Abstract
Human hearing loss is a common neurosensory disorder about which many basic research and clinically relevant questions are unresolved. This review on hereditary deafness focuses on three examples considered at first glance to be uncomplicated, however, upon inspection, are enigmatic and ripe for future research efforts. The three examples of clinical and genetic complexities are drawn from studies of (1) Pendred syndrome/DFNB4 (PDS, OMIM 274600), (2) Perrault syndrome (deafness and infertility) due to mutations of CLPP (PRTLS3, OMIM 614129), and (3) the unexplained extensive clinical variability associated with TBC1D24 mutations. At present, it is unknown how different mutations of TBC1D24 cause nonsyndromic deafness (DFNB86, OMIM 614617), epilepsy (OMIM 605021), epilepsy with deafness, or DOORS syndrome (OMIM 220500) that is characterized by deafness, onychodystrophy (alteration of toenail or fingernail morphology), osteodystrophy (defective development of bone), intellectual disability and seizures. A comprehensive understanding of the multifaceted roles of each gene associated with human deafness is expected to provide future opportunities for restoration as well as preservation of normal hearing.
Keywords: hereditary deafness, enlarged vestibular aqueduct, SLC26A4, Pendred syndrome, Perrault syndrome, CLPP, TBC1D24
Introduction and Background Information
For every 1000 neonates, one or two will have a permanent, significant hearing loss (Morton & Nance, 2006). Throughout adolescence, the prevalence of hearing loss markedly increases. A majority of eighty-year-olds has a variety of deficits including compromised ability to detect and discriminate between sounds, referred to as presbycusis. At any age, the foremost reasons for hearing loss are genetic and environmental factors. Acquired hearing loss can be caused by loud noise, cytomegalovirus infection and ototoxic drugs such as cisplatin and aminoglycoside antibiotics (Brock et al., 2012, Barbi et al., 2003). Environmental factors in combination with a specific genotype may cause deafness as well. As an example of a gene-environment interaction collectively causing deafness, the likelihood that head trauma or barotrauma will precipitate hearing loss is increased in subjects carrying mutations of the SLC26A4 gene (Usami et al., 1999). Similarly, approximately 15% of persons in the United States with aminoglycoside-associated hearing loss have the m.1555A>G mutation in their mitochondrial MTRNR1 gene, which is transmitted matrilineally and encodes the mitochondrial 12S ribosomal RNA (Prezant et al., 1993, Fischel-Ghodsian et al., 1997). This is a challenging medical predicament because aminoglycoside antibiotics are often inexpensive, widely available and sometimes a clinical necessity.
Nonsyndromic deafness
Genetic deafness is classified by the mode of inheritance of the mutation causing deafness and whether or not hearing loss is accompanied by other clinical relevant disorders as part of a syndrome. Approximately 60% to 70% of cases of inherited nonsyndromic deafness (i.e. absence of co-segregating extra-auditory features) are associated with autosomal recessive mutations (referred to as DFNB), 20% to 30% are autosomal dominant (DFNA), and a few percent are located on the X-chromosome (referred to as DFNX or X-linked). There are now approximately 100 different nonsyndromic deafness genes that have been reported (Griffith & Friedman, 2016). Some of the genes associated with human deafness have both dominant and recessive mutations. Examples include TMC1 (Kurima et al., 2002), MYO7A (Liu et al., 1997) and the TBC1D24 genes (Azaiez et al., 2014). Although there are nearly one hundred nonsyndromic deafness genes, less than 10 of them account for a majority of cases of nonsyndromic hearing loss inherited as a recessive trait (DFNB). Three of the most common genes underlying autosomal recessive nonsyndromic deafness are GJB2 at the DFNB1 locus on chromosome 13 (OMIM 220290), SLC26A4 at the DFNB4 locus on chromosome 7q22.3 (OMIM 600791) and MYO15A at the DFNB3 locus on chromosome 17p11.2 (OMIM 600316). Usually, but not always, the genotype of a deafness gene can accurately predict the phenotype. Some deafness-causing mutations resist the influence of environmental factors or modifiers in the genetic background, and thus are said to be fully penetrant. Although difficult to study and not yet well understood, there are examples of protective variants in our genetic backgrounds that can influence the severity and age of onset of hearing loss (Riazuddin et al., 2000, Bykhovskaya et al., 2000, Schultz et al., 2005).
Depending on the ethnicity, a particular pathogenic mutation may be a common cause of deafness. For example, in southern Europe, New Zealand, Australia and North America, a deletion of one G nucleotide (c.35delG) in GJB2 encoding connexin 26 accounts for an estimated 50% of recessive nonsyndromic deafness (Smith & Van Camp, 1993–2016). Among Ashkenazi Jews, the recessive c.167delT mutation of GJB2 is carried by approximately 4% of individuals (95 percent confidence interval, 2.5 to 6.0%) and explains a majority of nonsyndromic deafness in this population (Morell et al., 1998).
Is a genetic variant benign or pathogenic? Every human genome is assumed to have numerous mutations whose pathogenic potential is difficult to predict. An average human is heterozygous for approximately 100 loss-of-function recessive mutations and is homozygous for approximately 20 loss-of-function alleles (MacArthur et al., 2014, Tennessen et al., 2012). It is not uncommon to detect these mutations as coincidental variants in wide-scale DNA sequencing. Global encyclopedias of human genomic variation facilitate the interpretation of many variants as pathogenic or coincidental. Some variants initially considered to cause deafness have eventually been recognized as non-pathogenic. For example, variants of MYO1A encoding myosin 1A were reported to cause deafness but were subsequently found to be benign polymorphisms (Abou Tayoun et al., 2015, Donaudy et al., 2003, Eisenberger et al., 2014).
Genetic testing for many of the genes unequivocally demonstrated to be associated with prelingual (before the acquisition of speech)-onset deafness is commercially available and may establish a molecular diagnosis of childhood hearing loss. However, there are caveats. Notwithstanding a genotype predicted to cause a pathogenic phenotype, there are instances of nonpenetrance (i.e. nonappearance of the expected mutant phenotype) (Chen et al., 2016). Moreover, siblings may have a seemingly identical phenotype but the deafness may be caused by mutations of different genes (Rehman et al., 2015). When a molecular genetic cause of deafness is known in one sibling, it is not necessarily a safe assumption that the same genotype is responsible for deafness of a sibling or deaf relative (Rehman et al., 2015).
Syndromic deafness
Approximately one third of individuals with an inherited hearing loss also have additional abnormalities of other tissues or organs, which is referred to as syndromic deafness. There are at least 400 clinically distinct forms of syndromic deafness annotated in the Online Mendelian Inheritance in Man (http://www.omim.org). Different mutations of a single gene may be associated with nonsyndromic or syndromic hearing loss. For example, mutations of PCDH15 encoding protocadherin 15 can cause either type 1 Usher syndrome characterized by congenital profound deafness, vestibular areflexia and progressive loss of vision due to retinitis pigmentosa, or just nonsyndromic deafness DFNB23 (Ahmed et al., 2003, Ahmed et al., 2001). Similarly, mutations of CDH23 encoding cadherin 23 can also cause a type 1 Usher syndrome or nonsyndromic deafness DFNB12 (Bolz et al., 2001, Bork et al., 2001). Although the number of subjects was small, when a person had an Usher syndrome-associated mutation of one CDH23 allele in combination with a nonsyndromic deafness-associated allele of CDH23 (referred to as a compound heterozygosity), the resultant phenotype was nonsyndromic deafness DFNB12 (Schultz et al., 2011). These data indicate that a DFNB12-associated mutant CDH23 allele, hypothesized to have a low level of residual function, is phenotypically dominant to an USH1D (Usher syndrome type 1D, OMIM 601067)-associated mutant CDH23 allele. Perhaps a low level of CDH23 function is sufficient in the eye to maintain vision but is insufficient in the sensory epithelial cells of the inner ear to preserve hearing (Schultz et al., 2011). These observations have implications for therapies for retinitis pigmentosa in Usher syndrome. If only a small amount of partially functional PCDH15 and CDH23 protein is actually necessary to maintain vision, a therapy for Usher syndrome to maintain vision need not aim to achieve a normal level of these proteins to restore or preserve vision.
Pendred Syndrome, enlarged vestibular aqueduct and the mystery of missing mutations
Pendred syndrome (OMIM 274600) is inherited as an autosomal recessive disorder characterized by bilateral severe to profound prelingual- or perilingual-onset sensorineural hearing loss, enlargement of the vestibular aqueduct (EVA, OMIM 600791) and incompletely penetrant goiter with a typical onset around the time of sexual maturity (Phelps et al., 1998, Morgans & Trotter, 1958, Fraser, 1965). Pendred syndrome is caused by mutations of the SLC26A4 gene on chromosome 7q31 (Everett et al., 1997). SLC26A4, also called pendrin, is a transmembrane protein capable of ATP-independent exchange of chloride, iodide, bicarbonate and other anions. To date, SLC26A4 is the only gene with mutations reported to cause Pendred syndrome. Mutations of SLC26A4 were also reported to cause nonsyndromic deafness DFNB4 (Li et al., 1998). Different ethnic groups have their own distinct spectrum of mutations of SLC26A4. Depending on the population, Pendred syndrome accounts for as much as 10% of hereditary deafness and 7.5% of childhood deafness (Park et al., 2003, Fraser, 1965).
Most patients with two recessive mutations (biallelic mutations) of SLC26A4 develop Pendred syndrome although they initially present with nonsyndromic hearing loss and EVA. The most penetrant manifestation of Pendred syndrome is EVA. Computed tomography (CT) of the temporal bone reveals enlargement of the vestibular aqueduct while magnetic resonance imaging (MRI) shows enlargement of the soft tissue and fluid contents of the vestibular aqueduct: the endolymphatic duct and sac.
Only 25% of European or North American Caucasian individuals with EVA have two mutant alleles of SLC26A4. Fifty percent of subjects with EVA have no obvious mutations in the transcribed region of the SLC26A4 gene and 25% have only one mutant allele of SLC26A4. It has been hypothesized that EVA may result from digenic inheritance of one recessive SLC26A4 mutation and a second recessive mutation in another gene (Yang et al., 2009, Yang et al., 2007), but the evidence is not conclusive (Choi et al., 2009). Variants of KCNJ10 and FOXI1 have been associated with EVA although support for the involvement of these genes is not robust (Abou Tayoun et al., 2015, Yang et al., 2009, Yang et al., 2007) and has not been reproducible.
Other possibilities to explain only one or no mutant alleles in or near the exons of SLC26A4 in many EVA subjects include unidentified alterations of the regulatory sequences that remotely control SLC26A4 gene expression. However, efforts to identify such mutations have heretofore been unsuccessful. We note that enhancers of a gene need not even be linked to chromosome 7q31 (Kleinjan & van Heyningen, 2005, van Heyningen & Bickmore, 2013). An innovative experimental strategy will be required to identify the missing variants associated with the increased risk of EVA.
In mice, a null mutation (no functional SLC26A4 protein) of the Slc26a4 gene causes enlargement of endolymph-containing spaces, loss of the endocochlear potential, deafness, and vestibular dysfunction (Everett et al., 1997). There is also a “tet-on” mouse model where the only expression of SLC26A4 is from an Slc26a4 transgene introduced onto a Slc26a4 null genetic background. In the tet-on mouse model, induction of expression of Slc26a4 from a transgene requires the presence of doxycycline in drinking water (Choi et al., 2011). Data from this study indicated that Slc26a4 expression is required during development from embryonic day16.5 to postnatal day 2 for the initial acquisition of normal hearing at one month after birth (Choi et al., 2011). However, a low level of SLC26A4 expression is necessary to maintain normal hearing in the Slc26a4 “tet-on” mouse model (Choi et al., 2011, Li et al., 2013). This mouse model of EVA can be used to identify and test potential therapeutic options to prevent progression or fluctuation of hearing loss that is often associated with EVA in young children.
Perrault Syndrome and unresolved questions about male fertility
Perrault syndrome (PS, OMIM 233400) is a clinically and genetically heterogeneous sex-influenced rare recessive disorder that is characterized by sensorineural hearing loss of variable severity in men and women and ovarian disorders ranging from nonfunctional streak ovaries to premature ovarian insufficiency before the age of 40 years (Newman et al., 1993–2016, Jenkinson et al., 2013). As teenagers, young women with a normal karyotype and affected with PS may be hypoestrogenic and have an elevated level of follicle stimulating hormone and lutenizing hormone mimicking menopause (reviewed in Newman et al., 2014). Hypergonadotropic hypogonadism is a characteristic hormone profile of women with PS. Occasionally, other clinically relevant neurological signs have also been noted to co-occur and include ataxia, nystagmus, learning disability and progressive peripheral neuropathy (Pierce et al., 2010, Demain et al., 2016).
Recessive mutations of five genes are now known to be associated with PS, with many more PS genes remaining to be discovered (Demain et al., 2016, Jenkinson et al., 2012, Jenkinson et al., 2013, Pierce et al., 2011, Pierce et al., 2010). In one family, PS is associated with compound heterozygosity for two missense mutations of HARS2, encoding mitochondrial histidyl tRNA synthetase. These data suggested that PS is predominantly a disorder of mitochondrial proteostasis (Pierce et al., 2011). In support of this interpretation, a homozygous missense mutation of LARS2 was identified in a family with one deaf female and her two deaf brothers. Additionally, PS can also be caused by mutations of LARS2, encoding mitochondrial leucyl-tRNA synthetase (Pierce et al., 2013, Solda et al., 2015) and mutations of CLPP (Jenkinson et al., 2013). CLPP encodes a highly conserved chambered mitochondrial protease. Why are manifestations of dysfunctional mitochondrial proteostasis due to recessive mutations of CLPP, HARS2 and LARS2 limited largely to ears and ovaries? An answer to this question may emerge from investigations of animal models of PS mutations to explore the pathogenic mechanisms of hearing loss and ovarian impairment (Gispert et al., 2013).
In addition to CLPP, LARS2 and HARS2, mutations of HSD17B4 are associated with PS. HSD17B4 encodes a multifunctional peroxisomal pre-protein that is proteolytically cleaved into two separate enzymes, a 17-β-hydroxysteroid dehydrogenase type 4 and a hydratase, each catalyzing successive steps in fatty acid β-oxidation. Two hearing-impaired sisters were reported to be compound heterozygous for alleles of HSD17B4 and exhibited ovarian dysgenesis, short stature, compromised cognitive ability and peripheral neuropathy (Fiumara et al., 2004, McCarthy & Opitz, 1985, Pierce et al., 2010). More disabling mutations of HSD17B4 cause D-Bifunctional Protein deficiency (DBP deficiency; OMIM 261515). DBP deficiency is characterized by, but not limited to, perinatal hypotonia, seizures, craniofacial dysmorphology, hearing loss, vision impairment and a failure to thrive (Ferdinandusse et al., 2006, Lieber et al., 2014). Missense mutations of C10ORF2 encoding TWINKLE have recently extended the spectrum of mutant genes associated with PS (Morino et al., 2014, Demain et al., 2016).
Reduced fertility in PS women is documented comprehensively but there is much less data on the fecundity of deaf males with biallelic mutations of the PS genes. Consequently, there are limited data to support the assumption that deaf males have normal fertility when they have the same mutant genotype as a woman with PS. In a single family reported with a mutation of HARS2, there were three PS females and their two deaf male siblings (Pierce et al., 2011), one of whom was reported 32 years ago to have normal hearing children (Pallister & Opitz, 1979, Pierce et al., 2011). For the other PS genes, caution is suggested in assuming normal fertility in deaf males with PS genotypes for the LARS, HSD17B4 and CLPP genes. This matter is not resolved for three reasons. First, there are only a few deaf male siblings in all families reported to be segregating PS and fertility in only one male has been documented (Pallister & Opitz, 1979, Pierce et al., 2011). The scarcity of males in “Perrault families” may reflect an ascertainment bias towards families with deaf women with primary amenorrhea and a 46, XX karyotype (Newman et al., 1993–2016). Second, clinical data on sperm count and sperm motility for deaf males in Perrault families have not been reported. Third, and most vexing, a homozygous recessive null allele of Clpp in mouse causes deafness and sterility of both females and males (Gispert et al., 2013). A caveat is that in mouse a complete knockout of the Clpp gene does not precisely recapitulate the two human CLPP missense mutations associated with PS, which may have reduced, but not absent, proteolytic function in mitochondria (Jenkinson et al., 2013). Gene editing with TALENs or CRISPR/Cas9 system could be used to engineer the equivalent of the human PS p.Cys147Ser, p.Thr145Pro, and p.Cys144Arg missense mutations into the mouse Clpp gene (Demain et al., 2016, Jenkinson et al., 2013).
TBC1D24 Mutations Associated with Nonsyndromic Deafness, Epilepsy or DOORS
There is no shortage of human genes associated with deafness whose normal functions are not well understood. One such example is TBC1D24 located on chromosome 16p that belongs to a family of TBC (Tre-2, Bub2, Cdc16) domain containing RAB-specific GTPase-activating proteins. The eight annotated exons of the TBC1D24 (NM_001199107) encode a 559-amino acid protein that has an N-terminal TBC domain and a C-terminal TLDc domain (TBC, LysM, domain catalytic). Smaller isoforms of TBC1D24 are also expressed (see supplemental figure S1; (Rehman et al., 2014)). Some TLDc domain-containing proteins have a neuro-protective role against oxidative stress (Oliver et al., 2011, Durand et al., 2007, Finelli et al., 2016). TBC1D24 is expressed in various tissues including inner ear, brain, kidney, heart and liver. In the inner ear, TBC1D24 protein is expressed in the spiral ganglion neurons (Rehman et al., 2014) and also in the inner and outer hair cells (Azaiez et al., 2014). The question remains as to whether or not a normal function, or an additional normal function, of TBC1D24 is to diminish oxidative stress within spiral ganglions and in other inner ear cells types where it is expressed.
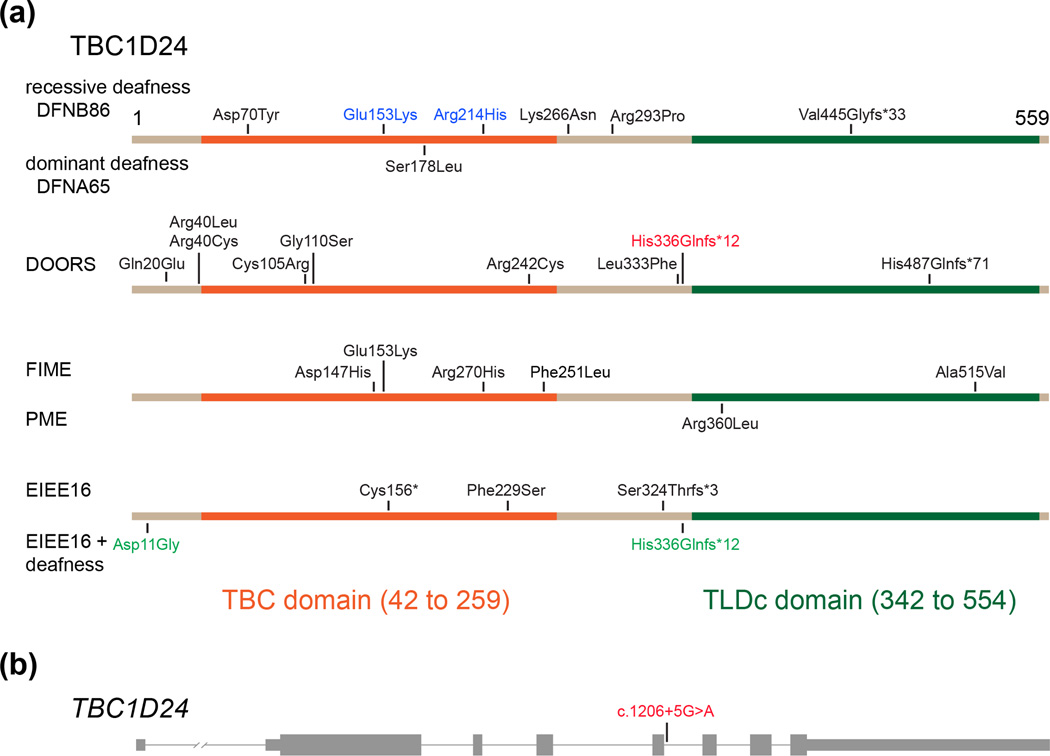
There are now 26 reported mutant alleles of TBC1D24 that are distributed across the gene, some of which are located in the sequences that encode the TBC and TLDc domains (Figure 1). Mutations of TBC1D24 are associated with nonsyndromic deafness segregating as an autosomal recessive trait (DFNB86, OMIM 614617) or as an autosomal dominant trait (DFNA65, OMIM 616044), familial infantile myoclonic epilepsy (FIME, OMIM 605021) with or without deafness, early infantile epileptic encephalopathy (EIEE16, OMIM 615338), progressive myoclonic epilepsy (PME, OMIM 310370), or deafness, onychodystrophy, osteodystrophy, mental retardation and seizures (DOORS, OMIM 220500). It is a conundrum as to how different homozygous or compound heterozygous mutations of TBC1D24 can give rise to such different phenotypes.
Figure 1.
Neurological disorders caused by mutations of TBC1D24. (a) Each distinct disorder is caused by different subset of twenty-six pathogenic mutations of TBC1D24, with two exceptions (p.Glu153Lys and p.His336Glnfs*12 shown in blue and red font, respectively). The p.Glu153Lys in homozygosity results in FIME (myoclonic epilepsy, familial infantile, OMIM 605021) while in compound heterozygosity with p.Arg214His causes nonsyndromic deafness DFNB86 (OMIM 614617). The p.Asp11Gly in compound heterozygosity with p.His336Glnfs*12 causes EIEE (epileptic encephalopathy, early infantile, 16 OMIM 615338) and deafness whereas in compound heterozygosity with an experimentally confirmed splice site mutation c.1206+5G>A causes DOORS syndrome (deafness, onychodystrophy, osteodystrophy, mental retardation and seizures, OMIM 220500). Mutation nomenclature is based on the full-length isoform of TBC1D24 (NM_001199107.1). The c.1333dupG(p.Val445Glyfs*33) was reported as c.1316insG(p.Val439Glyfs*32) in Bakhchane et al., 2015 based on a shorter isoform of TBC1D24 (NM_020705, personal communication with the last author). (b) TBC1D24 gene structure and location of a splice site mutation c.1206+5G>A that, in compound heterozygosity with p.His336Glnfs*12, causes DOORS syndrome. Thick and thin bars represent exons and introns, respectively. PME; progressive myoclonic epilepsy (OMIM 310370).
As illustrated in Figure 1, seven of 26 mutations of TBC1D24 are associated with nonsyndromic deafness (i.e no additional clinical anomalies) (Ali et al., 2012, Azaiez et al., 2014, Bakhchane et al., 2015, Rehman et al., 2014, Zhang et al., 2014). There are also ten mutant alleles of TBC1D24 reported in patients with epileptic syndromes (Corbett et al., 2010, Falace et al., 2010, Afawi et al., 2013, Guven & Tolun, 2013, Milh et al., 2013, Doummar et al., 2015, Muona et al., 2015, Poulat et al., 2015, Strazisar et al., 2015). With these observations in mind, two of the four available DFNB86 families segregating the nonsyndromic deafness-associated mutation p.Asp70Tyr of TBC1D24 were re-examined to determine if the subjects had epilepsy. A detailed family history of epilepsy was collected from 15 affected and 18 unaffected individuals from the two families. Magnetic resonance imaging (MRI) of the brain and electroencephalography (EEG) was also performed on selected affected and unaffected individuals. A history of seizures was reported in one of the 18 normal-hearing individuals. An 18-year-old deaf female had fever-associated seizures at the age of 8 years and reports no seizures since then. The remaining 17 normal-hearing individuals and 14 deaf individuals have never experienced a seizure. Given the high prevalence of epilepsy, co-occurrence of fever-associated seizures with hearing loss in one of the 15 deaf individuals could be coincidental (Rehman et al., 2014).
The association of TBC1D24 with nonsyndromic deafness was further solidified by identification of three missense and one frameshift mutation segregating in three Moroccan families (Bakhchane et al., 2015). Affected individuals from three families displayed severe to profound congenital sensorineural deafness and were compound heterozygous for two mutant alleles of TBC1D24. All deaf individuals shared the p.Arg214His allele and one of the following three variants as the second allele; p.Glu153Lys, p.Lys266Asn, or p.Val445Glyfs*33. The p.Arg214His mutation has an allele frequency of 2% in a presumably normal-hearing Moroccan control population. To date, no deaf individuals homozygous for p.Arg214His have been reported. One possible explanation is that p.Arg214His is a hypomorphic allele (residual normal function) and individuals homozygous for p.Arg214His have no obvious loss of hearing. The p.Arg214His may be associated with hearing loss only in compound heterozygosity with a more disabling mutation of TBC1D24.
With the exception of p.Ser178Leu, all other reported mutations of TBC1D24 are recessive. In a family of European descent and also in a Chinese family, p.Ser178Leu co-segregated with nonsyndromic dominant progressive hearing loss (Azaiez et al., 2014, Zhang et al., 2014). Individuals who are heterozygous (carriers) for truncating recessive mutations of TBC1D24 are phenotypically normal (Bakhchane et al., 2015, Guven & Tolun, 2013, Milh et al., 2013, Poulat et al., 2015). Therefore, the p.Ser178Leu allele is likely to cause hearing loss either through a gain-of-function or a dominant-negative mechanism and not due to haplo-insufficiency for TBC1D24 function (Zhang et al., 2014).
Recessive mutations of TBC1D24 are also associated with a range of epileptic disorders with or without hearing loss (Figure 1) or with DOORS syndrome. The diverse epileptic phenotypes include familial infantile myoclonic epilepsy without intellectual impairment, progressive myoclonus epilepsies, focal epilepsy with intellectual disability, early onset epileptic encephalopathy with hearing loss, and malignant migrating partial seizures of infancy (Corbett et al., 2010, Doummar et al., 2015, Falace et al., 2010, Guven & Tolun, 2013, Milh et al., 2013, Poulat et al., 2015, Strazisar et al., 2015, Muona et al., 2015). DOORS syndrome segregates as an autosomal recessive trait and presents extensive clinical variability. DOORS patients who have mutations of TBC1D24 exhibit five consistent features; (1) deafness, (2) intellectual disability or developmental delay, (3) seizures, (4) small distal phalanges, and (5) small or absent nails (Campeau et al., 2014a, Campeau et al., 2014b). In addition to these five main clinical features of DOORS, some patients have optic and peripheral neuropathy, visual impairment, triphalangeal thumbs, facial dysmorphisms and microcephaly. A cohort of 38 patients from 32 families segregating DOORS syndrome was screened to identify the underlying genetic basis. This effort led to identification of 10 recessive mutations of TBC1D24 in thirteen patients from 11 families. There are at least 23 DOORS syndrome patients with no mutations in TBC1D24 suggesting that there are other genes in which mutations can cause DOORS syndrome. Alternatively, there are unidentified variants associated with DOORS located in the regulatory elements of TBC1D24.
The location and type of mutations of TBC1D24 cannot currently be used to predict the associated phenotype. However, the same mutant allele in different individuals appears to cause a similar phenotype suggesting a minor role for genetic modifiers. The two exceptions to such a simplistic generalization are the mutations p.Glu153Lys and p.His336Glnfs*12. Homozygosity for the p.Glu153Lys allele results in familial infantile myoclonic epilepsy with moderate intellectual disability but, in compound heterozygosity with p.Arg214His, causes nonsyndromic deafness (Bakhchane et al., 2015, Poulat et al., 2015). Similarly, the p.His336Glnfs*12 allele in compound heterozygosity with an experimentally demonstrated splice site mutation, c.1206+5G>A, causes DOORS syndrome. In contrast, p.His336Glnfs*12 in compound heterozygosity with the missense allele p.Asp11Gly results in epilepsy, hearing loss, and developmental delay, but no onychodystrophy or osteodystrophy (Strazisar et al., 2015). As no obvious genotype-phenotype relationship has emerged for mutations of TBC1D24, predicting the phenotype associated with novel mutations of TBC1D24 is not likely to be straightforward. Identifying the spectrum of protein partners that interact with TBC1D24, studying the phenotypes of engineered mutant mouse models of Tbc1d24, or studying the physical structure of TBC1D24 protein in relationship to the locations of amino acid substitutions might reveal a conceptual framework for the correlation of TBC1D24 genotype and phenotype.
Conclusions
Additional genes associated with nonsyndromic and syndromic forms of hearing loss remain to be discovered. With time, molecular genetic screening will eventually include all of the deafness-causing genes as well as the cis-acting sequences necessary for regulating their expression. Improvements in genomic sequencing and its interpretation will continue to expand the benefits of genetic testing available for patients. These benefits include estimates of recurrence probability in a family, reducing misconceptions about the biological reason for hearing loss, and identification of deafness-associated medical conditions that require surveillance or management, and hopefully in the future, guidance for precision therapies.
Acknowledgments
We thank Sadaf Naz and Joan Guitart for their critiques of the manuscript. The authors are supported by NIDCD/NIH intramural research funds DC-000060-12 (A.J.G.) and DC-000039-19 (T.B.F.).
Footnotes
Author contributions
A. Rehman, T. Friedman and A. Griffith contributed to the drafting and editing of the manuscript and the figure.
Conflicts of interest: none to declare.
REFERENCES
- Abou Tayoun AN, Al Turki SH, Oza AM, Bowser MJ, Hernandez AL, Funke BH, Rehm HL, Amr SS. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2015 doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- Afawi Z, Mandelstam S, Korczyn AD, Kivity S, Walid S, Shalata A, Oliver KL, Corbett M, Gecz J, Berkovic SF, Jackson GD. TBC1D24 mutation associated with focal epilepsy, cognitive impairment and a distinctive cerebro-cerebellar malformation. Epilepsy Res. 2013;105:240–244. doi: 10.1016/j.eplepsyres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, Sieving P, Riazuddin S, Griffith AJ, Friedman TB, Belyantseva IA, Wilcox ER. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali RA, Rehman AU, Khan SN, Husnain T, Riazuddin S, Friedman TB, Ahmed ZM, Riazuddin S. DFNB86, a novel autosomal recessive non-syndromic deafness locus on chromosome 16p13.3. Clin Genet. 2012;81:498–500. doi: 10.1111/j.1399-0004.2011.01729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaiez H, Booth KT, Bu F, Huygen P, Shibata SB, Shearer AE, Kolbe D, Meyer N, Black-Ziegelbein EA, Smith RJ. TBC1D24 mutation causes autosomal-dominant nonsyndromic hearing loss. Hum Mutat. 2014;35:819–823. doi: 10.1002/humu.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhchane A, Charif M, Salime S, Boulouiz R, Nahili H, Roky R, Lenaers G, Barakat A. Recessive TBC1D24 Mutations Are Frequent in Moroccan Non-Syndromic Hearing Loss Pedigrees. PLoS One. 2015;10:e0138072. doi: 10.1371/journal.pone.0138072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbi M, Binda S, Caroppo S, Ambrosetti U, Corbetta C, Sergi P. A wider role for congenital cytomegalovirus infection in sensorineural hearing loss. Pediatr Infect Dis J. 2003;22:39–42. doi: 10.1097/00006454-200301000-00012. [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del CSCM, Vila MC, Molina OP, Gal A, Kubisch C. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Riazuddin S, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock PR, Knight KR, Freyer DR, Campbell KC, Steyger PS, Blakley BW, Rassekh SR, Chang KW, Fligor BJ, Rajput K, Sullivan M, Neuwelt EA. Platinum-induced ototoxicity in children: a consensus review on mechanisms, predisposition, and protection, including a new International Society of Pediatric Oncology Boston ototoxicity scale. J Clin Oncol. 2012;30:2408–2417. doi: 10.1200/JCO.2011.39.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykhovskaya Y, Estivill X, Taylor K, Hang T, Hamon M, Casano RA, Yang H, Rotter JI, Shohat M, Fischel-Ghodsian N. Candidate locus for a nuclear modifier gene for maternally inherited deafness. Am J Hum Genet. 2000;66:1905–1910. doi: 10.1086/302914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Hennekam RC group Dsc. DOORS syndrome: phenotype, genotype and comparison with Coffin-Siris syndrome. Am J Med Genet C Semin Med Genet. 2014a;166C:327–332. doi: 10.1002/ajmg.c.31412. [DOI] [PubMed] [Google Scholar]
- Campeau PM, Kasperaviciute D, Lu JT, Burrage LC, Kim C, Hori M, Powell BR, Stewart F, Felix TM, van den Ende J, Wisniewska M, Kayserili H, Rump P, Nampoothiri S, Aftimos S, Mey A, Nair LD, Begleiter ML, De Bie I, Meenakshi G, Murray ML, Repetto GM, Golabi M, Blair E, Male A, Giuliano F, Kariminejad A, Newman WG, Bhaskar SS, Dickerson JE, Kerr B, Banka S, Giltay JC, Wieczorek D, Tostevin A, Wiszniewska J, Cheung SW, Hennekam RC, Gibbs RA, Lee BH, Sisodiya SM. The genetic basis of DOORS syndrome: an exome-sequencing study. Lancet Neurol. 2014b;13:44–58. doi: 10.1016/S1474-4422(13)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, Sleiman P, Cheng WY, Chen W, Shah H, Shen Y, Fromer M, Omberg L, Deardorff MA, Zackai E, Bobe JR, Levin E, Hudson TJ, Groop L, Wang J, Hakonarson H, Wojcicki A, Diaz GA, Edelmann L, Schadt EE, Friend SH. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016 doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Choi BY, Alper SL, Griffith AJ. Response to: The c. −103T>C variant in the 5'-UTR of SLC26A4 gene: A pathogenic mutation or coincidental polymorphism? Hum Mutat. 2009;30:1471. doi: 10.1002/humu.21097. [DOI] [PubMed] [Google Scholar]
- Choi BY, Kim HM, Ito T, Lee KY, Li X, Monahan K, Wen Y, Wilson E, Kurima K, Saunders TL, Petralia RS, Wangemann P, Friedman TB, Griffith AJ. Mouse model of enlarged vestibular aqueducts defines temporal requirement of Slc26a4 expression for hearing acquisition. J Clin Invest. 2011;121:4516–4525. doi: 10.1172/JCI59353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett MA, Bahlo M, Jolly L, Afawi Z, Gardner AE, Oliver KL, Tan S, Coffey A, Mulley JC, Dibbens LM, Simri W, Shalata A, Kivity S, Jackson GD, Berkovic SF, Gecz J. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am J Hum Genet. 2010;87:371–375. doi: 10.1016/j.ajhg.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain LA, Urquhart JE, O'Sullivan J, Williams SG, Bhaskar SS, Jenkinson EM, Lourenco CM, Heiberg A, Pearce SH, Shalev SA, Yue WW, Mackinnon S, Munro KJ, Newbury-Ecob R, Becker K, Kim MJ, RT OK, Newman WG. Expanding the Genotypic Spectrum of Perrault syndrome. Clin Genet. 2016 doi: 10.1111/cge.12776. [DOI] [PubMed] [Google Scholar]
- Donaudy F, Ferrara A, Esposito L, Hertzano R, Ben-David O, Bell RE, Melchionda S, Zelante L, Avraham KB, Gasparini P. Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet. 2003;72:1571–1577. doi: 10.1086/375654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doummar D, Mignot C, Apartis E, Villard L, Rodriguez D, Chantot-Bastauraud S, Burglen L. A Novel Homozygous TBC1D24 Mutation Causing Multifocal Myoclonus With Cerebellar Involvement. Mov Disord. 2015;30:1431–1432. doi: 10.1002/mds.26303. [DOI] [PubMed] [Google Scholar]
- Durand M, Kolpak A, Farrell T, Elliott NA, Shao W, Brown M, Volkert MR. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol. 2007;8:13. doi: 10.1186/1471-2121-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberger T, Di Donato N, Baig SM, Neuhaus C, Beyer A, Decker E, Murbe D, Decker C, Bergmann C, Bolz HJ. Targeted and genomewide NGS data disqualify mutations in MYO1A, the "DFNA48 gene", as a cause of deafness. Hum Mutat. 2014;35:565–570. doi: 10.1002/humu.22532. [DOI] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, Sheffield VC, Green ED. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- Falace A, Filipello F, La Padula V, Vanni N, Madia F, De Pietri Tonelli D, de Falco FA, Striano P, Dagna Bricarelli F, Minetti C, Benfenati F, Fassio A, Zara F. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, Ylianttila MS, Gloerich J, Koski MK, Oostheim W, Waterham HR, Hiltunen JK, Wanders RJ, Glumoff T. Mutational spectrum of D-bifunctional protein deficiency and structure-based genotype-phenotype analysis. Am J Hum Genet. 2006;78:112–124. doi: 10.1086/498880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli MJ, Sanchez-Pulido L, Liu KX, Davies KE, Oliver PL. The Evolutionarily Conserved Tre2/Bub2/Cdc16 (TBC), Lysin Motif (LysM), Domain Catalytic (TLDc) Domain Is Neuroprotective against Oxidative Stress. J Biol Chem. 2016;291:2751–2763. doi: 10.1074/jbc.M115.685222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N, Prezant TR, Chaltraw WE, Wendt KA, Nelson RA, Arnos KS, Falk RE. Mitochondrial gene mutation is a significant predisposing factor in aminoglycoside ototoxicity. Am J Otolaryngol. 1997;18:173–178. doi: 10.1016/s0196-0709(97)90078-8. [DOI] [PubMed] [Google Scholar]
- Fiumara A, Sorge G, Toscano A, Parano E, Pavone L, Opitz JM. Perrault syndrome: evidence for progressive nervous system involvement. Am J Med Genet A. 2004;128A:246–249. doi: 10.1002/ajmg.a.20616. [DOI] [PubMed] [Google Scholar]
- Fraser GR. Association of Congenital Deafness with Goitre (Pendred's Syndrome) a Study of 207 Families. Ann Hum Genet. 1965;28:201–249. doi: 10.1111/j.1469-1809.1964.tb00479.x. [DOI] [PubMed] [Google Scholar]
- Gispert S, Parganlija D, Klinkenberg M, Drose S, Wittig I, Mittelbronn M, Grzmil P, Koob S, Hamann A, Walter M, Buchel F, Adler T, Hrabe de Angelis M, Busch DH, Zell A, Reichert AS, Brandt U, Osiewacz HD, Jendrach M, Auburger G. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum Mol Genet. 2013;22:4871–4887. doi: 10.1093/hmg/ddt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith AJ, Friedman TB. In: Ballenger’s otorhinolaryngology head and neck surgery, 18th edition. Wackym PA, Snow JB, editors. USA: People’s Medical Publishing House – USA; 2016. pp. 329–345. [Google Scholar]
- Guven A, Tolun A. TBC1D24 truncating mutation resulting in severe neurodegeneration. J Med Genet. 2013;50:199–202. doi: 10.1136/jmedgenet-2012-101313. [DOI] [PubMed] [Google Scholar]
- Jenkinson EM, Clayton-Smith J, Mehta S, Bennett C, Reardon W, Green A, Pearce SH, De Michele G, Conway GS, Cilliers D, Moreton N, Davis JR, Trump D, Newman WG. Perrault syndrome: further evidence for genetic heterogeneity. J Neurol. 2012;259:974–976. doi: 10.1007/s00415-011-6285-5. [DOI] [PubMed] [Google Scholar]
- Jenkinson EM, Rehman AU, Walsh T, Clayton-Smith J, Lee K, Morell RJ, Drummond MC, Khan SN, Naeem MA, Rauf B, Billington N, Schultz JM, Urquhart JE, Lee MK, Berry A, Hanley NA, Mehta S, Cilliers D, Clayton PE, Kingston H, Smith MJ, Warner TT, University of Washington Center for Mendelian G. Black GC, Trump D, Davis JR, Ahmad W, Leal SM, Riazuddin S, King MC, Friedman TB, Newman WG. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, Ghosh M, Menon PS, Deshmukh D, Oddoux C, Ostrer H, Khan S, Riazuddin S, Deininger PL, Hampton LL, Sullivan SL, Battey JF, Jr, Keats BJ, Wilcox ER, Friedman TB, Griffith AJ. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Li X, Sanneman JD, Harbidge DG, Zhou F, Ito T, Nelson R, Picard N, Chambrey R, Eladari D, Miesner T, Griffith AJ, Marcus DC, Wangemann P. SLC26A4 targeted to the endolymphatic sac rescues hearing and balance in Slc26a4 mutant mice. PLoS Genet. 2013;9:e1003641. doi: 10.1371/journal.pgen.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, Wilcox ER. A mutation in PDS causes non-syndromic recessive deafness. Nat Genet. 1998;18:215–217. doi: 10.1038/ng0398-215. [DOI] [PubMed] [Google Scholar]
- Lieber DS, Hershman SG, Slate NG, Calvo SE, Sims KB, Schmahmann JD, Mootha VK. Next generation sequencing with copy number variant detection expands the phenotypic spectrum of HSD17B4-deficiency. BMC Med Genet. 2014;15:30. doi: 10.1186/1471-2350-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Opitz JM. Perrault syndrome in sisters. Am J Med Genet. 1985;22:629–631. doi: 10.1002/ajmg.1320220324. [DOI] [PubMed] [Google Scholar]
- Milh M, Falace A, Villeneuve N, Vanni N, Cacciagli P, Assereto S, Nabbout R, Benfenati F, Zara F, Chabrol B, Villard L, Fassio A. Novel compound heterozygous mutations in TBC1D24 cause familial malignant migrating partial seizures of infancy. Hum Mutat. 2013;34:869–872. doi: 10.1002/humu.22318. [DOI] [PubMed] [Google Scholar]
- Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, Van Camp G, Berlin CI, Oddoux C, Ostrer H, Keats B, Friedman TB. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- Morgans ME, Trotter WR. Association of congenital deafness with goitre; the nature of the thyroid defect. Lancet. 1958;1:607–609. doi: 10.1016/s0140-6736(58)90866-3. [DOI] [PubMed] [Google Scholar]
- Morino H, Pierce SB, Matsuda Y, Walsh T, Ohsawa R, Newby M, Hiraki-Kamon K, Kuramochi M, Lee MK, Klevit RE, Martin A, Maruyama H, King MC, Kawakami H. Mutations in Twinkle primase-helicase cause Perrault syndrome with neurologic features. Neurology. 2014;83:2054–2061. doi: 10.1212/WNL.0000000000001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- Muona M, Berkovic SF, Dibbens LM, Oliver KL, Maljevic S, Bayly MA, Joensuu T, Canafoglia L, Franceschetti S, Michelucci R, Markkinen S, Heron SE, Hildebrand MS, Andermann E, Andermann F, Gambardella A, Tinuper P, Licchetta L, Scheffer IE, Criscuolo C, Filla A, Ferlazzo E, Ahmad J, Ahmad A, Baykan B, Said E, Topcu M, Riguzzi P, King MD, Ozkara C, Andrade DM, Engelsen BA, Crespel A, Lindenau M, Lohmann E, Saletti V, Massano J, Privitera M, Espay AJ, Kauffmann B, Duchowny M, Moller RS, Straussberg R, Afawi Z, Ben-Zeev B, Samocha KE, Daly MJ, Petrou S, Lerche H, Palotie A, Lehesjoki AE. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat Genet. 2015;47:39–46. doi: 10.1038/ng.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman WG, Friedman TB, Conway GS. Perrault Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993–2016. [Google Scholar]
- Oliver PL, Finelli MJ, Edwards B, Bitoun E, Butts DL, Becker EB, Cheeseman MT, Davies B, Davies KE. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS Genet. 2011;7:e1002338. doi: 10.1371/journal.pgen.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister PD, Opitz JM. The Perrault syndrome: autosomal recessive ovarian dysgenesis with facultative, non-sex-limited sensorineural deafness. Am J Med Genet. 1979;4:239–246. doi: 10.1002/ajmg.1320040306. [DOI] [PubMed] [Google Scholar]
- Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, Kim HN, Moon SK, Abe S, Tukamoto K, Riazuddin S, Kabra M, Erdenetungalag R, Radnaabazar J, Khan S, Pandya A, Usami SI, Nance WE, Wilcox ER, Riazuddin S, Griffith AJ. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40:242–248. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps PD, Coffey RA, Trembath RC, Luxon LM, Grossman AB, Britton KE, Kendall-Taylor P, Graham JM, Cadge BC, Stephens SG, Pembrey ME, Reardon W. Radiological malformations of the ear in Pendred syndrome. Clin Radiol. 1998;53:268–273. doi: 10.1016/s0009-9260(98)80125-6. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Chisholm KM, Lynch ED, Lee MK, Walsh T, Opitz JM, Li W, Klevit RE, King MC. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci U S A. 2011;108:6543–6548. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Gersak K, Michaelson-Cohen R, Walsh T, Lee MK, Malach D, Klevit RE, King MC, Levy-Lahad E. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet. 2013;92:614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, Fiumara A, Opitz JM, Levy-Lahad E, Klevit RE, King MC. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulat AL, Ville D, de Bellescize J, Andre-Obadia N, Cacciagli P, Milh M, Villard L, Lesca G. Homozygous TBC1D24 mutation in two siblings with familial infantile myoclonic epilepsy (FIME) and moderate intellectual disability. Epilepsy Res. 2015;111:72–77. doi: 10.1016/j.eplepsyres.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Rehman AU, Santos-Cortez RL, Drummond MC, Shahzad M, Lee K, Morell RJ, Ansar M, Jan A, Wang X, Aziz A, Riazuddin S, Smith JD, Wang GT, Ahmed ZM, Gul K, Shearer AE, Smith RJ, Shendure J, Bamshad MJ, Nickerson DA, University of Washington Center for Mendelian G. Hinnant J, Khan SN, Fisher RA, Ahmad W, Friderici KH, Riazuddin S, Friedman TB, Wilch ES, Leal SM. Challenges and solutions for gene identification in the presence of familial locus heterogeneity. Eur J Hum Genet. 2015;23:1207–1215. doi: 10.1038/ejhg.2014.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Santos-Cortez RL, Morell RJ, Drummond MC, Ito T, Lee K, Khan AA, Basra MA, Wasif N, Ayub M, Ali RA, Raza SI, University of Washington Center for Mendelian G. Nickerson DA, Shendure J, Bamshad M, Riazuddin S, Billington N, Khan SN, Friedman PL, Griffith AJ, Ahmad W, Riazuddin S, Leal SM, Friedman TB. Mutations in TBC1D24, a gene associated with epilepsy, also cause nonsyndromic deafness DFNB86. Am J Hum Genet. 2014;94:144–152. doi: 10.1016/j.ajhg.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Castelein CM, Ahmed ZM, Lalwani AK, Mastroianni MA, Naz S, Smith TN, Liburd NA, Friedman TB, Griffith AJ, Riazuddin S, Wilcox ER. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet. 2000;26:431–434. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, King KA, Ahmed ZM, Riazuddin S, Ahmad N, Hussain Z, Qasim M, Kahn SN, Meltzer MR, Liu XZ, Munisamy M, Ghosh M, Rehm HL, Tsilou ET, Griffith AJ, Zein WM, Brewer CC, Riazuddin S, Friedman TB. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J Med Genet. 2011;48:767–775. doi: 10.1136/jmedgenet-2011-100262. [DOI] [PubMed] [Google Scholar]
- Schultz JM, Yang Y, Caride AJ, Filoteo AG, Penheiter AR, Lagziel A, Morell RJ, Mohiddin SA, Fananapazir L, Madeo AC, Penniston JT, Griffith AJ. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N Engl J Med. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- Smith RJH, Van Camp G. Nonsyndromic Hearing Loss and Deafness, DFNB1. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993–2016. [Google Scholar]
- Solda G, Caccia S, Robusto M, Chiereghin C, Castorina P, Ambrosetti U, Duga S, Asselta R. First independent replication of the involvement of LARS2 in Perrault syndrome by whole-exome sequencing of an Italian family. J Hum Genet. 2015;61:295–300. doi: 10.1038/jhg.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazisar BG, Neubauer D, Paro Panjan D, Writzl K. Early-onset epileptic encephalopathy with hearing loss in two siblings with TBC1D24 recessive mutations. Eur J Paediatr Neurol. 2015;19:251–256. doi: 10.1016/j.ejpn.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO Project NES. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami S, Abe S, Weston MD, Shinkawa H, Van Camp G, Kimberling WJ. Non-syndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum Genet. 1999;104:188–192. doi: 10.1007/s004390050933. [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Bickmore W. Regulation from a distance: long-range control of gene expression in development and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120372. doi: 10.1098/rstb.2012.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Gurrola JG, 2nd, Wu H, Chiu SM, Wangemann P, Snyder PM, Smith RJ. Mutations of KCNJ10 together with mutations of SLC26A4 cause digenic nonsyndromic hearing loss associated with enlarged vestibular aqueduct syndrome. Am J Hum Genet. 2009;84:651–657. doi: 10.1016/j.ajhg.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Vidarsson H, Rodrigo-Blomqvist S, Rosengren SS, Enerback S, Smith RJ. Transcriptional control of SLC26A4 is involved in Pendred syndrome and nonsyndromic enlargement of vestibular aqueduct (DFNB4) Am J Hum Genet. 2007;80:1055–1063. doi: 10.1086/518314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hu L, Chai Y, Pang X, Yang T, Wu H. A dominant mutation in the stereocilia-expressing gene TBC1D24 is a probable cause for nonsyndromic hearing impairment. Hum Mutat. 2014;35:814–818. doi: 10.1002/humu.22558. [DOI] [PubMed] [Google Scholar]