Abstract
Pentatricopeptide repeat (PPR) proteins are extensive in all eukaryotes. Their functions remain as yet largely unknown. Mining potential stress responsive PPRs, and checking whether known PPR editing factors are affected in the stress treatments. It is beneficial to elucidate the regulation mechanism of PPRs involved in biotic and abiotic stress. Here, we explored the characteristics and origin of the 105 E subgroup PPRs in Arabidopsis thaliana. Phylogenetic analysis categorized the E subgroup PPRs into five discrete groups (Cluster I to V), and they may have a common origin in both A. thaliana and rice. An in silico expression analysis of the 105 E subgroup PPRs in A. thaliana was performed using available microarray data. Thirty-four PPRs were differentially expressed during A. thaliana seed imbibition, seed development stage(s), and flowers development processes. To explore potential stress responsive PPRs, differential expression of 92 PPRs was observed in A. thaliana seedlings subjected to different abiotic stresses. qPCR data of E subgroup PPRs under stress conditions revealed that the expression of 5 PPRs was responsive to abiotic stresses. In addition, PPR96 is involved in plant responses to salt, abscisic acid (ABA), and oxidative stress. The T-DNA insertion mutation inactivating PPR96 expression results in plant insensitivity to salt, ABA, and oxidative stress. The PPR96 protein is localized in the mitochondria, and altered transcription levels of several stress-responsive genes under abiotic stress treatments. Our results suggest that PPR96 may important function in a role connecting the regulation of oxidative respiration and environmental responses in A. thaliana.
Keywords: Arabidopsis thaliana, pentatricopeptide repeat (PPR) proteins, microarray analysis, seed imbibition, seed development stage(s), flower development processes, mitochondria, abiotic stresses
Introduction
Pentatricopeptide repeat (PPR) proteins are encoded by a large gene family in terrestrial plants. This family has 450 members in Arabidopsis thaliana, 477 members in rice, and 486 members in foxtail millet (Setaria italica) (Schmitz-Linneweber and Small, 2008; Liu et al., 2016). Non-plant organisms contain very few PPRs (Lurin et al., 2004; Andrés et al., 2007). For example, PPRs are virtually absent from prokaryotes (Pusnik et al., 2007), and the yeast, Drosophila and human genomes are predicted to contain only 5, 2, and 6 PPR genes, respectively, and it is clear that the family has expanded greatly in terrestrial plants (O'Toole et al., 2008; Liu et al., 2016). The PPR gene family was identified serendipitously over a decade ago as a result of bioinformatic analyses of the incomplete A. thaliana genome sequence (Small and Peeters, 2000). The higher plant PPR proteins are characterized by tandem arrays of a degenerate 35-amino acid repeat motif and have functions in RNA or DNA modification, acting through sequence-specific binding (Saha et al., 2007). In A. thaliana, this family can be split into two subfamilies based on the structure of the repeated motif: the P subfamily and the PLS subfamily. Members of the P subfamily contain the canonical P motif common to all eukaryotes, while members of the PLS subfamily contain the P motif as well as two P motif-derived variants, the short (S), and the long (L) motifs. Based on the presence of conserved domains in the C-terminal region, the PLS subfamily can be further divided into the PLS, E (E/E+), and DYW subgroups (Lurin et al., 2004; Andrés et al., 2007). The majority of plant PPR proteins are predicted to be localized to mitochondria or chloroplasts. PPR proteins characterized to date have functions in a wide range of physiological and developmental processes, including cytoplasmic male sterility, photosynthesis, respiration, and embryogenesis. A few of these proteins have been shown to play roles in post-transcriptional processes associated with RNA in plant organelles (Zsigmond et al., 2008; Liu et al., 2010; Sung et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Ye et al., 2012).
There have been very few functional studies of PPR proteins relating to biotic and abiotic stress-response mechanisms in higher plants. To date, of the 450 predicted PPR proteins, only GUN1, PPR40, ABO5, PGN, AHG11, SLG1, and MEF11/LOI1 have been implicated with A. thaliana defense biotic or abiotic stress response (Koussevitzky et al., 2007; Zsigmond et al., 2008; Liu et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Yuan and Liu, 2012). GUN1 is implicated with plastid-to-nucleus retrograde signaling, regulation of nuclear gene ABI4 expression, and photooxidative stress responses (Koussevitzky et al., 2007). A. thaliana PPR40 is implicated with mitochondrial oxidative respiration that also contributes to abiotic stress tolerance (Zsigmond et al., 2008). ROS accumulation was increased and some stress-responsive gene expression were altered in the ppr40 mutant grew in the culture medium contained ABA and salt (Zsigmond et al., 2008). ABO5 was required for cis-splicing of the mitochondrial nad2 intron 3, and altered the expression of several stress-responsive and nuclear-encoded genes to affect the ABA signaling pathway (Liu et al., 2010). The PGN functions in the regulation of reactive oxygen species (ROS) homeostasis by regulating RNA editing events in mitochondria during abiotic and biotic stress responses may occur through the regulation of mitochondria-nucleus retrograde signaling (Laluk et al., 2011). AHG11 regulates the nad4 transcriptional level and thus led to changes in oxidative levels by controlling RNA editing events in mitochondria and affecting plant responses to ABA (Murayama et al., 2012). SLG1 regulates the nad3 transcript by regulating RNA editing events in mitochondria and affecting the expression of genes involved in the alternative respiratory pathway (Yuan and Liu, 2012). MEF11/LOI1 is involved in mitochondrial RNA editing, and regulates biosynthesis of isoprenoids, metabolites known to affect defense gene expression in response to wounding and pathogen infection (Kobayashi et al., 2007; Tang et al., 2010). The prevalence of chloroplast- and mitochondrial-physiology in the findings of functional studies of many PPR proteins suggests that many of these proteins may play roles in regulating oxidation balance in cellular redox under different types of stress (Lurin et al., 2004; Andrés et al., 2007). Of the seven PPR proteins, GUN1, PPR40, and ABO5 belong to P subfamily, PGN, AHG11, and SLG1 belong to E subgroup, and MEF11/LOI1 belong to DYW subgroup which also contains E/E+ motif. Four members contain an E/E+ motif (MEF11/LOI1, PGN, AHG1, and SLG1), and those PPR proteins contain E/E+ motif (E and DYW subgroups) relating to biotic and abiotic stress-response by regulating RNA editing events in mitochondria and chloroplast (Kobayashi et al., 2007; Tang et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Yuan and Liu, 2012). It is thought that the E/E+ motif might have a common function among trans-factors of RNA editing in chloroplasts or mitochondria (Okuda et al., 2007). RNA editing is important for plants to maintain functional chloroplast and mitochondrial transcripts. As a vital component, PPR protein provides specificity of each editing event. However, very little has been done to study how editing is regulated under different stresses and whether it can be used as a coping mechanism. It would be interesting to the field to check whether known PPR editing factors are affected in the stress treatments. However, there have been very few functional studies of PPR proteins relating to biotic and abiotic stress response mechanisms in higher plants.
To explore potential stress responsive PPR proteins, and checking whether known PPR editing factors are affected in the stress treatments. Here, we explored the characteristics of 105 E subgroup PPR proteins in A. thaliana, including chromosomal location and phylogenetic relationships. A comprehensive in silico expression analysis of the E subgroup PPR genes at several development stages of A. thaliana, including seed imbibition, seed development, and flowers has been performed using available microarray data. The expression patterns under stress conditions showed that five E subgroup PPR genes are responsive to abiotic stresses, and finally confirmed that PPR96 is involved in abiotic stresses. In addition, we studied the function of A. thaliana PPR96 in finer detail. A fusion of the protein encoded by this gene with GFP was localized to the mitochondria. The expression of PPR96 was found to be responsive to salt stress, oxidative stress, and ABA treatments.
Materials and methods
Database searches for E subgroup PPR genes in A. thaliana
The gene and protein sequences of the A. thaliana E subgroup PPRs were acquired from TAIR (http://www.arabidopsis.org/); those of rice E subgroup PPRs were acquired from TIGR (http://rice.plantbiology.msu.edu/). The gene microarray data for A. thaliana were obtained from the NASCArrays server (http://bar.utoronto.ca/). Data for the following A. thaliana tissue/organs and developmental stages were analyzed: seed imbibition, roots, shoots and stems, leaves, flowers, and seed (Winter et al., 2007). Differential expression was defined according to the previous description: a gene as differentially expressed at a given stage only if the expression level of the gene at that stage was significantly higher (more than 2-fold) than the levels at all the other stages (Jain et al., 2007).
Identification of E subgroup PPR genes and evolutionary analyses
To explore the evolutionary relationship of E subgroup PPR genes in the flowering plants, we identify probable candidate eudicot model plant A. thaliana and monocots model plant rice E subgroup PPRs. The PPR domain (Pfam: PF01535) and the E/E+ motif (Lurin et al., 2004) were submitted as queries in BLASTP (E-value ≤ 10) searches of the A. thaliana and rice genome databases. A total of 105 A. thaliana and 138 rice putative E subgroup PPRs were obtained after manually filtering out repeated sequences. The amino acid sequences of the putative E subgroup PPRs were imported into MEGA5 (Tamura et al., 2011), and multiple sequence alignments were performed using ClustalW with a gap open penalty of 10 and a gap extension penalty of 0.1 (Thompson et al., 1997). The alignment was then used to produce an unrooted phylogenetic tree based on the neighbor-joining method (Saitou and Nei, 1987), and Poisson model was used as the amino acid substitution model; after bootstrap analysis with 500 replications, the final tree was generated.
Exon-intron structure and phylogenetic analysis
Manual curation and assessment of the numbers of introns and the exon-intron positions of the genes was based on comparing the full-length cDNA of the putative E subgroup PPR genes with their corresponding genomic sequences in A. thaliana. Physical mapping of the genes encoding the putative E subgroup PPRs onto the A. thaliana genome was performed by conducting BLASTP searches of the sequences against the Phytozome database adopting the default settings. The map was displayed using MapInspect software.
Reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qPCR)
Total RNA from leaves of A. thaliana (Col-0 and ppr96 mutants) was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and treated with RNase free DNase I (2 U μl−1, Invitrogen). The quality and purity of the RNA preparations were determined by measuring the OD260/OD280 absorption ratio (1.8–1.9), and the integrity of the preparations was determined according to the previous studies (Lata et al., 2014). DNase-treated RNA was examined by PCR to ensure complete DNA removal. Subsequently, cDNA synthesis, and RT-PCR were conducted as previously described (Xu et al., 2007), A. thaliana Act2 gene was used as an internal control. qPCR analysis for the examination of the expression of the putative E subgroup PPR genes was carried out according to the previous description (Liu et al., 2016), PCR was performed based on three technical replicates for each of the biological duplicates. Act2 was used as an internal control to normalize the expression data. qPCR data analysis was done according to previous studies (Livak and Schmittgen, 2001; Lata et al., 2014). All primers used in the study are listed in Table S1.
Plant materials and multiple stress treatments
Seeds of wild type A. thaliana (Columbia, Col-0) and mutant lines ppr96-1 (SALK_045553C) and ppr96-2 (SALK_121064) were surface sterilized with bleach and thoroughly washed three times with sterile water before incubation in a growth chamber following 3 days of cold treatment. For the stress treatments, 4-week-old plants were exposed to solutions containing, variously, 200 mM NaCl, 50 μM ABA, or 10 mM H2O2. Unstressed plants were maintained as controls. All plant materials were harvested and stored at −80°C.
Examination of T-DNA insertion mutants
The T-DNA insertion site in the ppr96 mutants was confirmed by PCR with the gene-specific primers in Table S1.
Seed germination and root growth assays
For the germination assays, Col-0 and ppr96 seeds were placed on ½ MS medium containing different contents of exogenous ABA (0.5–1 μM), or NaCl (0–200 mM). For the root growth assays, 7-day-old seedlings were grown on vertical agar plates in the presence or absence of ABA (5 or 10 μM) or NaCl (80 or 120 mM). Root lengths were measured after 14 days.
Stomatal opening assay and measurement of chlorophyll content
Stomatal opening assays were performed with epidermal peels from rosette leaves of 4- to 6-week-old plants according to the previous description (Leymarie et al., 1998). Stomatal pores were measured according to previously-described methods (Li et al., 2012). For the measurement of chlorophyll content, samples (0.05–0.1 g) of 3-week-old in vitro grown control and treated seedlings were homogenized in liquid nitrogen and extracted with 80% (v/v) acetone for 2 h. The homogenate was centrifuged at 15,000 × g for 10 min. Absorption of the extracts was measured at 663 and 645 nm and the chlorophyll A and chlorophyll B concentrations were calculated according to Lichtenthaler (1987).
Subcellular localization
The coding region of PPR96 was fused to the N-terminal end of GFP under control of the CaMV 35S promoter. For transient expression assays, ~4 × 104 A. thaliana mesophyll protoplasts were isolated and transfected with 15 μg of 35S::PPR96 plasmid. The empty GFP vector was used as a control. Transfected protoplasts were incubated in darkness at 25°C for 16 h (Xu et al., 2007; Liu et al., 2013). Transfected A. thaliana protoplasts were stained with a mitochondria-specific dye [MitoTracker Orange (Invitrogen catalog no. M7510)] (Jiang et al., 2006) and then observed with 488 and 543 nm illumination using a Zeiss LSM700 microscope.
Results
Identification of E subgroup PPR genes and phylogenetic analysis
In the A. thaliana genome, 450 putative PPR genes were identified, including 106 E subgroup PPR genes. (Lurin et al., 2004). However, At1g06145 was listed as obsolete in TAIR (http://www.arabidopsis.org/). Finally, a total of 105 E subgroup PPR genes in the genome of A. thaliana were identified using techniques described in the Materials and Methods section of this paper. The PHYTOZOME locus, subclass, open reading frame ORF length, predicted protein length, intron number, chromosomal location, and predicted protein targeting of each of these 105 E subgroup PPR genes is listed in Table S2. The predicted polypeptide lengths of E subgroup PPR proteins varied widely, ranging from 429 to 1028. EXPASY analysis suggested that the predicted E subgroup PPR protein sequences have great variations in both isoelectric point (pI) values (ranging from 5.23 to 9.11) and molecular weight (ranging from 46.921 to 111.541 kDa; Table S1).
To evaluate the phylogenetic relationships among the E subgroup PPR genes in A. thaliana, a phylogenetic analysis of 105 A. thaliana and 138 rice predicted E subgroup PPR sequences was performed by generating a neighbor-joining phylogenetic tree (Figure 1). The phylogenetic analysis categorized the E subgroup PPR proteins into five discrete groups (Cluster I to V), containing, respectively, 45, 16, 14, 7, and 23 predicted proteins (Figure 1). Many of the internal branches had high bootstrap values, indicating statistically reliable pairs of possible homologous proteins.
Figure 1.
NJ distance tree of E subgroup PPR proteins A. thaliana and rice. The tree is presented radially so that distances from the center represent cumulative branch lengths. Terminal branches and labels are colored to indicate: species (A. thaliana, blue; rice, orange) and predicted organelle targeting (mitochondria, red; plastids, green; and cytoplasm, gray).
Gene structure and chromosomal distribution
The intron numbers present within the ORF of each of the E subgroup PPR genes in A. thaliana were determined by analysis of their exon-intron organization. The large majority of the E subgroup PPR genes contain very few introns. Ninety-two E subgroup PPR genes contain no introns, 11 contain 1 intron, 3 and 4 introns were found in At3g50420 and At2g42920, respectively. It has been proposed that the gene families which are lack of intron can evolve rapidly, and the ways are either by gene duplication or reverse transcription/integration (Lecharny et al., 2003; Lurin et al., 2004; Jain et al., 2006).
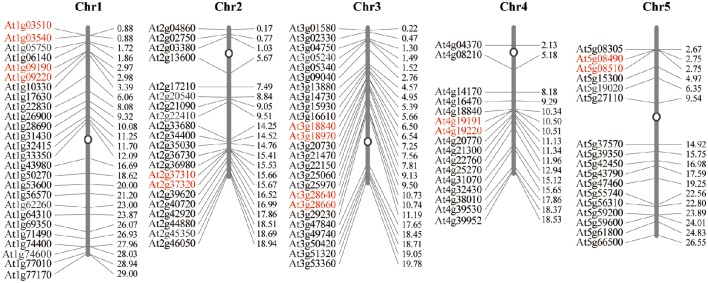
The 105 E subgroup PPR genes are unevenly distributed across all 5 of the A. thaliana chromosomes. Chromosome 1 of A. thaliana contains the highest number of the E subgroup PPR genes [26(24.76%)], while the lowest number are on chromosome 4 [16(15.23%)]. The exact position (in bp) of each E subgroup PPR gene on the A. thaliana chromosomes is available at PHYTOZOME and is represented diagrammatically in Figure 2 (the exact position in bp is given in Table S2).
Figure 2.
Distribution of the 105 E subgroup PPR genes onto the five A. thaliana chromosomes. Graphical (scaled) representation of physical locations for each E subgroup PPR gene on the five A. thaliana chromosomes (numbered 1–5). Tandem duplicated genes on a particular chromosome are depicted by scarlet letters. Chromosomal distances are given in Mb.
In silico microarray analysis of the E subgroup PPR genes during seed imbibition, seed, and flower development
To get gene expression profiling of E subgroup PPR genes in A. thaliana, microarray analysis was performed using available microarray data, which were downloaded from the NASCArrays server (http://bar.utoronto.ca/). The log signal values of A. thaliana corresponding tissues/organs and developmental stages of the 105 E subgroup PPR genes represented on the array were extracted.
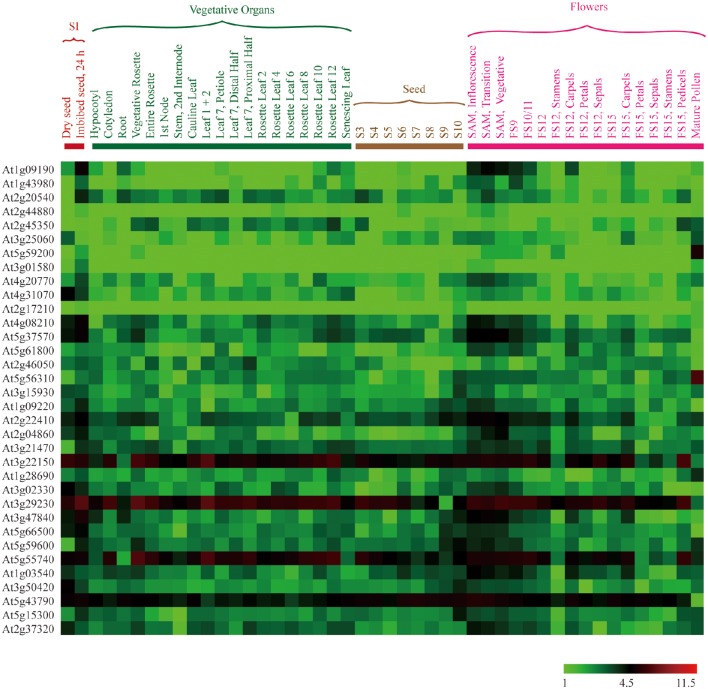
The average log signal values for all of the 105 E subgroup PPR genes are given in Table S3, and visualized in a hierarchical cluster in Figure S1. The results show that the majority of E subgroup PPR genes are expressed in at least one of the A. thaliana vegetative organs and/or stages of development that we downloaded the data for. Subsequently, differential expression analysis was performed to identify the E subgroup PPR genes with the highest expression among seed imbibition, the seed development stage(s), and flowers (Figure 3). Differential expression was defined according to the description of Jain et al. (2007). This analysis revealed that a total of 25, 23, and 34 E subgroup PPR genes were differentially expressed in at least one of the stages of seed imbibition, the seed development stage(s), and flowers, respectively (Figure 3, Table S4).
Figure 3.
Expression profiles of A. thaliana E subgroup PPR genes differentially expressed during seed imbibition, seed development stage(s), and flowers. The average log signal values of E subgroup PPR genes in various tissues/organs and developmental stages (mentioned at the top of each lane) are presented by cluster display. The color scale (representing log signal values) is shown at the bottom.
Microarray analysis of E subgroup PPR genes under abiotic stresses conditions
To examine expression patterns of A. thaliana E subgroup PPR genes under abiotic stress treatments, and mine potential stress responsive E subgroup PPR genes, microarray analysis was performed using available NASCArrays data (http://bar.utoronto.ca/) for RNA from A. thaliana seedlings subjected to salinity, cold, heat, drought, osmotic, oxidative, and wounding treatments for 0.5, 1, 6, and 12 h. We were able to identify 92 E subgroup PPR genes that were differentially expressed under one or more of these stress conditions (Figure 4 and Table S5). 84, 74, 88, 83, 62, 57, and 72 genes were differentially expressed under the salt, cold, heat, drought, osmotics, oxidative, and wounding treatments, respectively (Figure 4), and the differentially expressed genes were labeled by different background colors in Table S5. Twenty-seven E subgroup PPR genes were differentially expressed under all of the abiotic stress treatments (Table S5). Microarray analysis results showed that a large number of the E subgroup PPR genes were differentially expressed under the abiotic stress treatments, which may be due to the E/E+ motif of E subgroup PPR proteins which are implicated with the site-specific RNA editing events in plant mitochondria or chloroplasts (Okuda et al., 2007) to alter oxidation balance in vivo when plants grow under the abiotic and biotic stress conditions (Zsigmond et al., 2008; Liu et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Yuan and Liu, 2012).
Figure 4.
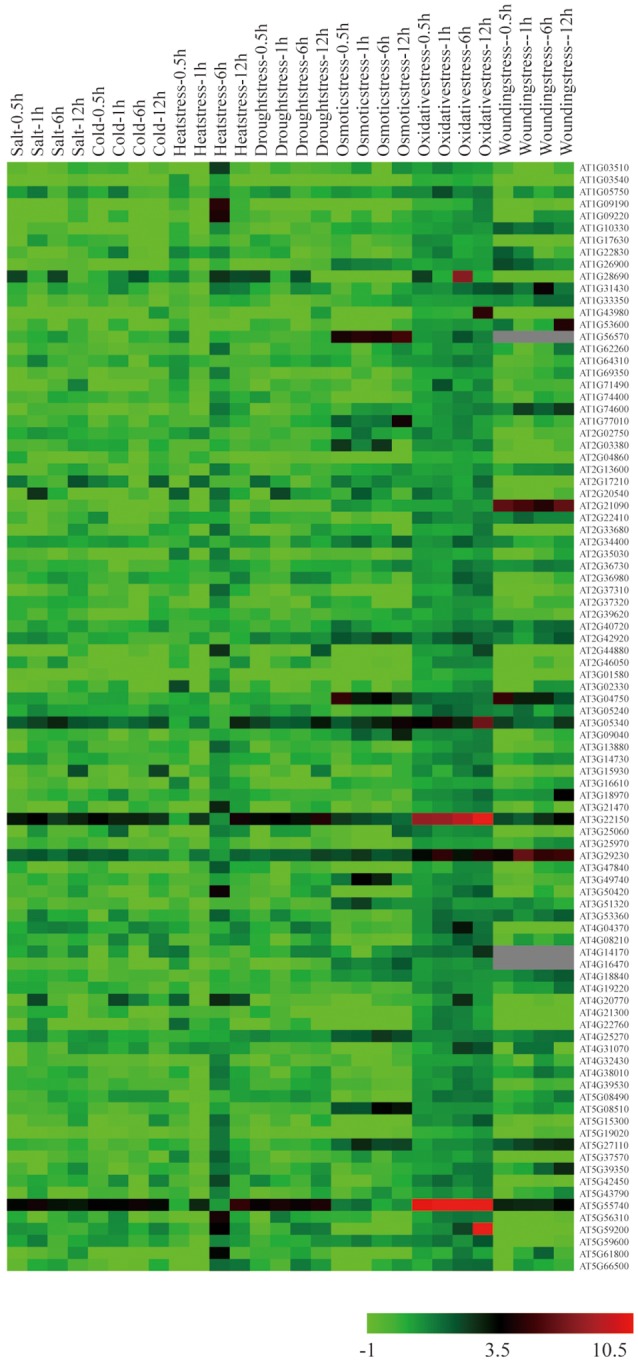
Expression profiles of E subgroup PPR genes differentially expressed under abiotic stress conditions. The values of E subgroup PPR genes under control (untreated) and various stress conditions (mentioned at the top of each lane) were presented by cluster display (values are given in Table S5). The color scale (representing signal values) is shown at the bottom.
Verification of microarray data using qPCR
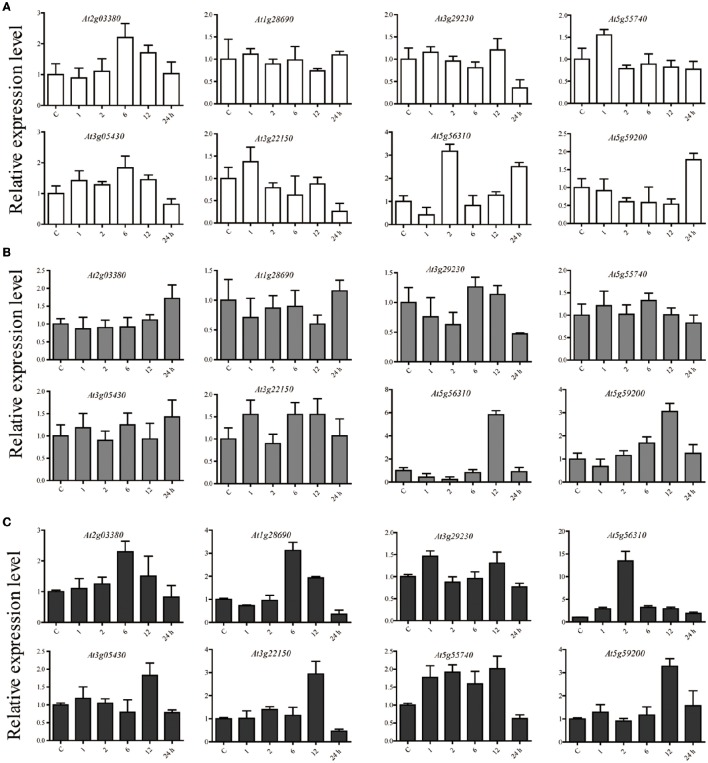
We used qPCR analysis to verify that the microarray results of interest. E subgroup PPR proteins were involved in biotic and abiotic stress-response by regulating RNA editing events in mitochondria and chloroplast, and many of these proteins may play roles in regulating oxidation balance in vivo elicited by different types of stress (Lurin et al., 2004; Andrés et al., 2007; Kobayashi et al., 2007; Tang et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Yuan and Liu, 2012). Considering this, 8 A. thaliana E subgroup PPR genes based on the expression level under oxidative stress in the microarray results were selected for further verification. A. thaliana plants were subjected to salinity, ABA, and oxidative stress for 1, 2, 6, 12, and 24 h, the unstressed plants were maintained as controls in each time point, and gene expression was analyzed with qPCR. In summary, the qPCR analysis showed that all of the candidate E subgroup PPR genes had variations in their expression patterns in response to one or more stresses relative to their expression in untreated control samples (Figure 5). The expression of 2 genes were up-regulated (>2-fold) by salt stress (At2g03380, and At5g59200), 2 were up-regulated by ABA (At5g56310, and At5g59200), and 5 were up-regulated by oxidative stress (At2g03380, At1g28690, At5g56310, At3g22150, and At5g59200). The expression of At2g03380 was up-regulated under both salt and oxidative stress treatments. At5g59200 expression was also up-regulated under both ABA and oxidative stress conditions. Notably, the expression of At5g56310 was up-regulated under salt, ABA, and oxidative stress conditions. We found that the expression of three A. thaliana E subgroup PPR genes (At3g05340, At3g29230, and At5g55740) was unchanged by either stress. The results of qPCR broadly consistent with the microarray analysis, and showed that At2g03380 (PPR96), At5g59200 (OTP80), and At5g56310 (MEF21) are responsive to abiotic stresses and ABA.
Figure 5.
The relative expression levels of 8 candidate E subgroup PPR genes analyzed using qPCR under (A) salinity stress, (B) ABA treatment, and (C) oxidative stress for 1, 2, 6, 12, and 24. The relative expression level of each gene was calculated relative to their expression in untreated control samples. Act2 was used as an internal control to normalize the expression data. The error bars represent the standard deviation calculated based on three technical replicates for each of the biological duplicates.
The expression of the PPR96 gene in A. thaliana and ppr96 mutants insensitivity to salt stress
According to the results of our qPCR analysis, At2g03380 (PPR96), At5g59200 (OTP80), and At5g56310 (MEF21) were selected for further study whether they respond to abiotic stresses. We obtained mutants for these genes from the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu) and verified that the ppr96 mutants seedlings were insensitive to salt, abscisic acid, and oxidative stress as compared to Col-0 plants. Mutants of the other two genes had no obvious phenotype to abiotic stresses treatment (data not shown). We confirmed that the ppr96 mutants by a tandem PCR, including ppr96-1 (SALK_045553) and ppr96-2 (SALK_121064). The ppr96-1 mutant has a T-DNA insertion caused a target site addition of 1 bp in the 66 bp downstream of the ATG codon, resulting in the presence of a stop codon in the 96 bp downstream of the predicted ATG codon (Figure 6A). The ppr96-2 mutant has a T-DNA insertion caused a target site deletion of 1 bp and was localized 1455 bp downstream of the predicted ATG codon (Figure 6A). The T-DNA insertion in ppr96 disrupts the expression of PPR96; RT-PCR analysis revealed that no PPR96 transcripts were detectable in ppr96 mutants (Figure 6B).
Figure 6.
Response of the ppr96 mutants to salt stress. (A) and (B) Verification of the T-DNA insertion in the ppr96 mutants. Positions of T-DNA insertions in the At2g03380 gene, and PPR96 expression was not detected in mutants plants. (C–F) Germination rates of seeds after 7 days in the presence or absence of NaCl. (G) Phenotypic comparison of root lengths of plants grown on MS medium with or without NaCl. Images were recorded on day 7 after the transfer of 7-day-old seedlings from ½ MS medium to plates containing NaCl. Bars = 1 cm. (H) Effect of different NaCl concentrations on root growth in Col-0 and mutant plants. Data represent means ± SD (n = 30). Student's t-tests were used to generate the P-values. *P < 0.05; **P < 0.01.
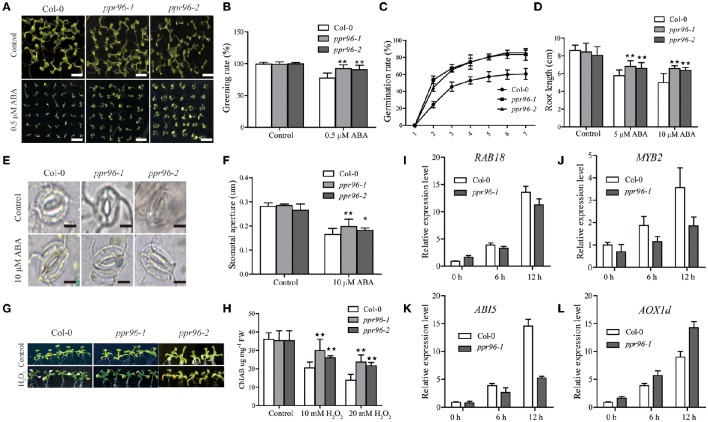
Under standard growth conditions, we observed no significant differences in the growth or morphology between mutant and wild type Col-0 plants. However, mutants seedlings displayed insensitivity to salt as compared to Col-0 (Figures 6C–E). In the presence of 80 mM and 150 mM NaCl, the germination rate of mutant was faster than that of Col-0. Mutants seedlings had enhanced salt stress tolerance and had longer root lengths compared to Col-0 (Figures 6F,G), suggesting that PPR96 may function in response to salt stress.
Mutant ppr96 plants are insensitive to ABA and oxidative stress
Assays were performed to examine the ABA insensitivity of ppr96-1 and ppr96-2. The germination and root length of ppr96 mutants seedings were not severely affected by exogenous ABA compared to Col-0 (Figures 7A–D). ABA regulates stomatal closure to avoid water loss during drought stress treatment (Leung and Giraudat, 1998). As shown in Figures 7E,F, treatment of ppr96-1 and ppr96-2 leaves with ABA did not cause pronounced stomatal closure, whereas ABA treatment caused obvious stomatal closure in Col-0 leaves. These results showed that the functional deficiency in ppr96 decreased exogenous ABA sensitivity in the process of stomatal closure, suggesting that PPR96 is a positive regulator of ABA responses and adjusts exogenous ABA responses in development and stress responses. We also found that ppr96-1 and ppr96-2 mutant plants are insensitive to oxidative stress. In the presence of sublethal contents of hydrogen peroxide, the ppr96-1 and ppr96-2 mutant displayed relatively slower bleaching and chlorophyll degradation responses as compared to Col-0 plants (Figures 7G,H).
Figure 7.
ABA and oxidative stress insensitivity of ppr96 mutants plants. (A) Growth of Col-0 and mutants plants on ½ MS medium containing zero or 0.5 μM ABA. Bars = 0.5 cm. (B) Comparison of green cotyledon percentages of Col-0 and mutants plants. (C) Germination rates of Col-0 and mutants. Seedlings were grown with or without 1 μM ABA. Germination rates were determined daily after stratificaction. Data represent means ± SD (n = 108). (D) Effects of ABA on root growth of Col-0 and mutants plants. Data represent means ± SD (n = 50) from three independent experiments. (E) Stomatal movement profiles of Col-0 and mutants plants. Stomatal guard cells were observed in the epidermal peels treated with a solution containing 25 mM KCl and 10 mM MES-Tris (pH 6.15) for 1 h in the light and subsequently treated with 10 μM ABA for 3 h. Bars = 10 μm. (F) Stomatal closure of guard cells resulting from ABA treatment. Data are the mean ratios of width to length ± SD of three independent experiments (n = 30). (G) 2-week old Col-0 and ppr96 mutants plants were treated with 10 mM H2O2 for 4 d. (H) Reduction of chlorophyll levels in response to H2O2 treatment during a 4 d period. (I–L) Expression levels of RAB18, MYB2, ABI5, and AOX1d in Col-0 and ppr96-1 mutant plants under normal conditions and under 1 μM ABA treatment during for 0, 6, and 12 h. Measurement was performed via qPCR. Values represent means ± SD with three biological replicates. Student's t-tests were used to generate the P-values. *P < 0.05; **P < 0.01.
Quantitative real-time PCR (qPCR) was performed to analyze the expression of stress-response marker genes (RAB18, MYB2, and ABI5) in Col-0 and ppr96-1 plants under ABA treatment (Figures 7I–L). The results showed that all the stress marker genes had up-regulated expression in Col-0 and ppr96-1 under ABA treatment at 6 and 12 h (Figures 7I–L). However, the expression levels of those marker genes in ppr96-1 plants were obviously lower than Col-0 plants at 6 and 12 h. Notably, ABA treatment enhanced AOX1d transcription in both Col-0 and ppr96-1 mutant plants at 0, 6, and 12 h, but the expression level of AOX1d gene in ppr96-1 plants was higher than Col-0 plants at each time point, and this trend was significantly increased at 12 h as compared to Col-0. AOXs proteins are known to prevent the accumulation of ROS during stress in plants (Navrot et al., 2007). Enhanced induction of AOX1d transcription suggested the activation of the compensatory AOX pathway in ppr96-1 mutant mitochondria. These results suggest that the PPR96 gene may important function in connecting the regulation of both oxidative respiration and environmental responses in A. thaliana.
PPR96 is localized in the mitochondria
The PPR96 gene has a single exon with an ORF of 2070 bp and putatively encodes a 77-kD PPR96 protein. Analysis of the PPR96 protein sequence by the TargetP program (http://www.cbs.dtu.dk/services/TargetP) (mitochondrial score 0.660), which suggested the PPR96 protein is targeted to mitochondria. The PPR96 protein carries a predicted mitochondrial targeting signal and 16 conserved PPR motifs, and shows the characteristics of the E subgroup of PPR proteins, having a M-63-S-P-S-S-P-P-S-P-S-4-S-P-P-3-S-P-L-S-5-E repeat sequence and a typical domain arrangement (numbers show the number of residues in between domains; Figure 8A). RT-PCR analysis with PPR96-specific primers was used to investigate the tissue specific expression pattern of PPR96. PPR96 was detected in roots, flowers, and shoots (Figure 8B). PPR96 orthologs sharing highly conserved domain structures have been identified in Brassica napus, Vitis vinifera, Populus trichocarpa, Glycine max, and rice (Figure 8C).
Figure 8.
Main features of the PPR96 protein and its subcellular localization. (A) Conserved domains of the PPR96 protein as defined by Lurin et al. (2004). (B) RT-PCR analysis of the expression pattern of PPR96. ACT2 was used as control. (C) Multiple sequence alignment of PPR96 and closely related proteins in other species. Black shading indicates conserved residues, and gray indicates residues identical to PPR96. Proteins were aligned using ClustalW with default gap penalties (Thompson et al., 1997). (D) PPR96 is localized to mitochondria. Green fluorescence indicates GFP, red fluorescence indicates stained protoplasts (Mitotracker Orange), and yellow fluorescence indicates images with the two types of fluorescence merged. Bars = 5 μm. Bna, Brassica napus; Vv, Vitis vinifera; Pt, Populus trichocarpa; Os, Oryza sativa; Rc, Ricinus communis.
To identify the subcellular localization of PPR96, the full-length PPR96 was cloned and inserted into a subcellular localization vector that included a GFP protein-encoding gene under the control of the 35S promoter; this construct was transformed into A. thaliana protoplasts. To confirm that the putative mitochondrion-targeting PPR96 protein was expressed in mitochondria, we stained transformed A. thaliana protoplasts with a mitochondria-specific dye (Jiang et al., 2006) and then observed the samples with 488 and 543 nm illumination. The green fluorescent signals co-localized with the MitoTracker Orange fluorescent signals (Figure 8D). This result revealed that the PPR96-GFP fusion was localized to mitochondria.
Discussion
RNA editing is a process of RNA maturation involved in the insertion, deletion, or modification of nucleotides (Smith et al., 1997). The RNA editing is more complex in plants than that of other organisms. In organellar transcripts of higher plants, specific cytidine residues are converted into uridine residues. The post-transcriptional conversion of specific cytosines to uracil in mitochondrial and plastid transcripts is unique to land plants (Steinhauser et al., 1999). Thirty-four sites are edited in Arabidopsis plastids (Chateigner-Boutin and Small, 2007), whereas more than 450 editing sites are edited in Arabidopsis mitochondria (Giegé and Brennicke, 1999; Bentolila et al., 2008; Zehrmann et al., 2008). In many cases, editing results in the restoration of conserved amino acid residues, a process that is essential for protein function in plastids (Bock et al., 1994; Sasaki et al., 2001). The A. thaliana genome contains more than 450 PPRs. Most of these are aimed at plastids and/or mitochondria (Lurin et al., 2004), and their functions remain largely sparse (Schmitz-Linneweber and Small, 2008). Of the 450 PPR proteins, the 105 E subgroup members may have a common functions as trans-factors of RNA editing in chloroplasts or mitochondria (Okuda et al., 2007). A. thaliana chloroplast editing trans-factors CRR4 and CRR21, for example, can regulate the activity of NAD(P)H dehydrogenase (Kotera et al., 2005; Okuda et al., 2007), and each is required for the editing of a different C target in the ndhD transcript. The PPR protein named CLB19 was required for editing of C targets in rpoA and clpP transcripts (Chateigner-Boutin et al., 2008). A. thaliana PPR protein SLO1 is required for RNA editing of nad4 and nad9 in mitochondria; its absence affects plant growth and development. In addition, several PPR proteins that contain E/E+ motif are known to be implicated with abiotic stresses in A. thaliana. These include: MEF11/LOI1, PGN, AHG1, and SLG1; all of these proteins have been implicated with A. thaliana biotic and/or abiotic stress tolerance (Sung et al., 2010; Laluk et al., 2011; Murayama et al., 2012; Yuan and Liu, 2012). The functional link of many PPRs in plastid and mitochondrial development and/or regulation suggests these proteins may play key roles in regulating oxidation balance in cellular redox that are elicited by different types of stress (Lurin et al., 2004; Andrés et al., 2007).
In this paper, the phylogenetic relationships of the E subgroup PPR proteins in eudicot model plant A. thaliana and monocots model plant rice were analyzed, revealing that all of the E subgroup PPR proteins can be categorized into five discrete groups (Cluster I to V) (Figure 1). E subgroup PPR proteins in both species showed there generally appears an even mix of genes from both species, excepting Cluster II. The genes in Cluster II may have expanded after the separation of monocots and dicots. Additionally, gene structure and chromosomal distribution analysis showed that 87.6% of the E subgroup PPR genes lacked intron(s) and 14 (13.3%) of the PPR genes were found to be tandem repeats (Figure 2). The A. thaliana genome has undergone genome-wide duplication events, including polyploidy, which has great impact on the amplification of members of a gene family in the genome (Seoighe, 2003). Recent research showed that the expansion of PPR gene family prior to the divergence of the euphyllophytes and the lycophytes in land plants, and tandem and segmental duplication are responsible for the expansion of the PPR gene family in vascular plants (Liu et al., 2016). These results suggested that the E subgroup PPR genes may have a common origin in both eudicot model plant A. thaliana and monocots model plant rice, and the expansion of this gene family occurred prior to the monocot/dicot divergence in land plants (O'Toole et al., 2008), and the expansion of E subgroup PPRs in A. thaliana may results from localized gene duplications.
Microarray analysis of the expression of the E subgroup PPR genes at several stages of development of A. thaliana revealed that 25, 23, and 34 of these genes were differentially expressed at least one of the stages of seed imbibition, seed development stage(s), and flowers, respectively (Figure 3, Table S4). Notably, 11, 18, and 17 genes were preferentially expressed in imbibing seeds, SAM, and mature pollen, respectively (Figure 3, Table S4). This suggested that these genes may play important roles in important roles in a wide range of physiological and developmental processes.
Recently, three E subgroup PPR proteins were found to function in the highly complex ABA signaling network. The functions of PGNs in the regulation of ROS homeostasis in mitochondria may occur through the regulation of mitochondria-nucleus retrograde signaling when plants grow under abiotic and biotic stress conditions. (Laluk et al., 2011). SLG1 regulates nad3 (mitochondrion complex I) transcription by regulating RNA editing events in mitochondria and affecting the expression of genes involved in the alternative respiratory pathway (Yuan and Liu, 2012). In the present study, analysis of publically-available microarray data suggested that several E subgroup PPR genes may participate in responses to abiotic stresses (Figure 4, Table S5). Mutant plants of the mitochondria-localized A. thaliana PPR96 gene were insensitive to salt, ABA, and oxidative stress. This is the first E subgroup PPR gene mutant identified to be associated with plant resistance to abiotic stresses. We performed a preliminary phenotypic characterization of ppr96 mutants plants under salt, ABA, and oxidative stress. It is unclear which specific RNA editing sites are deleteriously affected in the ppr96 mutants. It is not clear whether the loss of PPR96 function will potentially enhance or decrease ROS accumulation in seedlings in response to abiotic stress. qPCR showed that ABA treatment increased the AOX1d transcription level in the ppr96 mutant as compared to Col-0. AOX1d encodes an AOX protein; these proteins are known to capture the excess electrons from ubiquitin to prevent the accumulation of ROS during stress (Navrot et al., 2007). Whether the ppr96 mutant can induce changes in mitochondrial electron transport also requires further investigation. Enhanced induction of AOX1d transcription suggests the activation of the compensatory AOX pathway in ppr96 mutant mitochondria. These results suggest that PPR96 may play roles in connecting the regulation of both oxidative respiration and environmental adaptation in A. thaliana. Additional physiological and biochemical experiments will need to be performed to further explore this supposition.
Author contributions
ZX coordinated the project, conceived and designed experiments, and edited the manuscript; JL performed experiments, analyzed data, and wrote the first draft of the manuscript; JZ and PL analyzed data; MC provided analytical tools and managed reagents; CG contributed with valuable discussions; YM coordinated the project and contributed with valuable discussions. All authors have read and approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financially supported by the National Transgenic Key Project of the Chinese Ministry of Agriculture (2014ZX0800916B and 2016ZX08002-002) and the National Natural Science Foundation of China (31371620).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01825/full#supplementary-material
Hierarchial clustering display of 105 E subgroup PPR genes represented on NASCArrays A. thaliana genome array in various A. thaliana organs and developmental stages (mentioned at the top of each lane). The average log signal values were used for clustering. The color scale (representing log signal values) is shown at the bottom.
The sequences of primers used in the study. The sequences shown in lower case were added to generate a restriction enzyme site.
E-subgroup PPR genes in A. thaliana. Detailed genomic information including domain/class present, alias, ORF length, protein length, genomic locus (chromosomal location), number of introns within ORF, subcellular localization, isoelectric point, and molecular weight (kDa) of the PPR proteins for each PPR gene.
Average log signal values of 105 E-subgroup PPR genes from three biological replicates of each sample.
Average log signal values of 34 differentially expressed E-subgroup PPR genes from three biological replicates of each sample.
Average log signal values of 105 E-subgroup of PPR genes subjected to salt, cold, heat, drought, osmotic, oxidative, and wounding treatment. The background color indicates the differentially expressed genes under different stress treatments.
References
- Andrés C., Lurin C., Small I. D. (2007). The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129, 14–22. 10.1111/j.1399-3054.2006.00766.x [DOI] [Google Scholar]
- Bentolila S., Elliott L. E., Hanson M. R. (2008). Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178, 1693–1708. 10.1534/genetics.107.073585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., Kössel H., Maliga P. (1994). Introduction of a heterologous editing site into the tabacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 13, 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A. L., Small I. (2007). A rapid high-throughput method for the detection and quantification of RNA editing based of high-resolution melting of amplicons. Nucleic Acids Res. 35:e114. 10.1093/nar/gkm640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A. L., Ramos-Vega M., Guevara-García A., Andrés C., De La Luz Gutiérrez-Nava M., Cantero A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602. 10.1111/j.1365-313X.2008.03634.x [DOI] [PubMed] [Google Scholar]
- Giegé P., Brennicke A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. U.S.A. 96, 15324–15329. 10.1073/pnas.96.26.15324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., et al. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. 10.1104/pp.106.091900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Tyagi A. K., Khurana J. P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88, 360–371. 10.1016/j.ygeno.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Jiang K., Schwarzer C., Lally E., Zhang S., Ruzin S., Machen T., et al. (2006). Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 141, 397–403. 10.1104/pp.106.078246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Suzuki M., Tang J., Nagata N., Ohyama K., Seki H., et al. (2007). Lovastatin insensitive 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol. 48, 322–331. 10.1093/pcp/pcm005 [DOI] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. 10.1038/nature03229 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. 10.1126/science.1140516 [DOI] [PubMed] [Google Scholar]
- Laluk K., AbuQamar S., Mengiste T. (2011). The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 156, 2053–2068. 10.1104/pp.111.177501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Mishra A. K., Muthamilarasan M., Bonthala V. S., Khan Y., Prasad M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 9:e113092. 10.1371/journal.pone.0113092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharny A., Boudet N., Gy I., Aubourg S., Kreis M. (2003). Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J. Struct. Funct. Genomics 3, 111–116. [PubMed] [Google Scholar]
- Leung J., Giraudat J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Biol. 49, 199–222. 10.1146/annurev.arplant.49.1.199 [DOI] [PubMed] [Google Scholar]
- Leymarie J., Vavasseur A., Lascève G. (1998). CO2 sensing in stomata of abi1-1 and abi2-1 mutants of Arabidopsis thaliana. Plant Physiol. Biochem. 36, 539–543. 10.1016/S0981-9428(98)80180-0 [DOI] [Google Scholar]
- Li Z. Y., Xu Z. S., He G. Y., Yang G. X., Chen M., Li L. C., et al. (2012). A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 426, 522–527. 10.1016/j.bbrc.2012.08.118 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1987). Chlorophyll and carotenoids: pigments of photosynthetic biomembranes. Meth. Enzymol. 148, 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Liu J. M., Xu Z. S., Lu P. P., Li W. W., Chen M., Guo C. H., et al. (2016). Genome-wide investigation and expression analyses of the pentatricopeptide repeat protein gene family in foxtail millet. BMC Genomics 17:840. 10.1186/s12864-016-3184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Xu Z. S., Pan-Pan L., Hu D., Chen M., Li L. C., et al. (2013). A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J. Exp. Bot. 64, 2915–2927. 10.1093/jxb/ert133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He J., Chen Z., Ren X., Hong X., Gong Z. (2010). ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 63, 749–765. 10.1111/j.1365-313X.2010.04280.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. 10.1105/tpc.104.022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama M., Hayashi S., Nishimura N., Ishide M., Kobayashi K., Yagi Y., et al. (2012). Isolation of Arabidopsis ahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. J. Exp. Bot. 63, 5301–5310. 10.1093/jxb/ers188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrot N., Rouhier N., Gelhaye E., Jacquot J. P. (2007). Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol. Plant. 129, 185–195. 10.1111/j.1399-3054.2006.00777.x [DOI] [Google Scholar]
- Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. U.S.A. 104, 8178–8183. 10.1073/pnas.0700865104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128. 10.1093/molbev/msn057 [DOI] [PubMed] [Google Scholar]
- Pusnik M., Small I., Read L. K., Fabbro T., Schneider A. (2007). Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell. Biol. 27, 6876–6888. 10.1128/MCB.00708-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D., Prasad A. M., Srinivasan R. (2007). Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534. 10.1016/j.plaphy.2007.03.026 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Kozaki A., Ohmori A., Iguchi H., Nagano Y. (2001). Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 276, 3937–3940. 10.1074/jbc.M008166200 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. 10.1016/j.tplants.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Seoighe C. (2003). Turning the clock back on ancient genome duplication. Curr. Opin. Genet. Dev. 13, 636–643. 10.1016/j.gde.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Small I. D., Peeters N. (2000). The PPR motif–a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 45–47. 10.1016/S0968-0004(99)01520-0 [DOI] [PubMed] [Google Scholar]
- Smith H. C., Gott J. M., Hanson M. R. (1997). A guide to RNA editing. RNA 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- Steinhauser S., Beckert S., Capesius I., Malek O., Knoop V. (1999). Plant mitochondrial RNA editing. J. Mol. Evol. 48, 303–312. 10.1007/PL00006473 [DOI] [PubMed] [Google Scholar]
- Sung T. Y., Tseng C. C., Hsieh M. H. (2010). The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63, 499–511. 10.1111/j.1365-313X.2010.04258.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Kobayashi K., Suzuki M., Matsumoto S., Muranaka T. (2010). The mitochondrial PPR protein lovastatin insensitive 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through RNA editing. Plant J. 61, 456–466. 10.1111/j.1365-313X.2009.04082.x [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-S., Xia L.-Q., Chen M., Cheng X.-G., Zhang R.-Y., Li L.-C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. 10.1007/s11103-007-9237-9 [DOI] [PubMed] [Google Scholar]
- Ye J. W., Gong Z. Y., Chen C. G., Mi H. L., Chen G. Y. (2012). A mutation of OSOTP 51 leads to impairment of photosystem I complex assembly and serious photo-damage in rice. J. Integr. Plant Biol. 54, 87–98. 10.1111/j.1744-7909.2012.01094.x [DOI] [PubMed] [Google Scholar]
- Yuan H., Liu D. (2012). Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 70, 432–444. 10.1111/j.1365-313X.2011.04883.x [DOI] [PubMed] [Google Scholar]
- Zehrmann A., van der Merwe J. A., Verbitskiy D., Brennicke A., Takenaka M. (2008). Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion 8, 319–327. 10.1016/j.mito.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Zsigmond L., Rigó G., Szarka A., Székely G., Ötvös K., Darula Z., et al. (2008). Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 146, 1721–1737. 10.1104/pp.107.111260 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hierarchial clustering display of 105 E subgroup PPR genes represented on NASCArrays A. thaliana genome array in various A. thaliana organs and developmental stages (mentioned at the top of each lane). The average log signal values were used for clustering. The color scale (representing log signal values) is shown at the bottom.
The sequences of primers used in the study. The sequences shown in lower case were added to generate a restriction enzyme site.
E-subgroup PPR genes in A. thaliana. Detailed genomic information including domain/class present, alias, ORF length, protein length, genomic locus (chromosomal location), number of introns within ORF, subcellular localization, isoelectric point, and molecular weight (kDa) of the PPR proteins for each PPR gene.
Average log signal values of 105 E-subgroup PPR genes from three biological replicates of each sample.
Average log signal values of 34 differentially expressed E-subgroup PPR genes from three biological replicates of each sample.
Average log signal values of 105 E-subgroup of PPR genes subjected to salt, cold, heat, drought, osmotic, oxidative, and wounding treatment. The background color indicates the differentially expressed genes under different stress treatments.