Abstract
Background
Anaphoric references occur ubiquitously in clinical narrative text. However, the problem, still very much an open challenge, is typically less aggressively focused on in clinical text domain applications. Furthermore, existing research on reference resolution is often conducted disjointly from real-world motivating tasks.
Objective
In this paper, we present our machine-learning system that automatically performs reference resolution and a rule-based system to extract tumor characteristics, with component-based and end-to-end evaluations. Specifically, our goal was to build an algorithm that takes in tumor templates and outputs tumor characteristic, e.g. tumor number and largest tumor sizes, necessary for identifying patient liver cancer stage phenotypes.
Results
Our reference resolution system reached a modest performance of 0.66 F1 for the averaged MUC, B-cubed, and CEAF scores for coreference resolution and 0.43 F1 for particularization relations. However, even this modest performance was helpful to increase the automatic tumor characteristics annotation substantially over no reference resolution.
Conclusion
Experiments revealed the benefit of reference resolution even for relatively simple tumor characteristics variables such as largest tumor size. However we found that different overall variables had different tolerances to reference resolution upstream errors, highlighting the need to characterize systems by end-to-end evaluations.
Keywords: Natural language processing, Information extraction, Reference resolution, Radiology report, Cancer stages, Liver cancer
Graphical Abstract
1. Introduction
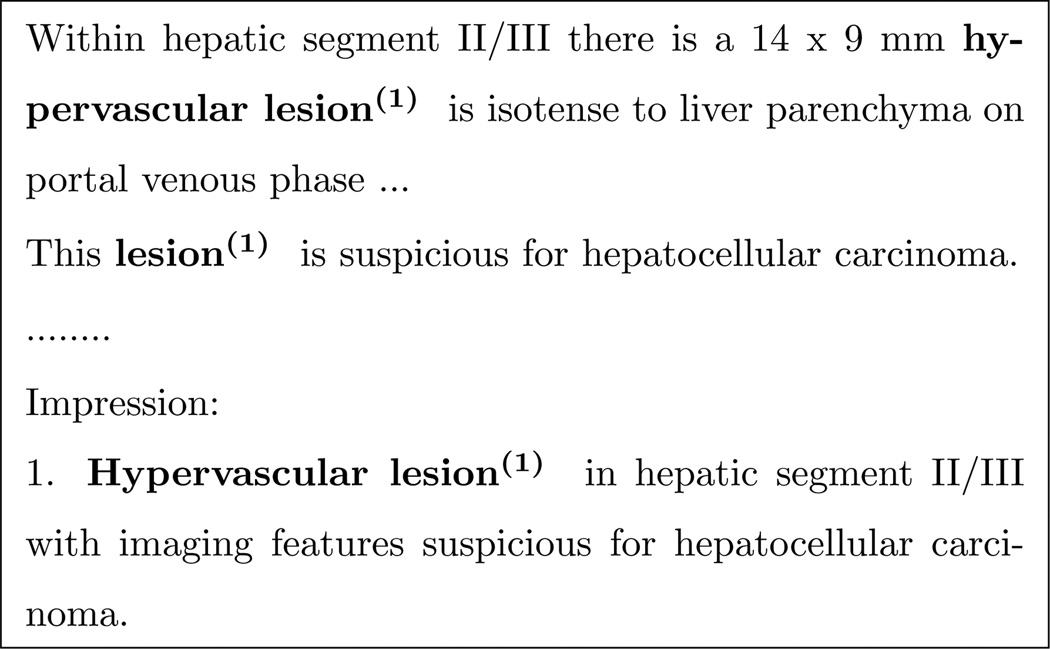
Reference resolution is the task of identifying expressions in text that refer to the same real-world entities. In natural discourse, humans readily employ reference resolution to thread together discrete pieces of information, creating a cohesive picture of discussed entities for both disseminating and processing information. For example, consider the following excerpt from a radiology report: The three mentions of the hypervascular lesion appear in separate sentences, yet the reader will naturally group them as one real world entity.
Solving reference resolution is imperative to unearthing the complex web of information trapped in clinical narrative text. Unfortunately, the state-of-the-art in reference resolution in the general domain is still limited; capabilities are even more modest in the clinical domain, in which there is a relative scarcity of annotated corpora. Furthermore, in the clinical domain, there are still well-known unsolved text processing problems such as ill-formed, ungrammatical, telegraphic, semi-structured, abbreviation-ridden narratives. This paper focuses Specifically on reference resolution for tumor references found in radiology reports.
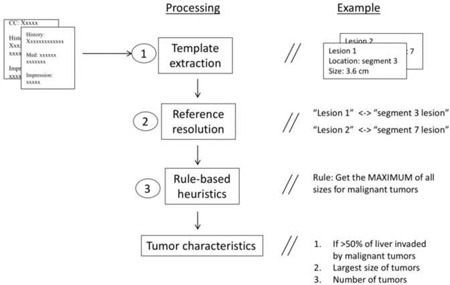
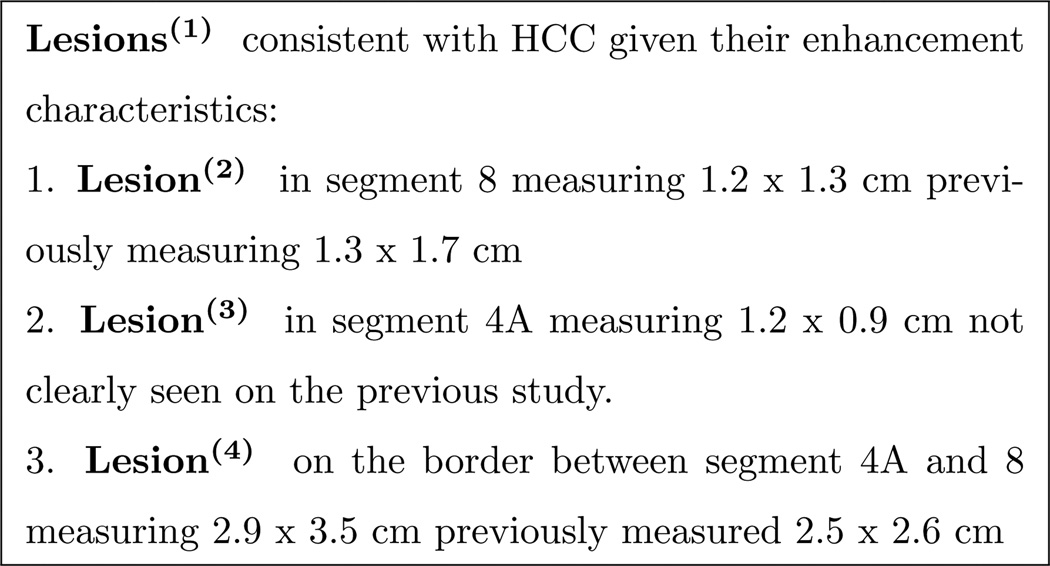
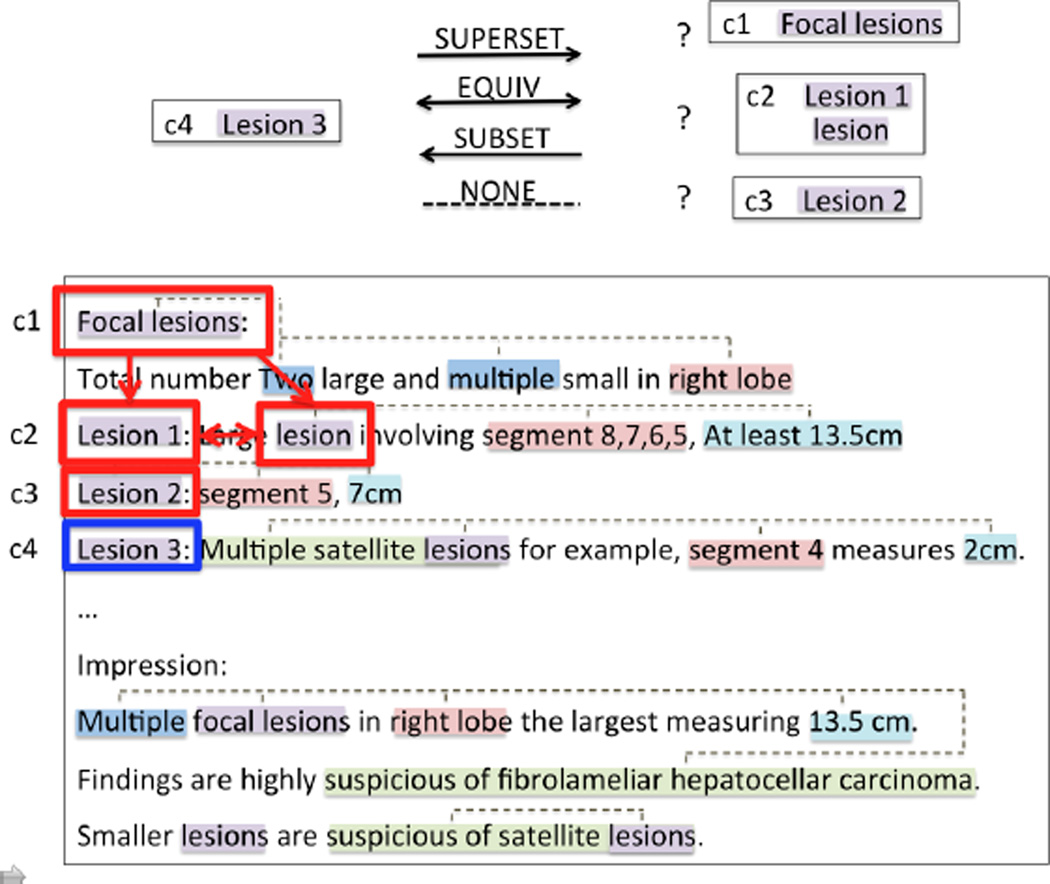
More precisely, our work targets coreference and particularization forms of anaphoric references, where coreference refers to instances in which two template heads are equivalent, as in Figure 1. We define a particularization relationship as a parent-child reference. For example, in Figure 2 Lesions(1) is a reference that represents a set of items, particularized by Lesion(2), Lesion(3), and Lesion(4).
Figure 1.
Radiology report excerpt
Figure 2.
Example of one reference and its particularizations
Additionally, we are interested in the real-world task of automatically categorizing patients into liver cancer staging phenotypes, for which reference resolution is a critical intermediate step. To this end, we are motivated to identify three summative tumor characteristic variables important for staging: (1) largest size of a malignant tumor, (2) tumor counts, and (3) whether 50% of the liver organ is invaded by tumors. These are relevant for three liver cancer staging algorithms: AJCC (American Joint Committee on Cancer), BCLC (Barcelona Cancer of the Liver Clinic), and CLIP (Cancer of the Liver Italian Program). Since these variables require aggregate knowledge of tumor-related attributes, an end-to-end evaluation, incorporating reference resolution, using these staging variables would provide a worthy perspective.
In this paper, we (a) detail our annotation for tumor reference resolution and tumor characteristics, (b) present our machine-learning reference resolution and rule-based tumor characteristics annotator approach for our tasks, and (c) report our reference resolution results, as well as an end-to-end result for the final tumor characteristics extraction.
2. Related work
Reference resolution is an active area of research in the natural language processing domain. General english NLP focus on reference resolution has primarily been on newswire text, with several notable information events such as the Message Understanding Conference (MUC) [1] and the Automatic Content Extraction (ACE) program [2]. The OntoNotes project includes coreference annotations across three languages (English, Chinese, and Arabic) for various text [3]. Similar to our goals, one previous work attempts to classify event, subevents, etc. using a pairwise logistic regression classifier [4].
In the biomedical domain, the BioNLP 2011 Shared Task featured anaphoric coreference of biomedical entities, e.g. biological entities, processes, and gene expressions [5].
In the clinical domain, annotation of a variety of concept types, e.g. person, tests, problems, for coreference, has been the focus of the 2011 i2b2/VA Cincinnati challenge [6]. Some difference between our task and that of the 2011 i2b2/VA Cincinnati challenge are the following: (a) we target very few specific mentions (tumor references instead of large classes such as person, test, or problems) and (b) our annotation is based on smaller noun phrase chunks. For example, the i2b2 challenge puts references between long noun phrases which includes descriptors such as: “’a left facial mass”, “a right parietal hyper dense and heterogeneously enhancing mass”, “an endobronchial tumor of the right upper lobe bronchus”, “a 5mm linear, focal area of enhancement in the left central semiovale”. In contrast, our references are between shorter phrases, e.g. “hypervascular lesion” or “tumor”. Similar to our task, the Ontology Development and Information Extraction (ODIE) part of the corpus has been annotated with anaphoric references, with identity, set/subset, and part/whole relations [7].
Related works on reference resolution relevant to tumors or clinical findings have been the subject of several works. Coden et al [8] identified coreferences in pathology reports using a rule-based system. Son et al [9] classified coreferrent tumor templates between documents with a MUC score of 0.72 precision and 0.63 recall. Sevenster et al [10] paired numerical finding measurements between documents.
Actual reference resolution tasks vary widely in scope. For example, nouns, pronouns, and noun phrases are common; however, coreference for nested noun phrases or nested named entities, (e.g. “America” in “Bank of America”), relative pronouns, and gerunds may not be annotated in a corpus [11]. Here our references are between the template heads of tumor templates. Our corpus does not include pronominal cases and nested references.
3. Methods
3.1. Dataset
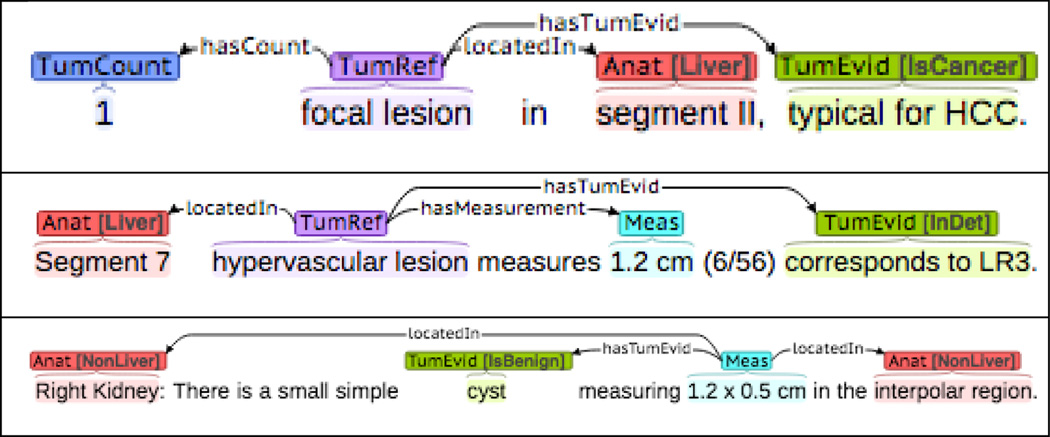
Our dataset is a set of 101 abdomen radiology reports drawn from 160 hepatocellular carcinoma (HCC) patients, annotated for 6 important entities, e.g. tumor reference and measurement entities, and 7 relations, e.g. hasSize. Several examples of the template annotations are shown in Figure 3.
Figure 3.
Three canonical template annotation examples. The last one is a case in which the template head is measurement entity.
Entities and entity attributes are described in Table 1, and relations described in Table 2. The total numbers of entities, relations, and strict templates for the were 3211, 2283 and 1006, respectively. The corpus is described in a previous work [12]. The number of relaxed templates which encode, isNegated, hadMeasurement, and hasTumorEvid relations as attributes, and re-attach nested relations to the highest head entity, is 999 (the number drop is due to tumor evidence and negation singletons being removed). The breakdown of relaxed templates by category is detailed in Table 3.
Table 1.
Entity description
| Label | Description | Freq. |
|---|---|---|
| Anatomy | Anatomic locations, e.g. “liver” with attributes (Liver, NonLiver) |
1043 |
| Measurement | Measurements findings, e.g. “2.4 cm” | 489 |
| Negation | Negation cue, e.g. “no” | 73 |
| Tumor count | Number of possible tumors, e.g. “2”, “multiple” |
174 |
| Tumorhood evidence |
Evidence indicating identity, type, or diagnostic information of a tumor referring expression, e.g. “cyst”, “suspicious for HCC”, with attributes (isCancer, isBenign, inDeterminate) |
630 |
| Tumor reference | References to possible tumors, e.g. “lesion” |
802 |
| ALL | 3211 |
Table 2.
Relation description
| Label | Description | Freq. |
|---|---|---|
| hadMeasurement | Past tense indication relation between tumor reference or measurement to another measurement |
32 |
| hasCount | Relates a tumor reference its corresponding tumor count |
171 |
| hasMeasurement | Relates a tumor reference its corresponding measurement |
334 |
| hasTumEvid | For a tumor reference or measurement, marks corresponding evidence to tumorhood evidence |
656 |
| isNegated | Relates a tumor reference to a negation cue |
75 |
| locatedIn | Identifies anatomy entity where a tumor reference or measurement is found |
955 |
| refersTo | Relates a measurement to an anatomy entity, indicating a measurement (rather than a locatedIn) |
60 |
| ALL | 2283 |
Table 3.
Relaxed template frequencies
| Label | Description | Freq. |
|---|---|---|
| AnatomyMeas | events with refersTo relations | 53 |
| Negation | events with isNegated relations | 75 |
| OtherSingleton | events with a single entity, which are not TumorSingleton events |
70 |
| TumorSingleton | events with a single entity, in which the entity is a tumor reference or measurement |
157 |
| Tumor | events not part of the previous event types |
644 |
| ALL | 999 |
3.2. Annotation for tumor reference resolution and tumor characteristics
Annotation for both reference resolution and tumor characteristics was performed on all 101 reports. 20 reports were used to measure inter-annotator agreement between a medical student and a biomedical informatics graduate student. The rest of the corpus was single-annotated by the biomedical informatics student.
3.2.1. Reference resolution annotation
Our reference resolution annotation are based on two types of relations for tumor-related templates (TumorSingleton and Tumor templates) heads:
coreference equivalence relations between mentions, e.g. Figure 1
particularization a directed relation in which the first argument represents a set of tumor reference(s) that contains the second argument tumor reference(s), e.g. Figure 2
Pronominal cases, e.g. “it”, “they”, and “these” are unmarked.
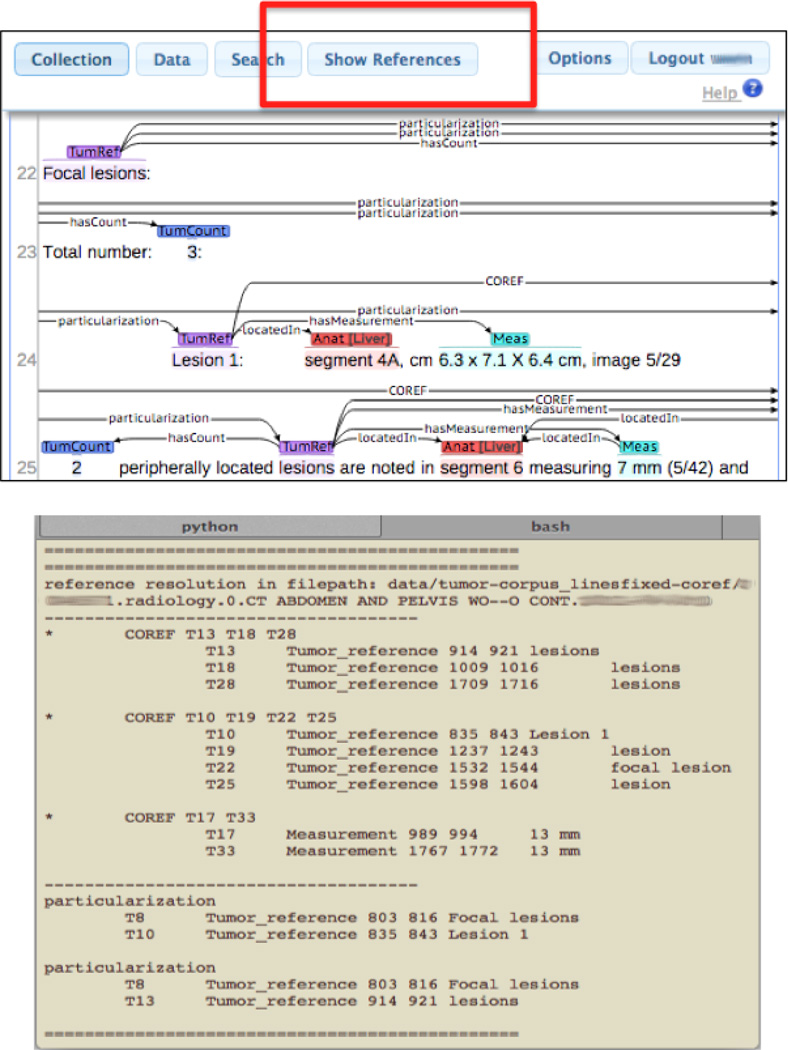
For our annotation software, we used brat[13], a web-based, annotation software for our reference resolution annotation. Since the number of coreference and particularization relations would visually render the annotations to be highly cluttered, we augmented the software to output text information regarding the clusters and particularizations annotated whenever the user selected a “show references” button, as shown in Figure 4.
Figure 4.
Brat annotation with augmentations
We measured inter-annotator agreement for coreference in terms of MUC [14], B-cubed [15], and CEAF [16] for tumor-related template heads. The agreements were at 0.956, 0.969 and 0.916 F1, respectively. A more detailed description of these metrics and our exact evaluation is described in Section 3.4. For annotator 2, there were 20 clusters (no singletons), 149 clusters (with singletons), with the average size of 2.7 entities per cluster. The cluster-normalized F1 measure for particularization relations was at 0.837.
Some ambiguities did occur between coreference and particularization, which accounted for some of the disparity in inter-annotator agreement. Mainly, as given in the examples of Figure 5, some mentions are singular but may be equivalent to the plural form of another mention.
Figure 5.
Examples of coreference relations that can be mistaken as particularizations
The final corpus has 210 clusters (no singletons), 479 cluster (with singletons), with an average of 2.60 mentions per cluster. Inferred particularization relations amounted to 573. The average and median number of sentences between the closest pairwise mentions in the same cluster are 10 and 6 sentences respectively. The large difference between mean and median suggests the existence of some very long-distance coreference relations. The mean proportion of mentions that are exact matches in a cluster is 37%, 43% if normalized for capitalizations. The average proportion of mentions found in the Findings and Impression section per cluster respectively, are 57% and 38%. The proportion of particularization relations that connect mentions in different sections is 47%.
3.2.2. Tumor characteristics annotation
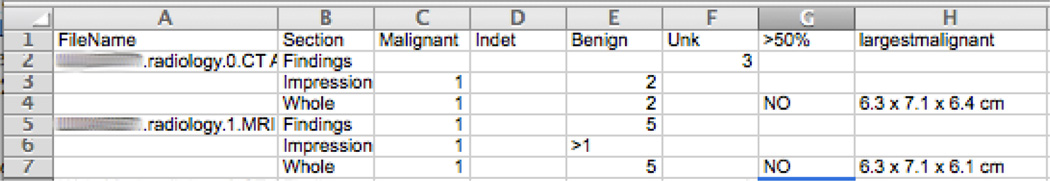
Our tumor characteristics annotation included a spreadsheet that referenced each document name and (1) the number of tumor counts by type (benign, indeterminate, unknown, and malignant), (2) the largest size for malignant tumors, and (3) whether or not more than 50% of the liver is invaded. We decided to mark inequalities, as at times the documents do not in fact give a clear number. Meanwhile, we also collected information regarding the various tumor counts for each of the Findings and Impression sections, as well as the entire document. A sample is of this is shown in Figure 6.
Figure 6.
Tumor characteristics annotation
Because the measurement for (3) is not readily quantifiable given the information in reports, we use a series of expert-created guidelines to determine the criteria for (3), as outlined in the Figure 7.
Figure 7.
Logic for >50% of liver invaded
The inter-annotator agreement is shown in Table 4. The explanation of our evaluation for inter annotator agreement is the same evaluation as those in our system. This is described more thoroughly in Section 3.4.3.
Table 4.
Tumor characteristics inter-annotator agreement
| Label | TP | F1 | F1 (relaxed) |
|---|---|---|---|
| Benign | 17 | 0.85 | 0.95 |
| Indet | 18 | 0.90 | 0.95 |
| Malignant | 17 | 0.85 | 0.95 |
| Unk | 20 | 1.00 | 1.00 |
| LargestSize | 17 | 0.85 | 0.95 |
| >50% | 20 | 1.00 | 1.00 |
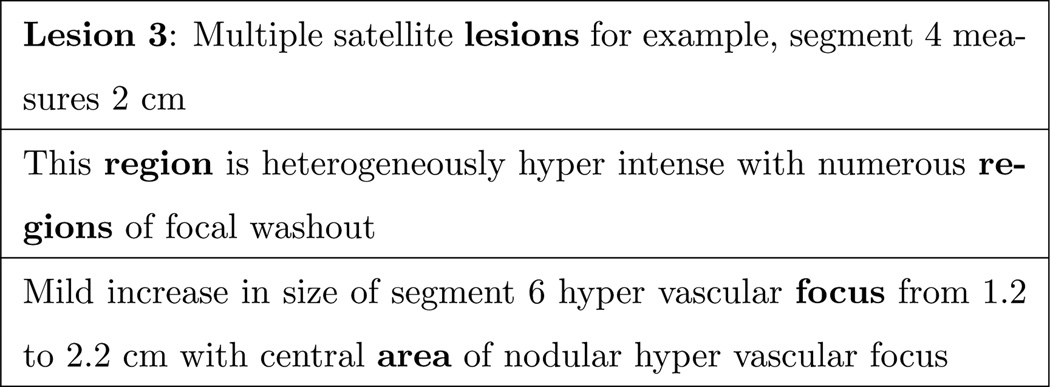
Tumor characteristics annotation were subject to various gray areas. For example for tumor counts, at times there were many ambiguous statements regarding the numbers. One example of this is in the case of conjunctions, several examples of which are shown in Figure 8.
Figure 8.
Conjunction ambiguities
The first statement can imply either one 5–6-mm focus in segment 6/7 and one in segment 5, or one 5–6-mm touching segments 6/7 and segment 5; or multiple 5–6-mm foci in the areas of segment 6/7 and 5. Similarly, “enhancing area,” in the latter statement, may be one large area inside segment 6 and 7 (which are adjacent) or separate areas in 6 and 7. Furthermore, to gather the most accurate number bounds for tumor counts, it was at times necessary to add multiple inequalities, e.g. if there are multiple (but unspecified or only partially specified) lesions in separate areas, which added to the cognitive load.
Annotating the largest size was the least controversial, though this too has some ambiguity. For example the same lesion may have two different measurements in a single report. For example in the Findings section, the largest size might be “2.5cm” but the same lesion is later referred to as “2.4cm” in the Impression section. In another example, one measurement mentioned may be specific, e.g. “6.3 × 6.1 × 9.8 cm”, and but later rounded, e.g. “6 × 6 × 10 cm”. Moreover, the amount of text and lengths of the documents, including many possible repetitions, could make it difficult to locate the best representable sizes.
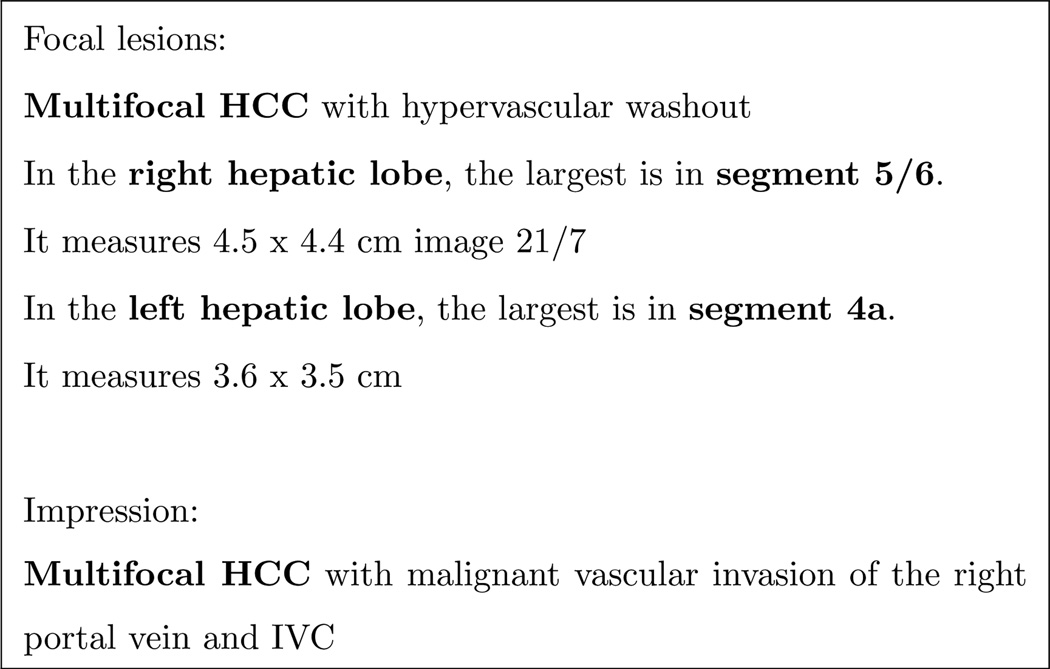
The >50% variable was at times still unclear, even with the guideline. Analyzing Figure 9 as an example, it is obvious that there are multiple tumors in both the right lobe and in the left lobe; however only 3 segments are Specifically mentioned. It is therefore not clear if the unmentioned numerous tumors may be all over the liver or only in those specific parts.
Figure 9.
Ambiguity in tumor invasion area
The distribution for the full corpus of the tumor characteristics annotation, binned along critical threshholds, is shown in Table 5.
Table 5.
Tumor characteristics annotation distributions, binned according to crucial staging values. The value of “[0, 1, 2–3, > 3]” was for a case in which the full number of lesions was given, but it was unclear how many were malignant, resulting in an unknown lesion inequality after subtraction < 5.
| Annotation categories | ||
| Tumor counts | Number | Freq. |
| Benign | 0 | 69 |
| 1 | 8 | |
| 2–3 | 10 | |
| > 3 | 8 | |
| [2–3, > 3] | 6 | |
| Indet | 0 | 62 |
| 1 | 19 | |
| 2–3 | 9 | |
| > 3 | 4 | |
| [2–3, > 3] | 7 | |
| Malig | 0 | 3.0 |
| 1 | 54 | |
| 2–3 | 25 | |
| > 3 | 13 | |
| [2–3, > 3] | 6 | |
| Unk | 0 | 89 |
| 1 | 5 | |
| 2–3 | 2 | |
| > 3 | 2 | |
| [2–3, > 3] | 2.0 | |
| [0, 1, 2–3, > 3] | 1.0 | |
| Largest size | Size (cm) | Freq. |
| [0,3) | 43 | |
| [3,5] | 26 | |
| (5–10) | 17 | |
| [10,) | 10 | |
| n/a | 5 | |
| >50% | Label | Freq. |
| n/a | 4 | |
| no | 83 | |
| yes | 14 | |
3.3. System
Our system consists of two distinct components: the reference resolution classifier and the tumor characteristics annotator. The reference resolution classifier takes structured tumor templates from a document and categorize which templates are equivalent or in a part-of relation. The tumor characteristics annotator receives grouped tumor templates and outputs (1) the number of tumor for each malignancy category, (2) the largest size malignant tumor, and (3) whether 50% of the liver is taken up by malignant tumors. The two components are further described in Section 4 and 5.
3.4. Evaluation metrics
Evaluation for our various information extraction goals were measured using F1:
| (1) |
where P = precision and R = recall used different definitions of instances, precision, and recall depending on each task. In the next sections, we detail the specific instance and F1 definitions for each evaluation task.
3.4.1. Coreference evaluation
For coreference evaluation, we use the F1 metrics for: MUC [14], B-cubed metric [15], and CEAF [16]. We also used the unweighted average of F1 between the three metrics as a separate measure.
3.4.2. Relation Evaluation
A relation is a labeled directed connection between two mentions. A correct relation requires the correct label (in this case particularization) and the correct identification of the first mention to the second mention. Here, (precision), (recall), TP is true positives, FP is false positives, and FN is false negatives.
3.4.3. Tumor characteristics evaluation
Our tumor characteristics evaluation is based on the correct label for each document and tumor characteristic variable: (1) tumor counts for benign, indeterminate, malignant, and unknown and (2) largest size for malignant tumors, and (3) whether > 50% of liver is invaded. Although we also labelled tumor counts for specific sections in a document (Findings and Impression) we only evaluate values for the entire document in this work.
We also introduced a relaxed match motivated by our specific extraction needs for liver cancer staging for AJCC, BCLC, and CLIP liver cancer algorithms. Based on staging criteria, there are only certain critical thresholds that affect the score. For example, given malignant tumor measurements all under 3cm, it does not make a difference if our algorithm cannot distinguish between 2 or 3 tumors, or if it cannot distinguish between 5 and 10 tumors; however, if the system cannot distinguish between a single tumor and multiple tumors, the cancer stage is changed drastically. The case is the same for certain sizes. Thus, our relaxed match measures based on the bins discretized from the critical values of our staging algorithms. The bin thresholds are the same as those summarizing our tumor characteristics annotation distribution in Table 5.
4. Reference resolution classifier
4.1. Approach
Our reference resolution classifier consists of a greedy algorithm which visits each template in the order of appearance in each document, and classifies the head of a template as EQUIV, SUBSETOF, SUPERSETOF, and NONE for each available cluster. If the template is EQUIV to one or more clusters, the template is added to the clusters and merged. For all other choices, the template forms a new cluster.
Figure 10 depicts the choice of a new potential cluster being being classified with one of the relation labels for each available existing cluster. At each round, classifications of the current template with existing clusters are done independently. Relations between clusters are updated at each round. When cycles emerge, all relevant clusters are merged. If there is a conflict due to a NONE classification and another label, the other labels take precedence. Classifications were trained using Lib-SVM and MALLET, for a support vector machine with a linear kernel with default settings. Feature values are scaled by the difference between the minimum and maximum values. All features (described in Section 4.2) were used for the classification.
Figure 10.
Reference resolution set up
After the assignment of EQUIV, SUBSETOF, SUPERSETOF, and NONE for each cluster in a document, the relations were translated back into coreference (EQUIV) or particularization (SUBSETOF or SUPERSETOF were converted back into a directed relation labeled as a particularization) relations for evaluation.
4.2. Reference resolution features
We detail several types of features shown in Table 6. Classes of these features are described in the following section.
Table 6.
Reference resolution features
| Feature Name | Feature Types | Description |
|---|---|---|
| closestTempDist | other | The distance of the closest template in a candidate cluster to the current template |
| containedIn | anatomic | If any of the anatomies in the current template are contained in the anatomy in the candidate cluster |
| containerOf | anatomic | If any of the anatomies in the candidate cluster are contained in the current template |
| header | static | If the sentence of the template looks like a section header |
| isSuperset | other | If the candidate cluster is already a superset of another cluster |
| malignancy | static | Malignancy status of template |
| malignancyOfCandCluster | static | Malignancy status of the cluster |
| nextBestSim | relative, similarity | L-2 norm of the next best similarity vector |
| ngrams | static | 1-, 2-, and 3- grams (using lemma) for sentences of template and a candidate cluster |
| ngramsMatching | other | Matching 1-, 2-, and 3- grams (using raw words) for sentences of template and a candidate cluster |
| nthTemplate | positional, static | The number template in the document |
| numOfCand | other | The number of candidate clusters |
| numOfMeas | static | The number of measurements |
| numOfTempInCluster | other | The number of templates in the candidate cluster |
| onlySameMal | relative | The only candidate cluster with matching malignancy as template |
| onlySameMeas | relative | The only candidate cluster with matching measurement malignancy as template |
| sameOrgan | anatomic | If the organ in the sentence matches organ in a cluster |
| sameLocations | anatomic | The matching locations of all |
| section | static | Section of the template |
| sim | similarity | The L2-norm of similarity vector |
| simvecfeats | similarity | This feature extends from the similarity vector features so that each individual similarity vector dimensions are each considered their own feature |
| summaryOf | static | If tumor reference is preceded with “the”, “this”, “these” |
| totalNumOfTemp | static | Total number of templates in the document |
| totalNumOfImpTemp | static | Total number of templates in the Impressions section |
| UMLS | other | Matching UMLS concept between the template and the cluster |
| Underheading | other | If there is a sentence belonging in the cluster that looks like a header of the current template |
Normalized anatomic location features
If anatomical entities are detected for a template, they are normalized to an anatomic concept. Based on this concept, we designed features based on anatomic hierarchy, e.g. “segment VIII” is contained in “liver”. The processing and normalization of anatomic entities is further described in Section 6. Normalization was based on Unified Medical Language System (UMLS) [18] concept names. Relevant related features are containedIn, containerOf, and sameLocations.
Positional features
Whether or not a template appears in the top or near the bottom of the template will affect how many options it will be clustered to and the threshold to what cluster similarity should be in order to be paired. We included several features related to the position of a template over all templates in a document. For example, nthTemplate gives both the absolute number and the ratio of the template position normalized to the number of all the templates.
Relative features
Relative features identify differences between candidate clusters. For example, onlySameMal is in the case of if a candidate cluster is the only one of the candidate clusters which has the same malignancy status. Another exists for same measurement.
Static features
Static features includes a variety of features, such as the section of the template, n-grams in the sentence, and number of measurements (numOfMeas). These features remain the same regardless of what candidate cluster a template head reference is being classified with.
Similarity features
Similarity features (simvecfeats and sim) are measured from the current template head to be classified to an existing candidate cluster. The similarity with the entire cluster is measured by taking the maximum of each similarity dimension among all the templates in the existing candidate clusters. For all dimension except for 0 and 1, subset candidate templates features are combined and normalized together.
Similarity features include the sentence similarity features, tumor reference similarity, as well as similarity between template attributes. For example, tumor reference similarity, measurement similarities, anatomy similarities, and anatomy similarities. The total of all similarity features combine to form a similarity vector of 9 dimensions. Each dimension is described in the Table 7.
Table 7.
Similarity features description
| Target | Description |
|---|---|
| sentence similarity |
jaccard proximity for sentence, word-tokenized |
| tumor reference similarity |
jarowinkler string proximity |
| number of measurements |
difference between number of measurement entities divided by the larger number of measurements |
| tumor count similarity |
difference in tumor count divided by the larger tumor count |
| matching measurement1 |
The number of matching measurements divided by the total number of measurements in template 1 (Measurements considered matching if within 0.1 cm) |
| matching measurement2 |
The number of matching measurements divided by the total number of measurements in template 2 |
| anatomy1 | Sum of pairwise jarowinkler proximity for all anatomy entity combinations between template 1 and 2, divided over the number of anatomy entities in template 1 |
| anatomy2 | Sum of pairwise jarowinkler proximity for all anatomy entity combinations between template 1 and 2, divided over the number of anatomy entities in template 2 |
| malignancy1 | The number of matching malignancy status (combined malignancy status’ get broken up, e.g. “INDET-BENIGN” becomes “INDET” and “BENIGN”), divided by the total number of malignancy status for template 1 |
| malignancy2 | The number of matching malignancy status, divided by the total number of malignancy status for template 1 |
5. Tumor characteristics annotator
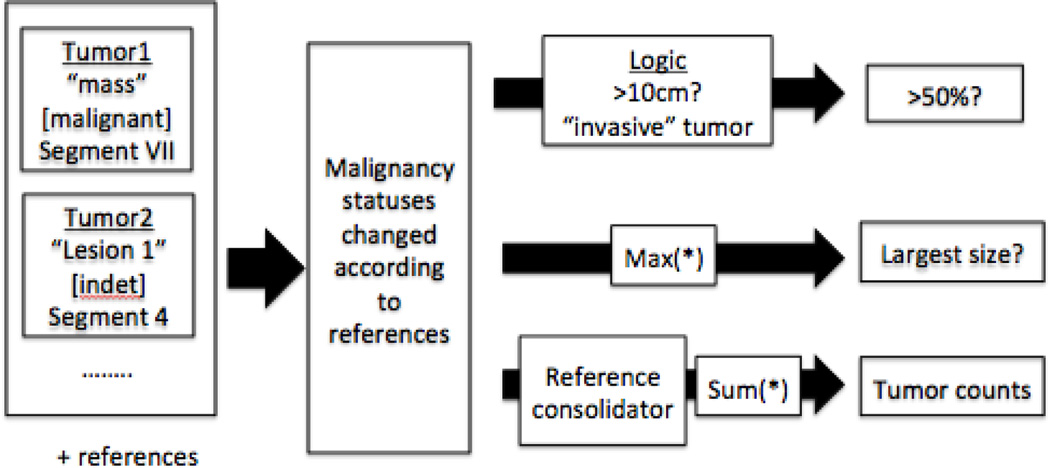
The tumor characteristics annotator takes in tumor templates and reference resolution information and outputs the three types of variables for our liver cancer staging (size, number, and whether > 50% of liver is invaded), using a series of heuristic rule-based algorithms. The various system components parts are shown in Figure 11. First the templates are updated to a new malignancy status depending on their coreference and particularization relations to other templates, next the templates are sent through several various pipelines depending on the chosen variable. In the following sections, we describe several of the non-obvious components in the pipeline: the module for updating malignancy status, the module for classifying whether >50% of liver is invaded, and the module for consolidating referenced tumors.
Figure 11.
Tumor characteristics annotator
5.1. Updating malignancy status
The malignancy statuses for related tumor templates are updated in the following way. The malignancy status for coreferrent templates are updated to the most critical case. Thus, anything coreferrent to a malignant tumor template is also malignant; if the most critical status is indeterminate then all templates are updated to indeterminate. In regards to particularizations (superset/subset relations), we take a top-to-bottom approach. The status of the superset is transferred down to the templates in the subset. After this top-down-transfer, the inter-cluster malignancy status is updated once more. Extension of this updating algorithm continuously is left for future work.
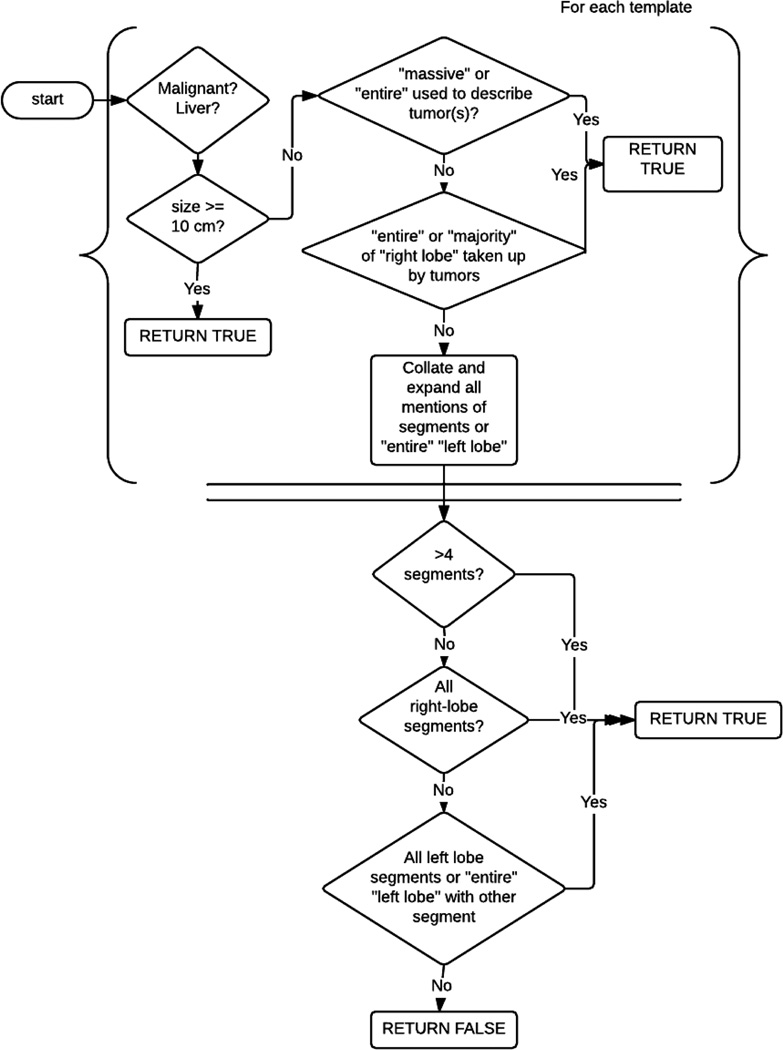
5.2. Invasion of >50% of liver logic
The logic for deciding whether or not >50% of the liver is invaded, as shown in Figure 12. The algorithm is based on the expert guidelines introduced in Figure 7.
Figure 12.
Algorithm for >50% liver is invaded
Concepts such as “right lobe”, “left lobe”, and “liver”, as well as decisions on which lines are segments are involved, are based on the anatomy normalizations from the anatomy normalization module, described in Section 6.
5.3. Reference consolidator
The reference consolidator is responsible for updating templates to the most current set of information and removing extraneous other templates. The premise is to be able to refine all the given information to a few representative templates. For example, if a reference in “Several liver lesions, suspicious for HCC” has the particularizations of “Lesion 1: segment 8, 3.0cm” and “Lesion 2: segment 5: 2.1×1.1 cm” then the template associated with the first passage will be (1) updated with measurements of “3.0cm” and “2.1×1.1 cm”, (2) updated with anatomies of “segment 8” and “segment 5”, and (3) updated to have a number of “2” for tumor count. Furthermore, if the particularization templates match the malignancy status of its superset template then those are deleted. The final result should yield a set of tumor templates with updated count, measurement, anatomy, and malignancy attributed that can be easily summed to determine the number of each type of tumors found in the radiology report.
Our exact algorithm includes heuristics for deciding for unambiguous cases, for example:
If the tumor count is set to 3 what happens if there are more than 3 measurements?
If the tumor count is not reliably determinable, how should it be decided based on the number of associated measurements?
Both coreference and particularization relations are used in the decisions.
6. Anatomy Normalizer Module
Even with properly marked anatomy entities, concept normalization requires both conjunction normalization as well as concept disambiguation. For example, “segment 2, 4A/B, and 5” must be normalized to “segment 2”, “segment 4A”, “segment 4B”, and “segment 5”. Furthermore, “left lobe” may refer to “lung” or “liver”.
Anatomy named entity clauses are normalized to discrete concepts by first, determining the organ dictionary to use using the organ context of the sentence. Afterwards, text-spans adjusted to account for missed endings for system entities, e.g. “segments VIII and V/IVb”, and terms are conjunction-normalized. Finally, concepts are matched based on the lowest score of summing together the matching edit distance with any leftover substrings.
In the following sections, we describe our rule-based algorithms for how to map sentences to an organ context and how to normalized for conjunctions; as well as our automatic creation of organ-specific hierarchal dictionaries using the Foundation Model of Human Anatomy (FMA) ontology [19].
6.1. Mapping sentences to Organ Scope
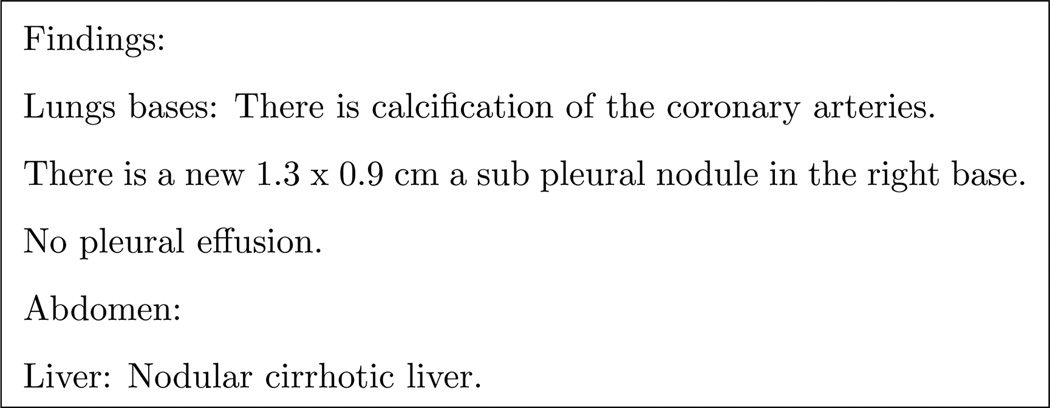
In order to differentiate between ambiguous anatomic locations, e.g. “left lobe” the organ context for a sentence must be understood. However, this information is not always available within a sentence, requiring external information. An example of this is shown in Figure 13.
Figure 13.
Different parts of the report have anatomical context not necessarily immediately available in the same sentence or not explicitly clear. In the third sentence, “right base” can be inferred to be part of the lungs by the reference to “Lungs bases” in the previous sentence or the mention of “pleural” in the same sentence.
Our algorithm is detailed as follows. From starting at the beginning of a document to the end, each sentence, previously tagged with UMLS concepts using MetaMap [20], was categorized as related to one or more organ concepts, if these two conditions were met: (1) an anatomic location semantic type was found and (2) the corresponding matched string was matched to the organ dictionary. The list of semantic type abbreviations included in the anatomic location list are: anst, bdsy, blor, bpoc, bsoj, and tisu. The dictionary of organ-related UMLS concept identifiers was created by recursively identifying “is-a” relations starting from the top (non-inclusive) concepts listed in Figure 14.
Figure 14.
Starting organ concept identifiers
Our algorithm also assigns organ context by matching to organ-related adjectives, e.g. “hepatic” refers to the liver. The mapping from a organ-related adjective to an organ was created by taking pertainyms from WordNet [21] which point to a MetaMap-matched organ. Examples of the resulting dictionary is shown in Table 8. If no match occurs, the previous line’s organ is set for the current line. At the start of each section, the assigned organ is reset to a default state. In our case, the default organ concept was set to the liver.
Table 8.
Organ adjectives identified using WordNet pertainyms. As bones are considered organs in the FMA, adjective forms of specific bones were also captured (tibial).
| Organ | Adjective forms |
|---|---|
| kidney | nephritic, renal, adrenal |
| liver | hepatic |
| lung | pulmonic, lung-like, pulmonary, pneumogastric, pneumonic, cardiopul- monary, intrapulmonary |
| prostate | prostatic, prostate |
| spleen | lienal, splenetic, splenic |
| tibia | tibial |
A manual review of 5 randomly drawn documents (215 sentences) revealed a precision of 94% for this procedure.
6.2. Normalizing for conjunctions
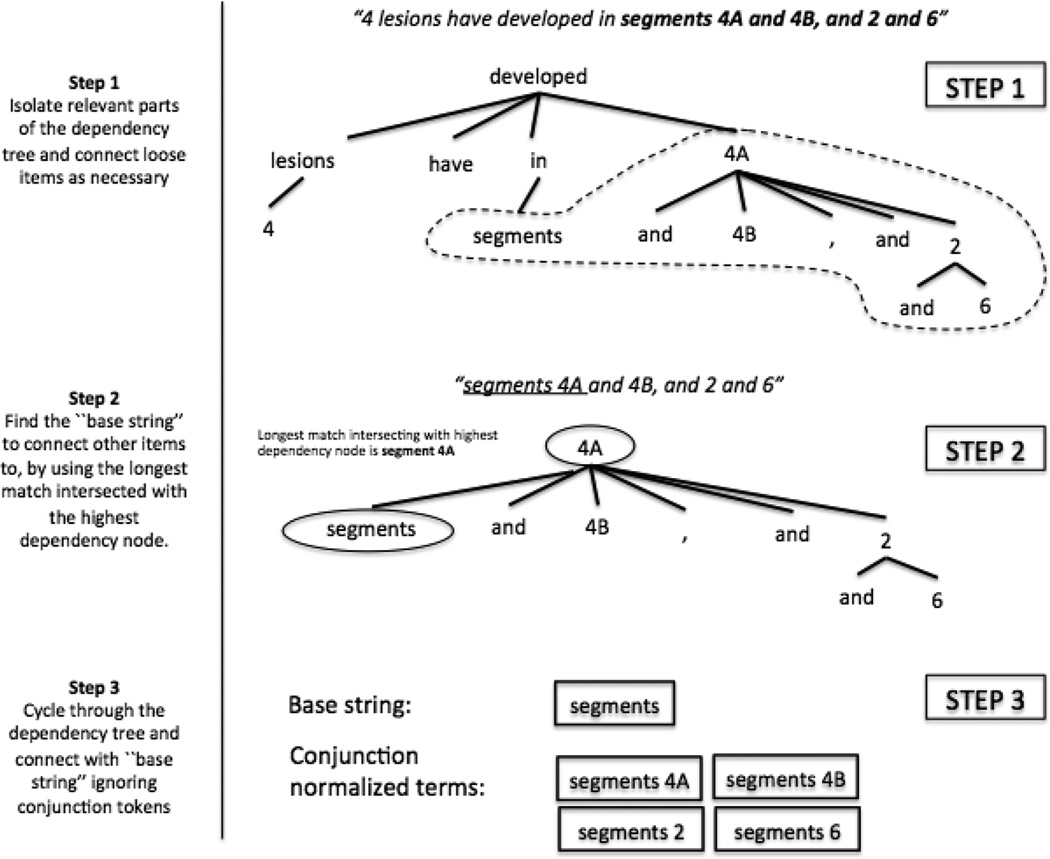
Conjunctions were normalized by first finding the longest match from organ-specific dictionaries. The automatic creation of these hierarchal organ-specific dictionaries is detailed in Section 6.3. The overlap of the longest match was then intersected with the highest node of the sentence dependency tree. The longest match was determined by finding the terms with the lowest edit distance. Starting from this match, the center-most word is popped off. Then, each unused word from the anatomy entity is paired with the match, ignoring terms such as “and”, “or”, “/”, “-” and “.”. The construction of the pairings for “segments 4A and 4B, and 2 and 6”, as shown in Figure 15. The same algorithm is designed to also be used for cases such as “Tumor thrombus within main, right and proximal left portal veins”.
Figure 15.
Conjunction normalization process. Step 1: Isolate relevant parts of the dependency tree and connect loose items as necessary. Step 2: Find the “base string” to connect other items to, by using the longest match intersected with the highest dependency node. Step 3: Cycle through the dependency tree and connect with “base string” ignoring conjunction tokens.
Our aim here was to provide a way to capture both types of conjunction problems that we encounter for our anatomy entities, such as the right-branching conjunctions of “segments x, x, and x” as well as the left-branching conjunctions of “x, x and x portal veins” in the least assuming way possible. Thus, the generalization of this heuristic for other cases is left for future investigation.
6.3. Organ-specific hierarchal dictionary creation
Portions of each organ’s hierarchal constituent structures were extracted starting from the organ concept identifiers listed in the previous section. The concepts were collected by recursively following relations: has regional part, has constitutional part, and has attributed part.
Synonym dictionaries for each concept was augmented by adding synonyms in which roman numerals were replaced with numbers (1–12), e.g. “segment II” would be duplicated with variant “segment 2”. Synonyms that required mentions of the specific organ, e.g. “right lobe of the liver”, were also duplicated with the removal the organ mention to allow better matching, e.g. “right lobe”. The following regular expressions were used to identify portions of synonyms to be augmented: “ of [organ]$”, “[^organ]”, “[^organ-adjective]”. These regular expressions were created after studying the naming conventions of the FMA. As a caveat, this may not generalizable to all ontologies and is subject to changes of the FMA terminology.
7. Results
Tables 9 show coreference and particularization classifications results, using gold standard tumor reference and templates, with a simple baseline of ngrams and ngrams-matching features compared with our system with the full set of features. Though there was little improvement for particularizations, the coreference performance increased sizably.
Table 9.
Reference resolution results.
| Evaluation | N-grams + Ngrams matching |
All Features | |||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ||
| Coreference classification | |||||||
| MUC | 0.49 | 0.35 | 0.41 | 0.63 | 0.54 | 0.58 | |
| B-cubed | 0.82 | 0.72 | 0.77 | 0.84 | 0.78 | 0.81 | |
| CEAF | 0.61 | 0.36 | 0.45 | 0.68 | 0.52 | 0.59 | |
| 0.54 | 0.66 | ||||||
| Particularization relation | |||||||
| particularization | 0.51 | 0.39 | 0.44 | 0.42 | 0.44 | 0.43 | |
(P=precision, R=recall, F1=F1-score)
In order to quantify how well our tumor characteristics annotator works, we experiment with using no reference resolution information, using gold standard reference resolution, and, finally, system reference resolution using gold standard templates. The results are shown in Table 10. Given system reference resolution annotations, the tumor characteristics significantly dropped, however the performance remained high for the > 50% variable, and dropped less drastically for the largest size variable, compared to those for the tumor count variables.
Table 10.
Tumor characteristics annotation results (gold standard templates)
| No Ref. Res. | Gold Ref. Res. |
System Ref. Res. |
||||
|---|---|---|---|---|---|---|
| TP | F1 | TP | F1 | TP | F1 | |
| >50% | 94 | 0.93 | 95 | 0.94 | 95 | 0.94 |
| #benign | 72 | 0.71 | 80 | 0.79 | 76 | 0.75 |
| #indet | 67 | 0.66 | 78 | 0.77 | 76 | 0.75 |
| #malig | 14 | 0.14 | 70 | 0.69 | 56 | 0.55 |
| #unk | 12 | 0.12 | 60 | 0.59 | 52 | 0.51 |
| largest size |
80 | 0.79 | 94 | 0.93 | 87 | 0.86 |
We were also interested in knowing how the two components affect our entire system end-to-end. That is given, system produced templates, what is our tumor characteristics annotation results? The comparison results are shown in Table 11. From these results we see the > 50% variable remains high, suggesting that it is a variable that is more robust to changes in reference resolution errors as well as template extraction problems. The tumor count variables for all types of malignancies are shown once again to drop substantially. However, this makes sense as even with perfect gold reference resolution, our annotation logic would not get past 0.80 exact F1; furthermore, figuring out the number of tumors requires very exact reference resolution information, making the tolerance for errors very much lower. The largest size variable was the least affected using both the system references and system templates; this accounts to the ability of the metric to absorb errors (it uses a maximum function).
Table 11.
Tumor characteristics annotation results (system templates)
| No Ref. Res. | System Ref. Res. |
System Ref. Res. (relaxed) |
||||
|---|---|---|---|---|---|---|
| TP | F1 | TP | F1 | TP | F1 | |
| >50% | 89 | 0.90 | 90 | 0.90 | 90 | 0.90 |
| #benign | 67 | 0.67 | 66 | 0.66 | 68 | 0.68 |
| #indet | 64 | 0.64 | 68 | 0.68 | 72 | 0.72 |
| #malig | 18 | 0.18 | 50 | 0.50 | 62 | 0.62 |
| #unk | 2 | 0.02 | 34 | 0.34 | 34 | 0.34 |
| largest size |
63 | 0.63 | 77 | 0.77 | 79 | 0.79 |
We also allowed our system to only process certain sections of the document, e.g. Findings only, Impression only, or both (default). We present our results of doing so for our three important variables in Table 12, with different combinations of gold and system reference resolution and template annotations.
Table 12.
Tumor characteristics annotation results restricted by section measured in accuracy (gold-templates, gold references / gold-templates, system references / system-templates, system references)
| Section | |||
|---|---|---|---|
| Findings | Impression | Both | |
| >50% | 0.81/ 0.80/0.69 | 0.89 /0.89/0.78 | 0.94/0.93/0.90 |
| #malig | 0.67/0.56/0.40 | 0.69/0.61/0.56 | 0.69/0.50/0.50 |
| Largest size |
0.76/0.70/0.57 | 0.43/0.39/0.37 | 0.93/0.86/0.77 |
While the tumor count variable for malignant tumors did better using only the Impression section, the other two variables benefitted from having information across both the Findings and Impression section. Interestingly, the largest size variable is much lower for the Impression section compared to the Findings section, which reinforces the observation we have found that more detailed information are often kept in the Findings section, with more summary information in the Impression section.
8. Error analysis and discussion
8.1. Tumor reference resolution classification
Analyzing the misclassification of relations, we found that of the 358 FP particularizations, 350 were represented in the gold standard except with the opposites direction (supersetof/subsetof switch) and 27 corresponded to equivalent relations in the gold standard (the may overlap with the supersetof/subsetof switch since they are not mutually exclusive). Similarly, for the 253 FN, 217 were reversed and 34 were related to equivalent relations in the system.
There are many areas for improvement with this classification. Firstly, the greedy merge approach for all coreference and particularization loops is simplistic. An algorithm that resolves this issue by ranking probabilities of each individual relation may do better to resolve the loop without causing large chain reactions. In general english, there are constraints such as pronoun agreement (“John” and “he” vs “her”) that are used for coreference systems. We did not implement any such constraints, partly because of our small corpus. Some ideas in this vein could be constraints against different “named lesions” being in the same cluster, e.g. “Lesion 1” and “Lesion 2”. Our classification for each template to all candidate clusters were done individually, though perhaps joint classification could yield marginally better results. Finally, our system aggregated clusters from top to bottom in a greedy fashion, allowing the possibility of cascading errors.
8.2. Tumor characteristics annotation
Analysis of the tumor characteristics annotator using gold standard templates and referencer resolution annotations revealed some interesting phenomenon.
While the tumor count errors was partly due to our system not producing inequalities (which is required in the gold standard under strict evaluation), it was also due to the heuristic rules of changing malignancy status (only if coreferrent or top-down) and in merging. Furthermore, while particularization hierarchies may go down several levels, we limited our number, measurement, and anatomy update rules to a scope to 3 levels.
In the case of > 50% invasion of the liver, there were only a handful of mistakes. One false positive was due to a possible typo in the report (listed as 24 cm in Findings but 24 mm in the Impression), one false negative in which no template was attached to a malignancy evidence finding (it was outside the Findings/Impression section), and one case which was labelled “n/a” due to no size or anatomy in the report at all. The remaining cases included one false negative in which “both segments” was not converted to mean segments 1–8 in the liver and a false positive in which “majority” was not meant to modify “liver” in the sentence. The performance for this was quite high regardless of reference resolution for two reasons. There was a skew in population towards < 50% invasion of the liver, which was the default. Secondly, for positive cases, only some documents required reference resolution. Of those that required reference resolution, it was not necessary to be as precise as for calculating tumor counts. For example, a lesion greater than 10 cm may only be known to be malignant through a reference in another sentence. As long as the the measurement can be labeled as malignant through either a coreference or particularization relation (regardless which one is true), then the overall < 50% invasion of the liver would be easily determined.
For the largest size variable, two errors were due to no malignancy evidence attached to templates, four errors were due to either differences in reported measurements (mistakes or simply precision differences, e.g. 2.5 cm vs. 2.4 cm). Finally, one error was due to malignancy status not being updated in a down-up fashion. This variable improved even with modest reference resolution performance because it only required reference resolution for better assignment of malignancy status; therefore as in the previous case, it requires less precise reference resolution classification. Afterwards, if other measurements with their malignancy statuses are correctly identified, the largest size could be calculated easily.
9. Conclusions and future work
In this work, we present our annotation as well as our system design for tumor reference resolution and tumor characteristics annotation. Although our reference resolution and tumor count results are modest, our experiments demonstrated that improvements in reference resolution will also lead to improvements in downstream tasks.
Finding the number of tumors proved to be the most difficult variable, as it requires very precise reference annotations. Meanwhile the other variables, > 50% invasion of the liver and largest size, were more tolerant to errors.
Some limitations to this work is that our dataset is from a single institution, our corpus size is small, and our annotations are mostly single-annotated. Our corpus annotations are also specific for tumors and not generalizable towards general medical concepts.
In future work, we will incorporate our system into our overall patient liver cancer staging system.
Highlights.
-
-
Reference resolution is essential for understanding the content in free text radiology reports.
-
-
For tumor-related entities and specific tumor characteristics, e.g. largest size and tumor count, reference resolution is shown to improve predictive performance.
-
-
Different information extraction application tasks have different thresholds for reference resolution errors; therefore reference resolution must be measured using end-to-end experiments.
Acknowledgments
We thank Tyler Denman for his help with annotation. This project was partially funded by the National Institutes of Health, National Center for Advancing Translational Sciences (KL2 TR000421) and the UW Institute of Translational Health Sciences (UL1TR000423).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grishman R, Sundheim B. Message understanding conference-6: A brief history. COLING. 1996;96:466–471. [Google Scholar]
- 2.Doddington GR, Mitchell A, Przybocki MA, Ramshaw LA, Strassel S, Weischedel RM. The automatic content extraction (ace) program-tasks, data, and evaluation. LREC. 2004;2:1. [Google Scholar]
- 3.OntoNotes Release 5.0 - Linguistic Data Consortium. URL https://catalog.ldc.upenn.edu/LDC2013T19. [Google Scholar]
- 4.Araki J, Liu Z, Hovy E, Mitamura T. Detecting Subevent Structure for Event Coreference Resolution. URL http://citeseerx.ist.psu.edu/viewdoc/citations;jsessionid=AC6C5BDE654DDC3D6C1940135817B1A7?doi=10.1.1.650.8871. [Google Scholar]
- 5.Kim J-D, Nguyen N, Wang Y, Tsujii J, Takagi T, Yonezawa A. The Genia Event and Protein Coreference tasks of the BioNLP Shared Task 2011. BMC Bioinformatics. 2012;13(11):1–12. doi: 10.1186/1471-2105-13-S11-S1. URL http://dx.doi.org/10.1186/1471-2105-13-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. Journal of the American Medical Informatics Association: JAMIA. 2012;19(5):786–791. doi: 10.1136/amiajnl-2011-000784. URL http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3422835/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman WW, Savova GK, Zheng J, Tharp M, Crowley R. Anaphoric reference in clinical reports: characteristics of an annotated corpus. Journal of Biomedical Informatics. 2012;45(3):507–521. doi: 10.1016/j.jbi.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Coden A, Savova G, Sominsky I, Tanenblatt M, Masanz J, Schuler K, Cooper J, Guan W, de Groen PC. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. Journal of Biomedical Informatics. 2009;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Son RY, Taira RK, Kangarloo H. Inter-document coreference resolution of abnormal findings in radiology documents. Studies in Health Technology and Informatics. 2004;107(Pt 2):1388–1392. [PubMed] [Google Scholar]
- 10.Sevenster M, Bozeman J, Cowhy A, Trost W. A natural language processing pipeline for pairing measurements uniquely across free-text CT reports. Journal of Biomedical Informatics. 2015;53:36–48. doi: 10.1016/j.jbi.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Stoyanov V, Gilbert N, Cardie C, Riloff E. Conundrums in Noun Phrase Coreference Resolution: Making Sense of the State-of-the-art. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2- Volume 2, ACL ’09, Association for Computational Linguistics; Stroudsburg, PA, USA. 2009. pp. 656–664. URL http://dl.acm.org/citation.cfm?id=1690219.1690238. [Google Scholar]
- 12.Yim W-w, Denman T, Kwan S, Yetisgen M. Proceedings of AMIA 2016 Joint Summits on Translational Science. San Francisco, USA: 2016. Tumor information extraction in radiology reports for hepatocellular carcinoma patients. [PMC free article] [PubMed] [Google Scholar]
- 13.Stenetorp P, Pyysalo S, Topi G, Ohta T, Ananiadou S, Tsujii J. BRAT: A Web-based Tool for NLP-assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics, EACL ’12, Association for Computational Linguistics; Stroudsburg, PA, USA. 2012. pp. 102–107. URL http://dl.acm.org/citation.cfm?id=2380921.2380942. [Google Scholar]
- 14.Vilain M, Burger J, Aberdeen J, Connolly D, Hirschman L. A model-theoretic coreference scoring scheme. Sixth Message Understanding Conference (MUC-6): Proceedings of a Conference Held in Columbia; November 6–8, 1995; Maryland. 1995. pp. 45–52. URL http://www.aclweb.org/anthology/M95-1005. [Google Scholar]
- 15.Bagga A, Baldwin B. Algorithms for Scoring Coreference Chains; In The First International Conference on Language Resources and Evaluation Workshop on Linguistics Coreference; 1998. pp. 563–566. [Google Scholar]
- 16.Luo X. On Coreference Resolution Performance Metrics. Proceedings of the Conference on Human Language Technology and Empirical Methods in Natural Language Processing, HLT ’05, Association for Computational Linguistics; Stroudsburg, PA, USA. 2005. pp. 25–32. URL http://dx.doi.org/10.3115/1220575.1220579. [Google Scholar]
- 17.Kuhn HW. The hungarian method for the assignment problem. Naval Research Logistics Quarterly. 1955;2(1–):83–97. URL http://dx.doi.org/10.1002/nav.3800020109. [Google Scholar]
- 18.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32(Database issue):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosse C, VM JL., Jr . The Foundational Model of Anatomy Ontology. In: B AB MSc, D D BSc, B R BSc, editors. Anatomy Ontologies for Bioinformatics, no. 6 in Computational Biology. Springer London: 2008. pp. 59–117. URL http://link.springer.com/chapter/10.1007/978-1-84628-885-2_4. [Google Scholar]
- 20.Aronson AR. Proceedings / AMIA … Annual Symposium. AMIA Symposium; 2001. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 21.Bentivogli L, Forner P, Magnini B, Pianta E. Revising theWordnet Domains Hierarchy: Semantics, Coverage and Balancing. Proceedings of the Workshop on Multilingual Linguistic Ressources, MLR ’04, Association for Computational Linguistics; Stroudsburg, PA, USA. 2004. pp. 101–108. URL http://dl.acm.org/citation.cfm?id=1706238.1706254. [Google Scholar]