seropositivity, need to be used to further confirm these findings.
Acknowledgments
Dr Calin is The Alan M Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr Calin's laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, The UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE (2P50CA127001), the Leukemia SPORE (5P50CA100632), the Center for radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot project, a 2014 Knowledge GAP MDACC grant, a CLL Moonshot pilot project, The UT MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson. Dr Giza was supported in part by Ministry of National Education, CNCS-UEFISCDI project number 22 from 28/08/2013 (PN-II-ID-PCE-2012-4-0018). Dr Fuentes-Mattei were supported in part by the NIH Loan Repayment Program for Clinical Research, (LRP-CR), US Department of Health and Human Services. Dr Bullock was supported by the 2015 Fulbright-RCS research fellowship. Dr Lupu's laboratory is supported by NIH grants R01GM116184, R01GM097747, U19AI062629 and R21AI113020. Parts of the figures were prepared by using ChemBioDraw version 15.0 (Perkin Elmer).
Glossary
- miRNA
microRNA
- mRNA
messenger RNA
- KSHV
Kaposi sarcoma herpesvirus
- SIRS
systemic inflammation response syndrome
- TLR
Toll-like receptor
The authors declare no conflicts of interest.
Footnotes
Edited by E Fuentes-Mattei
References
- Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MM, Artigas A, Phillips GS, Rhodes A, Beale R, Osborn T et al. Outcomes of the Surviving Sepsis Campaign in intensive care units in the USA and Europe: a prospective cohort study. Lancet Infect Dis 2012; 12: 919–924. [DOI] [PubMed] [Google Scholar]
- Angus DC, Wax RS. Epidemiology of sepsis: an update. Crit Care Med 2001; 29(Suppl): S109–S116. [DOI] [PubMed] [Google Scholar]
- Jawad I, Luksic I, Rafnsson SB. Assessing available information on the burden of sepsis: global estimates of incidence, prevalence and mortality. J Glob Health 2012; 2: 010404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001; 29: 1303–1310. [DOI] [PubMed] [Google Scholar]
- Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med 2013; 369: 840–851. [DOI] [PubMed] [Google Scholar]
- Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2003; 348: 1546–1554. [DOI] [PubMed] [Google Scholar]
- Pfuntner A, Wier LM, Steiner C. Costs for Hospital Stays in the United States, 2011: Statistical Brief #168. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs, Rockville, MD, 2013..
- Ranieri VM, Thompson BT, Barie PS, Dhainaut JF, Douglas IS, Finfer S et al. Drotrecogin alfa (activated) in adults with septic shock. N Engl J Med 2012; 366: 2055–2064. [DOI] [PubMed] [Google Scholar]
- Opal SM, Garber GE, LaRosa SP, Maki DG, Freebairn RC, Kinasewitz GT et al. Systemic host responses in severe sepsis analyzed by causative microorganism and treatment effects of drotrecogin alfa (activated). Clin Infect Dis 2003; 37: 50–58. [DOI] [PubMed] [Google Scholar]
- Lagu T, Rothberg MB, Shieh MS, Pekow PS, Steingrub JS, Lindenauer PK. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med 2012; 40: 754–761. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009; 302: 2323–2329. [DOI] [PubMed] [Google Scholar]
- van der Poll T, van Deventer SJ. Cytokines and anticytokines in the pathogenesis of sepsis. Infect Dis Clin North Am 1999; 13: 413–426 ix. [DOI] [PubMed] [Google Scholar]
- Muenzer JT, Davis CG, Chang K, Schmidt RE, Dunne WM, Coopersmith CM et al. Characterization and modulation of the immunosuppressive phase of sepsis. Infect Immun 2010; 78: 1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, Kurosawa S, Stepien D, Valentine C et al. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiol Rev 2013; 93: 1247–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D et al2001. SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med 2003; 29: 530–538. [DOI] [PubMed] [Google Scholar]
- Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D et al2001. SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med 2003; 31: 1250–1256. [DOI] [PubMed] [Google Scholar]
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34: 1589–1596. [DOI] [PubMed] [Google Scholar]
- Seymour CW, Rosengart MR. Septic shock: advances in biagnosis and treatment. JAMA 2015; 314: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One 2009; 4: e7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Strmsek Z, Kunej T. MicroRNA silencing by DNA methylation in human cancer: a literature analysis. Non-Coding RNA 2015; 1: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarsky Y et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res 2001; 61: 6640–6648. [PubMed] [Google Scholar]
- Essandoh K, Fan GC. Role of extracellular and intracellular microRNAs in sepsis. Biochim Biophys Acta 2014; 1842: 2155–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor S, Giza DE, Lin HY, Fabris L, Yoshiaki K, D'Abundo L et al. Cellular and Kaposi's sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis 2014; 5: e1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem 2010; 285: 20940–20951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007; 179: 5082–5089. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006; 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H et al. A genomic storm in critically injured humans. J Exp Med 2011; 208: 2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-kappaB activation. Innate Immun 2012; 18: 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham E. Nuclear factor-kappaB and its role in sepsis-associated organ failure. J Infect Dis 2003; 187(Suppl 2): S364–S369. [DOI] [PubMed] [Google Scholar]
- Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol 2011; 8: 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Gong H, Li Y, Jie K, Ding C, Shao Q et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res 2013; 39: 275–282. [DOI] [PubMed] [Google Scholar]
- Konecny FA. Review of cellular and molecular pathways linking thrombosis and innate immune system during sepsis. J Res Med Sci 2010; 15: 348–358. [PMC free article] [PubMed] [Google Scholar]
- Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics 2011; 43: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820. [DOI] [PubMed] [Google Scholar]
- Toubiana J, Courtine E, Pene F, Viallon V, Asfar P, Daubin C et al. IRAK1 functional genetic variant affects severity of septic shock. Crit Care Med 2010; 38: 2287–2294. [DOI] [PubMed] [Google Scholar]
- Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem 2007; 282: 28929–28938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X et al. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J Cell Biochem 2014; 115: 919–927. [DOI] [PubMed] [Google Scholar]
- Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y et al. MyD88 as a target of microRNA-203 in regulation of lipopolysaccharide or Bacille Calmette-Guerin induced inflammatory response of macrophage RAW264.7 cells. Mol Immunol 2013; 55: 303–309. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 2010; 11: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 2008; 27: 4712–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging 2009; 1: 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MY, Calin GA. The mix of two worlds: non-coding RNAs and hormones. Nucleic Acid Ther 2013; 23: 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 2011; 306: 2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata JM, Krajewska M, Morse HC III, Choi Y, Reed JC. TNF receptor-associated factor (TRAF) domain and Bcl-2 cooperate to induce small B cell lymphoma/chronic lymphocytic leukemia in transgenic mice. Proc Natl Acad Sci USA 2004; 101: 16600–16605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Scholten D, Frey N et al. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 2013; 8: e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Meng K, Chen W, Feng D, Jia Y, Xie L. Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock 2012; 37: 263–267. [DOI] [PubMed] [Google Scholar]
- Li Y, Dalli J, Chiang N, Baron RM, Quintana C, Serhan CN. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by microRNA and proresolving mediators. Immunity 2013; 39: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke F, Roderburg C, Benz F, Cardenas DV, Luedde M, Hippe HJ et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit Care Med 2014; 42: 1096–1104. [DOI] [PubMed] [Google Scholar]
- Huang J, Sun Z, Yan W, Zhu Y, Lin Y, Chen J et al. Identification of microRNA as sepsis biomarker based on miRNAs regulatory network analysis. Biomed Res Int 2014; 2014: 594350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH et al. Reactivation of multiple viruses in patients with sepsis. PLoS One 2014; 9: e98819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007; 131: 146–159. [DOI] [PubMed] [Google Scholar]
- Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 2010; 11: 141–147. [DOI] [PubMed] [Google Scholar]
- Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. Kaposi's sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol 2012; 86: 11663–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 2007; 81: 12836–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E. Kaposi's sarcoma-associated herpesvirus microRNAs. Front Microbiol 2012; 3: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S. Update on the role of Toll-like receptors during bacterial infections and sepsis. Wien Med Wochenschr 2010; 160: 107–111. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 2012; 109: E2110–E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther 2007; 7: 1009–1019. [DOI] [PubMed] [Google Scholar]
- Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst 2015; 107. pii: djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang HC, Chen C, Zeng J, Wang Q, Zheng L et al. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp Ther Med 2013; 5: 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS One 2012; 7: e38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Vilanova D, Atalar K, Delfour O, Edgeworth J, Ostermann M et al. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS One 2013; 8: e75918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Dai QC, Shen HL, Zhang XW. Diagnostic value of elevated serum miRNA-143 levels in sepsis. J Int Med Res 2016; 44: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bei Y, Shen S, Huang P, Shi J, Zhang J et al. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J Mol Cell Cardiol 2016; 94: 43–53. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu C, Wang Z, Huang J, Zeng Q. microRNA-23a-5p acts as a potential biomarker for sepsis-induced acute respiratory distress syndrome in early stage. Cell Mol Biol (Noisy-le-grand) 2016; 62: 31–37. [PubMed] [Google Scholar]
- Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheumat Dis 2008; 67(Suppl 3): iii50–iii55. [DOI] [PubMed] [Google Scholar]
- Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest 2007; 117: 3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009; 457: 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonisch T, Fowler PA, Hombach-Klonisch S. Molecular and genetic regulation of testis descent and external genitalia development. Dev Biol 2004; 270: 1–18. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 2009; 8: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Yu Y, Li M, Wang G, Chen R, Fan GC et al. Inhibition of microRNA 195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J Infect Dis 2016; 213: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov 2010; 9: 293–307. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol 2005; 6: 127–138. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Carthew RW. Silence from within: endogenous siRNAs and miRNAs. Cell 2005; 122: 9–12. [DOI] [PubMed] [Google Scholar]
- Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun 2010; 394: 184–188. [DOI] [PubMed] [Google Scholar]
- Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L et al. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest 2012; 122: 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med 2014; 24: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie LX. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin Chem Lab Med 2012; 50: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Zhang PJ, Chen WJ, Feng D, Jia YH, Xie LX. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J Trauma Acute Care Surg 2012; 73: 850–854. [DOI] [PubMed] [Google Scholar]
- Wang H, Yu B, Deng J, Jin Y, Xie L. Serum miR-122 correlates with short-term mortality in sepsis patients. Crit Care 2014; 18: 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Deng J, Wang JY, Zhang PJ, Xin Z, Xiao K et al. Serum miR-122 levels are related to coagulation disorders in sepsis patients. Clin Chem Lab Med 2014; 52: 927–933. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li C, He Y, Li Q, Wang G, Wen P et al. [Relationship between expression of microRNA and inflammatory cytokines plasma level in pediatric patients with sepsis]. Zhonghua Er Ke Za Zhi 2014; 52: 28–33. [PubMed] [Google Scholar]
- Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int 2015; 35: 1172–1184. [DOI] [PubMed] [Google Scholar]
- Puskarich MA, Nandi U, Shapiro NI, Trzeciak S, Kline JA, Jones AE. Detection of microRNAs in patients with sepsis. J Acute Dis 2015; 4: 101–106. [Google Scholar]
- How CK, Hou SK, Shih HC, Huang MS, Chiou SH, Lee CH et al. Expression profile of MicroRNAs in gram-negative bacterial sepsis. Shock 2015; 43: 121–127. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X, Liu X, Wang X, Xu J, Hou S et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med 2015; 8: 5683–5690. [PMC free article] [PubMed] [Google Scholar]
- Yao L, Liu Z, Zhu J, Li B, Chai C, Tian Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int J Clin Exp Pathol 2015; 8: 7675–7684. [PMC free article] [PubMed] [Google Scholar]
- Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 2011; 3: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AJ, Guo C, Cook JA, Wolf B, Halushka PV, Fan H. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit Care 2015; 19: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Lovat F, Romano G, Calore F, Acunzo M, Bell EH et al. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J Biol Chem 2014; 289: 26406–26416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. microRNAs are ligands of Toll-like receptors. RNA 2013; 19: 737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Gu JT, Yi B, Tang ZZ, Tao GC. microRNA-23b regulates the expression of inflammatory factors in vascular endothelial cells during sepsis. Exp Ther Med 2015; 9: 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gong X, Tian K, Chen D, Sun J, Wang G et al. miR-25 promotes glioma cell proliferation by targeting CDKN1C. Biomed Pharmacother 2015; 71: 7–14. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep 2012; 27: 594–598. [DOI] [PubMed] [Google Scholar]
- Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol 2009; 326: 431–443. [DOI] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J 2012; 31: 2207–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Takata A, Otsuka M, Kishikawa T, Kojima K, Yoshida H et al. Silencing of microRNA-122 enhances interferon-alpha signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep 2012; 2: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Wu D, Xu B, Qian W, Li P, Lu Q et al. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep 2012; 27: 1967–1975. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour Biol 2015; 36: 9779–9788. [DOI] [PubMed] [Google Scholar]
- Kopp F, Schnoedt M, Haase R, Wagner E, Roidl A, Ogris M. De-targeting by miR-143 decreases unwanted transgene expression in non-tumorigenic cells. Gene Ther 2013; 20: 1104–1109. [DOI] [PubMed] [Google Scholar]
- Hou C, Yang Z, Kang Y, Zhang Z, Fu M, He A et al. MiR-193b regulates early chondrogenesis by inhibiting the TGF-beta2 signaling pathway. FEBS Lett 2015; 589: 1040–1047. [DOI] [PubMed] [Google Scholar]
- Zheng D, Yu Y, Li M, Wang G, Chen R, Fan GC et al. Inhibition of miR-195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J Infect Dis 2015; 213: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn EM, Wang J, Redmond HP. The emerging role of microRNA in regulation of endotoxin tolerance. J Leuk Biol 2012; 91: 721–727. [DOI] [PubMed] [Google Scholar]
- Essandoh K, Li Y, Huo J, Fan GC. Mirna-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 2016; 46: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ma N, Zhao R, Wu G, Zhang Y, Qiao Y et al. Overexpression of miR-483-5p/3p cooperate to inhibit mouse liver fibrosis by suppressing the TGF-beta stimulated HSCs in transgenic mice. J Cell Mol Med 2014; 18: 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang YW, Tong XH, Liu YS. Characterization of microRNA profile in human cumulus granulosa cells: identification of microRNAs that regulate Notch signaling and are associated with PCOS. Mol Cell Endocrinol 2015; 404: 26–36. [DOI] [PubMed] [Google Scholar]
- Pan T, Liu Z, Yin J, Zhou T, Liu J, Qu H. Notch signaling pathway was involved in regulating programmed cell death 1 expression during sepsis-induced immunosuppression. Mediat Inflamm 2015; 2015: 539841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mees ST, Mardin WA, Sielker S, Willscher E, Senninger N, Schleicher C et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol 2009; 16: 2339–2350. [DOI] [PubMed] [Google Scholar]
- Li Q, Li X, Guo Z, Xu F, Xia J, Liu Z et al. MicroRNA-574-5p was pivotal for TLR9 signaling enhanced tumor progression via down-regulating checkpoint suppressor 1 in human lung cancer. PLoS One 2012; 7: e48278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitas G, Burt BM, Nguyen HM, Bamboat ZM, DeMatteo RP. Toll-like receptor 9 inhibition reduces mortality in polymicrobial sepsis. J Exp Med 2008; 205: 1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fu S, Wang J. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Arch Virol 2014; 159: 2997–3003. [DOI] [PubMed] [Google Scholar]