Abstract
The global scale-up of antiretroviral (ARV) therapy (ART) has led to dramatic reductions in HIV-1 mortality and incidence. However, HIV drug resistance (HIVDR) poses a potential threat to the long-term success of ART and is emerging as a threat to the elimination of AIDS as a public health problem by 2030. In this review we describe the genetic mechanisms, epidemiology, and management of HIVDR at both individual and population levels across diverse economic and geographic settings. To describe the genetic mechanisms of HIVDR, we review the genetic barriers to resistance for the most commonly used ARVs and describe the extent of cross-resistance between them. To describe the epidemiology of HIVDR, we summarize the prevalence and patterns of transmitted drug resistance (TDR) and acquired drug resistance (ADR) in both high-income and low- and middle-income countries (LMICs). We also review to two categories of HIVDR with important public health relevance: (i) pre-treatment drug resistance (PDR), a World Health Organization-recommended HIVDR surveillance metric and (ii) and pre-exposure prophylaxis (PrEP)-related drug resistance, a type of ADR that can impact clinical outcomes if present at the time of treatment initiation. To summarize the implications of HIVDR for patient management, we review the role of genotypic resistance testing and treatment practices in both high-income and LMIC settings. In high-income countries where drug resistance testing is part of routine care, such an understanding can help clinicians prevent virological failure and accumulation of further HIVDR on an individual level by selecting the most efficacious regimens for their patients. Although there is reduced access to diagnostic testing and to many ARVs in LMIC, understanding the scientific basis and clinical implications of HIVDR is useful in all regions in order to shape appropriate surveillance, inform treatment algorithms, and manage difficult cases.
Keywords: HIV-1, drug resistance, antiretroviral therapy, diagnostic test, surveillance, mutations
1. Introduction
The world has embarked on a fast-track strategy to achieve the Joint United Nations Programme on HIV/AIDS 90-90-90 target and reduce the number of new HIV infections to 500,000 per year by 2020. Achieving the 90-90-90 target requires that 90% of all people living with HIV will have been diagnosed, 90% of all people with known HIV infection will be receiving antiretroviral (ARV) therapy (ART), and 90% of all people receiving ART will have a suppressed viral load. To meet the targeted reduction in new infections, the incidence must decline over four-fold from 2.1 million in 2015 (UNAIDS, 2014, 2016). To help achieve these goals, the World Health Organization (WHO) in 2016 recommended that all HIV-infected individuals should begin ART as soon after diagnosis as possible (Treat All) and that pre-exposure prophylaxis (PrEP) should be considered for persons at high risk of HIV infection (World Health Organization, 2016b). While the successful implementation of these recommendations will reduce the number of new HIV infections, the dramatic increase in ARV use will likely increase the prevalence of acquired drug resistance (ADR) in treated individuals and transmitted drug resistance (TDR) in newly infected individuals (Phillips et al., 2014b).
Whereas the principles of drug resistance are similar in all populations, differences in access to diagnostic testing and to many ARVs in low- and middle-income countries (LMICs) necessitate different approaches to the management of TDR and ADR. In most LMICs, HIV drug resistance (HIVDR) testing is neither feasible nor routinely recommended for patients receiving ART. Nonetheless, understanding the principles and mechanisms of HIVDR is useful to developing appropriate surveillance, informing treatment algorithms, and managing particularly difficult clinical cases. In high-income countries where drug resistance testing is part of routine care, such an understanding can also help clinicians prevent virological failure (VF) and accumulation of further HIVDR by selecting the most efficacious regimens for individual patients infected with drug-resistant viruses.
Although twenty-seven ARVs belonging to six classes have been approved for HIV-1 treatment, eleven of these ARVs are no longer available, no longer recommended, or rarely used, and thus will not be reviewed. We will instead focus on the 16 more commonly used ARVs belonging to 5 drug classes: 5 nucleoside reverse transcriptase inhibitors (NRTIs), 4 non-nucleoside reverse transcriptase inhibitors (NNRTIs), 3 protease inhibitors (PIs), 3 integrase strand transfer inhibitors (INSTIs) and one CCR5 antagonist (Table 1). In this review, we will not distinguish between ritonavir (RTV) and cobicistat pharmacoenhancement nor between the tenofovir prodrugs, tenofovir alafenamide fumarate (TAF) and tenofovir disoproxil fumarate (TDF), as their patterns of cross-resistance are similar (Gallant et al., 2015; Margot et al., 2015). Additionally, specific fixed dose combinations (FDCs) will not be addressed due to the rapid evolution of generic ARV formulations that currently dominate the ARV marketplace in LMICs (Waning et al., 2010).
Table 1.
Commonly Used Antiretrovirals with Typical Use in LMICs and High-Income Countries.
| Drug Class | Drug Name | Abbreviation | LMIC Use | High-income Country Use |
|---|---|---|---|---|
| NRTIs | Lamivudine | 3TC | First-line, second-line | First-line, second-line |
| Emtricitabine | FTC | First-line, second-line | First-line, second-line | |
| Tenofovir | TDF | First-line, second-line | First-line, second-line | |
| Abacavir | ABC | Not commonly used (HLA testing required) | First-line, second-line | |
| Zidovudine | AZT | Alternative first-line, second-line | Salvage | |
|
| ||||
| NNRTIs | Efavirenz | EFV | First-line | Alternative first-line |
| Rilpivirine | RPV | Not commonly used | Alternative first-line | |
| Etravirine | ETR | Not commonly used | Second-line, salvage | |
| Nevirapine | NVP | Alternative first-line | Not commonly used | |
|
| ||||
| PIs | Atazanavir | ATV | Second-line | Alternative first-line |
| Darunavir | DRV | Alternative second-line, third-line | First-line, second-line, salvage | |
| Lopinavir | LPV | Second-line | Not commonly used | |
|
| ||||
| INSTIs | Elvitegravir | EVG | Not commonly used | First-line |
| Raltegravir | RAL | Alternative second-line, third-line | First-line | |
| Dolutegravir | DTG | Alternative first-line | First-line, second-line, salvage | |
|
| ||||
| CCR5 Antagonist | Maraviroc | MVC | Not commonly used | Salvage |
Abbreviations: NRTIs: nucleoside reverse transcriptase inhibitors; NNRTIs: non-nucleoside reverse transcriptase inhibitors; PIs: protease inhibitors; INSTIs: integrase strand transfer inhibitors; LMIC: low- and middle-income countries (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016; World Health Organization, 2015a).
2. Mechanisms of HIV-1 Drug Resistance
HIV-1 genetic variability results from the high rate of HIV-1 reverse transcriptase (RT) processing errors, recombination when more than one viral variants infect the same cell, and the accumulation of proviral variants during the course of infection (Abram et al., 2010; Coffin, 1995; Levy et al., 2004; Mansky, 1996). Although most HIV-1 infections are initiated by a single viral variant (Keele et al., 2008), innumerable variants (or quasispecies) related to the initial transmitted virus emerge within weeks following infection (Coffin, 1995; Perelson and Ribeiro, 2013).
The selection of drug-resistant variants depends on the extent to which viral replication continues during incompletely suppressive therapy, the ease of acquisition of a particular drug resistance mutation (DRM), and the effect of DRMs on drug susceptibility and virus replication. Although naturally occurring drug-resistant viruses arise every day in untreated patients (Perelson and Ribeiro, 2013), these variants rarely rise to detectable levels because they are less fit than drug-susceptible viruses in the absence of selective drug pressure. Indeed, nearly all clinically significant DRMs arise only as a result of selective drug pressure and are otherwise non-polymorphic.
For some ARVs, multiple DRMs are required to reduce susceptibility, while for others a single DRM is sufficient. The number of DRMs required and the effect of each DRM on virus fitness contribute to the ARV’s genetic barrier to resistance. DRMs can be categorized as primary DRMs that directly reduce drug susceptibility and accessory DRMs that enhance fitness of variants containing primary DRMs or that contribute further reductions in susceptibility. The extent to which an ARV reduces plasma HIV-1 RNA levels is known as its antiviral potency. An ARV’s intrinsic antiviral potency combined with its genetic barrier to resistance influences its ability to protect an ART regimen from VF (Figure 1).
Figure 1.
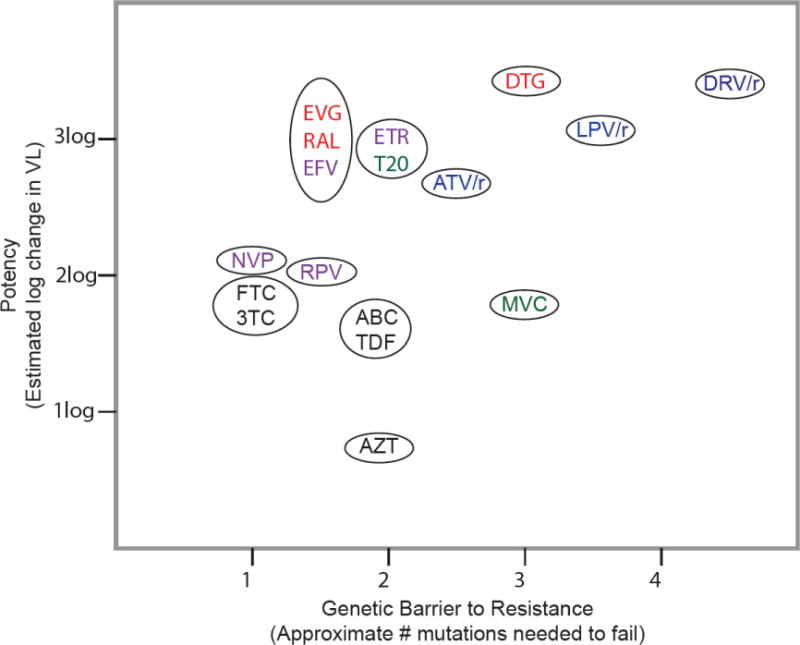
ARV potency versus genetic barrier to resistance.
Abbreviations: ARV: antiretroviral; VL: viral load; ABC: abacavir; ATV/r: boosted atazanavir; DRV/r: boosted darunavir; DTG: dolutegravir; EFV: efavirenz; FTC: emtricitabine; EVG: elvitegravir; T20: enfuvirtide; ETR: etravirine; 3TC: lamivudine; LPV/r: boosted lopinavir; MVC: maraviroc; NVP: nevirapine; RAL: raltegravir; RPV: rilpivirine; and TDF: tenofovir. ARVs in black font are nucleoside or nucleotide reverse transcriptase inhibitors (NRTIs), those in purple font are non-NRTIs (NNRTIs), those in blue font are protease inhibitors (PIs), those in red font are integrase strand transfer inhibitors (INSTIs), and those in green font are entry inhibitors. ARVs appearing together in the same ellipse should be considered to have roughly equivalent potencies and genetic barriers to resistance.
There is essentially no cross-resistance between drug classes. Viruses that are highly-resistant to drugs in one ARV class are completely susceptible to ARVs from unused classes (Larder, 1994). In contrast, significant cross-resistance within a drug class is common because most DRMs reduce susceptibility to multiple ARVs of the same class (Melikian et al., 2012; Melikian et al., 2014; Rhee et al., 2010; Tang and Shafer, 2012; Whitcomb et al., 2003). However, there are important exceptions in that several DRMs increase susceptibility to other ARVs of the same class (Whitcomb et al., 2002). Knowledge of ARV cross-resistance profiles is therefore essential when using more than one drug from the same ARV class either in combination or in sequence.
Most ART regimens used for first-line therapy are sufficiently potent to completely block HIV-1 replication and have a genetic barrier to resistance high enough to maintain long-term virological suppression. As a result, most cases of VF and drug-resistance arise from incomplete adherence, which exposes a patient’s virus to the incompletely suppressive ARV levels capable of exerting drug selective pressure. Accordingly, HIVDR appears to be less common in patients receiving FDCs containing ARVs that have similar half-lives because incomplete adherence to these combinations is less likely to expose a virus to selective drug pressure (Chi et al., 2007; Geretti et al., 2013; Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). Lower rates of HIVDR have also been associated with routine viral load monitoring programs in which early detection of virological rebound provides the opportunity for adherence counseling or regimen modification as necessary, prior to the evolution of multiple DRMs (Charest et al., 2014; Gupta et al., 2009; TenoRes Study, 2016).
3. HIV-1 Drug Resistance Testing
HIV-1 drug resistance testing can be performed phenotypically or genotypically. Phenotypic in vitro susceptibility assays measure ARV susceptibility in cell culture. Susceptibility is usually reported as the ARV concentration that inhibits HIV-1 replication by 50% (IC50). The IC50 of a patient’s virus is compared to that of a drug-susceptible reference strain, and expressed as a ratio, referred to as fold change, of the IC50 of the patient’s virus relative to the reference control. Nearly all susceptibility assays use recombinant viruses created by inserting PCR-amplified patient virus gene segments (protease/RT, integrase, envelope) into the backbone of a wild type laboratory clone (Petropoulos et al., 2000). Due to cost and lengthy turnaround time, phenotypic resistance testing is typically reserved for drug development, drug resistance research, or complex clinical cases.
The clinical significance of the reductions in in vitro susceptibility reported from phenotypic testing varies markedly among ARVs, owing to both pharmacokinetic and pharmacodynamic factors. For example, the NRTIs must be triphosphorylated to their active form, a process that often occurs at different rates in vivo compared with the cell lines typically used for drug-resistance testing. Additionally, the dynamic susceptibility range, or the fold change in IC50 between wild type and the most NRTI-resistant isolates, is as low as 5-fold for TDF and more than 200-fold for zidovudine (AZT), lamivudine (3TC), and emtricitabine (FTC) (Melikian et al., 2012; Whitcomb et al., 2003). Similar but less pronounced differences in dynamic susceptibility ranges exist within the NNRTI, PI, and INSTI classes.
Standard genotypic resistance testing (SGRT) involves the use of dideoxyterminator Sanger sequencing of protease, RT, and/or integrase to identify established clinically significant DRMs. SGRT is performed on PCR products directly amplified from cDNA that has been reverse-transcribed from plasma RNA. Because the virus population in an individual is typically heterogeneous, it is common for more than one nucleotide (i.e. a mixture) to be present in a given position on Sanger sequence electropherograms. Standardization between laboratories for studies of drug resistance has been facilitated through the use of software tools that apply a consistent approach to identify mixed bases (Woods et al., 2012) and to assess sequence quality (Shafer et al., 2000). Indeed, the information present in an electropherogram provides intrinsic quality control data including the level of background “noise” as indicated by low signal intensities and numerous poorly defined peaks. In addition, a high number of electrophoretic mixtures, atypical mutations, and APOBEC mutations observed in a nucleotide sequence can signal quality control problems (Rhee et al., 2016; Shafer et al., 2000). Furthermore, phylogenetic analysis and distance measurements of sequences can be used to detect possible laboratory contamination or specimen mislabeling at the point of collection.
Although the lower limit of SGRT detection is generally agreed to be around 20%, both point mutation assays (PMAs) and next generation deep sequencing (NGS) technologies often detect drug-resistant variants at levels well below a 20% threshold. However, the frequency with which these ultra-sensitive PMA and NGS assays reliably detect variants present at levels less than 20% varies widely between studies. Some of the low-abundance DRMs detected by highly sensitive assays may arise from PCR or sequencing errors (Jabara et al., 2011; Keys et al., 2015). Although the use of Primer IDs, random sequence tags incorporated into primers to generate templates with unique primer identification sequences, can reduce such errors (Jabara et al., 2011), this approach is used primarily in research settings. Low-abundance DRMs have been associated with a reduced response to NNRTI-based ART regimens, and the effect is greatest when the DRM-bearing variants are present at frequencies ≥1% (Li et al., 2011). Despite the potential clinical implications of low-abundance DRMs in patients initiating certain regimens, guidelines for the use of these more sensitive technologies have not yet been developed.
In high-income countries, SGRT, which has a retail cost of 400–500 USD, is considered cost-effective at time of diagnosis or treatment initiation and at VF. However, in LMIC settings, models have produced conflicting results regarding the cost-effectiveness of SGRT (Levison et al., 2013; Phillips et al., 2014a; Phillips et al., 2014b; Rosen et al., 2011). Platforms that are more economical than SGRT may play a role in expanding access to genotypic drug resistance testing, which is currently limited or non-existent in most LMICs.
Due to the high number of sequences obtained with NGS, the cost per sequence obtained with this platform is lower than with SGRT. In NGS deep sequencing the depth of coverage refers to the number of viral variants sequenced, typically several hundred. Researchers have shown that harnessing the cost savings of NGS using a “wide sequencing” strategy, wherein many samples are processed in a single run with a lesser depth of coverage, could be a viable method of genotypic testing in LMICs (Dudley et al., 2012; Lapointe et al., 2015). Cost-effectiveness models using NGS platforms have not been published, but the estimated cost per sample is much lower than SGRT at 1 to 20 USD (Dudley et al., 2012; Lapointe et al., 2015).
Whereas wide sequencing with NGS would require processing batched samples in few centralized laboratories, point-of-care (POC) assays aim to be deployable in a variety of settings, from hospitals to remote clinics, and to deliver actionable results during a single clinical encounter. The platforms in development that appear the most amenable to POC or near-POC use in LMICs are PMAs capable of detecting key DRMs at costs below $5 USD per locus (MacLeod et al., 2015; Mutsvangwa et al., 2014; Panpradist et al., 2016; Zhang et al., 2016; Zhang et al., 2013). A major challenge to the successful development of a PMA is the extreme genetic variability of HIV-1 (Clutter et al., 2016b). The variability surrounding DRM positions will necessitate extensive validation studies using diverse subtypes and may require multiple primers, thus driving up the cost of assay development and production. However, recent efforts to mitigate the effect of genetic variability on the success of a PMA have shown promising results (MacLeod et al., 2015). Notably PMAs do not produce sequences; thus they cannot be used for phylogenetic analyses or the discovery of novel DRMs.
4. Drug Resistance Test Interpretation
Understanding the results of drug resistance tests is one of the most difficult tasks facing HIV care providers. There are many DRMs and they arise in complex patterns that cause varying levels of drug resistance. Some DRMs are responsible for just low levels of resistance to a drug but would not be a contraindication to using that drug if the remaining components of a patient’s regimen were fully active. Further complicating genotypic resistance test interpretation is the recognition that SGRT and even NGS deep sequencing may not detect all of the resistant variants within an individual patient and that inference into the possible presence of these variants will depend on a patient’s past treatment regimens.
A DRM can be characterized according to whether it is selected by therapy and whether it is polymorphic or nonpolymorphic in untreated individuals. The development of a mutation while on ART suggests that the mutation is associated with reduced susceptibility to an ARV in the regimen. This type of evidence is particularly important for DRMs associated with drugs that exhibit only subtle in vitro susceptibility changes. Most major DRMs are non-polymorphic in that they do not occur naturally in the absence of selective drug pressure. An important corollary of this observation is that HIVDR to the current NRTIs, NNRTIs, PIs, and INSTIs rarely occurs in the absence of prior selective drug pressure.
DRMs can also be characterized by their effects on in vitro susceptibility and in vivo response to therapy. The difference in ARV susceptibility between a wild type laboratory clone and a clone bearing a DRM allows for direct assessment of that DRM’s phenotypic effect. The reduction in susceptibility caused by a particular DRM can also be studied in clinical isolates, in which multiple DRMs may be present, through the use of regression analyses where the presence of the DRM is an explanatory variable and the fold reduction in susceptibility is the outcome variable. The regression coefficients obtained from these models indicate the relative contribution of a DRM to reduced ARV susceptibility while controlling for other DRMs in a virus sequence (Melikian et al., 2012; Melikian et al., 2014; Rhee et al., 2010).
The clinical significance of DRMs for ART-naïve individuals was first demonstrated by the poor responses to therapy observed in the earliest reports of patients with TDR (Grant et al., 2002; Little et al., 2002). It was then re-enforced in subsequent studies in which initial therapy was selected without the availability of genotypic resistance testing but for which the presence of resistance mutations prior to initiation of therapy was detected retrospectively through the analysis of cryopreserved blood samples (Borroto-Esoda et al., 2007; Chaix et al., 2007; Chung et al., 2014; Hamers et al., 2012; Kuritzkes et al., 2008; Lee et al., 2014; Little et al., 2002; Wittkop et al., 2011).
The predictive value of specific mutations on the response to a new ARV has been shown most convincingly in large clinical trials in which individuals were randomly assigned to receiving a new ARV or placebo in combination with an optimized background regimen (Castagna et al., 2014a; de Meyer et al., 2008; Eron et al., 2013; King et al., 2007; Lanier et al., 2004; Miller et al., 2004; Vingerhoets et al., 2010). Additionally, observational studies have also shown that the predicted activity of a new ARV regimen (often called the genotypic susceptibility score) is a strong independent predictor of the response to that regimen (Rhee et al., 2009; Wittkop et al., 2011).
The Stanford HIV Drug Resistance Database (HIVDB) provides an online genotypic resistance interpretation program to help clinicians and laboratories interpret HIV-1 genotypic resistance tests (http://hivdb.stanford.edu). The program accepts RT, protease and/or integrase sequences submitted by the user and returns a list of penalty scores for each DRM and an estimate of reduced susceptibility for each ARV. The estimate of reduced susceptibility for each ARV is obtained by adding each of the DRMs’ penalty scores for the ARV. The DRM penalty scores (http://hivdb.stanford.edu/DR/) are based upon the DRM characteristics and the consensus on the clinical significance of a DRM among experts in the field, such as the IAS-USA Drug Resistance Mutations Group (Wensing et al., 2014).
5. NRTI Resistance Mutations
There are two genetic mechanisms of NRTI resistance: (i) discriminatory mutations that enable RT to distinguish between dideoxy-NRTI chain terminators and the cell’s own dNTPs, thus preventing NRTIs from being incorporated into viral DNA; and (ii) primer unblocking mutations that facilitate the phosphorylytic excision of an NRTI-triphosphate from viral DNA. Unblocking mutations are also referred to as thymidine analogue mutations (TAMs) because they are selected by the thymidine analogues AZT and stavudine (d4T) (Tang and Shafer, 2012). Even though d4T has not been used for years in high-income countries and AZT’s use has declined, TAMs have persisted in the circulating viruses this setting due to the relatively higher virus fitness associated with these mutations compared to the discriminatory DRMs. This higher degree of fitness slows reversion to wild type, and allows for ongoing transmission of these DRMs.
The most common NRTI-resistance mutations are M184V/I (Figure 2). M184V/I are selected by and cause high-level (>200-fold) reduced susceptibility to the cytosine analog NRTIs, 3TC and FTC. M184V/I is also selected by, and causes low-level resistance to abacavir (ABC). During VF on a regimen containing either 3TC or FTC, M184I often emerges before M184V. However, in most patients M184V will outcompete M184I within several weeks (Frost et al., 2000; Keulen et al., 1996). Despite the high-levels of 3TC and FTC phenotypic resistance associated with M184V/I, it is common practice to continue to use these NRTIs to treat viruses with M184V/I because maintenance of these DRMs reduces virological fitness (Campbell et al., 2005; Castagna et al., 2006; Diallo et al., 2003; Eron et al., 1995) and increases susceptibility to AZT and TDF (Kuritzkes et al., 1996; Melikian et al., 2012; Miller et al., 2004; Nijhuis et al., 1997; Whitcomb et al., 2003). In addition, 3TC and FTC are well tolerated with few associated side effects or toxicities.
Figure 2.
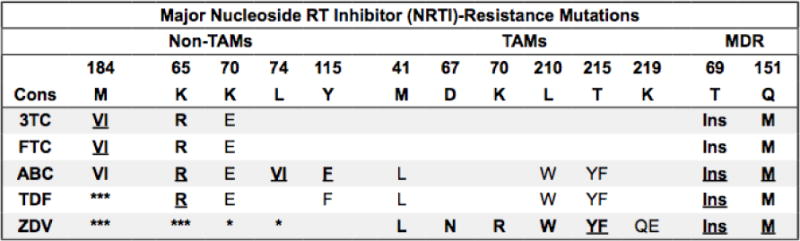
Summary of nucleoside/nucleotide reverse transcriptase inhibitor drug resistance mutations
Bold/underline: High-level reduced susceptibility or virological response. Bold: reduced suceptibility or virological response. Plain text: reduced susceptibility in combination with other NRTI-resistance mutations. Asterisk: increased susceptibility. Additional NRTIs: Stavudine (d4T) and didanosine (ddI) are no longer recommended. TAMs: Thymidine analog mutations. Selected by AZT and d4T and facilitate primer unblocking. Non-TAMs prevent NRTI incorporation. MDR: Multidrug resistance mutations. T69 insertions occur with TAMs. Q151M occurs with non-TAMs and accessory mutations A62V, V75I, F77L, and F116Y. M184VI: Although they cause high-level in vitro resistance to 3TC/FTC, they are not contraindications to 3TC/FTC because they increase TDF and AZT susceptibility and decrease viral replication fitness. Additional mutations: K65N is similar but weaker than K65R. K70GQ is similar to K70E. T69D and V75MT reduce susceptibility to d4T and ddI. T215SCDEIV (T215 revertants) evolve from T215YF in the absence of NRTIs. E40F, E44DA, D67GE, V118I, and K219NR are accessory TAMs. T69 deletions occur in combination with K65R and/or Q151M. With K65R (but not Q151M) they increase AZT susceptibility. References: http://hivdb.stanford.edu/DR/NRTIResiNote.html.
The next most common discriminatory mutations include K65R, K70E/G/Q, L74V/I, Y115F and the Q151M complex of mutations. K65R is selected primarily by TDF and to a lesser extent by ABC and d4T. K65R increases susceptibility to AZT. Although K65R reduces TDF susceptibility in vitro by only two-fold, it is, after M184V, the most common DRM in patients with VF on a TDF-containing regimen.
L74V/I and Y115F are selected primarily by ABC. However, L74I and Y115F are also often selected by TDF, particularly in LMICs where resistance tests are often performed after prolonged VF (Skhosana et al., 2015; Van Zyl et al., 2013). K70E/G/Q are also selected by TDF and ABC-containing regimens and are associated with minimal reductions on in vitro susceptibility to these NRTIs (TenoRes Study, 2016). Q151M is an uncommon DRM that usually occurs only in heavily treated patients with prolonged VF. It usually occurs in combination with several accessory DRMs (A62V, V75I, F77L and F116Y) and is associated high-level resistance to AZT and ABC and intermediate resistance to TDF, 3TC, and FTC.
The TAMs include M41L, D67N, K70R, L210W, T215F/Y and K219Q/E. Although they are selected only by AZT and d4T, they confer lower levels of cross-resistance to TDF and ABC. TAMs occur in two distinct but overlapping patterns denoted as type I and type II (Marcelin et al., 2004; Tang and Shafer, 2012). The type I TAMs, which include M41L, L210W and T215Y, cause higher levels of phenotypic and clinical cross resistance to TDF and ABC than the type II TAMs (D67N, K70R, T215F and K219Q/E). The presence of all three type I TAMs markedly worsens clinical response to TDF- and ABC-containing regimens (Lanier et al., 2004; Miller et al., 2004).
The TAMs and discriminatory mutations other than M184V/I occur through two separate, opposing pathways (Boucher et al., 2006; Parikh et al., 2006; Rhee et al., 2007). Viruses already containing multiple TAMs are more likely to develop additional TAMs than K65R when treated with TDF or ABC. Some viruses with multiple TAMs will also develop a double-amino acid insertion at RT position 69 referred to as T69S_SS (indicating a wildtype threonine replaced by three serines) or simply T69insertion. In combination with multiple TAMs this DRM causes high-level resistance to AZT, ABC, and TDF, and intermediate resistance to 3TC and FTC (Masquelier et al., 2001; Winters et al., 1998).
6. NNRTI Resistance Mutations
The NNRTIs have a relatively low genetic barrier to resistance. To develop high-level resistance, one DRM in the case of nevirapine (NVP), one to two DRMs in the case of efavirenz (EFV), and two DRMs in the case of etravirine (ETR), are required (Melikian et al., 2014; Sluis-Cremer and Tachedjian, 2008; Vingerhoets et al., 2010). Although rilpivirine (RPV) has a similar structure to ETR, its genetic barrier to clinically significant resistance is lower than ETR because it is administered at a much lower dose (Cohen et al., 2011; Rimsky et al., 2012).
There is a high level of cross-resistance within the NNRTI class as a result of two mechanisms: (i) most NNRTI-resistance mutations reduce susceptibility to two or more NNRTIs (Melikian et al., 2014); and (ii) the low genetic barrier to NNRTI resistance allows for the emergence of multiple independent NNRTI-resistant lineages, even if not all are detectible by SGRT (Antinori et al., 2002; Ruxrungtham et al., 2008; Tang and Shafer, 2012; Varghese et al., 2009).
The most common NNRTI mutations are L100I, K101E/P, K103N/S, V106A/M, Y181C/I/V, Y188C/H/L, G190A/S/E, and M230L (Figure 3). Each of these cause intermediate or high-level phenotypic resistance to NVP. With the exception of Y181C/I/V, they cause intermediate- or high-level phenotypic resistance to EFV (Melikian et al., 2014). Although Y181C alone reduces EFV susceptibility by only 2-fold, it has been associated with a reduced response to EFV in patients with virological failure on an NVP-containing regimen possibly as a result of minority variant mutations 15608522. With the exception of K103N/S, V106A/M, Y188C/H, and G190A/S, they cause phenotypic ETR and RPV resistance (Melikian et al., 2014).
Figure 3.
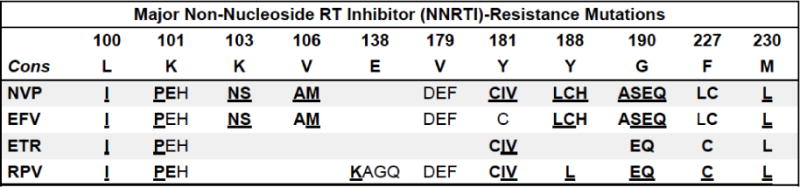
Summary of non-nucleoside reverse transcriptase inhibitor drug resistance mutations.
Bold/underline: High-level reduced susceptibility or virological response. Bold: reduced suceptibility or virological response. Plain text: reduced susceptibility in combination with other NNRTI-resistance mutations. Asterisk: increased susceptibility. Abbreviations: nevirapine (NVP), efavirenz (EFV), etravirine (ETR), rilpivirine (RPV). Synergistic combinations: V179D+K103R reduce NVP and EFV susceptibility >10-fold. Y181C+V179F cause high-level ETR and RPV resistance. ETR genotypic susceptibility score (GSS): Y181IV (3.0); L100I, K101P, Y181C, M230L (2.5); V90I, E138A, V179F, G190S (1.5); A98G, K101EH, V106I, V179DT, G190A (1.0); 3.0 high-level. V90I, A98G, V106I, E138A, V179DT, G190A/S have little effect on ETR susceptibility unless they occur with a bolded mutations. Additional accessory mutations: V90I (ETR), A98G (NVP, EFV, ETR, RPV), V108I, V179T (ETR), V179L (RPV), P225H (EFV), K238T (NVP, EFV), L318F (NVP). References: http://hivdb.stanford.edu/DR/NNRTIResiNote.html.
In addition, the previously unrecognized mutations E138K/G/Q/A are among the most common mutations to emerge in patients receiving RPV (Rimsky et al., 2012). Although E138K reduces RPV susceptibility only minimally, it is among the most common DRMs to emerge in patients with VF on a first-line RPV-containing regimen. E138A is a notable DRM for RPV because it occurs in up to 5% of ARV-naïve patients, depending on virus subtype (Sluis-Cremer et al., 2014), and was an exclusionary criterion for enrollment in the clinical trials of Complera (TDF/FTC/RPV).
Because it has the highest genetic barrier to resistance in its class, ETR is the most commonly used NNRTI in patients previously failing other NNRTIs and is often combined with other drugs to which the virus is likely to be susceptible (e.g. boosted PIs). In studies of treatment-experienced patients initiating boosted darunavir (DRV/r), NRTIs, and ETR, a scoring system based on the number and weight of ETR DRMs was shown to be useful in predicting VF (Vingerhoets et al., 2010).
7. PI Resistance Mutations
Boosted lopinavir (LPV/r), boosted atazanavir (ATV/r), and DRV/r are the three most commonly used PIs. ATV/r and DRV/r are the two most commonly recommended PIs in high-income countries (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). LPV/r is widely used in second-line regimens in LMICs. LPV/r and DRV/r have high genetic barriers to resistance in that multiple DRMs are required before antiviral activity is compromised (de Meyer et al., 2008; King et al., 2007). LPV/r and DRV/r have also been shown to be effective at maintaining virological suppression in ARV-experienced patients undergoing regimen simplification (Bartlett et al., 2012; Collins et al., 2016; Delfraissy et al., 2008; Llibre et al., 2013; Pulido et al., 2010). Several lines of evidence suggest that ATV/r has a lower genetic barrier to resistance than LPV/r and DRV/r: (i) fewer mutations are needed for reductions in in vitro susceptibility; (ii) smaller reductions in in vitro susceptibility are required to increase rates of clinical virological failure (Monogram Biosciences, 2016; Naeger and Struble, 2006); (iii) ATV/r monotherapy has also been less effective than LPV/r and DRV/r for regimen simplification (Castagna et al., 2014b; Guiguet et al., 2012; Karlstrom et al., 2007).
PI DRMs develop much less frequently in patients receiving an initial LPV/r, ATV/r, or DRV/r-containing regimen than do NRTI and NNRTI DRMs in patients receiving NRTI/NNRTI-containing regimens (Barber et al., 2012; Dolling et al., 2013; El Bouzidi et al., 2014; Lathouwers et al., 2011; Molina et al., 2008; Mollan et al., 2012; Van Zyl et al., 2013). The narrow drug concentration range over which PI levels are both low enough to allow viral replication yet high enough to exert selective drug pressure likely accounts for the reduced risk of resistance associated with boosted PIs (Rosenbloom et al., 2012; Siliciano and Siliciano, 2013). Indeed, most patients without PI DRMs who experience VF while on an initial PI-containing regimen achieve virological suppression with improved adherence (Lopez-Cortes et al., 2016; Zheng et al., 2014). Nonetheless, the possibility that mutations outside of protease may also cause VF is actively being investigated (Fun et al., 2012; Rabi et al., 2013). Specifically, it has been proposed that mutations in gag matrix (MA) and the gp41 cytoplasmic domain (gp41-CD) contribute to PI resistance but none consistently responsible for PI resistance have been identified (Gupta et al., 2010; Jinnopat et al., 2009; Sutherland et al., 2014).
Although PI-associated DRMs rarely develop in PI-naïve patients receiving one of the currently recommended boosted PIs, the historical use of unboosted PIs in high-income countries has led to the evolution of many highly PI-resistant viruses. Indeed, more than 80 non-polymorphic PI-selected mutations have been reported (Rhee et al., 2016; Shahriar et al., 2009). The majority of these reduce in vitro susceptibility to one or more PIs (Rhee et al., 2010). The DRMs with the greatest impact on susceptibility are in the substrate cleft including V32I, G48V/M, I50V/L, V82A/T/L/F/S/C/M, and I84V/A/C (Figure 4). In addition, several mutations in the enzyme flap including M46I/L and I54V/M/L/T/S/A, and in the enzyme core L33F, L76V, and N88S also markedly reduce susceptibility. Most PI-resistant viruses also require one or more accessory protease mutations and one or more compensatory gag cleavage-site mutations (Foulkes-Murzycki et al., 2007; Fun et al., 2012). The requirement for multiple accessory mutations outside of protease almost certainly contributes to the high genetic barrier to resistance of boosted PIs.
Figure 4.
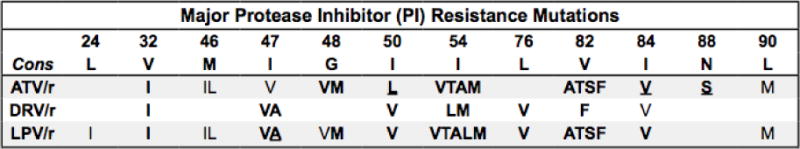
Summary of protease inhibitor drug resistance mutations.
Bold/underline: High-level reduced susceptibility or virological response. Bold: reduced suceptibility or virological response. Plain text: reduced susceptibility in combination with other PI-resistance mutations. Abbreviations: atazanavir (ATV), darunavir (DRV), lopinavir (LPV). Administered with ritonavir for pharmacokinetic boosting (/r). Additional PIs: Fosamprenavir (FPV), indinavir (IDV), saquinavir (SQV), and tipranavir (TPV) are rarely used. Nelfinavir (NFV) is no longer recommended. FPV/r and IDV/r are never more active than DRV/r and rarely if ever more active than LPV/r vs resistant viruses. TPV/r is occasionally useful for salvage therapy as it can be active vs LPV/r and DRV/r-resistant viruses with mutations that increase TPV susceptibility. Expert consultation +/− phenotypic testing should be obtained prior to using FPV, FPV/r, IDV/r, SQV/r, and TPV/r. Additional mutations: D30N and N88D are major NFV-resistance mutations. L10F, V11I, K20TV, L23I, K43T, F53L, Q58E, A71IL, G73STCA, T74P, N83D, and L89V are common nonpolymorphic accessory mutations. L10RY, V11L, L24F, M46V, G48ASTLQ, F53Y, I54S, V82CM, I84AC, N88TG are rare nonpolymorphic variants. Hypersusceptibility: I50L (each PI except ATV); L10F, L24I, I50V, I54L (TPV); L76V (ATV, SQV, TPV); I47A (SQV); N88S (FPV). References: http://hivdb.stanford.edu/DR/PIResiNote.html.
The ATV-selected DRM, I50L, is notable for causing high-level resistance to just one PI. Each of the remaining major mutations reduces susceptibility to two or more PIs. Due to the large number of PI DRMs and the fact that different DRMs at the same position can have markedly different effects on PI susceptibility, PI cross resistance is complex (Tang and Shafer, 2012). This is particularly the case for DRMs at positions 50, 54 and 82 (Rhee et al., 2010).
Due to their high genetic barriers to resistance, DRV/r and LPV/r are often used in patients failing therapy, even those who are PI-experienced. In such patients, multiple models incorporating information from 10 or more PI DRM positions have been shown to predict virological responses to LPV/r (Kempf et al., 2002; King et al., 2007). In studies evaluating the effect of baseline DRMs on response to DRV/r in PI-experienced patients, reduced responses were associated with 3 to 5 of the following DRV DRMs: V11I, V32I, L33F, I47V, I50V, I54L/M, T74P, L76V, I84V, and L89V (de Meyer et al., 2008; Pozniak et al., 2008).
8. INSTI Resistance Mutations
Figure 5 shows the most common INSTI-associated DRMs. More information is known about the DRMs selected by raltegravir (RAL) because it was the first FDA-approved INSTI. RAL resistance occurs by three main, often overlapping mutational pathways with the following signature DRMs: N155H±E92Q; Q148H/R/K±G140S/A; and Y143C/R (Blanco et al., 2011; Geretti et al., 2012). The presence of two of these signature DRMs is usually associated with >150-fold reduced RAL susceptibility. Although each of these DRMs alone is associated with much lower levels of phenotypic resistance, they are likely a harbinger of high-level resistance.
Figure 5.
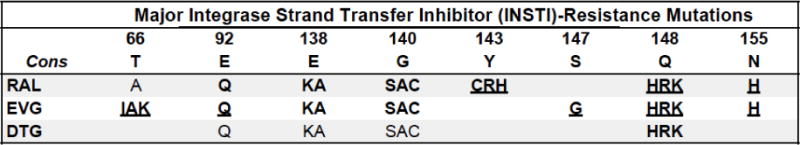
Summary of integrase strand transfer inhibitor drug resistance mutations.
Bold/underline: High-level reduced susceptibility or virological response. Bold: reduced suceptibility or virological response. Plain text: reduced susceptibility in combination with other INSTI-resistance mutations. Asterisk: increased susceptibility. Abbreviations: raltegravir (RAL), elvitegravir (EVG), dolutegravir (DTG). Additional mutations: H51Y, L74M, T97A, S153YF, G163RK, S230R, and R263K are relatively nonpolymorphic INSTI-selected accessoryresistance mutations. E92GV, E138T, Y143KSGA, Q148N, and N155ST are unusual variants at the positions listed above. P145S and Q146P are rare EVG-resistance mutations. G118R and F121Y are rare nonpolymorphic INSTI resistance mutations. References: http://hivdb.stanford.edu/DR/INIResiNote.html.
Whereas the published data on RAL-resistant isolates are from patients receiving first-line RAL-containing regimens and from patients receiving RAL for late salvage therapy, most published experience with elvitegravir (EVG) resistance is from virus isolates obtained during first-line EVG-containing regimens. EVG shares two mutational pathways with RAL: N155H±E92Q and Q148H/R/K±G140SA. E92Q alone is particularly common in patients receiving EVG and is often the most common EVG-resistance DRM in clinical trials (White et al., 2014a). There are also two non-polymorphic DRMs – T66I/K and S147G – that occur more commonly with EVG and cause lower-levels of cross-resistance to RAL (Abram et al., 2013).
Considering the extensive overlap between the genetic mechanisms of resistance to RAL and EVG, there are no scenarios in which patients with VF while receiving one of these INSTIs should be switched to the other INSTI. Moreover, even viruses with DRMs that do not appear to cause resistance to both INSTIs when they occur in isolation (e.g. Y143C/R for RAL) may be associated with higher than expected levels of cross-resistance if one or more of the accessory DRMs in the Figure 5 legend are present (Huang et al., 2013).
In contrast to RAL and EVG, multiple DRMs are required for clinically significant dolutegravir (DTG) resistance. Most data on the genetic mechanisms of DTG resistance have been obtained from RAL-experienced patients who received DTG salvage therapy in the VIKING trials (Castagna et al., 2014a; Eron et al., 2013). In these trials, patients with viruses containing a Q148 mutation in combination with E138 ± G140 mutations were at increased risk of VF. A number of accessory INSTI-resistance DRMs and N155H also appear to increase the risk of DTG VF particularly when these DRMs are present in combination with Q148. There are two reports of treatment-experienced but INSTI-naïve patients developing VF with R263K, a relatively weak DTG-associated resistance mutation which is associated with 2–5-fold reduced DTG susceptibility and which has also been selected by DTG in vitro. There have also been two reports of G118R in patients receiving DTG monotherapy. This mutation has also been selected in vitro by DTG. In vitro susceptibility data have been highly inconsistent possibly because viruses with this mutation replicate poorly.
9. HIV-1 Tropism
Maraviroc allosterically inhibits gp120 Env of CCR5 (R5)-tropic HIV-1 strains from binding to the seven-transmembrane G protein-coupled R5 receptor (Tan et al., 2013). Whereas HIV-1 gp120 binds to the N-terminus and second extracellular loop region of R5, maraviroc binds to a pocket formed by the transmembrane helices (Tan et al., 2013). In patients receiving R5 inhibitors, the most common mechanism of VF is the expansion of pre-existing CXCR4 (X4) tropic viruses that are intrinsically resistant to R5 inhibitors (Gulick et al., 2008; Westby et al., 2006). HIV-1 can also develop maraviroc resistance mutations that allow HIV-1 gp120 to bind to an inhibitor-bound R5 receptor. However, reports of such resistance have been documented primarily in vitro and in only a small number of clinical viral isolates (Jiang et al., 2015; Westby et al., 2007).
More than 80% of patients are initially infected with exclusively R5 tropic viruses. X4 tropic viruses usually emerge during later stages of infection, and about 50% of patients chronically infected with HIV-1 harbor X4 tropic viruses (Brumme et al., 2005; Hunt et al., 2006; Melby et al., 2006; Poveda et al., 2006). X4-tropic viruses are often present in low proportions relative to R5-tropic viruses when they emerge (Brumme et al., 2005; Wilkin et al., 2007).
A tropism assay should be performed to confirm that a patient is not infected with X4-tropic variants before using maraviroc. Tropism can be detected phenotypically or genotypically. The phenotypic Trofile test assesses the tropism of complete env genes amplified from patient samples using reporter cell lines expressing R5 or X4 (Whitcomb et al., 2007). Given a sufficiently high plasma virus level, it has the sensitivity to detect X4 variants at populations less than 1% (Su et al., 2009).
Genotypic testing for tropism involves sequencing the envelope V3 loop and using algorithms to predict tropism from the sequence. Positively charged residues at positions 11 and 25 of the V3 loop of gp120 and several less common combinations of mutations primarily but not exclusively within the V3 loop are associated with X4 tropism (Hartley et al., 2005). SGRT has a specificity of ~85% and sensitivity of 50–70% for detecting the presence of X4 tropic variants. The low sensitivity of SGRT arises from the fact that X4 tropic variants are often present at levels below the 20% sensitivity of SGRT and because some of the determinants of tropism lie outside of the V3 loop. However, sequencing multiple independent aliquots of viral cDNA or using NGS deep sequencing technology can increase the sensitivity of genotypic tropism testing (Archer et al., 2012; Swenson et al., 2010). Several clinical laboratories in high-income countries offer one or more genotypic approaches for detecting minor X4 tropic viruses.
10. Effect of Subtype on HIV-1 Drug Resistance
Subtype B viruses comprise more than 95% of HIV-1 strains in the U.S. and more than 60% of strains in Europe but only about 10% of strains worldwide. Subtype B viruses played an outsized role in the earliest ARV development efforts and in the earliest studies of ARV resistance. Nonetheless, existing ARVs are equally effective at treating subtype B and nonsubtype B viruses (Bannister et al., 2006; Geretti et al., 2009; Scherrer et al., 2011). Moreover, each of the mutations that cause resistance in subtype B viruses also cause resistance in non-B subtypes.
However, several DRMs preferentially occur in certain HIV-1 subtypes. The majority of these subtype-specific differences in DRM distribution are attributed to differences in codon usage. The NNRTI DRM V106M occurs more often in subtype C viruses from patients treated with NVP or EFV because V106M requires a single base-pair change in subtype C viruses – GTG (V) => ATG (M) – but a two base-pair change in all other subtypes – GTA (V) => ATG (M) (Brenner et al., 2003). CRF01_AE viruses preferentially develop the NRTI DRM V75M (Ariyoshi et al., 2003), subtype G viruses preferentially develop the PI DRM V82M (Palma et al., 2012), and subtype A viruses from the former Soviet Union (AFSU) preferentially develop the NNRTI DRM G190S (Kolomeets et al., 2014) by a similar mechanism.
In subtype B viruses, integrase G140 is encoded primarily by GGT or GGC whereas for nearly all other subtypes, G140 is encoded primarily by GGG or GGA (Doyle et al., 2015). Therefore, a single G to A change at the first position of the G140 codon results in a serine in subtype B viruses, but not in other subtypes. G140S is an important compensatory mutation for Q148 mutations and this may explain why Q148 mutations occur significantly more commonly among subtype B patients compared to non-B patients with RAL VF (Boyd et al., 2015; Doyle et al., 2015; Fourati et al., 2015).
Several lines of evidence suggest that K65R is more likely to emerge in subtype C viruses than viruses belonging to other subtypes. First, biochemical studies have shown that the unique subtype C sequence in the region of K65R, specifically five consecutive adenosines preceding the adenosine at the second position in the K65 codon, facilitates mutation during reverse transcription (Coutsinos et al., 2009; Invernizzi et al., 2009). Second, K65R emerges more rapidly during in vitro passage of subtype C viruses than during passage of subtype B viruses in the presence of tenofovir (Brenner et al., 2006). Third, three retrospective studies have suggested that in patients who develop VF on a TDF-containing regimen are at a somewhat higher risk of developing K65R if they are infected with a subtype C virus (TenoRes Study, 2016; Theys et al., 2013; White et al., 2016). Despite the reported tendency to acquire the K65R DRM, a recent study showed that when demographic and clinical factors are controlled for among patients receiving at TDF-containing regimen, infection with a subtype C virus does not increase the risk of developing VF (White et al., 2016).
11. Transmitted Drug Resistance
TDR is a type of HIVDR that occurs when primary HIV infection is caused by a DRM-bearing virus. The prevalence of TDR by SGRT is about 12% to 24% in the U.S., and 10% in Europe (Boden et al., 1999; Booth and Geretti, 2007; Kassaye et al., 2016; Li et al., 2016; Little et al., 2002; Parker et al., 2007; Rhee et al., 2015a; Shet et al., 2006; Simon et al., 2002; Weinstock et al., 2000; Weinstock et al., 2004). While limited data suggest that the prevalence is below 5% in most of the LMICs in sub-Saharan Africa and South/Southeast Asia (Frentz et al., 2012; Rhee et al., 2015a; Wheeler et al., 2010), intermediate TDR rates of 5 to 10% have been reported from Latin America, Eastern Europe, and high-income Asian countries (Frentz et al., 2012; Rhee et al., 2015a). Upward temporal trends in TDR continue to be reported from many regions, largely attributable to increases in NNRTI TDR (Bertagnolio et al., 2016; Gupta et al., 2012; Jordan et al., 2016; Rhee et al., 2015a).
Because DRMs reduce viral fitness, many transmitted drug-resistant viruses gradually revert to wild type over a period of several years (Brenner et al., 2002; Castro et al., 2013; Jain et al., 2011; Little et al., 2008; Pao et al., 2004). DRMs associated with the greatest reductions in viral fitness revert to wild type more quickly than those with less effect on viral fitness. Conversely, those DRMs with a low fitness cost have been shown to persist for a prolonged period following transmission (Mourad et al., 2015).
In the absence of selective drug pressure, PI and NNRTI DRMs gradually revert to wild type after an average of approximately 3 years. Within the NRTI class, the rate of reversion is more variable: M184V usually fades within one year; T215FY also fades rapidly; but T215 revertants persist for more than ten years (Castro et al., 2013; Jain et al., 2011). It is because of this tendency to fade to lower levels over time that the rates of detectable TDR are higher in recently compared with chronically infected patients, and that studies using more sensitive methods for detecting low-abundance drug-resistant viruses (see section on Diagnosis and Management of Drug Resistance) will detect higher rates of TDR than those using SGRT (Buckton et al., 2011; Cozzi-Lepri et al., 2015; Ellis et al., 2009; Goodman et al., 2011; Metzner et al., 2014).
The most common transmitted NNRTI-associated DRMs include K103N, Y181C, G190A, and K101E which combined accounted for >80% of NNRTI-associated TDR in a recent global meta-analysis. The most common NRTI-associated DRMs include M184V, the TAMs, the T215 revertants, T69D, and F77L (Rhee et al., 2015a). Not surprisingly, M184V occurs more commonly in recently infected patients compared with chronically infected patients. The most common PI SDRMs were L90M, M46I/L, I85V, I54V, N88D, I84V, D30N, G73S, and they occurred less frequently (Bertagnolio et al., 2016; Rhee et al., 2015a). Although transmitted INSTI-associated resistance has been practically absent in ongoing surveillance efforts (Casadella et al., 2015; Doyle et al., 2015; Stekler et al., 2015), there have been at least four case reports of INSTI-associated TDR (Boyd et al., 2011; Varghese et al., 2016; Volpe et al., 2015; Young et al., 2011).
TDR arises from two distinct sources: (i) from patients failing therapy who have developed ADR, or (ii) from an ART-naïve person with TDR, a phenomenon termed “onward transmission”. With a greater number of intervening transmission events in the case of onward transmission, the prevalence of DRMs with high fitness costs, such as M184V, is likely to decrease. Onward transmission is therefore responsible for established drug-resistant lineages that typically contain fit drug-resistant strains. Such established lineages are often the major sources of TDR in European cohorts (Mourad et al., 2015; Yerly et al., 2009).
In high-income countries, SGRT is recommended at the time of a patient’s initial diagnosis. If therapy is delayed, SGRT may be repeated at the time of ART initiation if the patient is considered to have been at risk for superinfection (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). If transmitted INSTI drug resistance is suspected, SGRT of the integrase gene should also be performed (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016).
Despite its increasing frequency, there are few published guidelines for treating patients with TDR. In patients with isolated NNRTI resistance, multiple studies indicate that boosted PIs are highly effective for initial treatment (Chaix et al., 2007; Clutter et al., 2016a; Oette et al., 2006; Wittkop et al., 2011; Zu Knyphausen et al., 2014) and several studies have indicated that standard first-line INSTI-containing regimens are also likely to be highly effective (Clutter et al., 2016a; Kulkarni et al., 2014). In patients with transmitted NRTI resistance, the selection of first-line therapy should be based on the extent of NRTI resistance. Both LPV/r and DRV/r-containing regimens have been shown to be highly effective for treating patients with multiple NRTI and NNRTI-associated DRMs following the failure of a first-line NRTI/NNRTI-containing regimen (Ajose et al., 2012; Bartlett et al., 2012; Madruga et al., 2007; Paton et al., 2014). Therefore, they are each also likely to be highly effective at treating ARV-naïve patients harboring multiple NRTI-resistance mutations. However, first-line ATV/r, RAL, or EVG-containing regimens should be used only for treating patients with minimal transmitted NRTI resistance. The use of DTG-containing regimens in this scenario may be useful but needs further study.
Due to historically lower levels of TDR in LMICs and cost-effectiveness considerations, individual-level baseline SGRT is not recommended by the WHO in LMICs (Phillips et al., 2014a). However population-level HIVDR surveillance programs are strongly recommended and they have played a key role in tracking TDR trends and in shaping first-line ARV regimen recommendations (see Section 14).
In LMICs, ART programs are facing important challenges such as ARV stock outs and suboptimal retention in care (World Health Organization, 2016c). Patients experiencing HIV treatment interruptions are more likely to carry DRMs. When these patients re-engage into care, they most frequently re-initiate treatment with the same first-line regimen with which they were previously treated, despite the increased risk of resistance relative to the truly ART-naive population (Hamers et al., 2012). Therefore, to assess the predicted activity of first-line ART, the WHO recommends monitoring pre-treatment resistance (PDR) not only among individuals initiating ART for the first time but also among individuals re-initiating first-line ART (World Health Organization, 2014a). PDR is indeed the result of either TDR or ADR, the later due to prior ARV treatment among re-starters, prior exposure to PMTCT among women, or prior ARV prophylaxis.
PDR surveys will be critical given the more recently observed signals of high level HIVDR in several LMIC settings, including Angola (16%), Cuba (22%), Mexico (12%), Papua New Guinea (16%), Botswana (10%) and South Africa (14%) (Afonso et al., 2012; Avila-Rios et al., 2016; Lavu et al., 2015; National Institute for Communicable Diseases, 2016; Perez et al., 2013). In the absence of SGRT-guided therapy, national PDR surveillance among individuals starting ART is critical to predict the activity of available first-line ART and inform the composition of these regimens. Where PDR levels remain low, patients are anticipated to do well on the WHO-recommended first-line regimen of EFV + TDF/3TC or FTC (World Health Organization, 2015a). Where PDR levels are high, alternative first-line regimens may become necessary for all starters or for a subset of those reporting prior ARV drug exposure (that is, people who defaulted from ART and restart, women exposed to PMTCT and perhaps those infected despite ARV drug prophylaxis [i.e. post-exposure prophylaxis and PrEP]). Although these surveys are strongly recommended by WHO and are essential to address data gaps in LMIC, their implementation has been slow due to lack of awareness of HIVDR as a public health threat, limited resources, and competing national and donor priorities. As discussed in Section 14, WHO is developing a coordinated and resourced global action plan for HIVDR.
12. Acquired Drug Resistance
In most studies, more than 70–80% of patients with VF develop ADR (Jordan et al., 2016; Kumarasamy et al., 2015; Paton et al., 2014; Prosperi et al., 2011; Schultze et al., 2015; Second-Line Study Group et al., 2013; TenoRes Study, 2016). M184V/I is usually selected as the initial NRTI DRM for cytidine analog (3TC or FTC) containing regimens. Among patients receiving an NNRTI-containing regimen, one or more NNRTI DRMs are also typically seen early in VF. Other NRTI DRMs usually occur after M184V/I and the NNRTI DRMs (Jordan et al., 2016; Rhee et al., 2015b). INSTI DRMs may develop early in treatment, but PI DRMs are rare and are generally a late occurrence. Resistance is detected more often in patients with infrequent viral load testing because VF is usually detected later, after DRMs have had time to accumulate (Gupta et al., 2009). Through this mechanism of prolonged VF, infrequent viral load testing may jeopardize the efficacy of second-line options, particularly NNRTI- and INSTI-based regimens.
Patterns of ADR at VF vary according to regimen and patient adherence levels. Indeed, the lack of any ADR in the setting of treatment failure usually indicates non-adherence. Because all recommended first-line regimens contain a cytidine analog, M184V is the most commonly acquired DRM in both high-income countries and LMICs (Rhee et al., 2015b). The later NRTI DRMs to develop depend on the second NRTI in the regimen. Although both d4T and AZT are thymidine analogs, they differ somewhat in the mutations they select. AZT selects almost exclusively TAMs whereas d4T selects primarily TAMs but may also select discriminatory mutations including K65R. TDF selects primarily for K65R and less commonly for K70E/G/Q and Y115F (Jordan et al., 2016; TenoRes Study, 2016; Van Zyl et al., 2013). Abacavir selects primarily L74V/I, Y115F, and less commonly K65R.
NNRTI DRMs are selected early and often by NNRTI-based regimens, occurring in the vast majority of patients with VF (Delaugerre et al., 2001; Molina et al., 2011; Second-Line Study Group et al., 2013; TenoRes Study, 2016). In both high-income countries and LMICs, K103N followed by Y181C, G190A/S, Y188L, and V106M are the most commonly observed acquired NNRTI DRMs (Rhee et al., 2015b). After failing a first-line NNRTI-based regimen containing EFV or NVP, cross-resistance to the second generation NNRTIs RPV and ETR is common and has been associated with VF (Ruxrungtham et al., 2008; Teeranaipong et al., 2016; Theys et al., 2015).
In previously PI-naïve patients, PI DRMs often occur in approximately 10% or fewer patients failing PI-based therapy (Barber et al., 2012; Dolling et al., 2013; El Bouzidi et al., 2014; Mollan et al., 2012). As noted previously, a high proportion of patients with VF on an initial PI-based regimen but without PI DRMs will achieve virological suppression with improved adherence alone (Zheng et al., 2014).
In patients with VF on a first-line RAL- or EVG-based regimen, INSTI resistance is not uncommon (Kulkarni et al., 2014; Rockstroh et al., 2013). In contrast, treatment-emergent INSTI resistance is rarely observed in patients failing first-line DTG-based regimens (Raffi et al., 2013; White et al., 2014b).
In high-income countries, SGRT is recommended in patients with VF when the viral load is >1000 copies/mL (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016), and should be considered when the viral load is >50 copies/mL (Gonzalez-Serna et al., 2014; Santoro et al., 2014). In patients with VF, the ability to detect ADR is much higher, if SGRT is performed while a patient is on the failing regimen or within four weeks of discontinuation (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). Genotypic testing can indicate whether VF is due primarily to non-adherence – i.e., if no DRMs are present – and can also be used to help design second-line therapy when ADR is the cause of VF. Patients who develop VF without DRMs can often be treated successfully with adherence counseling alone (Bonner et al., 2013; Hoffmann et al., 2014). In contrast, patients with VF who have developed DRMs on a first-line regimen usually require a change in ARVs.
In high-income countries where SGRT is routinely performed in the setting of VF, a second-line regimen should include at least two, preferably three, fully active agents. As a general principle, adding a single fully active ARV to a failing regimen is not recommended due to the risk of selecting for resistance to the new ARV. A fully active agent is defined as an ARV which is expected to have activity based on the patient’s treatment history and SGRT results (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). Managing patients with ADR who are failing second-line regimens is more challenging, and often requires expert guidance. With complex patterns of resistance, phenotypic testing may be useful to determine the activity of various ARVs (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016). The number of available fully active ARVs diminishes with each successive treatment failure. Therefore, a salvage regimen in patients who have experienced multiple VFs and who have multiclass resistant viruses is likely to be complicated and to require multiple ARVs that may have only partial residual activity.
Late salvage therapy ARVs can be placed into the following categories: (i) ARVs belonging to a previously unused drug class. Enfuvirtide is a highly active treatment option for late salvage therapy. However, it is not a popular treatment choice among clinicians and patients because of its requirement for parenteral intramuscular administration. Although maraviroc is well tolerated, it is often not an option because patients with multiple VFs tend to have more advanced immunosuppression and an increased likelihood of harboring X4-tropic viruses; (ii) ARVs belonging to a previously used drug class that retain significant antiviral activity including the PIs DRV/r and tipranavir/r, the INSTI dolutegravir, and in some instances the NNRTI ETR; (iii) NRTI combinations, which often retain in vivo virological activity even in the presence of reduced in vitro NRTI susceptibility; (iv) ARVs associated with previous VF episodes and treatment-emergent drug resistance that may have regained their activity as a result of viral reversion to wild type; and (v) investigational ARVs (Tang and Shafer, 2012).
In most LMICs, a diagnosis of ADR is made empirically and the selection of second-line therapy is guided by WHO recommendations. A patient is presumed to have ADR when two viral loads performed 3 months apart are both above 1,000 copies/mL, and a switch to second-line therapy is indicated (World Health Organization, 2015b). The preferred second-line options include a boosted PI, namely ATV/r or LPV/r, and two NRTIs. Subsequent options include DRV/r plus two NRTIs or LPV/r plus RAL. These alternatives are equally efficacious, but due to higher costs they are not currently preferred options in second-line regimens. Another consideration is that, as previously noted, PI-based regimens rarely select for PI DRMs, but failure of a RAL-containing regimen often selects for INSTI DRMs that may jeopardize INSTI activity in subsequent regimens. Second-line NRTI selection is directed by the first-line NRTIs used: if VF developed on TDF/FTC or 3TC, then AZT/3TC is recommended; if VF developed on AZT/3TC, then TDF/FTC or 3TC is recommended (World Health Organization, 2015a). In some LMICs with limited access to SGRT, resistance testing is recommended at VF of second-line therapy (Republic of South Africa Department of Health, 2014) and is managed according to the resistance detected and regimens available.
13. PrEP and Drug Resistance
As of September 2015, the World Health Organization has recommended TDF-containing pre-exposure prophylaxis (PrEP) for men who have sex with men, heterosexually-active men and women, and intravenous drug users considered at substantial risk for HIV-1 infection (World Health Organization, 2015c). Although the risk of HIV-1 infection is markedly reduced in patients receiving PrEP, those who become infected are theoretically at high risk of developing drug resistance. In a systematic review and meta-analysis of PrEP trials containing TDF, 11 (0.1%) of 9,222 PrEP users developed FTC or TDF DRMs. However, because 7 of these 11 infections actually had acute HIV-1 infection at the time of PrEP initiation, the rate of breakthrough infection with resistance is even lower (Baeten et al., 2012; Fonner et al.; Liegler et al., 2014; Thigpen et al., 2012; World Health Organization, 2015c). It has been estimated that for every case of FTC resistance attributed to starting PrEP in the setting of unrecognized acute HIV-1 infection, between 8 to 50 HIV-1 infections are prevented (Fonner et al.; World Health Organization, 2015c). The most commonly reported PrEP-associated DRMs are the FTC-associated M184V/I mutations followed by the TDF-associated K65R mutation (Baeten et al., 2012; Liegler et al, 2014; Thigpen et al., 2012). The risk of PrEP-associated ADR can likely by mitigated through routine frequent laboratory testing to identify incident HIV-1 infections and initiate ART in any incident cases detected. Despite the very low levels of resistance observed in clinical trials, resistance may increase as PrEP interventions are more widely implemented, particularly in settings where unrecognized HIV infections may be exposed to PrEP for longer periods.
14. HIV Drug Resistance Surveillance
Between 2004 and 2013, the World Health Organization recommended the surveillance of TDR amongst recently infected populations (Bennett et al., 2008; Myatt and Bennett, 2008; Shafer et al., 2008). The WHO-recommended method for surveillance of TDR limited enrollment to ARV-naïve populations likely to have been recently infected, and assessed for the presence of 93 non-polymorphic DRMs found in all subtypes, termed surveillance DRMs (SDRMs) (Bennett et al., 2009; Shafer et al., 2008).
In 2015, WHO revised the recommendations and prioritized the surveillance of drug resistance among all populations initiating ART, regardless of duration of infection or prior ARV drug exposure(s), primarily because of its programmatic impact in guiding recommended first-line regimens and feasibility. The WHO’s HIVDR Surveillance and Monitoring approach consists of surveillance of PDR (World Health Organization, 2014a), ADR (World Health Organization, 2014b), HIVDR in infants naïve to ART, and annual monitoring of early warning indicators (EWIs) of HIVDR, defined as clinic- and program-related factors associated with the emergence of HIVDR. The surveys provide nationally representative estimates of HIVDR and aim to guide the selection of first-,second-, and third-line regimens (World Health Organization, 2015d).
The wide implementation of the newly released WHO HIV Consolidated Guidelines recommending initiation of ART to all and scale up of PrEP are expected to lead to a marked decrease in HIV incidence. However, among those patients who become infected a higher proportion of individuals will be infected primarily with a drug-resistant strain and a higher proportion of all HIV-infected patients will be at risk for ADR. To address this concern and ensure a coordinated action to monitor and respond to HIVDR, the WHO is currently developing a Global Action Plan on HIV Drug Resistance. The development of this five-year plan reflects a global consensus that HIVDR in LMICs requires a coordinated and resourced response. The Global Action Plan is an agreement by key partners about their respective roles in preventing, monitoring, and responding to HIVDR (World Health Organization, 2016a).
15. Summary and Conclusions
An unprecedented scale-up of ART has been observed in the last 10 years: at the end of 2015, 17 million people were receiving antiretroviral therapy. However, the HIVDR emergence associated with increased ART use can compromise the effectiveness of antiretroviral drugs. Resistance is an inevitable consequence of any interventions that, through the use of ARVs, aim to reduce mortality, morbidity and HIV transmission. While concern for resistance should never limit ARV access for all HIV-infected individuals, the long-term implications of earlier initiation on adherence and drug resistance need to be closely monitored. Knowledge of the mechanisms, implications, and patterns of HIVDR can be harnessed to both select effective SGRT-guided therapy for individuals and/or to adapt national treatment guidelines according to surveillance findings following the public health approach to HIV treatment.
Highlights.
We review the patterns, genetic mechanisms, and management of HIV-1 drug resistance
Rates of transmitted drug resistance are increasing
Acquired drug resistance is common during treatment failure
Genetic barriers to resistance and cross-resistance of antiretrovirals predict efficacy
HIV drug resistance testing can guide individual therapy where available
HIV drug resistance surveillance can guide programmatic recommendations
Acknowledgments
Funding: This work was supported by a KL2 Mentored Career Development Award of the Stanford Clinical and Translational Science Award to Spectrum [grant number NIH KL2 TR 001083 and NIH UL1 TR 001085 to DSC], by the National Institutes of Health [grant number R01 AI068581 to RWS], by CFAR [grant number 1P30A142853 to MRJ] and by the Christine E. Driscoll O’Neill and James M. Driscoll, Driscoll-O’Neill Charitable Foundation [support for MRJ].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest: DSC receives research funding through the Bristol-Myers Squibb Virology Fellows Research Program. RWS is a consultant for Celera, and receives research funding from Roche Molecular, Gilead Sciences, Bristol-Myers Squibb, and Merck. The other authors have no potential conflicts of interest to disclose.
Disclaimer:
The opinions and conclusions in this article are those of the authors and do not reflect those of the respective institutions, including the World Health Organization.
Contributor Information
Dana S Clutter, Email: dclutter@stanford.edu.
Michael R Jordan, Email: mjordan@tuftsmedicalcenter.org.
Silvia Bertagnolio, Email: bertagnolios@who.int.
Robert W Shafer, Email: rshafer@stanford.edu.
References
- Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. Journal of virology. 2010;84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram ME, Hluhanich RM, Goodman DD, Andreatta KN, Margot NA, Ye L, Niedziela-Majka A, Barnes TL, Novikov N, Chen X, Svarovskaia ES, McColl DJ, White KL, Miller MD. Impact of Primary Elvitegravir Resistance-Associated Mutations in HIV-1 Integrase on Drug Susceptibility and Viral Replication Fitness. Antimicrobial agents and chemotherapy. 2013 doi: 10.1128/AAC.02568-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso JM, Bello G, Guimaraes ML, Sojka M, Morgado MG. HIV-1 genetic diversity and transmitted drug resistance mutations among patients from the North, Central and South regions of Angola. PloS one. 2012;7:e42996. doi: 10.1371/journal.pone.0042996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Zaccarelli M, Cingolani A, Forbici F, Rizzo MG, Trotta MP, Di Giambenedetto S, Narciso P, Ammassari A, Girardi E, De Luca A, Perno CF. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS research and human retroviruses. 2002;18:835–838. doi: 10.1089/08892220260190308. [DOI] [PubMed] [Google Scholar]
- Archer J, Weber J, Henry K, Winner D, Gibson R, Lee L, Paxinos E, Arts EJ, Robertson DL, Mimms L, Quinones-Mateu ME. Use of Four Next-Generation Sequencing Platforms to Determine HIV-1 Coreceptor Tropism. PloS one. 2012;7 doi: 10.1371/journal.pone.0049602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi K, Matsuda M, Miura H, Tateishi S, Yamada K, Sugiura W. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. Journal of acquired immune deficiency syndromes. 2003;33:336–342. doi: 10.1097/00126334-200307010-00007. [DOI] [PubMed] [Google Scholar]
- Avila-Rios S, Garcia-Morales C, Tapia-Trejo D, Matias-Florentino M, Quiroz-Morales V, Casillas-Rodriguez J, Sierra-Madero J, Leon E, Magis-Rodriguez C, Reyes-Teran G for the HIVDR MexNet Group. HIV Pre-Treatment Drug Resistance in Mexico: A Nationally Representative WHO Survey. XXV International HIV Drug Resistance Workshop; Boston, USA: 2016. Abstract 64. [Google Scholar]
- Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, Tappero JW, Bukusi EA, Cohen CR, Katabira E, Ronald A, Tumwesigye E, Were E, Fife KH, Kiarie J, Farquhar C, John-Stewart G, Kakia A, Odoyo J, Mucunguzi A, Nakku-Joloba E, Twesigye R, Ngure K, Apaka C, Tamooh H, Gabona F, Mujugira A, Panteleeff D, Thomas KK, Kidoguchi L, Krows M, Revall J, Morrison S, Haugen H, Emmanuel-Ogier M, Ondrejcek L, Coombs RW, Frenkel L, Hendrix C, Bumpus NN, Bangsberg D, Haberer JE, Stevens WS, Lingappa JR, Celum C, Partners Pr, E.P.S.T Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. The New England journal of medicine. 2012;367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister WP, Ruiz L, Loveday C, Vella S, Zilmer K, Kjaer J, Knysz B, Phillips AN, Mocroft A, Euro, S.S.G HIV-1 subtypes and response to combination antiretroviral therapy in Europe. Antiviral therapy. 2006;11:707–715. [PubMed] [Google Scholar]
- Barber TJ, Harrison L, Asboe D, Williams I, Kirk S, Gilson R, Bansi L, Pillay D, Dunn D. Frequency and patterns of protease gene resistance mutations in HIV-infected patients treated with lopinavir/ritonavir as their first protease inhibitor. The Journal of antimicrobial chemotherapy. 2012;67:995–1000. doi: 10.1093/jac/dkr569. [DOI] [PubMed] [Google Scholar]
- Bartlett JA, Ribaudo HJ, Wallis CL, Aga E, Katzenstein DA, Stevens WS, Norton MR, Klingman KL, Hosseinipour MC, Crump JA, Supparatpinyo K, Badal-Faesen S, Kallungal BA, Kumarasamy N. Lopinavir/ritonavir monotherapy after virologic failure of first-line antiretroviral therapy in resource-limited settings. Aids. 2012;26:1345–1354. doi: 10.1097/QAD.0b013e328353b066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS one. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antiviral therapy. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- Bertagnolio S, Barcarolo J, Davis DH, Gregson J, Sigaloff KC, Hamers RL, Rinke de Wit TF, Parkin N, Jordan MR, Group, R.T.S.W Assessment of Transmitted HIV Drug Resistance in Resource Limitied Countries, 2004–2010. 2016 Unpublished. [Google Scholar]
- Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. HIV-1 integrase inhibitor resistance and its clinical implications. The Journal of infectious diseases. 2011;203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Jones E, Tsay J, Ip J, Farthing C, Limoli K, Parkin N, Markowitz M. HIV-1 drug resistance in newly infected individuals. Jama. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- Bonner K, Mezochow A, Roberts T, Ford N, Cohn J. Viral load monitoring as a tool to reinforce adherence: a systematic review. Journal of acquired immune deficiency syndromes. 2013;64:74–78. doi: 10.1097/QAI.0b013e31829f05ac. [DOI] [PubMed] [Google Scholar]
- Booth CL, Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. The Journal of antimicrobial chemotherapy. 2007;59:1047–1056. doi: 10.1093/jac/dkm082. [DOI] [PubMed] [Google Scholar]
- Borroto-Esoda K, Waters JM, Bae AS, Harris J, Hinkle JE, Quinn JB, Rousseau FS. Baseline genotype as a predictor of virological failure to emtricitabine or stavudine in combination with didanosine and efavirenz. AIDS research and human retroviruses. 2007;23:998–995. doi: 10.1089/aid.2006.0310. [DOI] [PubMed] [Google Scholar]
- Boucher S, Recordon-Pinson P, Ragnaud JM, Dupon M, Fleury H, Masquelier B. HIV-1 reverse transcriptase (RT) genotypic patterns and treatment characteristics associated with the K65R RT mutation. HIV medicine. 2006;7:294–298. doi: 10.1111/j.1468-1293.2006.00379.x. [DOI] [PubMed] [Google Scholar]
- Boyd MA, Moore CL, Molina JM, Wood R, Madero JS, Wolff M, Ruxrungtham K, Losso M, Renjifo B, Teppler H, Kelleher AD, Amin J, Emery S, Cooper DA, group, S.-L.s Baseline HIV-1 resistance, virological outcomes, and emergent resistance in the SECOND-LINE trial: an exploratory analysis. The lancet. HIV. 2015;2:e42–51. doi: 10.1016/S2352-3018(14)00061-7. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Maldarelli F, Sereti I, Ouedraogo GL, Rehm CA, Boltz V, Shoemaker D, Pau AK. Transmitted raltegravir resistance in an HIV-1 CRF_AG-infected patient. Antiviral therapy. 2011;16:257–261. doi: 10.3851/IMP1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, Marlink RG, Schapiro J, Roger M, Wainberg MA. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. Aids. 2003;17:F1–5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- Brenner BG, Oliveira M, Doualla-Bell F, Moisi DD, Ntemgwa M, Frankel F, Essex M, Wainberg MA. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. Aids. 2006;20:F9–13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- Brenner BG, Routy JP, Petrella M, Moisi D, Oliveira M, Detorio M, Spira B, Essabag V, Conway B, Lalonde R, Sekaly RP, Wainberg MA. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. Journal of virology. 2002;76:1753–1761. doi: 10.1128/JVI.76.4.1753-1761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JS, Harrigan PR. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. The Journal of infectious diseases. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- Buckton AJ, Harris RJ, Pillay D, Cane PA. HIV type-1 drug resistance in treatment-naive patients monitored using minority species assays: a systematic review and meta-analysis. Antiviral therapy. 2011;16:9–16. doi: 10.3851/IMP1687. [DOI] [PubMed] [Google Scholar]
- Campbell TB, Shulman NS, Johnson SC, Zolopa AR, Young RK, Bushman L, Fletcher CV, Lanier ER, Merigan TC, Kuritzkes DR. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2005;41:236–242. doi: 10.1086/430709. [DOI] [PubMed] [Google Scholar]
- Casadella M, van Ham PM, Noguera-Julian M, van Kessel A, Pou C, Hofstra LM, Santos JR, Garcia F, Struck D, Alexiev I, Bakken Kran AM, Hoepelman AI, Kostrikis LG, Somogyi S, Liitsola K, Linka M, Nielsen C, Otelea D, Paraskevis D, Poljak M, Puchhammer-Stockl E, Stanekova D, Stanojevic M, Van Laethem K, Zidovec Lepej S, Clotet B, Boucher CA, Paredes R, Wensing AM, programme, S Primary resistance to integrase strand-transfer inhibitors in Europe. The Journal of antimicrobial chemotherapy. 2015;70:2885–2888. doi: 10.1093/jac/dkv202. [DOI] [PubMed] [Google Scholar]
- Castagna A, Danise A, Menzo S, Galli L, Gianotti N, Carini E, Boeri E, Galli A, Cernuschi M, Hasson H, Clementi M, Lazzarin A. Lamivudine monotherapy in HIV-1-infected patients harbouring a lamivudine-resistant virus: a randomized pilot study (E-184V study) Aids. 2006;20:795–803. doi: 10.1097/01.aids.0000218542.08845.b2. [DOI] [PubMed] [Google Scholar]
- Castagna A, Maggiolo F, Penco G, Wright D, Mills A, Grossberg R, Molina JM, Chas J, Durant J, Moreno S, Doroana M, Ait-Khaled M, Huang J, Min S, Song I, Vavro C, Nichols G, Yeo JM, Group, V.-S Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. The Journal of infectious diseases. 2014a;210:354–362. doi: 10.1093/infdis/jiu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagna A, Spagnuolo V, Galli L, Vinci C, Nozza S, Carini E, D’Arminio Monforte A, Montella F, Antinori A, Di Biagio A, Rusconi S, Lazzarin A, Group, M.O.S Simplification to atazanavir/ritonavir monotherapy for HIV-1 treated individuals on virological suppression: 48-week efficacy and safety results. Aids. 2014b;28:2269–2279. doi: 10.1097/QAD.0000000000000407. [DOI] [PubMed] [Google Scholar]
- Castro H, Pillay D, Cane P, Asboe D, Cambiano V, Phillips A, Dunn DT, Resistance, U.K.C.G.o.H.D Persistence of HIV-1 transmitted drug resistance mutations. The Journal of infectious diseases. 2013;208:1459–1463. doi: 10.1093/infdis/jit345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix ML, Desquilbet L, Descamps D, Costagliola D, Deveau C, Galimand J, Goujard C, Signori-Schmuck A, Schneider V, Tamalet C, Pellegrin I, Wirden M, Masquelier B, Brun-Vezinet F, Rouzioux C, Meyer L, French, P.C.S.G., French, A.A.C.R.S.G Response to HAART in French patients with resistant HIV-1 treated at primary infection: ANRS Resistance Network. Antiviral therapy. 2007;12:1305–1310. [PubMed] [Google Scholar]
- Charest H, Doualla-Bell F, Cantin R, Murphy DG, Lemieux L, Brenner B, Hardy I, Moisi D, Lo E, Baril JG, Wainberg MA, Roger M, Tremblay C. A significant reduction in the frequency of HIV-1 drug resistance in Quebec from 2001 to 2011 is associated with a decrease in the monitored viral load. PloS one. 2014;9:e109420. doi: 10.1371/journal.pone.0109420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BH, Sinkala M, Mbewe F, Cantrell RA, Kruse G, Chintu N, Aldrovandi GM, Stringer EM, Kankasa C, Safrit JT, Stringer JS. Single-dose tenofovir and emtricitabine for reduction of viral resistance to non-nucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for perinatal HIV prevention: an open-label randomised trial. Lancet. 2007;370:1698–1705. doi: 10.1016/S0140-6736(07)61605-5. [DOI] [PubMed] [Google Scholar]
- Chung MH, Beck IA, Dross S, Tapia K, Kiarie JN, Richardson BA, Overbaugh J, Sakr SR, John-Stewart GC, Frenkel LM. Oligonucleotide Ligation Assay Detects HIV Drug Resistance Associated With Virologic Failure Among Antiretroviral-Naive Adults in Kenya. Journal of acquired immune deficiency syndromes. 2014;67:246–253. doi: 10.1097/QAI.0000000000000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutter DS, Fessel WJ, Rhee SY, Hurley LB, Klein DB, Ioannidis JP, Silverberg MJ, Shafer RW. Response to Therapy in Antiretroviral Therapy-Naive Patients with Isolated Nonnucleoside Reverse-Transcriptase Inhibitor-Associated Transmitted Drug Resistance. Journal of acquired immune deficiency syndromes. 2016a doi: 10.1097/QAI.0000000000000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutter DS, Rojas Sanchez P, Rhee SY, Shafer RW. Genetic Variability of HIV-1 for Drug Resistance Assay Development. Viruses. 2016b;8 doi: 10.3390/v8020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Andrade-Villanueva J, Clotet B, Fourie J, Johnson MA, Ruxrungtham K, Wu H, Zorrilla C, Crauwels H, Rimsky LT, Vanveggel S, Boven K, group, T.s Rilpivirine versus efavirenz with two background nucleoside or nucleotide reverse transcriptase inhibitors in treatment-naive adults infected with HIV-1 (THRIVE): a phase 3, randomised, non-inferiority trial. Lancet. 2011;378:229–237. doi: 10.1016/S0140-6736(11)60983-5. [DOI] [PubMed] [Google Scholar]
- Collins SE, Grant PM, Shafer RW. Modifying Antiretroviral Therapy in Virologically Suppressed HIV-1-Infected Patients. Drugs. 2016;76:75–98. doi: 10.1007/s40265-015-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutsinos D, Invernizzi CF, Xu H, Moisi D, Oliveira M, Brenner BG, Wainberg MA. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. Journal of virology. 2009;83:2029–2033. doi: 10.1128/JVI.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, Schuurman R, Daumer M, Aitken S, Ceccherini-Silberstein F, D’Arminio Monforte A, Geretti AM, Booth CL, Kaiser R, Michalik C, Jansen K, Masquelier B, Bellecave P, Kouyos RD, Castro E, Furrer H, Schultze A, Gunthard HF, Brun-Vezinet F, Paredes R, Metzner KJ, Group, C.M.H.-V.W Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. The Journal of antimicrobial chemotherapy. 2015;70:930–940. doi: 10.1093/jac/dku426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meyer S, Vangeneugden T, van Baelen B, de Paepe E, van Marck H, Picchio G, Lefebvre E, de Bethune MP. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS research and human retroviruses. 2008;24:379–388. doi: 10.1089/aid.2007.0173. [DOI] [PubMed] [Google Scholar]
- Delaugerre C, Rohban R, Simon A, Mouroux M, Tricot C, Agher R, Huraux JM, Katlama C, Calvez V. Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. Journal of medical virology. 2001;65:445–448. [PubMed] [Google Scholar]
- Delfraissy JF, Flandre P, Delaugerre C, Ghosn J, Horban A, Girard PM, Norton M, Rouzioux C, Taburet AM, Cohen-Codar I, Van PN, Chauvin JP. Lopinavir/ritonavir monotherapy or plus zidovudine and lamivudine in antiretroviral-naive HIV-infected patients. Aids. 2008;22:385–393. doi: 10.1097/QAD.0b013e3282f3f16d. [DOI] [PubMed] [Google Scholar]
- Diallo K, Gotte M, Wainberg MA. Molecular impact of the M184V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrobial agents and chemotherapy. 2003;47:3377–3383. doi: 10.1128/AAC.47.11.3377-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolling DI, Dunn DT, Sutherland KA, Pillay D, Mbisa JL, Parry CM, Post FA, Sabin CA, Cane PA, Database, U.H.D.R., Study, U.K.C.H.C Low frequency of genotypic resistance in HIV-1-infected patients failing an atazanavir-containing regimen: a clinical cohort study. The Journal of antimicrobial chemotherapy. 2013;68:2339–2343. doi: 10.1093/jac/dkt199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T, Dunn DT, Ceccherini-Silberstein F, De Mendoza C, Garcia F, Smit E, Fearnhill E, Marcelin AG, Martinez-Picado J, Kaiser R, Geretti AM, Group, C.S Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. The Journal of antimicrobial chemotherapy. 2015;70:3080–3086. doi: 10.1093/jac/dkv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DM, Chin EN, Bimber BN, Sanabani SS, Tarosso LF, Costa PR, Sauer MM, Kallas EG, O’Connor DH. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PloS one. 2012;7:e36494. doi: 10.1371/journal.pone.0036494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bouzidi K, White E, Mbisa JL, Phillips A, Mackie N, Pozniak A, Dunn D. Protease mutations emerging on darunavir in protease inhibitor-naive and experienced patients in the UK. Journal of the International AIDS Society. 2014;17:19739. doi: 10.7448/IAS.17.4.19739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis GM, Page LC, Burman BE, Buskin S, Frenkel LM. Increased detection of HIV-1 drug resistance at time of diagnosis by testing viral DNA with a sensitive assay. Journal of acquired immune deficiency syndromes. 2009;51:283–289. doi: 10.1097/QAI.0b013e3181a9972c. [DOI] [PubMed] [Google Scholar]
- Eron JJ, Benoit SL, Jemsek J, MacArthur RD, Santana J, Quinn JB, Kuritzkes DR, Fallon MA, Rubin M. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. The New England journal of medicine. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- Eron JJ, Clotet B, Durant J, Katlama C, Kumar P, Lazzarin A, Poizot-Martin I, Richmond G, Soriano V, Ait-Khaled M, Fujiwara T, Huang J, Min S, Vavro C, Yeo J. Safety and Efficacy of Dolutegravir in Treatment-Experienced Subjects With Raltegravir-Resistant HIV Type 1 Infection: 24-Week Results of the VIKING Study. The Journal of infectious diseases. 2013;207:740–748. doi: 10.1093/infdis/jis750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonner G, Grant R, Baggaley R. Oral pre-exposure prophylaxis (PrEP) for all populations: a systematic review and meta-analysis of effectiveness, safety, and sexual and reproductive health outcomes. Unpublished. [Google Scholar]
- Foulkes-Murzycki JE, Scott WR, Schiffer CA. Hydrophobic sliding: a possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure. 2007;15:225–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S, Charpentier C, Amiel C, Morand-Joubert L, Reigadas S, Trabaud MA, Delaugerre C, Nicot F, Rodallec A, Maillard A, Mirand A, Jeulin H, Montes B, Barin F, Bettinger D, Le Guillou-Guillemette H, Vallet S, Signori-Schmuck A, Descamps D, Calvez V, Flandre P, Marcelin AG, Group, A.A.R.S Cross-resistance to elvitegravir and dolutegravir in 502 patients failing on raltegravir: a French national study of raltegravir-experienced HIV-1-infected patients. The Journal of antimicrobial chemotherapy. 2015;70:1507–1512. doi: 10.1093/jac/dku535. [DOI] [PubMed] [Google Scholar]
- Frentz D, Boucher CA, van de Vijver DA. Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS reviews. 2012;14:17–27. [PubMed] [Google Scholar]
- Frost SD, Nijhuis M, Schuurman R, Boucher CA, Brown AJ. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. Journal of virology. 2000;74:6262–6268. doi: 10.1128/jvi.74.14.6262-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fun A, Wensing AM, Verheyen J, Nijhuis M. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012;9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JE, Koenig E, Andrade-Villanueva JF, Chetchotisakd P, DeJesus E, Antunes F, Arasteh K, Rizzardini G, Fehr J, Liu HC, Abram ME, Cao H, Szwarcberg J. Brief Report: Cobicistat Compared With Ritonavir as a Pharmacoenhancer for Atazanavir in Combination With Emtricitabine/Tenofovir Disoproxil Fumarate: Week 144 Results. Journal of acquired immune deficiency syndromes. 2015;69:338–340. doi: 10.1097/QAI.0000000000000598. [DOI] [PubMed] [Google Scholar]
- Geretti AM, Armenia D, Ceccherini-Silberstein F. Emerging patterns and implications of HIV-1 integrase inhibitor resistance. Current opinion in infectious diseases. 2012;25:677–686. doi: 10.1097/QCO.0b013e32835a1de7. [DOI] [PubMed] [Google Scholar]
- Geretti AM, Fox Z, Johnson JA, Booth C, Lipscomb J, Stuyver LJ, Tachedjian G, Baxter J, Touloumi G, Lehmann C, Owen A, Phillips A, Group, I.S.f.M.o.A.T.S Sensitive assessment of the virologic outcomes of stopping and restarting non-nucleoside reverse transcriptase inhibitor-based antiretroviral therapy. PloS one. 2013;8:e69266. doi: 10.1371/journal.pone.0069266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti AM, Harrison L, Green H, Sabin C, Hill T, Fearnhill E, Pillay D, Dunn D, U.K. Collaborative Group on HIV Drug Resistance Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009;48:1296–1305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Serna A, Min JE, Woods C, Chan D, Lima VD, Montaner JS, Harrigan PR, Swenson LC. Performance of HIV-1 drug resistance testing at low-level viremia and its ability to predict future virologic outcomes and viral evolution in treatment-naive individuals. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58:1165–1173. doi: 10.1093/cid/ciu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DD, Zhou Y, Margot NA, McColl DJ, Zhong L, Borroto-Esoda K, Miller MD, Svarovskaia ES. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. Aids. 2011;25:325–333. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ, Hellmann NS, Chesney M, Busch MP, Kahn JO. Time trends in primary HIV-1 drug resistance among recently infected persons. Jama. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- Guiguet M, Ghosn J, Duvivier C, Meynard JL, Gras G, Partisani M, Teicher E, Mahamat A, Rodenbourg F, Launay O, Costagliola D, Fhdh-Anrs CO. Boosted protease inhibitor monotherapy as a maintenance strategy: an observational study. Aids. 2012;26:2345–2350. doi: 10.1097/QAD.0b013e32835646e0. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, Montana JB, McHale M, Sullivan J, Ridgway C, Felstead S, Dunne MW, van der Ryst E, Mayer H, Teams, M.S Maraviroc for previously treated patients with R5 HIV-1 infection. The New England journal of medicine. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Hill A, Sawyer AW, Cozzi-Lepri A, von Wyl V, Yerly S, Lima VD, Gunthard HF, Gilks C, Pillay D. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. The Lancet infectious diseases. 2009;9:409–417. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, Sawyer AW, Hamers RL, Ndembi N, Pillay D, Bertagnolio S. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012;380:1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Kohli A, McCormick AL, Towers GJ, Pillay D, Parry CM. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in vitro assays. Aids. 2010;24:1651–1655. doi: 10.1097/qad.0b013e3283398216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers RL, Schuurman R, Sigaloff KC, Wallis CL, Kityo C, Siwale M, Mandaliya K, Ive P, Botes ME, Wellington M, Osibogun A, Wit FW, van Vugt M, Stevens WS, de Wit TF. Effect of pretreatment HIV-1 drug resistance on immunological, virological, and drug-resistance outcomes of first-line antiretroviral treatment in sub-Saharan Africa: a multicentre cohort study. The Lancet infectious diseases. 2012;12:307–317. doi: 10.1016/S1473-3099(11)70255-9. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS research and human retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Hoffmann CJ, Charalambous S, Grant AD, Morris L, Churchyard GJ, Chaisson RE. Durable HIV RNA resuppression after virologic failure while remaining on a first-line regimen: a cohort study. Tropical medicine & international health: TM & IH. 2014;19:236–239. doi: 10.1111/tmi.12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Frantzell A, Fransen S, Petropoulos CJ. Multiple genetic pathways involving amino acid position 143 of HIV-1 integrase are preferentially associated with specific secondary amino acid substitutions and confer resistance to raltegravir and cross-resistance to elvitegravir. Antimicrobial agents and chemotherapy. 2013;57:4105–4113. doi: 10.1128/AAC.00204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Harrigan PR, Huang W, Bates M, Williamson DW, McCune JM, Price RW, Spudich SS, Lampiris H, Hoh R, Leigler T, Martin JN, Deeks SG. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. The Journal of infectious diseases. 2006;194:926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- Invernizzi CF, Coutsinos D, Oliveira M, Moisi D, Brenner BG, Wainberg MA. Signature nucleotide polymorphisms at positions 64 and 65 in reverse transcriptase favor the selection of the K65R resistance mutation in HIV-1 subtype C. The Journal of infectious diseases. 2009;200:1202–1206. doi: 10.1086/605894. [DOI] [PubMed] [Google Scholar]
- Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Sucupira MC, Bacchetti P, Hartogensis W, Diaz RS, Kallas EG, Janini LM, Liegler T, Pilcher CD, Grant RM, Cortes R, Deeks SG, Hecht FM. Differential persistence of transmitted HIV-1 drug resistance mutation classes. The Journal of infectious diseases. 2011;203:1174–1181. doi: 10.1093/infdis/jiq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Feyertag F, Meehan CJ, McCormack GP, Travers SA, Craig C, Westby M, Lewis M, Robertson DL. Characterizing the Diverse Mutational Pathways Associated with R5-Tropic Maraviroc Resistance: HIV-1 That Uses the Drug-Bound CCR5 Coreceptor. Journal of virology. 2015;89:11457–11472. doi: 10.1128/JVI.01384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnopat P, Isarangkura-na-ayuthaya P, Utachee P, Kitagawa Y, de Silva UC, Siripanyaphinyo U, Kameoka Y, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. Impact of amino acid variations in Gag and protease of HIV type 1 CRF01_AE strains on drug susceptibility of virus to protease inhibitors. Journal of acquired immune deficiency syndromes. 2009;52:320–328. doi: 10.1097/QAI.0b013e3181b4b18c. [DOI] [PubMed] [Google Scholar]
- Jordan MR, Barcarolo J, Hamers RL, Rinke de Wit TF, Sigaloff KC, Davis DH, Gregson J, Parkin N, Bertagnolio S, Group, H.R.A.S Surveillance of HIV Drug Resistance in Resource Limited Countries Before and After Initiation of First-line Antiretroviral Traetment, 2006-1010. 2016 Unpublished. [Google Scholar]
- Karlstrom O, Josephson F, Sonnerborg A. Early virologic rebound in a pilot trial of ritonavir-boosted atazanavir as maintenance monotherapy. Journal of acquired immune deficiency syndromes. 2007;44:417–422. doi: 10.1097/QAI.0b013e31802e2940. [DOI] [PubMed] [Google Scholar]
- Kassaye SG, Grossman Z, Balamane M, Johnston-White B, Liu C, Kumar P, Young M, Sneller MC, Sereti I, Dewar R, Rehm C, Meyer W, 3rd, Shafer R, Katzenstein D, Maldarelli F. Transmitted HIV Drug Resistance Is High and Longstanding in Metropolitan Washington, DC. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2016 doi: 10.1093/cid/ciw382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf DJ, Isaacson JD, King MS, Brun SC, Sylte J, Richards B, Bernstein B, Rode R, Sun E. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antiviral therapy. 2002;7:165–174. [PubMed] [Google Scholar]
- Keulen W, Boucher C, Berkhout B. Nucleotide substitution patterns can predict the requirements for drug-resistance of HIV-1 proteins. Antiviral research. 1996;31:45–57. doi: 10.1016/0166-3542(96)00944-8. [DOI] [PubMed] [Google Scholar]
- Keys JR, Zhou S, Anderson JA, Eron JJ, Jr, Rackoff LA, Jabara C, Swanstrom R. Primer ID Informs Next-Generation Sequencing Platforms and Reveals Preexisting Drug Resistance Mutations in the HIV-1 Reverse Transcriptase Coding Domain. AIDS research and human retroviruses. 2015;31:658–668. doi: 10.1089/aid.2014.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MS, Rode R, Cohen-Codar I, Calvez V, Marcelin AG, Hanna GJ, Kempf DJ. Predictive genotypic algorithm for virologic response to lopinavir-ritonavir in protease inhibitor-experienced patients. Antimicrobial agents and chemotherapy. 2007;51:3067–3074. doi: 10.1128/AAC.00388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomeets AN, Varghese V, Lemey P, Bobkova MR, Shafer RW. A uniquely prevalent nonnucleoside reverse transcriptase inhibitor resistance mutation in Russian subtype A HIV-1 viruses. Aids. 2014;28:F1–8. doi: 10.1097/QAD.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R, Abram ME, McColl DJ, Barnes T, Fordyce MW, Szwarcberg J, Cheng AK, Miller MD, White KL. Week 144 resistance analysis of elvitegravir/cobicistat/emtricitabine/tenofovir DF versus atazanavir+ritonavir+emtricitabine/tenofovir DF in antiretroviral-naive patients. HIV clinical trials. 2014;15:218–230. doi: 10.1310/hct1505-218. [DOI] [PubMed] [Google Scholar]
- Kumarasamy N, Aga E, Ribaudo HJ, Wallis CL, Katzenstein DA, Stevens WS, Norton MR, Klingman KL, Hosseinipour MC, Crump JA, Supparatpinyo K, Badal-Faesen S, Bartlett JA. Lopinavir/ritonavir Monotherapy As Second-line Antiretroviral Treatment in Resource-limited Settings - Week 104 Analysis of ACTG A5230. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2015 doi: 10.1093/cid/civ109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes DR, Lalama CM, Ribaudo HJ, Marcial M, Meyer WA, 3rd, Shikuma C, Johnson VA, Fiscus SA, D’Aquila RT, Schackman BR, Acosta EP, Gulick RM. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. The Journal of infectious diseases. 2008;197:867–870. doi: 10.1086/528802. [DOI] [PubMed] [Google Scholar]
- Kuritzkes DR, Quinn JB, Benoit SL, Shugarts DL, Griffin A, Bakhtiari M, Poticha D, Eron JJ, Fallon MA, Rubin M. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. Aids. 1996;10:975–981. doi: 10.1097/00002030-199610090-00007. [DOI] [PubMed] [Google Scholar]
- Lanier ER, Ait-Khaled M, Scott J, Stone C, Melby T, Sturge G, St Clair M, Steel H, Hetherington S, Pearce G, Spreen W, Lafon S. Antiviral efficacy of abacavir in antiretroviral therapy-experienced adults harbouring HIV-1 with specific patterns of resistance to nucleoside reverse transcriptase inhibitors. Antiviral therapy. 2004;9:37–45. doi: 10.1177/135965350400900102. [DOI] [PubMed] [Google Scholar]
- Lapointe HR, Dong W, Lee GQ, Bangsberg DR, Martin JN, Mocello AR, Boum Y, Karakas A, Kirkby D, Poon AF, Harrigan PR, Brumme CJ. HIV drug resistance testing by high-multiplex “wide” sequencing on the MiSeq instrument. Antimicrob Agents Chemother. 2015;59:6824–6833. doi: 10.1128/AAC.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder BA. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. The Journal of general virology. 1994;75(Pt 5):951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- Lathouwers E, De Meyer S, Dierynck I, Van de Casteele T, Lavreys L, de Bethune MP, Picchio G. Virological characterization of patients failing darunavir/ritonavir or lopinavir/ritonavir treatment in the ARTEMIS study: 96-week analysis. Antiviral therapy. 2011;16:99–108. doi: 10.3851/IMP1719. [DOI] [PubMed] [Google Scholar]
- Lavu E, Dala N, Gurung A, Kave E, Mosoro E, Markby J, Aleksic E, Gare J, Elsum I, Nano G, Myatt M, Kiama P, Crowe S, Bertagnolio S, Hearps A, Jordan MR. Transmitted HIV drug resistance survey in two provinces in Papau New Guinea. International AIDS Society Conference on HIV Pathogenesis, Treatment & Prevention; Vancouver, Canada. 2015. Poster: MOPED723. [Google Scholar]
- Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, Bennett J, Hunt PW, Knapp D, Brumme CJ, Harrigan PR, Martin JN. Prevalence and virologic consequences of transmitted HIV-1 drug resistance in Uganda. AIDS research and human retroviruses. 2014;30:896–906. doi: 10.1089/aid.2014.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levison JH, Wood R, Scott CA, Ciaranello AL, Martinson NA, Rusu C, Losina E, Freedberg KA, Walensky RP. The clinical and economic impact of genotype testing at first-line antiretroviral therapy failure for HIV-infected patients in South Africa. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2013;56:587–597. doi: 10.1093/cid/cis887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Linley L, Kline R, Ziebell R, Heneine W, Johnson JA. Sensitive sentinel mutation screening reveals differential underestimation of transmitted HIV drug resistance among demographic groups. Aids. 2016;30:1439–1445. doi: 10.1097/QAD.0000000000001099. [DOI] [PubMed] [Google Scholar]
- Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. Jama. 2011;305:1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegler T, Abdel-Mohsen M, Bentley LG, Atchison R, Schmidt T, Javier J, Mehrotra M, Eden C, Glidden DV, McMahan V, Anderson PL, Li P, Wong JK, Buchbinder S, Guanira JV, Grant RM, iPrEx Study, T HIV-1 drug resistance in the iPrEx preexposure prophylaxis trial. The Journal of infectious diseases. 2014;210:1217–1227. doi: 10.1093/infdis/jiu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. Journal of virology. 2008;82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, Conway B, Kilby M, Wang L, Whitcomb JM, Hellmann NS, Richman DD. Antiretroviral-drug resistance among patients recently infected with HIV. The New England journal of medicine. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- Llibre JM, Imaz A, Clotet B. From TMC114 to darunavir: five years of data on efficacy. AIDS reviews. 2013;15:112–121. [PubMed] [Google Scholar]
- Lopez-Cortes LF, Castano MA, Lopez-Ruz MA, Rios-Villegas MJ, Hernandez-Quero J, Merino D, Jimenez-Aguilar P, Marquez-Solero M, Terron-Pernia A, Tellez-Perez F, Viciana P, Orihuela-Canadas F, Palacios-Baena Z, Vinuesa-Garcia D, Fajardo-Pico JM, Romero-Palacios A, Ojeda-Burgos G, Pasquau-Liano J. Effectiveness of Ritonavir-Boosted Protease Inhibitor Monotherapy in Clinical Practice Even with Previous Virological Failures to Protease Inhibitor-Based Regimens. PloS one. 2016;11:e0148924. doi: 10.1371/journal.pone.0148924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod IJ, Rowley CF, Essex M. Pan Degenerate Amplification and Adaptation for Highly Sensitive Detection of ARV Drug Resistance. Conference on Retroviruses and Opportunistic Infections 2016; IAS-USA, Boston. 2015. [Google Scholar]
- Mansky LM. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS research and human retroviruses. 1996;12:307–314. doi: 10.1089/aid.1996.12.307. [DOI] [PubMed] [Google Scholar]
- Marcelin AG, Delaugerre C, Wirden M, Viegas P, Simon A, Katlama C, Calvez V. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. Journal of medical virology. 2004;72:162–165. doi: 10.1002/jmv.10550. [DOI] [PubMed] [Google Scholar]
- Margot NA, Johnson A, Miller MD, Callebaut C. Characterization of HIV-1 Resistance to Tenofovir Alafenamide In Vitro. Antimicrobial agents and chemotherapy. 2015;59:5917–5924. doi: 10.1128/AAC.01151-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquelier B, Race E, Tamalet C, Descamps D, Izopet J, Buffet-Janvresse C, Ruffault A, Mohammed AS, Cottalorda J, Schmuck A, Calvez V, Dam E, Fleury H, Brun-Vezinet F. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrobial agents and chemotherapy. 2001;45:1836–1842. doi: 10.1128/AAC.45.6.1836-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T, DeSpirito M, DeMasi R, Heilek-Snyder G, Greenberg ML, Graham N. HIV-1 coreceptor use in triple-class treatment experienced patients: Baseline prevalence, correlates, and relationship to enfuvirtide response. Journal of Infectious Diseases. 2006;194:238–246. doi: 10.1086/504693. [DOI] [PubMed] [Google Scholar]
- Melikian GL, Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia-Cancio PV, Zolopa A, Robbins GK, Kagan R, Israelski D, Shafer RW. Standardized comparison of the relative impacts of HIV-1 reverse transcriptase (RT) mutations on nucleoside RT inhibitor susceptibility. Antimicrobial agents and chemotherapy. 2012;56:2305–2313. doi: 10.1128/AAC.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikian GL, Rhee SY, Varghese V, Porter D, White K, Taylor J, Towner W, Troia P, Burack J, Dejesus E, Robbins GK, Razzeca K, Kagan R, Liu TF, Fessel WJ, Israelski D, Shafer RW. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. The Journal of antimicrobial chemotherapy. 2014;69:12–20. doi: 10.1093/jac/dkt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzner KJ, Scherrer AU, von Wyl V, Boni J, Yerly S, Klimkait T, Aubert V, Furrer H, Hirsch HH, Vernazza PL, Cavassini M, Calmy A, Bernasconi E, Weber R, Gunthard HF, Swiss, H.I.V.C.S Limited clinical benefit of minority K103N and Y181C-variant detection in addition to routine genotypic resistance testing in antiretroviral therapy-naive patients. Aids. 2014;28:2231–2239. doi: 10.1097/QAD.0000000000000397. [DOI] [PubMed] [Google Scholar]
- Miller MD, Margot N, Lu B, Zhong L, Chen SS, Cheng A, Wulfsohn M. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. The Journal of infectious diseases. 2004;189:837–846. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- Molina JM, Andrade-Villanueva J, Echevarria J, Chetchotisakd P, Corral J, David N, Moyle G, Mancini M, Percival L, Yang R, Thiry A, McGrath D, Team, C.S Once-daily atazanavir/ritonavir versus twice-daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral-naive HIV-1-infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet. 2008;372:646–655. doi: 10.1016/S0140-6736(08)61081-8. [DOI] [PubMed] [Google Scholar]
- Molina JM, Cahn P, Grinsztejn B, Lazzarin A, Mills A, Saag M, Supparatpinyo K, Walmsley S, Crauwels H, Rimsky LT, Vanveggel S, Boven K, group, E.s Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial. Lancet. 2011;378:238–246. doi: 10.1016/S0140-6736(11)60936-7. [DOI] [PubMed] [Google Scholar]
- Mollan K, Daar ES, Sax PE, Balamane M, Collier AC, Fischl MA, Lalama CM, Bosch RJ, Tierney C, Katzenstein D, Team, A.C.T.G.S.A HIV-1 amino acid changes among participants with virologic failure: associations with first-line efavirenz or atazanavir plus ritonavir and disease status. The Journal of infectious diseases. 2012;206:1920–1930. doi: 10.1093/infdis/jis613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monogram Biosciences. PhenoSense HIV Drug Resistance Assay 2016 [Google Scholar]
- Mourad R, Chevennet F, Dunn DT, Fearnhill E, Delpech V, Asboe D, Gascuel O, Hue S, Database, U.H.D.R., the Collaborative Hiv, A.H.I.V.D.R.N A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. Aids. 2015;29:1917–1925. doi: 10.1097/QAD.0000000000000768. [DOI] [PubMed] [Google Scholar]
- Mutsvangwa J, Beck IA, Gwanzura L, Manhanzva MT, Stranix-Chibanda L, Chipato T, Frenkel LM. Optimization of the oligonucleotide ligation assay for the detection of nevirapine resistance mutations in Zimbabwean Human Immunodeficiency Virus type-1 subtype C. Journal of virological methods. 2014;210C:36–39. doi: 10.1016/j.jviromet.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt M, Bennett DE. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antiviral therapy. 2008;13(Suppl 2):37–48. [PubMed] [Google Scholar]
- Naeger LK, Struble KA. Effect of baseline protease genotype and phenotype on HIV response to atazanavir/ritonavir in treatment-experienced patients. Aids. 2006;20:847–853. doi: 10.1097/01.aids.0000218548.77457.76. [DOI] [PubMed] [Google Scholar]
- National Institute for Communicable Diseases, D.o.t.N.H.L.S. Prospective sentinel surveillance of human immunodeficiency virus related drug resistance. Communicable Disease Communiqué. 2016:10–11. [Google Scholar]
- Nijhuis M, Schuurman R, de Jong D, van Leeuwen R, Lange J, Danner S, Keulen W, de Groot T, Boucher CA. Lamivudine-resistant human immunodeficiency virus type 1 variants (184V) require multiple amino acid changes to become co-resistant to zidovudine in vivo. The Journal of infectious diseases. 1997;176:398–405. doi: 10.1086/514056. [DOI] [PubMed] [Google Scholar]
- Oette M, Kaiser R, Daumer M, Petch R, Fatkenheuer G, Carls H, Rockstroh JK, Schmaloer D, Stechel J, Feldt T, Pfister H, Haussinger D. Primary HIV drug resistance and efficacy of first-line antiretroviral therapy guided by resistance testing. Journal of acquired immune deficiency syndromes. 2006;41:573–581. doi: 10.1097/01.qai.0000214805.52723.c1. [DOI] [PubMed] [Google Scholar]
- Palma AC, Covens K, Snoeck J, Vandamme AM, Camacho RJ, Van Laethem K. HIV-1 protease mutation 82M contributes to phenotypic resistance to protease inhibitors in subtype G. The Journal of antimicrobial chemotherapy. 2012;67:1075–1079. doi: 10.1093/jac/dks010. [DOI] [PubMed] [Google Scholar]
- Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; 2016. Available at: http://aidsinfo.nih.gov/guidelines. [Google Scholar]
- Panpradist N, Beck IA, Chung MH, Kiarie JN, Frenkel LM, Lutz BR. Simplified Paper Format for Detecting HIV Drug Resistance in Clinical Specimens by Oligonucleotide Ligation. PloS one. 2016;11:e0145962. doi: 10.1371/journal.pone.0145962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao D, Andrady U, Clarke J, Dean G, Drake S, Fisher M, Green T, Kumar S, Murphy M, Tang A, Taylor S, White D, Underhill G, Pillay D, Cane P. Long-term persistence of primary genotypic resistance after HIV-1 seroconversion. Journal of acquired immune deficiency syndromes. 2004;37:1570–1573. doi: 10.1097/00126334-200412150-00006. [DOI] [PubMed] [Google Scholar]
- Parikh UM, Bacheler L, Koontz D, Mellors JW. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. Journal of virology. 2006;80:4971–4977. doi: 10.1128/JVI.80.10.4971-4977.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MM, Gordon D, Reilly A, Horowitz HW, Waters M, Bennett R, Hallack R, Smith J, Lamson D, Aydemir A, Dvali N, Agins BD, Drusano GL, Taylor J, Resistance Study, G Prevalence of drug-resistant and nonsubtype B HIV strains in antiretroviral-naive, HIV-infected individuals in New York State. AIDS patient care and STDs. 2007;21:644–652. doi: 10.1089/apc.2006.0172. [DOI] [PubMed] [Google Scholar]
- Paton NI, Kityo C, Hoppe A, Reid A, Kambugu A, Lugemwa A, van Oosterhout JJ, Kiconco M, Siika A, Mwebaze R, Abwola M, Abongomera G, Mweemba A, Alima H, Atwongyeire D, Nyirenda R, Boles J, Thompson J, Tumukunde D, Chidziva E, Mambule I, Arribas JR, Easterbrook PJ, Hakim J, Walker AS, Mugyenyi P, Team, E.T Assessment of Second-Line Antiretroviral Regimens for HIV Therapy in Africa. The New England journal of medicine. 2014;371:234–247. doi: 10.1056/NEJMoa1311274. [DOI] [PubMed] [Google Scholar]
- Perelson AS, Ribeiro RM. Modeling the within-host dynamics of HIV infection. BMC biology. 2013;11:96. doi: 10.1186/1741-7007-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L, Kouri V, Aleman Y, Abrahantes Y, Correa C, Aragones C, Martinez O, Perez J, Fonseca C, Campos J, Alvarez D, Schrooten Y, Dekeersmaeker N, Imbrechts S, Beheydt G, Vinken L, Soto Y, Alvarez A, Vandamme AM, Van Laethem K. Antiretroviral drug resistance in HIV-1 therapy-naive patients in Cuba. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;16:144–150. doi: 10.1016/j.meegid.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrobial agents and chemotherapy. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A, Cambiano V, Nakagawa F, Mabugu T, Miners A, Ford D, Pillay D, De Luca A, Lundgren J, Revill P. Cost-effectiveness of HIV drug resistance testing to inform switching to second line antiretroviral therapy in low income settings. PloS one. 2014a;9:e109148. doi: 10.1371/journal.pone.0109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AN, Cambiano V, Miners A, Revill P, Pillay D, Lundgren JD, Bennett D, Raizes E, Nakagawa F, De Luca A, Vitoria M, Barcarolo J, Perriens J, Jordan MR, Bertagnolio S. Effectiveness and cost-effectiveness of potential responses to future high levels of transmitted HIV drug resistance in antiretroviral drug-naive populations beginning treatment: modelling study and economic analysis. The lancet. HIV. 2014b;1:e85–93. doi: 10.1016/S2352-3018(14)70021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda E, Briz V, Quinones-Mateu M, Soriano V. HIV tropism: diagnostic tools and implications for disease progression and treatment with entry inhibitors. Aids. 2006;20:1359–1367. doi: 10.1097/01.aids.0000233569.74769.69. [DOI] [PubMed] [Google Scholar]
- Pozniak A, Opravil M, Beatty G, Hill A, de Bethune MP, Lefebvre E. Effect of baseline viral susceptibility on response to darunavir/ritonavir versus control protease inhibitors in treatment-experienced HIV type 1-infected patients: POWER 1 and 2. AIDS research and human retroviruses. 2008;24:1275–1280. doi: 10.1089/aid.2007.0275. [DOI] [PubMed] [Google Scholar]
- Prosperi MC, Mackie N, Di Giambenedetto S, Zazzi M, Camacho R, Fanti I, Torti C, Sonnerborg A, Kaiser R, Codoner FM, Van Laethem K, Bansi L, van de Vijver DA, Geretti AM, De Luca A, consortium, S Detection of drug resistance mutations at low plasma HIV-1 RNA load in a European multicentre cohort study. The Journal of antimicrobial chemotherapy. 2011;66:1886–1896. doi: 10.1093/jac/dkr171. [DOI] [PubMed] [Google Scholar]
- Pulido F, Matarranz M, Rodriguez-Rivera V, Fiorante S, Hernando A. Boosted protease inhibitor monotherapy. What have we learnt after seven years of research? AIDS reviews. 2010;12:127–134. [PubMed] [Google Scholar]
- Rabi SA, Laird GM, Durand CM, Laskey S, Shan L, Bailey JR, Chioma S, Moore RD, Siliciano RF. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. The Journal of clinical investigation. 2013;123:3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffi F, Rachlis A, Stellbrink HJ, Hardy WD, Torti C, Orkin C, Bloch M, Podzamczer D, Pokrovsky V, Pulido F, Almond S, Margolis D, Brennan C, Min S, Group, S.-S Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet. 2013;381:735–743. doi: 10.1016/S0140-6736(12)61853-4. [DOI] [PubMed] [Google Scholar]
- Republic of South Africa Department of Health. National consolidated guidelines for the prevention of mother-to-child transmission of HIV (PMTCT) and the management of HIV in children, adolescents, and adults 2014 [Google Scholar]
- Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, Hamers RL, Bertagnolio S, Rinke de Wit TF, Aghokeng AF, Albert J, Avi R, Avila-Rios S, Bessong PO, Brooks JI, Boucher CA, Brumme ZL, Busch MP, Bussmann H, Chaix ML, Chin BS, D’Aquin TT, De Gascun CF, Derache A, Descamps D, Deshpande AK, Djoko CF, Eshleman SH, Fleury H, Frange P, Fujisaki S, Harrigan PR, Hattori J, Holguin A, Hunt GM, Ichimura H, Kaleebu P, Katzenstein D, Kiertiburanakul S, Kim JH, Kim SS, Li Y, Lutsar I, Morris L, Ndembi N, Ng KP, Paranjape RS, Peeters M, Poljak M, Price MA, Ragonnet-Cronin ML, Reyes-Teran G, Rolland M, Sirivichayakul S, Smith DM, Soares MA, Soriano VV, Ssemwanga D, Stanojevic M, Stefani MA, Sugiura W, Sungkanuparph S, Tanuri A, Tee KK, Truong HH, van de Vijver DA, Vidal N, Yang C, Yang R, Yebra G, Ioannidis JP, Vandamme AM, Shafer RW. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS medicine. 2015a;12:e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Fessel WJ, Liu TF, Marlowe NM, Rowland CM, Rode RA, Vandamme AM, Van Laethem K, Brun-Vezinet F, Calvez V, Taylor J, Hurley L, Horberg M, Shafer RW. Predictive value of HIV-1 genotypic resistance test interpretation algorithms. The Journal of infectious diseases. 2009;200:453–463. doi: 10.1086/600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Jordan MR, Raizes E, Chua A, Parkin N, Kantor R, Van Zyl GU, Mukui I, Hosseinipour MC, Frenkel LM, Ndembi N, Hamers RL, Rinke de Wit TF, Wallis CL, Gupta RK, Fokam J, Zeh C, Schapiro JM, Carmona S, Katzenstein D, Tang M, Aghokeng AF, De Oliveira T, Wensing AM, Gallant JE, Wainberg MA, Richman DD, Fitzgibbon JE, Schito M, Bertagnolio S, Yang C, Shafer RW. HIV-1 Drug Resistance Mutations: Potential Applications for Point-of-Care Genotypic Resistance Testing. PloS one. 2015b;10:e0145772. doi: 10.1371/journal.pone.0145772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Liu TF, Holmes SP, Shafer RW. HIV-1 subtype B protease and reverse transcriptase amino acid covariation. PLoS computational biology. 2007;3:e87. doi: 10.1371/journal.pcbi.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Sankaran K, Varghese V, Winters M, Hurt CB, Eron JJ, Parkin N, Holmes SP, Holodniy M, Shafer RW. HIV-1 Protease, Reverse Transcriptase, and Integrase Variation. Journal of virology. 2016 doi: 10.1128/JVI.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrobial agents and chemotherapy. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. Journal of acquired immune deficiency syndromes. 2012;59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- Rockstroh JK, DeJesus E, Lennox JL, Yazdanpanah Y, Saag MS, Wan H, Rodgers AJ, Walker ML, Miller M, DiNubile MJ, Nguyen BY, Teppler H, Leavitt R, Sklar P, Investigators, S Durable efficacy and safety of raltegravir versus efavirenz when combined with tenofovir/emtricitabine in treatment-naive HIV-1-infected patients: final 5-year results from STARTMRK. Journal of acquired immune deficiency syndromes. 2013;63:77–85. doi: 10.1097/QAI.0b013e31828ace69. [DOI] [PubMed] [Google Scholar]
- Rosen S, Long L, Sanne I, Stevens WS, Fox MP. The net cost of incorporating resistance testing into HIV/AIDS treatment in South Africa: a Markov model with primary data. Journal of the International AIDS Society. 2011;14:24. doi: 10.1186/1758-2652-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom DI, Hill AL, Rabi SA, Siliciano RF, Nowak MA. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nature medicine. 2012;18:1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruxrungtham K, Pedro RJ, Latiff GH, Conradie F, Domingo P, Lupo S, Pumpradit W, Vingerhoets JH, Peeters M, Peeters I, Kakuda TN, De Smedt G, Woodfall B, group, T.C.s Impact of reverse transcriptase resistance on the efficacy of TMC125 (etravirine) with two nucleoside reverse transcriptase inhibitors in protease inhibitor-naive, nonnucleoside reverse transcriptase inhibitor-experienced patients: study TMC125-C227. HIV medicine. 2008;9:883–896. doi: 10.1111/j.1468-1293.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- Santoro MM, Fabeni L, Armenia D, Alteri C, Di Pinto D, Forbici F, Bertoli A, Di Carlo D, Gori C, Carta S, Fedele V, D’Arrigo R, Berno G, Ammassari A, Pinnetti C, Nicastri E, Latini A, Tommasi C, Boumis E, Petrosillo N, D’Offizi G, Andreoni M, Ceccherini-Silberstein F, Antinori A, Perno CF. Reliability and clinical relevance of the HIV-1 drug resistance test in patients with low viremia levels. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58:1156–1164. doi: 10.1093/cid/ciu020. [DOI] [PubMed] [Google Scholar]
- Scherrer AU, Ledergerber B, von Wyl V, Boni J, Yerly S, Klimkait T, Burgisser P, Rauch A, Hirschel B, Cavassini M, Elzi L, Vernazza PL, Bernasconi E, Held L, Gunthard HF, Swiss, H.I.V.C.S Improved virological outcome in White patients infected with HIV-1 non-B subtypes compared to subtype B. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2011;53:1143–1152. doi: 10.1093/cid/cir669. [DOI] [PubMed] [Google Scholar]
- Schultze A, Phillips AN, Paredes R, Battegay M, Rockstroh JK, Machala L, Tomazic J, Girard PM, Januskevica I, Gronborg-Laut K, Lundgren JD, Cozzi-Lepri A, Euro, S.i.E HIV resistance testing and detected drug resistance in Europe. Aids. 2015;29:1379–1389. doi: 10.1097/QAD.0000000000000708. [DOI] [PubMed] [Google Scholar]
- Second-Line Study Group. Boyd MA, Kumarasamy N, Moore CL, Nwizu C, Losso MH, Mohapi L, Martin A, Kerr S, Sohn AH, Teppler H, Van de Steen O, Molina JM, Emery S, Cooper DA. Ritonavir-boosted lopinavir plus nucleoside or nucleotide reverse transcriptase inhibitors versus ritonavir-boosted lopinavir plus raltegravir for treatment of HIV-1 infection in adults with virological failure of a standard first-line ART regimen (SECOND-LINE): a randomised, open-label, non-inferiority study. Lancet. 2013;381:2091–2099. doi: 10.1016/S0140-6736(13)61164-2. [DOI] [PubMed] [Google Scholar]
- Shafer RW, Jung DR, Betts BJ. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nature medicine. 2000;6:1290–1292. doi: 10.1038/81407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer RW, Rhee SY, Bennett DE. Consensus drug resistance mutations for epidemiological surveillance: basic principles and potential controversies. Antiviral therapy. 2008;13(Suppl 2):59–68. [PMC free article] [PubMed] [Google Scholar]
- Shahriar R, Rhee SY, Liu TF, Fessel WJ, Scarsella A, Towner W, Holmes SP, Zolopa AR, Shafer RW. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrobial agents and chemotherapy. 2009;53:4869–4878. doi: 10.1128/AAC.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shet A, Berry L, Mohri H, Mehandru S, Chung C, Kim A, Jean-Pierre P, Hogan C, Simon V, Boden D, Markowitz M. Tracking the prevalence of transmitted antiretroviral drug-resistant HIV-1: a decade of experience. Journal of acquired immune deficiency syndromes. 2006;41:439–446. doi: 10.1097/01.qai.0000219290.49152.6a. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. Recent trends in HIV-1 drug resistance. Current opinion in virology. 2013;3:487–494. doi: 10.1016/j.coviro.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V, Vanderhoeven J, Hurley A, Ramratnam B, Louie M, Dawson K, Parkin N, Boden D, Markowitz M. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. Aids. 2002;16:1511–1519. doi: 10.1097/00002030-200207260-00008. [DOI] [PubMed] [Google Scholar]
- Skhosana L, Steegen K, Bronze M, Lukhwareni A, Letsoalo E, Papathanasopoulos MA, Carmona SC, Stevens WS. High Prevalence of the K65R Mutation in HIV-1 Subtype C Infected Patients Failing Tenofovir-Based First-Line Regimens in South Africa. PloS one. 2015;10:e0118145. doi: 10.1371/journal.pone.0118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis-Cremer N, Jordan MR, Huber K, Wallis CL, Bertagnolio S, Mellors JW, Parkin NT, Harrigan PR. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral research. 2014;107:31–34. doi: 10.1016/j.antiviral.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus research. 2008;134:147–156. doi: 10.1016/j.virusres.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekler JD, McKernan J, Milne R, Tapia KA, Mykhalchenko K, Holte S, Maenza J, Stevens CE, Buskin SE, Mullins JI, Frenkel LM, Collier AC. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007–2013. Antiviral therapy. 2015;20:77–80. doi: 10.3851/IMP2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su ZH, Gulick RM, Krambrink A, Coakley E, Hughes MD, Han D, Flexner C, Wilkin TJ, Skolnik PR, Greaves WL, Kuritzkes DR, Reeves JD, Tea, A.C.T.G.A Response to Vicriviroc in Treatment-Experienced Subjects, as Determined by an Enhanced-Sensitivity Coreceptor Tropism Assay: Reanalysis of AIDS Clinical Trials Group A5211. Journal of Infectious Diseases. 2009;200:1724–1728. doi: 10.1086/648090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KA, Mbisa JL, Cane PA, Pillay D, Parry CM. Contribution of Gag and protease to variation in susceptibility to protease inhibitors between different strains of subtype B human immunodeficiency virus type 1. The Journal of general virology. 2014;95:190–200. doi: 10.1099/vir.0.055624-0. [DOI] [PubMed] [Google Scholar]
- Swenson LC, Moores A, Low AJ, Thielen A, Dong WN, Woods C, Jensen MA, Wynhoven B, Chan D, Glascock C, Harrigan PR. Improved Detection of CXCR4-Using HIV by V3 Genotyping: Application of Population-Based and “Deep” Sequencing to Plasma RNA and Proviral DNA. Jaids-J Acq Imm Def. 2010;54:506–510. doi: 10.1097/QAI.0b013e3181d0558f. [DOI] [PubMed] [Google Scholar]
- Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs. 2012;72:e1–25. doi: 10.2165/11633630-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeranaipong P, Sirivichayakul S, Mekprasan S, Ohata PJ, Avihingsanon A, Ruxrungtham K, Putcharoen O. Role of Rilpivirine and Etravirine in Efavirenz and Nevirapine-Based Regimens Failure in a Resource-Limited Country: A Cross-Sectional Study. PloS one. 2016;11:e0154221. doi: 10.1371/journal.pone.0154221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TenoRes Study, G. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. The Lancet infectious diseases. 2016 doi: 10.1016/S1473-3099(15)00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theys K, Camacho RJ, Gomes P, Vandamme AM, Rhee SY, Portuguese, H.I.V.R.S.G Predicted residual activity of rilpivirine in HIV-1 infected patients failing therapy including NNRTIs efavirenz or nevirapine. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21:607 e601–608. doi: 10.1016/j.cmi.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Theys K, Vercauteren J, Snoeck J, Zazzi M, Camacho RJ, Torti C, Schulter E, Clotet B, Sonnerborg A, De Luca A, Grossman Z, Struck D, Vandamme AM, Abecasis AB. HIV-1 subtype is an independent predictor of reverse transcriptase mutation K65R in HIV-1 patients treated with combination antiretroviral therapy including tenofovir. Antimicrobial agents and chemotherapy. 2013;57:1053–1056. doi: 10.1128/AAC.01668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thigpen MC, Kebaabetswe PM, Paxton LA, Smith DK, Rose CE, Segolodi TM, Henderson FL, Pathak SR, Soud FA, Chillag KL, Mutanhaurwa R, Chirwa LI, Kasonde M, Abebe D, Buliva E, Gvetadze RJ, Johnson S, Sukalac T, Thomas VT, Hart C, Johnson JA, Malotte CK, Hendrix CW, Brooks JT, Group, T.D.F.S Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. The New England journal of medicine. 2012;367:423–434. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- UNAIDS. Fast-Track: Ending the AIDS Epidemic by 2030. Geneva, Switzerland: 2014. Available at: http://www.unaids.org/sites/default/files/media_asset/JC2686_WAD2014report_en.pdf. [Google Scholar]
- UNAIDS. Fact Sheet 2016. Geneva, Switzerland: 2016. Available at: http://www.unaids.org/en/resources/fact-sheet. [Google Scholar]
- Van Zyl GU, Liu TF, Claassen M, Engelbrecht S, de Oliveira T, Preiser W, Wood NT, Travers S, Shafer RW. Trends in Genotypic HIV-1 Antiretroviral Resistance between 2006 and 2012 in South African Patients Receiving First- and Second-Line Antiretroviral Treatment Regimens. PloS one. 2013;8:e67188. doi: 10.1371/journal.pone.0067188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese V, Pinsky BA, Smith DS, Klein D, Shafer RW. Q148N, a Novel Integrase Inhibitor Resistance Mutation Associated with Low-Level Reduction in Elvitegravir Susceptibility. AIDS research and human retroviruses. 2016 doi: 10.1089/aid.2016.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese V, Shahriar R, Rhee SY, Liu T, Simen BB, Egholm M, Hanczaruk B, Blake LA, Gharizadeh B, Babrzadeh F, Bachmann MH, Fessel WJ, Shafer RW. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. Journal of acquired immune deficiency syndromes. 2009;52:309–315. doi: 10.1097/QAI.0b013e3181bca669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingerhoets J, Tambuyzer L, Azijn H, Hoogstoel A, Nijs S, Peeters M, de Bethune MP, De Smedt G, Woodfall B, Picchio G. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. Aids. 2010;24:503–514. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- Volpe JM, Ward DJ, Napolitano L, Phung P, Toma J, Solberg O, Petropoulos CJ, Walworth CM. Five Antiretroviral Drug Class-Resistant HIV-1 in a Treatment-Naive Patient Successfully Suppressed with Optimized Antiretroviral Drug Selection. Journal of the International Association of Providers of AIDS Care. 2015;14:398–401. doi: 10.1177/2325957415596229. [DOI] [PubMed] [Google Scholar]
- Waning B, Diedrichsen E, Moon S. A lifeline to treatment: the role of Indian generic manufacturers in supplying antiretroviral medicines to developing countries. Journal of the International AIDS Society. 2010;13:35. doi: 10.1186/1758-2652-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock H, Respess R, Heneine W, Petropoulos CJ, Hellmann NS, Luo CC, Pau CP, Woods T, Gwinn M, Kaplan J. Prevalence of mutations associated with reduced antiretroviral drug susceptibility among human immunodeficiency virus type 1 seroconverters in the United States, 1993–1998. The Journal of infectious diseases. 2000;182:330–333. doi: 10.1086/315686. [DOI] [PubMed] [Google Scholar]
- Weinstock HS, Zaidi I, Heneine W, Bennett D, Garcia-Lerma JG, Douglas JM, Jr, LaLota M, Dickinson G, Schwarcz S, Torian L, Wendell D, Paul S, Goza GA, Ruiz J, Boyett B, Kaplan JE. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. The Journal of infectious diseases. 2004;189:2174–2180. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2014 Update of the drug resistance mutations in HIV-1. Topics in antiviral medicine. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, van der Ryst E. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. Journal of virology. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. Journal of virology. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, Mahle KC, Heneine W, Johnson JA, Hall HI, Variant A, Resistant, H.I.V.S.G Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. Aids. 2010;24:1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, Chappey C, Kiss LD, Paxinos EE, Petropoulos CJ. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrobial agents and chemotherapy. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb JM, Huang W, Limoli K, Paxinos E, Wrin T, Skowron G, Deeks SG, Bates M, Hellmann NS, Petropoulos CJ. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: clinical, phenotypic and genotypic correlates. Aids. 2002;16:F41–47. doi: 10.1097/00002030-200210180-00002. [DOI] [PubMed] [Google Scholar]
- Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. The Journal of infectious diseases. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- White E, Smit E, Churchill D, Collins SE, Booth C, Tostevin A, Sabin C, Pillay D, Dunn D, on behalf the, U.H.D.R.D.a.U.C.H.C.S No evidence that HIV-1 subtype C infection compromises the efficacy of tenofovir-containing regimens: cohort study in the United Kingdom. Journal of Infectious Diseases. 2016 doi: 10.1093/infdis/jiw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KL, Kulkarni R, McColl DJ, Rhee MS, Szwarcberg J, Cheng AK, Miller MD. Week 144 resistance analysis of elvitegravir/cobicistat/emtricitabine/tenofovir DF versus efavirenz/emtricitabine/tenofovir DF in antiretroviral-naive patients. Antiviral therapy. 2014a doi: 10.3851/IMP2885. [DOI] [PubMed] [Google Scholar]
- White KL, Raffi F, Miller MD. Resistance analyses of integrase strand transfer inhibitors within phase 3 clinical trials of treatment-naive patients. Viruses. 2014b;6:2858–2879. doi: 10.3390/v6072858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin TJ, Su ZH, Kuritzkes DR, Hughes M, Flexner C, Gross R, Coakley E, Greaves W, Godfrey C, Skolnik PR, Timpone J, Rodriguez B, Gulick RM. HIV type 1 chemokine coreceptor use among antiretroviral-experienced patients screened for a clinical trial of a CCR5 inhibitor: AIDS Clinical Trial Group A5211. Clinical Infectious Diseases. 2007;44:591–595. doi: 10.1086/511035. [DOI] [PubMed] [Google Scholar]
- Winters MA, Coolley KL, Girard YA, Levee DJ, Hamdan H, Shafer RW, Katzenstein DA, Merigan TC. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. The Journal of clinical investigation. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkop L, Gunthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, Kucherer C, Obel N, von Wyl V, Masquelier B, Stephan C, Torti C, Antinori A, Garcia F, Judd A, Porter K, Thiebaut R, Castro H, van Sighem AI, Colin C, Kjaer J, Lundgren JD, Paredes R, Pozniak A, Clotet B, Phillips A, Pillay D, Chene G, EuroCoord, C.s.g Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. The Lancet infectious diseases. 2011;11:363–371. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- Woods CK, Brumme CJ, Liu TF, Chui CK, Chu AL, Wynhoven B, Hall TA, Trevino C, Shafer RW, Harrigan PR. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. Journal of clinical microbiology. 2012;50:1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Concept note: Surveillance of HIV drug resistance in adults initiating antiretroviral therapy (pre-treatment HIV drug resistance) HIV/AIDS. Geneva, Switzerland: 2014a. [Google Scholar]
- World Health Organization. Concept note: Surveillance of HIV drug resistance in adults receiving ART, HIV/AIDS. Geneva, Switzerland: 2014b. [Google Scholar]
- World Health Organization. Fact Sheet: What’s New in HIV Treatment. HIV Treatment and Care; Geneva, Switzerland: 2015a. Available at: http://apps.who.int/iris/bitstream/10665/204347/1/WHO_HIV_2015.44_eng.pdf?ua=1. [Google Scholar]
- World Health Organization. Fact Sheet: What’s New in Monitoring. HIV Treatment and Care; 2015b. Available at: http://apps.who.int/iris/bitstream/10665/204345/1/WHO_HIV_2015.42_eng.pdf?ua=1. [Google Scholar]
- World Health Organization. Guideline on when to start antiretroviral tharapy and on pre-exposure prophylaxis for HIV. Geneva, Switzerland: 2015c. Available at: http://apps.who.int/iris/bitstream/10665/186275/1/9789241509565_eng.pdf?ua=1. [PubMed] [Google Scholar]
- World Health Organization. HIV drug resistance surveillance guidance. Geneva, Switzerland: 2015d. Available at: http://www.who.int/hiv/pub/drugresistance/hiv-drug-resistance-2015-update/en/ [Google Scholar]
- World Health Organization. Brief: Global action plan for HIV drug resistance 2016–2021. HIV Drug Resistance; Geneva, Switzerland: 2016a. Available at: http://apps.who.int/iris/bitstream/10665/204518/1/WHO_HIV_2016.02_eng.pdf?ua=1. [Google Scholar]
- World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach - Second edition. Geneva, Switzerland: 2016b. Available at: http://www.who.int/hiv/pub/arv/arv-2016/en/ [PubMed] [Google Scholar]
- World Health Organization. Global report on early warning indicators of HIV drug resistance. Geneva, Switzewrland: 2016c. Available at: http://www.who.int/hiv/pub/drugresistance/ewi-hivdr-2016/en/ [Google Scholar]
- Yerly S, Junier T, Gayet-Ageron A, Amari EB, von Wyl V, Gunthard HF, Hirschel B, Zdobnov E, Kaiser L, Swiss, H.I.V.C.S The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. Aids. 2009;23:1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- Young B, Fransen S, Greenberg KS, Thomas A, Martens S, St Clair M, Petropoulos CJ, Ha B. Transmission of integrase strand-transfer inhibitor multidrug-resistant HIV-1: case report and response to raltegravir-containing antiretroviral therapy. Antiviral therapy. 2011;16:253–256. doi: 10.3851/IMP1748. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cai F, de Rivera IL, Zhou Z, Zhang J, Nkengasong J, Gao F, Yang C. Simultaneous Detection of Major Drug Resistance Mutations of HIV-1 Subtype B Viruses from Dried Blood Spot Specimens by Multiplex Allele-Specific Assay. Journal of clinical microbiology. 2016;54:220–222. doi: 10.1128/JCM.02833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cai F, Zhou Z, DeVos J, Wagar N, Diallo K, Zulu I, Wadonda-Kabondo N, Stringer JS, Weidle PJ, Ndongmo CB, Sikazwe I, Sarr A, Kagoli M, Nkengasong J, Gao F, Yang C. Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype C by use of a multiplex allele-specific assay. Journal of clinical microbiology. 2013;51:3666–3674. doi: 10.1128/JCM.01669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Hughes MD, Lockman S, Benson CA, Hosseinipour MC, Campbell TB, Gulick RM, Daar ES, Sax PE, Riddler SA, Haubrich R, Salata RA, Currier JS. Antiretroviral therapy and efficacy after virologic failure on first-line boosted protease inhibitor regimens. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;59:888–896. doi: 10.1093/cid/ciu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Knyphausen F, Scheufele R, Kucherer C, Jansen K, Somogyi S, Dupke S, Jessen H, Schurmann D, Hamouda O, Meixenberger K, Bartmeyer B. First line treatment response in patients with transmitted HIV drug resistance and well defined time point of HIV infection: updated results from the German HIV-1 seroconverter study. PloS one. 2014;9:e95956. doi: 10.1371/journal.pone.0095956. [DOI] [PMC free article] [PubMed] [Google Scholar]


