Abstract
The International Committee for the Taxonomy and Nomenclature of Viruses does not rule on virus classifications below the species level. The definition of species for viruses cannot be clearly defined for all types of viruses. The complex and interesting epidemiology of Human Immunodeficiency Viruses demands a detailed and informative nomenclature system, while at the same time it presents challenges such that many of the rules need to be flexibly applied or modified over time. This review outlines the nomenclature system for primate lentiviruses and provides an update on new findings since the last review was written in 2000.
Keywords: Immunodeficiency, HIV, Lentivirus, Nomenclature, Classification
Introduction
1.1 Primate Immunodeficiency Virus nomenclature history
The first complete genome of a human immunodeficiency virus, named HTLV-III at that time, was sequenced in 1984(Ratner et al., 1985). By 1992 it was clear that there was considerable diversity between HIV-1 M group viruses, that HIV-2 was also infecting humans, that chimpanzees carried viruses similar to HIV-1(Huet et al., 1990), and that sooty mangabeys carried viruses similar to HIV-2(Sanchez-Pescador et al., 1985)(Myers et al., 1992)(Hirsch et al., 1989). In subsequent years analyses of tens of thousands of virus genomic sequences have provided great insight into the origins and spread of these viruses. The rules for naming viruses, established and overseen by the International Committee for the Taxonomy and Classification of Viruses (ICTV), only go to the species level, with nomenclatures of subspecies, strains, isolates, serotypes and similar subspecies designations left up to the research community that studies each virus(Fauquet and Fargette, 2005). The HIV research community worked on names informally until a formal meeting was held in September 1999 (Robertson et al., 2000), where a set of rules for naming primate lentiviruses (HIVs and SIVs) was agreed upon.
1.2 Lentiviruses and Primate Immunodeficiency Viruses
The broad picture of lentiviruses is that they have only been detected in a small subset of mammals, and that the phylogeny of the viruses is similar enough to the phylogeny of the host species to suggest a very long time of co-evolution of these viruses with their hosts. However, there are many exceptions where the virus phylogeny disagrees with host phylogeny due to recent and/or ancient cross species transfer events, and also due to ancient or recent recombination events between different virus lineages. The non-primate lentiviruses include bovine immunodeficiency viruses found in both domestic cattle and wild buffalo, Equine Infectious Anemia Virus, Small Ruminant Lentiviruses of goats and sheep, and feline immunodeficiency viruses. The feline immunodeficiency viruses have been extensively studied and have been found in domestic housecats, New World felines including pumas, bobcats and lynx, and Old World felines including lions, leopards and Pallas’ cats. Outside the primate immunodeficiency viruses (PIVs), less effort has been made to standardize the classification and nomenclature, but in cases where there has been some effort made, it generally follows a similar set of criteria as have been used for the PIVs. This review will only cover the nomenclature of PIVs, with a special emphasis on HIV-1.
1.3 PIV species specificity, coevolution
For the most part, the lentiviruses do not easily cross species boundaries, and there are several virus-host protein interactions which either partially or completely block the ability of lentivirus from one host to infect another host species. Because of this, it makes sense to name each lineage of simian lentivirus for the host it infects, such as SIV_Gorilla (SIVgor) for simian immunodeficiency viruses isolated from gorillas. Short sequence or isolate names are nice for labeling phylogenetic trees and other purposes, so the convention is to use a 3-letter code for the primate species from which the virus was isolated. The exact line between species and subspecies of primates, as with most other organisms, is in some cases difficult to define. For some groups, the geographic range is more important than genetic distances, while for others the genetics is more important. For example, zoo keepers have reported that Pan troglodytes schweinfurthi, Pan troglodytes verus and Pan troglodytes troglodytes can produce fertile offspring with Pan paniscus and thus the species (paniscus vs troglodytes) and subspecies of chimpanzees are better defined by their geographic ranges. A similar situation exists for baboons and African green monkeys. In other cases such as the greater spot-nosed monkey (Cercopithecus nictitans) and lesser spot-nosed monkey (Cercopithecus petaurista) the species ranges overlap but mating preferences and genetic distances justify separate species designations(Tosi et al., 2005). The three letter code chosen to represent each primate species or subspecies from which a PIV has been isolated is usually derived from the common name (cpz for chimpanzee, for example) but sometimes derived from the Latin name, ex. SIVasc from Cercopithecus ascanius (red tailed guenon). In some cases a single species or subspecies of primate can harbor two or more very distinct lineages of SIV, in which case sequential numbers are added to the name (HIV-1 and HIV-2, SIVmnd-1, SIVmnd-2). Table 1 is a comprehensive list of primate species which have been tested by serology or virus sequencing. The HIV Databases at LANL maintain a listing of codes used for the sequenced viruses here: http://www.hiv.lanl.gov/content/sequence/HelpDocs/subtypes.html
Table 1.
| Virus species | Primate species | Comments | Primate&species |
|---|---|---|---|
| ASC | Red-tailed Guenon | Also dubbed SIVrtg. There are at least 3 SIVasc species infecting the two subspecies of Red-tailed Guenons, but complete genomes not yet available for all 3. | Cercopithecus ascanius schmidti |
| BAB | Baboon | Baboon infected in the wild with vervet SIV. | Papio spp. |
| BLC | Bioko Black Colobus | Cercopithecus,satanas,satanas | |
| BKM | Black Mangabey | Lophocebus aterrimus | |
| BLU | Blue Monkey | Cercopithecus,mitis | |
| COL | Colobus'Monkey | Colobus guereza | |
| CPZ | Chimpanzee | SIVcpz infects Pan troglodytes troglodytes and Pan troglodytes schweinfurthi subspecies. Pan paniscus does not seem to have a SIV. | Pan troglodytes troglodytes or P. t. schweinfurthii |
| DEB | DeBrazza’ s Monkey | Cercopithecus neglectus | |
| DEN | Dent s Monkey | Cercopithecus denti | |
| DRL | Drill Monkey | Mandrillus leucophaeus | |
| GOR | Gorilla | Gorilla gorilla | |
| GRV | Grivet | Formerly a subspecies of African Green Monkey. | Chlorocebus aethiops |
| GSN | Greater Spotnosed | Monkey Cercopithecus nictitans | |
| KRC | Kibale Red Colobus | Procolobus [Piliocolobus] rufomitratus tephrosceles | |
| LST | L’Hoest ’ s Monkey | Cercopithecus lhoesti | |
| MAC | Macaque | Rhesus Macaque naturally infected by SIVsmm. | Macaca mulatta |
| MND-1 | Mandrill | Mandrills are infected with two types of SIV nearly as diverse from each other as HIV-1 and HIV-2 are from each other. | Mandrillus sphinx |
| MNE | Pig-tailed Macaque | Pig-tailed Macaque infected in captivity by SIVsmm. | Macaca nemestrina |
| MON | Mona Monkey | Cercopithecus mona | |
| MUS-1 | |||
| MUS-2 | |||
| MUS-3 | Mustached Monkey | At least 3 distinct SIV species infect Mustached Monkeys. | Cercopithecus cephus |
| OLC | Olive Colobus | Procolobus verus | |
| PAT | Patas Monkey | Erythrocebus patas | |
| PRG | Preuss s Guenon | Preuss ’ s Red-eared Guenon from Bioko. | Cercopithecus preussi insularis |
| RCM | Red-capped Mangabey | Cercocebus torquatus torquatus | |
| REG | Red-eared Guenon | Red-eared Guenon from Bioko. Also dubbed SIVery. | Cercopithecus erythrotis erythrotis |
| SAB | Sabaeus Monkey | Also called Green Monkey. | Chlorocebus sabaeus |
| SMM | Sooty Mangabey | SMM is the species for both Sooty Mangabey and macaques experimentally infected with SMM. | Cercocebus atys atys |
| STM | Stump-tailed Macaque | Stump-tailed Macaque infected in captivity by SIVsmm. | Macaca arctoides |
| SUN | Sun-tailed Monkey | Cercopithecus solatus | |
| SYK | Sykes Monkey | Cercopithecus albogularis | |
| TAL | Talapoin Monkey | Miopithecus ogouensis or Miopithecus talapoin | |
| TAN | Tantalus Monkey | Formerly a subspecies of African Green Monkey. | Chlorocebus tantalus |
| TRC | Tshuapa Red Colobus | Piliocolobus tholloni | |
| VER | Vervet | Formerly a subspecies of African Green Monkey. | Chlorocebus pygerythrus |
| WCM | White-crowned | ||
| Mangabey | White-crowned Mangabey infected in captivity by SIVver. | Cercocebus torquatus lunulatus | |
| WOL | Wolf s Monkey | Cercopithecus wolfi | |
| WRC | Western Red Colobus | Procolobus verus |
1.4 Human Immunodeficiency Viruses
There have been several transfers of nonhuman primate immunodeficiency viruses into humans, from chimpanzees, gorillas and sooty mangabeys. Viruses in the HIV-1 M and N groups are most closely related to viruses found in wild chimpanzees of the Pan troglodytes troglodytes subspecies from south eastern Cameroon(Keele et al., 2006, Van Heuverswyn et al 2007). HIV-1 O and P group viruses are most closely related to viruses found in western lowland gorillas in Cameroon(D’arc et al., 2015). Each group of HIV-1 viruses seems to be the result of a separate cross-species transmission event from chimpanzee or gorilla into humans, followed by different degrees of epidemic spread within humans. The HIV-1 M group spread far more extensively in humans than did N, O, or P group viruses. The AIDS pandemic is primarily caused by viruses in HIV-1 M group.
HIV-2 viruses similarly have been introduced into humans several times, from sooty mangabeys in West Africa, of which only two transfers resulted in significant spread within humans in West Africa. The viruses from each of the sooty mangabey to human transfers were initially named as “subtypes” of HIV-2 and the subtype (A, B, C) designations remain in use today even though they are equivalent to the “groups” of HIV-1, and not equivalent to the subtypes which have been defined within the HIV-1 M group (http://www.hiv.lanl.gov/content/sequence/HelpDocs/subtypes-more.html accessed May 26, 2016). The HIV-2 clades should be referred to as “groups” and not “subtypes”.
2 HIV-1, SIVcpz, SIVgor
Because the AIDS pandemic is primarily caused by viruses in the HIV-1 M group, far more research has been conducted on this group of viruses. The global spread of HIV-1 M group viruses has also resulted in a much more complex and interesting epidemiological history, than those of the other HIV-1 groups or HIV-2. Likewise the extra interest in this group has also resulted in increased searching and sampling of chimpanzees and gorillas aimed at tracing the origins of the HIV-1 viruses and a better understanding of the natural history of these viruses than any of the other primate lentiviruses (Keele 2006 (D’arc 2015) (Li 2012).
It is now clear that the HIV-1 M and N groups came from chimpanzees in Cameroon, while the HIV-1 O and P groups came from western lowland gorillas in Cameroon (D’arc et al., 2015; Keele et al., 2006; Li et al., 2012; Sharp and Hahn, 2010). The diversity of HIV-1 O group viruses in humans is nearly equivalent to the diversity of HIV-1 M group viruses, and one of the earliest proven AIDS cases in humans was with O group virus infecting a Norwegian sailor and his family in 1970(Wertheim and Worobey, 2009) (Froeland 1988). Despite apparently similar timing of the origins, the O group has not spread around the world in humans and evolved into subtypes, while the HIV-1 M group has. It is possible that the reason for the limited spread of group O versus M was due to stochastics associated with the ignition and generation of HIV-1 epidemic at the early stage(Faria et al., 2014)(Hogan et al., 2016).
2.1 HIV-1 M group
2.1.1 Subtypes, Subsubtypes and local outbreak clusters
Very early in the study of HIV and AIDS it was noted that there was very great diversity between viruses isolated from patients sampled in the developed world, and patients sampled in Africa (Smith 1988). Although the very first HIV-1 isolates to be cloned and sequenced were from the developed world, they were named “subtype B” in the 1992 HIV Database Compendium (Myers 1992). The 1992 Database Compendium named subtypes A through D for gag and A though E for envelope sequences, noting that subtype E viruses were classified as A in the gag gene. Although this publication established the subtype nomenclature for the HIV-1 M group, there is no discussion of how the letters were assigned to each clade. Researchers studying serological reactions to various isolates in human infections and in animals experimentally infected with cloned isolates had been using the term “subtype” for at least 4 years (Cheng-Mayer 1988)(Neurath 1990) but those serological groups do not all coincide with the genotypes.
By 1994 subtype F had been detected among orphans infected in an outbreak in Romania, and in Brazil more subtype F, as well as B/F recombinant virus was found (Cernescu et al., 1994)(Morgado et al., 1994). Subtype G was also described in 1994 (Bobkov 1994).
Between 1992 and 2000 subtypes A though K were established and many circulating recombinant forms were described (Triques et al., 2000). Although there have been a few nearly complete genomes of HIV-1 M group viruses sequenced since 2000 which are unique from the established subtypes, and thus hint that other subtypes of HIV-1 exist, in no case have 3 or more of the same clade been identified to qualify for naming a new subtype. Conversely, a set of related viruses which was initially designated as subtype I was renamed as a circulating recombinant form because several regions of its genome fell within the established A–H clades even though other regions were unique enough to warrant a new subtype designation (Gao et al., 1998; Kostrikis et al., 1995)(Paraskevis et al., 2001)(Nasioulas et al., 1999). Indeed these unique regions were later found to be more closely related to subytpes J and K once those subtypes were described (Montavon et al., 2002). Although it is not completely incorrect to refer to the unique regions in CRF01_AE as “E” or the unique regions in CRF04_cpx as “I”, it is highly recommended to refer to these lineages, and subgenomic regions derived from these lineages, as CRF01_AE and CRF04_cpx.
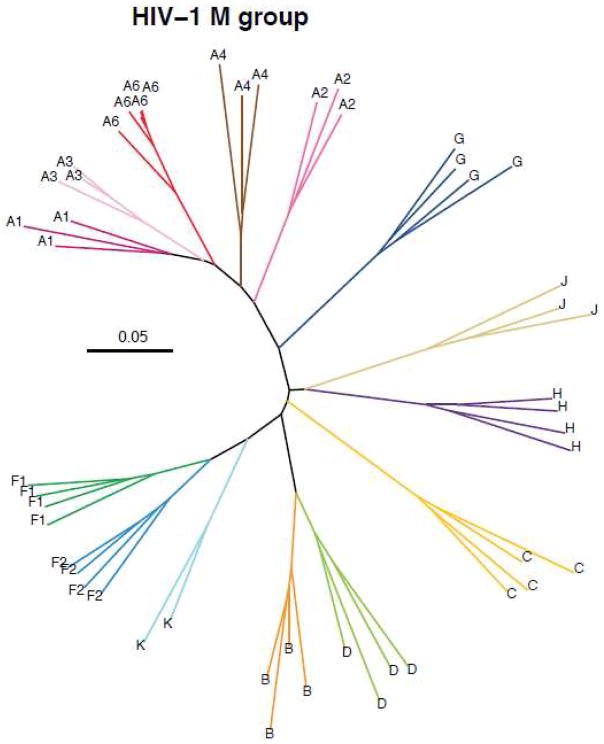
Differences between the subtypes of the HIV-1 M group include many other attributes in addition to the DNA and amino acid sequence diversity and distances. Some subtypes account for millions of infections in dozens of countries while other subtypes have a very limited epidemiology. Some of the subtypes apparently began spreading earlier than others and are thus more diverse, while others followed different epidemiological trajectories. Subtype A is very diverse and six major subclades have been designated subsubtypes A1 through A6(Vidal et al., 2009, 2006). Subsubtype A1 spread considerably across Africa especially in eastern Africa and A2 to some extent in central Africa. The A3, A4 and A5 subsubtypes each have few sequences described and no pure A5 strains have been identified, A5 is only found in large regions of the CRF26_A5U genomes (Meloni et al., 2004)(Vidal et al., 2006)(Vidal et al., 2009). Another group of divergent subtype A viruses, labeled as subsubtype A6 in figure 3, has spread quite widely among IDU in the regions of the former Soviet Union and surrounding areas(Bobkov et al., 1994; Lapovok et al., 2014). This group of viruses have been also named as AFSU corresponding the geographic area (former Soviet Union countries) of their endemicity. Subtype B is closely related to subtype D, suggesting that these clades share a more recent common ancestor than the other subtypes. However subtype B was defined and named as its own subtype long before the addition of subsubtypes to the nomenclature scheme and is grandfathered in as a separate subtype. A similar relationship, although deeper, exists between subtypes F and K.
Figure 3.
Maximum likelihood phylogenetic tree constructed using nearly complete HIV-1 M group genomes (gag through nef genes). Sequences were aligned with the HIV Databases GeneCutter tool (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html) and a maximum likelihood phylogenetic tree was constructed using the GTR model of evolution plus site-specific rates of evolution using IQ-tree(Nguyen et al., 2015).
The assignment of subsubtypes and of other subdivisions within a subtype creates some difficult issues. Soon after HIV-1 subtype B was detected in Thailand in the 1990s, it was noted that the Thai epidemic seemed to be spreading from one introduction of virus, and this became known as the B’ (B-prime) or Thai-B strain(Kalish et al., 1995). Numerous other such founder effects have resulted in similar subclades or local epidemic strains in other parts of the world. It is acceptable and indeed good practice to give labels to any subclades or local strains in order to clearly convey concepts about the epidemiology of a subset of viruses. However, it is obvious that there can be no clear definitions put in place to standardize the nomenclature of these lineages. It would be useful to name these subclades according to the geographic area(s) and/or risk groups where they are endemic.
The Subtyping Distance tool (SUDI; http://www.hiv.lanl.gov/content/sequence/SUDI/sudi.html) and the PhyloPlace tool (http://www.hiv.lanl.gov/content/sequence/phyloplace/PhyloPlace.html) were created in order to assist researchers in determining whether or not a group of related sequences represents a true “sub-subtype”, a new subtype, or just some variants of an already labeled subtype.
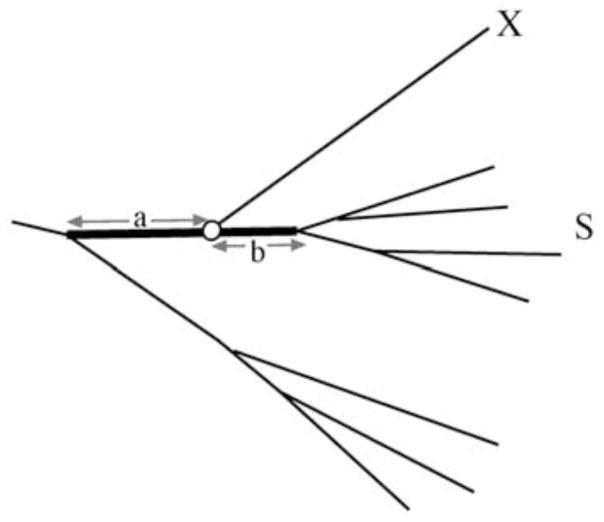
The HIV and SIV nomenclature committee has not defined a cut-off rule on the branching index value for determining when a single virus isolate sequence or group of sequences should be named as a subtype or variant (Fig 4). However, a subtype specific branching index evaluation was reported in Hraber et al (Hraber et al., 2008). While an overall cut-off at 0.66 can be used as a rule of thumb, the cut-off for certain subtype associations varies across the genome and between subtypes. Applying a universal cut-off is complicated by the inclusion of new subtype reference sequences, and thus relies on a fixed set of reference sequences for meaningful results. For instance, we may have a small region of a complex recombinant genome that is “H-like” or falls close to the “crown group” of subtype H sequences that were chosen as the subtype H reference set of sequences, in phylogenetic analysis. If the branching index is greater than 0.66 it makes a lot of sense to label this small region of the genome as “H” or “H-like” rather than labeling it as “U” or “undetermined” subtype. As another example, we may have a group of related viruses such as the virus isolates now labeled as subtype K and we note that in some regions of the genome they are F-like and in other regions of the genome they are C-like. In this case we are concerned with whether we should name the viruses as a new subtype (subtype K) or name them as a circulating recombinant form. Even though at least one region of the subtype K genomes fall in between the F1 and F2 subsubtypes, in no place do the subtype K genomes have a branching index of more than 0.66 for either F1 or F2, so they were named as subtype K. An important point in this decision, is that the genomic region where subtype K sequences fall in between F1 and F2 is a region where F1 and F2 are very diverse from one another, a region of low signal to noise ratio. Historically different clades have been named as subtypes based on partial genomic analysis. Moreover, at the early time of subtype assignment, currently existing tools and nomenclature proposal were not available. Thus some of the existing classification rules are not fulfilled by some subtypes (e.g. subtypes D and K). No matter the previous issues, our recommendation for current assignments is that they should be according to the criteria specified in the existing nomenclature proposal.
Figure 4.
Schematic picture of the branching index. Here the association of sequence X to the subtype cluster S is investigated. The letters a and b represent genetic distances that depend on the position of the node of sequence X (white circle) at the bold branch. The branching index is defined as a/(a + b) and can take values between 0 and 1 , where 0 means no support and 1 means solid support for belonging to the subtype(Wilbe et al., 2003).
Isolates of HIV-1 M group virus which are not related to any one of the defined subtypes or circulating forms are labeled as “U” for unique or unsubtyped. Because the HIV-1 M group pandemic began with a Chimpanzee cross-species transmission to human in central Africa it is rather common to find such unique viruses in or near central Africa(Mokili et al., 2002). To date, the sampling and sequencing of nearly complete genomes of HIV-1 isolated from central Africa has been rather sparse in comparison to the number of infections there, and this contributes to a lack of identifying more subtypes. A recent paper studying published genomes suggests that more subtypes may be defined in the future(Tongo et al., 2015). The ongoing PANGEA_HIV project (http://www.pangea-hiv.org/) may also make contributions in this area(Pillay et al., 2015).
In recent years several research groups have named local subclades of CRF01_AE viruses found in various cities, or risk groups within cities in China, with sequential numbers(Zeng et al., 2016)(Feng et al., 2013). It is difficult to ascertain whether the authors of different papers are using the same numbering system such that “cluster 1” in one paper is the same local epidemic as “clade 1” in another publication. The papers most often do not provide a table clearly linking each virus isolate to each cluster or subclade, and the GenBank entries also lack clear annotation, so it is very difficult to determine which sequences belong to each subclade. Most of the analyses are based on small genomic regions rather than complete genomes in order to obtain enough isolate sequences to find the clusters. Using those same genomic regions and subtype B from the USA, similar clusters or subclades are notable, although they are not always geographically ordered. Another difficulty with this fine classification within a subtype or CRF, is that the geographical location is often not specified below the country level, so it is not possible to search GenBank or the HIV database for, for example, “New York” or “California” isolate sequences. Although it can be useful to give names to local clusters in order to discuss them in a paper, no expectations can be made for those names to be useful beyond the one study being published. Authors of papers describing sequences are encouraged to provide annotation in GenBank that clearly identifies the virus sample date, genotype, location and any other pertinent information.
2.1.2 Circulating Recombinant Forms
Whenever an individual is infected by more than one HIV-1 subtype, whether it is a dual infection before an effective immune response to the first virus is mounted, or a superinfection after the first virus has been partially suppressed by the host immune response, recombinants between the two virus lineages may soon be formed. The recombination is not due to strand breakage and re-joining as in most dsDNA recombination events. It is due to template switching during the reverse transcription process. Two copies of the viral RNA genome are packaged in each virus particle, so a single cell must be dually infected such that one copy of each of the two genomes is packaged, and the reverse transcriptase switches between the two templates during the reverse transcription process. This process leads to a recombination rate on par with the high substitution rate (Neher and Leitner, 2010)
Quite early in the studies of complete genome sequences from isolates of the HIV-1 M group viruses, it was noted that viruses from Thailand formed a clade with some of the viruses from Africa in gag and pol regions of the genome but formed their own distinct clade in the env gene region (Carr et al., 1996)(McCutchan et al., 1992). The env gene genotype had already been named “subtype E” (Myers et al., 1992). Some debate was carried out as to whether the envelope gene of these viruses had just evolved very rapidly compared to the gag-pol region of the genome, or alternatively if recombination had been the cause (Anderson et al., 2000)(Robertson et al., 2000). The name CRF01_AE was adopted by the majority vote of the HIV and SIV nomenclature committee(Robertson et al., 2000). The CRF01_AE genotype was, and remains, somewhat problematic because no full length non-recombinant “genotype E” virus has yet been sampled and sequenced. Thus the env region could be called “U” for “untyped” if it was not already grandfathered in as “subtype E”. Moreover it has been shown that it contains a small genomic fragment closely related to subtype G suggesting that CRF01_AE has a complicated pattern of mosaicism consisting of subtypes A, G, and E.(Magiorkinis et al., 2002).
The CRF02_AG genomes are somewhat similar to CRF01_AE with regard to both of these circulating recombinant forms originating many decades ago. It is clear that these viruses had already spread quite substantially before HIV was discovered. Careful analyses of subregions of HIV-1 genomes suggests that recombinations were ongoing in the early years before the subtypes each expanded, and it has been suggested that for example CRF02_AG may predate the origin of subtype G(Abecasis et al., 2007)(Zhang et al., 2010). As stated in the HIV nomenclature proposal, the “pure subtypes” were not intended to imply that no recombinations had contributed to their formation, but only that they had spread extensively and have a relatively uniform phylogenetic signal or origin across their genomes (Robertson et al., 2000). The initial HIV classification reflects in fact sampling history rather than evolutionary history of the virus. The situation is different in recent epidemics. For example, the third recombinant form to be discovered, CRF03_AB from the Ukraine, was the result of a very recent recombination event that took place very shortly before it was discovered (Liitsola et al., 1998)(Liitsola et al., 2000). Although the exact individual who was dual infected with subtypes A and B to create the CRF03_AB virus that then spread among IDU in the region was never identified, isolates of viruses very closely related to the parental strains were found among IDU in the same geographical region and sequenced(Zarandia et al., 2006).
By the time of the 1999 HIV and SIV Nomenclature Committee meeting, four circulating recombinant forms had been identified and published and it was clear that more were in press or in the process of being characterized(Robertson et al., 2000)(http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/nomenclature/Nomen.html accessed May 26, 2016). The committee established and agreed that new subtypes and CRFs would require at least three independent isolates of the same form. The complete genome sequence must be determined for at least two of the isolates. The third isolate has to be sequenced in gene regions which prove that it belongs to the same new subtype or recombinant form.
2.1.3 Unique Intersubtype Recombinants
Overall, dual infections with more than one subtype of virus are rather rare except in certain regions of the world where two or more subtypes or CRFs co-circulate, in population groups with high HIV prevelance and high risk behavior, for example within an IDU community, or an MSM network. In central and western Africa many intersubtype recombinant viruses are assumed to have arisen via sexual transmissions, because opioid injection drug use is not very common.
The number of recombinant viruses that are sequenced is influenced not only by the number of dual-infected people per year, but also by the length of time that two or more subtypes have been circulating in the local area, and the percentage of infected people who are sampled and sequenced.
2.1.4 Intrasubtype Recombinants
With more than 30 million people infected with HIV today, and less than 100,000 infected people sampled for DNA sequencing (excluding drug resistance testing, where the sequences are not made public), a considerable proportion of sequences are unique enough to fall on their own branch within a subtype, rather than being part of a subclade within the subtype. Even when studies have sampled infections in rather small villages the infections have most often been found to be surprisingly diverse, with most transmissions coming into the villages from surrounding cities(Grabowski et al., 2014)(Carrel et al., 2014).
2.1.5 Small Genome Regions
In a recent paper, Marcel Tongo et al. propose using lowercase letters to indicate regions of a virus genome that are somewhat similar to, but fall outside the known diversity of a given subtype(Tongo et al., 2015). These regions are currently listed as “U” for untyped or unclassified. One of the problems with this proposal, is that there can be many different reasons why a small region is not classifiable. In many cases, it is very likely that recombination has confounded our ability to reconstruct an accurate history. On the other side of the same issue, some researchers have argued that only regions of the genome which give 100% bootstrap support for being in the crown group of a given subtype should be labeled with that subtype, and other regions should be listed as “U” for uncertain or untyped.
For most small regions of the genome, less than 100 or in some cases 200 bases long, obtaining solid bootstrap support for the correct classification is problematic. Indeed, the length of the genomic fragment that gives proper subtype signal varies across the genome.
3 HIV-2, SIV-SMM, SIV-Mac
HIV-2 is the result of cross species transmissions from sooty mangabeys (Cercocebus atys) to humans. To date at least nine different lineages representing nine different cross species transfers to humans have been described, resulting in HIV-2 groups A–I(Ayouba et al., 2013)(Santiago et al., 2005). HIV-2 groups A and B have spread in western Africa and have also been detected in India and other countries. Dual infections with HIV-2 groups A and B have been reported, and at least one HIV-2 A/B recombinant virus has spread to several individuals to become listed as HIV-2 CRF01_AB(Ibe et al., 2010). Dual infections with HIV-1 and HIV-2 have been reported many times, but not recombination between these two distantly related viruses, and for most reports of dual infection there is only serological and not genetic data for confirming the dual infection.
Although HIV-2 groups A and B have spread to many humans, the groups C through I of HIV-2 have mostly been detected and sequenced in just one or two individuals and technically do not meet the nomenclature criteria to be named as a group. However, because of the relative importance of cross species transmission events, it is worthwhile to note each event, even when it does not result in significant human to human spread of the virus.
The SIVsmm also crossed species to infect macaques in captivity in the USA several times in the 1960s and 1970s(Apetrei et al., 2005). Several lineages of the virus then evolved in captive macaques and/or sooty mangabeys co-housed with macaques in different primate research centers in the USA. A few of these viruses were isolated in the 1980s and became very important research reagents for studying immune deficiency in macaques and testing vaccines in a nonhuman primate model(Fischer et al., 2012)(http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/KUIKEN2000/Kuiken.html accessed June 7, 2016).
For virus lineages where the transfer from sooty mangabeys to captive macaques occurred unintentionally they have been labeled as SIVstm for stump tailed macaque(Novembre et al., 1992), SIVmac for rhesus macaque(Apetrei et al., 2005), and SIVmne for Macaca nemestrema (pig tailed macaque)(Kimata et al., 1998). Laboratory made chimeras between SIVmac and HIV-1, most often with the envelope gene derived from HIV-1, are common in vaccine research, and these are called SHIVs for simian-human immunodeficiency virus.
4 PIV Details and peculiarities
The number of primate immunodeficiency viruses isolated and sequenced from nonhuman primates has increased dramatically in recent years since the development of protocols for isolating viruses from freshly collected fecal samples allowed non-invasive sampling of wild animals(Santiago et al., 2003). It has long been assumed that the ancient virus host coevolution of these viruses has resulted in a decrease in pathogenicity over time to the point where the viruses are nonpathogenic to their natural hosts. Indeed there is evidence that many of the viruses are far less pathogenic to the natural host than HIV-1 is to humans or SIVsmm and SIVmac are to macaques. However, it was always known that the chimpanzee named Marilyn, from which the first SIVcpz was isolated, had died with infections in 1985 after giving birth to stillborn twins(Gao et al., 1999). More recent studies have determined that at least some of the natural infections are pathogenic but typically with long latency periods not unlike humans infected with HIV-1 or HIV-2(Keele et al., 2009).
Several studies specifically focused on developing highly pathogenic SIVs and SHIVs in order to produce vaccine challenge viruses such that large improvements in the clinical outcome of vaccinated animals could be seen in short periods of time(Haddrick et al., 2001)(McCormick-Davis et al., 1998). Although these viruses prove that there can be great differences in pathogenicity between viruses that are very similar to each other, the majority of studies of rates of disease progression in humans infected with HIV-1 M group viruses have shown that it is human genotype rather than viral genotype that is most often the determining factor(McLaren and Carrington, 2015)(Limou and Zagury, 2013). Highly active antiretroviral therapies in humans have made studies of disease progression rates in untreated humans unethical(INSIGHT START Study Group et al., 2015)(Wagner et al., 2016).
Estimating the length of time that PIVs have coevolved with their natural hosts is problematic due to the lack of solid fossil record or datable events deep in the phylogenies. Molecular phylogenetic methods are hampered by the problem of saturation(Duchêne et al., 2015). Although no New World monkeys are known to be infected with PIVs, felines around the world are infected with feline immunodeficiency viruses with no greater diversity than is found in the African PIVs(Troyer et al., 2011)(O’Brien et al., 2012). Endogenous lentiviruses have been found in lemurs, rabbits, ferrets and weasels and estimated to have become endogenous as much as ten million years ago(Han and Worobey, 2012). Although other estimates of PIV origins have been made with populations of primates separated for long periods of time, these methods can be confounded by not knowing that the animals which became isolated from each other were carrying just one lineage of virus at the time of isolation(Wertheim and Worobey, 2009)(Worobey et al., 2010)(Ayouba et al., 2015).
Cross species transmissions of PIVs are not limited to primate to human transfers. It is clear for example that baboons have repeatedly been infected in the wild with SIVver from the vervet species of African Green Monkeys, Chlorocebus pygerythrus(van Rensburg et al., 1998)(Jin et al., 1994). The SIV lineage infecting chimanzees and gorillas is apparently the result of a very ancient recombination event, with the pol gene of SIVcpz somewhat related to virus from red capped mangabeys while the env gene is more closely related to virus from greater spot-nosed monkeys(Etienne et al., 2013)(Bailes, 2003) (Leitner et al., 2007). Fighting and predation between primate species is rather common, so the relative lack of cross species transmission events is a testament to how well the primate APOBEC system, and other innate and adaptive immune functions protect against these viruses(Puvvada and Patel, 2013)(Bibollet-Ruche et al., 2004)(Etienne et al., 2015)(D’arc et al., 2015).
5 Conclusions
Although there are some inconsistencies and confusing issues in the nomenclature of HIVs and PIVs, these problems are mostly a reflection either of changes in the state of knowledge about the diversity of these viruses over time, or inherent biological quirks such as recombination and cross species transmissions. The goals of the classification and nomenclature system are both to make biological sense and to make it possible for humans and man-made databases to organize and understand the data. Moreover classification of viral sequences is crucial to understand the complex epidemiology of HIV-1 epidemic and also to monitor temporal and spatial changes on a global and regional scale. There have been proposals to better organize the CRFs such as grouping related recombinants together(Zhang et al., 2010). The future seems certain to bring challenges to the existing system. One confusing issue may come up if more subtypes in the HIV-1 M group are discovered and classified, such that the A, B, C, D naming of subtypes overtakes the M, N, O, P naming of groups.
Despite some complexities or difficulties with naming primate lentiviruses, it is very clear that the current system works well in comparison to the classification of many or even most other viruses(Fauquet and Fargette, 2005). For many viruses, the lack of standards set by the International Committee for the Taxonomy of Viruses (ICTV) below the species level is a barrier to accurate classification. One problem is that there is no biologically solid way to define what constitutes a “species” of virus. For the lentiviruses a classification based partly on host makes good sense because in general the cross species events are quite rare in comparison to coevolution within a virus-host pair. But for other viruses such as the influenza A virus it is very common for a virus that is very clearly a human virus to be isolated from a pig, or vise versa.
In cases where the current rules for nomenclature and classification of lentiviruses are violated by names that were “grandfathered in” before these rules were standardized, the problem of rules broken is less severe than the confusion which would be caused by renaming well-known groups now. Most challenges with the current system are local in nature, such as describing the epidemiology of local outbreaks, and do not have an impact on the HIV-1 M group pandemic. It is completely acceptable to use names for local strains as has already been done many times, but authors should not expect those names to become incorporated into the official nomenclature.
Supplementary Material
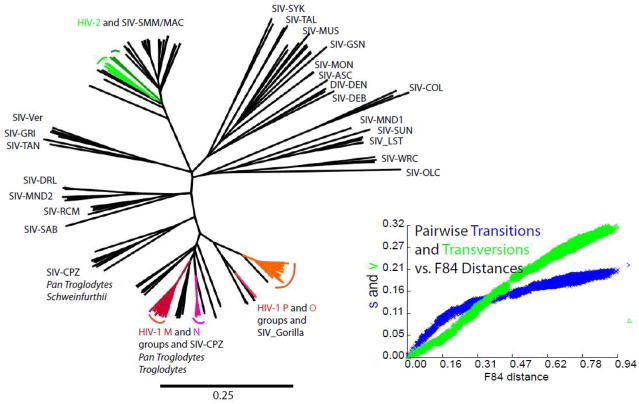
Figure 1.
A maximum likelihood phylogenetic tree constructed from the nearly complete genomes (gag through nef genes) of primate immunodeficiency viruses. The plot of transitions and transversions vs estimated evolutionary distances shows that the distances across the alignment are far beyond saturation of silent site mutations and thus underestimated in comparison to distances within a recent expansion such as the HIV-1 M group of viruses. Clades in color represent viruses identified in humans after relatively recent transfer from chimpanzee, gorilla, and sooty mangabey reservoirs.
Figure 2.
The cover of the 1992 HIV Database Compendium illustrated the diversity of HIV-1 M group viruses that had been sequenced to date, with a phylogenetic tree built from envelope gene sequences including 3 sequences from each subtype, plus one from “subtype E” which later became known as the CRF01_AE circulating recombinant form.
Acknowledgments
Funding: This work was supported by the National Institutes of Health, NIH Contract AAI12007-001-01001
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abecasis AB, Lemey P, Vidal N, de Oliveira T, Peeters M, Camacho R, Shapiro B, Rambaut A, Vandamme AM. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J Virol. 2007;81:8543–51. doi: 10.1128/JVI.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, Coon M, Girard M, Osmanov S, Hood L, Mullins JI. Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J Virol. 2000;74:10752–65. doi: 10.1128/jvi.74.22.10752-10765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol. 2005;79:8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, Li Y, Hahn BH, Delaporte E, Leendertz FH, Peeters M. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural Côte d’Ivoire. AIDS. 2013;27:2488–91. doi: 10.1097/01.aids.0000432443.22684.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayouba A, Njouom R, Chia JE, Ahuka-Mundeke S, Kfutwah A, Ngole EM, Nerrienet E, Delaporte E, Peeters M. Molecular characterization of a new mosaic Simian Immunodeficiency Virus in a naturally infected tantalus monkey (Chlorocebus tantalus) from Cameroon: a challenge to the virus-host co-evolution of SIVagm in African green monkeys. Infect Genet Evol. 2015;30:65–73. doi: 10.1016/j.meegid.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes E. Hybrid Origin of SIV in Chimpanzees. Science (80- ) 2003;300:1713–1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, Mwenda JM, Langat DK, Chege GK, McClure HM, Mpoudi-Ngole E, Delaporte E, Peeters M, Shaw GM, Sharp PM, Hahn BH. New simian immunodeficiency virus infecting De Brazza’s monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J Virol. 2004;78:7748–62. doi: 10.1128/JVI.78.14.7748-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkov A, Cheingsong-Popov R, Garaev M, Rzhaninova A, Kaleebu P, Beddows S, Bachmann MH, Mullins JI, Louwagie J, Janssens W. Identification of an env G subtype and heterogeneity of HIV-1 strains in the Russian Federation and Belarus. AIDS. 1994;8:1649–55. doi: 10.1097/00002030-199412000-00002. [DOI] [PubMed] [Google Scholar]
- Carr JK, Salminen MO, Koch C, Gotte D, Artenstein AW, Hegerich PA, St Louis D, Burke DS, McCutchan FE. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–43. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel M, Eron JJ, Emch M, Hurt CB. Spatial epidemiology of recently acquired HIV infections across rural and urban areas of North Carolina. PLoS One. 2014;9:e88512. doi: 10.1371/journal.pone.0088512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernescu CE, Tardei G, Necula A, Ruta SM, Pau CP. The serologic significance of F viral genotype for human immunodeficiency virus type 1 epidemic. J Infect Dis. 1994;170:1043–4. doi: 10.1093/infdis/170.4.1043. [DOI] [PubMed] [Google Scholar]
- D’arc M, Ayouba A, Esteban A, Learn GH, Boué V, Liegeois F, Etienne L, Tagg N, Leendertz FH, Boesch C, Madinda NF, Robbins MM, Gray M, Cournil A, Ooms M, Letko M, Simon VA, Sharp PM, Hahn BH, Delaporte E, Mpoudi Ngole E, Peeters M. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A. 2015;112:E1343–52. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne S, Ho SYW, Holmes EC. Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evol Biol. 2015;15:36. doi: 10.1186/s12862-015-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Bibollet-Ruche F, Sudmant PH, Wu LI, Hahn BH, Emerman M. The Role of the Antiviral APOBEC3 Gene Family in Protecting Chimpanzees against Lentiviruses from Monkeys. PLoS Pathog. 2015;11:e1005149. doi: 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Hahn BH, Sharp PM, Matsen FA, Emerman M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe. 2013;14:85–92. doi: 10.1016/j.chom.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pépin J, Posada D, Peeters M, Pybus OG, Lemey P. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Fargette D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J. 2005;2:64. doi: 10.1186/1743-422X-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He X, Hsi JH, Li F, Li X, Wang Q, Ruan Y, Xing H, Lam TTY, Pybus OG, Takebe Y, Shao Y. The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS. 2013;27:1793–802. doi: 10.1097/QAD.0b013e328360db2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Apetrei C, Santiago ML, Li Y, Gautam R, Pandrea I, Shaw GM, Hahn BH, Letvin NL, Nabel GJ, Korber BT. Distinct evolutionary pressures underlie diversity in simian immunodeficiency virus and human immunodeficiency virus lineages. J Virol. 2012;86:13217–31. doi: 10.1128/JVI.01862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–41. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Gao F, Robertson DL, Carruthers CD, Li Y, Bailes E, Kostrikis LG, Salminen MO, Bibollet-Ruche F, Peeters M, Ho DD, Shaw GM, Sharp PM, Hahn BH. An isolate of human immunodeficiency virus type 1 originally classified as subtype I represents a complex mosaic comprising three different group M subtypes (A, G, and I) J Virol. 1998;72:10234–41. doi: 10.1128/jvi.72.12.10234-10241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski MK, Lessler J, Redd AD, Kagaayi J, Laeyendecker O, Ndyanabo A, Nelson MI, Cummings DAT, Bwanika JB, Mueller AC, Reynolds SJ, Munshaw S, Ray SC, Lutalo T, Manucci J, Tobian AAR, Chang LW, Beyrer C, Jennings JM, Nalugoda F, Serwadda D, Wawer MJ, Quinn TC, Gray RH. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med. 2014;11:e1001610. doi: 10.1371/journal.pmed.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrick M, Brown CR, Plishka R, Buckler-White A, Hirsch VM, Ginsberg H. Biologic studies of chimeras of highly and moderately virulent molecular clones of simian immunodeficiency virus SIVsmPBj suggest a critical role for envelope in acute AIDS virus pathogenesis. J Virol. 2001;75:6645–59. doi: 10.1128/JVI.75.14.6645-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GZ, Worobey M. Endogenous lentiviral elements in the weasel family (Mustelidae) Mol Biol Evol. 2012;29:2905–8. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–92. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- Hogan CA, Iles J, Frost EH, Giroux G, Cassar O, Gessain A, Dion M-J, Ilunga V, Rambaut A, Yengo-Ki-Ngimbi A-É, Behets F, Pybus OG, Pépin J. Epidemic History and Iatrogenic Transmission of Blood-borne Viruses in Mid-20th Century Kinshasa. J Infect Dis. 2016 doi: 10.1093/infdis/jiw009. [DOI] [PubMed] [Google Scholar]
- Hraber P, Kuiken C, Waugh M, Geer S, Bruno WJ, Leitner T. Classification of hepatitis C virus and human immunodeficiency virus-1 sequences with the branching index. J Gen Virol. 2008;89:2098–107. doi: 10.1099/vir.0.83657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–9. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Ibe S, Yokomaku Y, Shiino T, Tanaka R, Hattori J, Fujisaki S, Iwatani Y, Mamiya N, Utsumi M, Kato S, Hamaguchi M, Sugiura W. HIV-2 CRF01_AB: first circulating recombinant form of HIV-2. J Acquir Immune Defic Syndr. 2010;54:241–7. doi: 10.1097/QAI.0b013e3181dc98c1. [DOI] [PubMed] [Google Scholar]
- INSIGHT START Study Group. Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, Avihingsanon A, Cooper DA, Fätkenheuer G, Llibre JM, Molina J-M, Munderi P, Schechter M, Wood R, Klingman KL, Collins S, Lane HC, Phillips AN, Neaton JD. Initiation of Antiretroviral Therapy in Early Asymptomatic HIV Infection. N Engl J Med. 2015;373:795–807. doi: 10.1056/NEJMoa1506816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MJ, Rogers J, Phillips-Conroy JE, Allan JS, Desrosiers RC, Shaw GM, Sharp PM, Hahn BH. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–60. doi: 10.1128/jvi.68.12.8454-8460.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish ML, Baldwin A, Raktham S, Wasi C, Luo CC, Schochetman G, Mastro TD, Young N, Vanichseni S, Rübsamen-Waigmann H. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS. 1995;9:851–7. doi: 10.1097/00002030-199508000-00004. [DOI] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, Wroblewski E, Mosser A, Raphael J, Kamenya S, Lonsdorf EV, Travis DA, Mlengeya T, Kinsel MJ, Else JG, Silvestri G, Goodall J, Sharp PM, Shaw GM, Pusey AE, Hahn BH. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–9. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JFY, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–6. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata JT, Mozaffarian A, Overbaugh J. A lymph node-derived cytopathic simian immunodeficiency virus Mne variant replicates in nonstimulated peripheral blood mononuclear cells. J Virol. 1998;72:245–56. doi: 10.1128/jvi.72.1.245-256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrikis LG, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho DD. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–30. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapovok I, Kazennova E, Laga V, Vasilyev A, Utegenova A, Abishev A, Dzissyuk N, Tukeev M, Bobkova M. Short communication: molecular epidemiology of HIV type 1 infection in Kazakhstan: CRF02_AG prevalence is increasing in the southeastern provinces. AIDS Res Hum Retroviruses. 2014;30:769–74. doi: 10.1089/AID.2013.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T, Dazza MC, Ekwalanga M, Apetrei C, Saragosti S. Sequence diversity among chimpanzee simian immunodeficiency viruses (SIVcpz) suggests that SIVcpzPts was derived from SIVcpzPtt through additional recombination events. AIDS Res Hum Retroviruses. 2007;23:1114–8. doi: 10.1089/aid.2007.0071. [DOI] [PubMed] [Google Scholar]
- Li Y, Ndjango JB, Learn GH, Ramirez MA, Keele BF, Bibollet-Ruche F, Liu W, Easlick JL, Decker JM, Rudicell RS, Inogwabini BI, Ahuka-Mundeke S, Leendertz FH, Reynolds V, Muller MN, Chancellor RL, Rundus AS, Simmons N, Worobey M, Shaw GM, Peeters M, Sharp PM, Hahn BH. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol. 2012;86:10776–91. doi: 10.1128/JVI.01498-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liitsola K, Holm K, Bobkov A, Pokrovsky V, Smolskaya T, Leinikki P, Osmanov S, Salminen M. An AB recombinant and its parental HIV type 1 strains in the area of the former Soviet Union: low requirements for sequence identity in recombination. UNAIDS Virus Isolation Network. AIDS Res Hum Retroviruses. 2000;16:1047–53. doi: 10.1089/08892220050075309. [DOI] [PubMed] [Google Scholar]
- Liitsola K, Tashkinova I, Laukkanen T, Korovina G, Smolskaja T, Momot O, Mashkilleyson N, Chaplinskas S, Brummer-Korvenkontio H, Vanhatalo J, Leinikki P, Salminen MO. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS. 1998;12:1907–19. doi: 10.1097/00002030-199814000-00023. [DOI] [PubMed] [Google Scholar]
- Limou S, Zagury JF. Immunogenetics: Genome-Wide Association of Non-Progressive HIV and Viral Load Control: HLA Genes and Beyond. Front Immunol. 2013;4:118. doi: 10.3389/fimmu.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorkinis G, Paraskevis D, Magiorkinis E, Vandamme AM, Hatzakis A. Reanalysis of the HIV-1 circulating recombinant form A/E (CRF01_AE): evidence of A/E/G recombination. J Acquir Immune Defic Syndr. 2002;30:124–9. doi: 10.1097/00042560-200205010-00017. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Zhao LJ, Mukherjee S, Leung K, Sheffer D, Joag SV, Narayan O, Stephens EB. Chronology of genetic changes in the vpu, env, and Nef genes of chimeric simian-human immunodeficiency virus (strain HXB2) during acquisition of virulence for pig-tailed macaques. Virology. 1998;248:275–83. doi: 10.1006/viro.1998.9300. [DOI] [PubMed] [Google Scholar]
- McCutchan FE, Hegerich PA, Brennan TP, Phanuphak P, Singharaj P, Jugsudee A, Berman PW, Gray AM, Fowler AK, Burke DS. Genetic variants of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1887–95. doi: 10.1089/aid.1992.8.1887. [DOI] [PubMed] [Google Scholar]
- McLaren PJ, Carrington M. The impact of host genetic variation on infection with HIV-1. Nat Immunol. 2015;16:577–83. doi: 10.1038/ni.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni ST, Kim B, Sankalé JL, Hamel DJ, Tovanabutra S, Mboup S, McCutchan FE, Kanki PJ. Distinct human immunodeficiency virus type 1 subtype A virus circulating in West Africa: sub-subtype A3. J Virol. 2004;78:12438–45. doi: 10.1128/JVI.78.22.12438-12445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JLK, Rogers M, Carr JK, Simmonds P, Bopopi JM, Foley BT, Korber BT, Birx DL, McCutchan FE. Identification of a novel clade of human immunodeficiency virus type 1 in Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2002;18:817–23. doi: 10.1089/08892220260139567. [DOI] [PubMed] [Google Scholar]
- Montavon C, Toure-Kane C, Nkengasong JN, Vergne L, Hertogs K, Mboup S, Delaporte E, Peeters M. CRF06-cpx: a new circulating recombinant form of HIV-1 in West Africa involving subtypes A, G, K, and J. J. Acquir. Immune Defic Syndr. 2002;29:522–30. doi: 10.1097/00126334-200204150-00014. [DOI] [PubMed] [Google Scholar]
- Morgado MG, Sabino EC, Shpaer EG, Bongertz V, Brigido L, Guimaraes MD, Castilho EA, Galvão-Castro B, Mullins JI, Hendry RM. V3 region polymorphisms in HIV-1 from Brazil: prevalence of subtype B strains divergent from North American/European prototype and detection of subtype F. AIDS Res Hum Retroviruses. 1994;10:569–76. doi: 10.1089/aid.1994.10.569. [DOI] [PubMed] [Google Scholar]
- Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;8:373–86. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- Nasioulas G, Paraskevis D, Magiorkinis E, Theodoridou M, Hatzakis A. Molecular analysis of the full-length genome of HIV type 1 subtype I: evidence of A/G/I recombination. AIDS Res Hum Retroviruses. 1999;15:745–58. doi: 10.1089/088922299310836. [DOI] [PubMed] [Google Scholar]
- Neher RA, Leitner T. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput Biol. 2010;6:e1000660. doi: 10.1371/journal.pcbi.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre FJ, Hirsch VM, McClure HM, Fultz PN, Johnson PR. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology. 1992;186:783–7. doi: 10.1016/0042-6822(92)90047-s. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Troyer JL, Brown MA, Johnson WE, Antunes A, Roelke ME, Pecon-Slattery J. Emerging viruses in the Felidae: shifting paradigms. Viruses. 2012;4:236–57. doi: 10.3390/v4020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Magiorkinis M, Vandamme AM, Kostrikis LG, Hatzakis A. Re-analysis of human immunodeficiency virus type 1 isolates from Cyprus and Greece, initially designated “subtype I”, reveals a unique complex A/G/H/K/? mosaic pattern. J Gen Virol. 2001;82:575–80. doi: 10.1099/0022-1317-82-3-575. [DOI] [PubMed] [Google Scholar]
- Pillay D, Herbeck J, Cohen MS, de Oliveira T, Fraser C, Ratmann O, Brown AL, Kellam P, PANGEA-HIV Consortium. PANGEA-HIV: phylogenetics for generalised epidemics in Africa. Lancet Infect Dis. 2015;15:259–61. doi: 10.1016/S1473-3099(15)70036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvvada MP, Patel SS. Role of trim5α in the suppression of cross-species transmission and its defence against human immunodeficiency virus. Curr HIV Res. 2013;11:601–9. doi: 10.2174/1570162x12666140306124543. [DOI] [PubMed] [Google Scholar]
- Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–84. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–6. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pescador R, Power MD, Barr PJ, Steimer KS, Stempien MM, Brown-Shimer SL, Gee WW, Renard A, Randolph A, Levy JA. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227:484–92. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- Santiago ML, Bibollet-Ruche F, Bailes E, Kamenya S, Muller MN, Lukasik M, Pusey AE, Collins DA, Wrangham RW, Goodall J, Shaw GM, Sharp PM, Hahn BH. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J Virol. 2003;77:2233–42. doi: 10.1128/JVI.77.3.2233-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noë R, Peeters M, Brookfield JFY, Shaw GM, Sharp PM, Hahn BH. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Taï Forest, Côte d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–27. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci. 2010;365:2487–94. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongo M, Dorfman JR, Martin DP. High Degree of HIV-1 Group M (HIV-1M) Genetic Diversity within Circulating Recombinant Forms: Insight into the Early Events of HIV-1M Evolution. J Virol. 2015;90:2221–9. doi: 10.1128/JVI.02302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi AJ, Detwiler KM, Disotell TR. X-chromosomal window into the evolutionary history of the guenons (Primates: Cercopithecini) Mol Phylogenet Evol. 2005;36:58–66. doi: 10.1016/j.ympev.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Triques K, Bourgeois A, Vidal N, Mpoudi-Ngole E, Mulanga-Kabeya C, Nzilambi N, Torimiro N, Saman E, Delaporte E, Peeters M. Near-full-length genome sequencing of divergent African HIV type 1 subtype F viruses leads to the identification of a new HIV type 1 subtype designated K. AIDS Res. Hum Retroviruses. 2000;16:139–51. doi: 10.1089/088922200309485. [DOI] [PubMed] [Google Scholar]
- Troyer JL, Roelke ME, Jespersen JM, Baggett N, Buckley-Beason V, MacNulty D, Craft M, Packer C, Pecon-Slattery J, O’Brien SJ. FIV diversity: FIV Ple subtype composition may influence disease outcome in African lions. Vet Immunol Immunopathol. 2011;143:338–46. doi: 10.1016/j.vetimm.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rensburg EJ, Engelbrecht S, Mwenda J, Laten JD, Robson BA, Stander T, Chege GK. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J Gen Virol. 1998;79(Pt 7):1809–14. doi: 10.1099/0022-1317-79-7-1809. [DOI] [PubMed] [Google Scholar]
- Vidal N, Bazepeo SE, Mulanga C, Delaporte E, Peeters M. Genetic characterization of eight full-length HIV type 1 genomes from the Democratic Republic of Congo (DRC) reveal a new subsubtype, A5, in the A radiation that predominates in the recombinant structure of CRF26_A5U. AIDS Res Hum Retroviruses. 2009;25:823–32. doi: 10.1089/aid.2008.0283. [DOI] [PubMed] [Google Scholar]
- Vidal N, Mulanga C, Bazepeo SE, Lepira F, Delaporte E, Peeters M. Identification and molecular characterization of subsubtype A4 in central Africa. AIDS Res Hum Retroviruses. 2006;22:182–7. doi: 10.1089/aid.2006.22.182. [DOI] [PubMed] [Google Scholar]
- Wagner GJ, Linnemayr S, Ghosh-Dastidar B, Currier JS, Hoffman R, Schneider S. Supporting Treatment Adherence Readiness through Training (START) for patients with HIV on antiretroviral therapy: study protocol for a randomized controlled trial. Trials. 2016;17:162. doi: 10.1186/s13063-016-1287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol. 2009;5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbe K, Salminen M, Laukkanen T, McCutchan F, Ray SC, Albert J, Leitner T. Characterization of novel recombinant HIV-1 genomes using the branching index. Virology. 2003;316:116–25. doi: 10.1016/j.virol.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Worobey M, Telfer P, Souquière S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, Roques P, Apetrei C, Kazanji M, Marx PA. Island biogeography reveals the deep history of SIV. Science. 2010;329:1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- Zarandia M, Tsertsvadze T, Carr JK, Nadai Y, Sanchez JL, Nelson AK. HIV-1 genetic diversity and genotypic drug susceptibility in the Republic of Georgia. AIDS Res Hum Retroviruses. 2006;22:470–6. doi: 10.1089/aid.2006.22.470. [DOI] [PubMed] [Google Scholar]
- Zeng H, Li T, Wang Y, Sun B, Yang R. The Epidemic Dynamics of Four Major Lineages of HIV-1 CRF01_AE Strains After Their Introduction into China. AIDS Res Hum Retroviruses. 2016;32:420–6. doi: 10.1089/AID.2015.0212. [DOI] [PubMed] [Google Scholar]
- Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.