Abstract
Phenotypic (co)variation is a prerequisite for evolutionary change, and understanding how (co)variation evolves is of crucial importance to the biological sciences. Theoretical models predict that under directional selection, phenotypic (co)variation should evolve in step with the underlying adaptive landscape, increasing the degree of correlation among co-selected traits as well as the amount of genetic variance in the direction of selection. Whether either of these outcomes occurs in natural populations is an open question and thus an important gap in evolutionary theory. Here, we documented changes in the phenotypic (co)variation structure in two separate natural populations in each of two chipmunk species (Tamias alpinus and T. speciosus) undergoing directional selection. In populations where selection was strongest (those of T. alpinus), we observed changes, at least for one population, in phenotypic (co)variation that matched theoretical expectations, namely an increase of both phenotypic integration and (co)variance in the direction of selection and a re-alignment of the major axis of variation with the selection gradient.
Keywords: quantitative genetics, adaptive landscape, P-matrix, genotype–phenotype map, chipmunks, Tamias
1. Introduction
In order to persist over evolutionary time, species must have the ability to respond in the direction of selection. As organisms are formed by a combination of multiple traits organized into a coherent whole (a multidimensional system), understanding the interaction between the available phenotypic (co)variation and selection is crucial to understand species' responses to selection [1] and, consequently, species' persistence over time. For instance, if a species lacks phenotypic (co)variation in a certain direction it can quickly become extinct when directional selection operates along that trajectory [2]. How the available phenotypic (co)variation shapes species evolution is therefore an important area of research in evolutionary biology, and is relatively well appreciated theoretically, empirically and computationally (e.g. [1,3,4]). On the other hand, our understanding of how directional selection shapes the evolution of available phenotypic (co)variation, although equally important, is only just beginning. Most studies addressing this issue have been simulations derived from theory [5,6], although some experimental evidence has emerged [6–9].
In traditional evolutionary thinking, directional selection is thought to deplete genetic variation, leading to a decrease in phenotypic (co)variation [2,10]. Therefore, establishing plausible mechanisms that account for the widespread phenotypic (co)variation observed in nature became a priority in evolutionary biology [10]. Despite the inherent difficulties in pursuing answers to this issue [5], recent advances in theoretical and computational studies have provided some benchmarks for how available phenotypic (co)variation is expected to evolve under directional selection [11–15]. These models are based on the evolution of the genotype–phenotype map (GP-map), which describes how genetic (co)variation is translated into phenotypic (co)variation. If different genotypes differ in genetic (co)variation among traits (i.e. the degree of pleiotropy and epistasis among traits), this leads to the possibility that genetic (co)variation among traits can evolve in response to selection [13,14]. In these models, selection might actually drive the evolution of the mutational machinery to align available phenotypic (co)variation with selection [16]. Consequently, the impact of directional selection on the GP-map organization is thought to be substantial and can occur at a rapid pace [12,13]. Such models allow us to make predictions about how change occurs in the available phenotypic (co)variation that are easily testable by empirical data. First, we would expect an increase in genetic variation in the direction of selection (figure 1). Second, the degree to which traits are correlated is expected to change; specifically, we expect increased correlation among traits that are being co-selected [6]. Lastly, we expect a re-orientation of the pattern of available phenotypic (co)variation to match the expected direction of selection [12] (figure 1).
Figure 1.
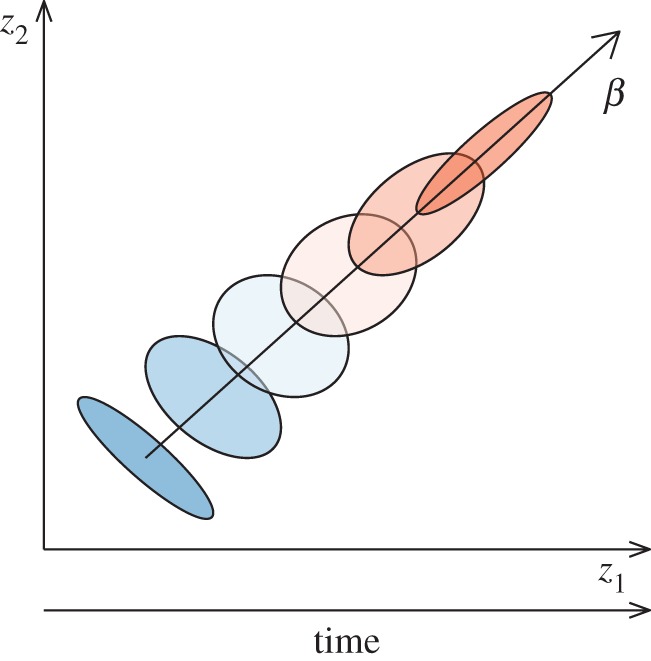
Expected change in the (co)variation pattern between two traits (z1 and z2) under a scenario of sustained directional selection (β). In this hypothetical example, prior to selection, both traits were tightly negatively correlated. Each ellipse represents the (co)variation pattern of one generation, with blue representing ancestral and red representing derived populations. As selection was acting to increase both traits, we observe a change in the magnitude and pattern of covariation between both traits due to selection. More specifically, we observe, first, an increase in the total amount of variation in the direction of selection; second, a change in the pattern of correlation among traits in order to mirror the selective regime, in this case resulting in a positive tight correlation between both traits; and third, a re-orientation of the (co)variation pattern to match the expected direction of selection. (Online version in colour.)
Validation of these predictions in multidimensional traits from natural populations is essential to further develop our understanding of evolution itself and of how species adapt to new selective pressures. It is especially relevant in a world where most natural environments are under some kind of stress due to anthropogenic pressures, either direct (e.g. land use change) or indirect (e.g. climate change). However, three main problems hinder the gathering of empirical data to assess these questions in natural populations. First, sampled populations should be separated by many generations, given that such changes are not expected in a short period of time. Second, populations must be well sampled in order to properly estimate statistical parameters. Third, the populations in question should have experienced directional selection. Here, we used a unique sample set that matches the first and second requirements, and which thus permits us to test the third requirement.
Our sample is composed of chipmunk specimens of two species, Tamias alpinus and T. speciosus, collected almost a century apart. These come from two independent transects separated by approximately 180 km along the Sierra Nevada of California. The first of these is in the central part of the Sierras, in Yosemite National Park; the other is in the Southern Sierras, within or just east of Sequoia-Kings Canyon National Parks. These species are phylogenetically close, but have responded in strikingly different manner to a century of climatic and associated environmental changes. While the alpine chipmunk, T. alpinus, has shifted its elevational distribution upwards and decreased its genetic diversity, the lodgepole chipmunk, T. speciosus, has changed neither its elevational distribution nor genetic diversity [17–19].
In this study, we tested for both species whether evolution between the historical and modern periods was driven by directional selection, and because we found evidence favouring a directional selection scenario, we tested how the selective regime changed the available phenotypic (co)variation over time. To this end, we analysed 35 skull traits in those populations using a quantitative genetics framework. We then compared the phenotypic (co)variation matrices between periods (historical and modern) for each species and transect in order to assess whether the specific selective regimes in each population had an impact on the overlying phenotypic (co)variation patterns.
2. Material and methods
(a). Samples
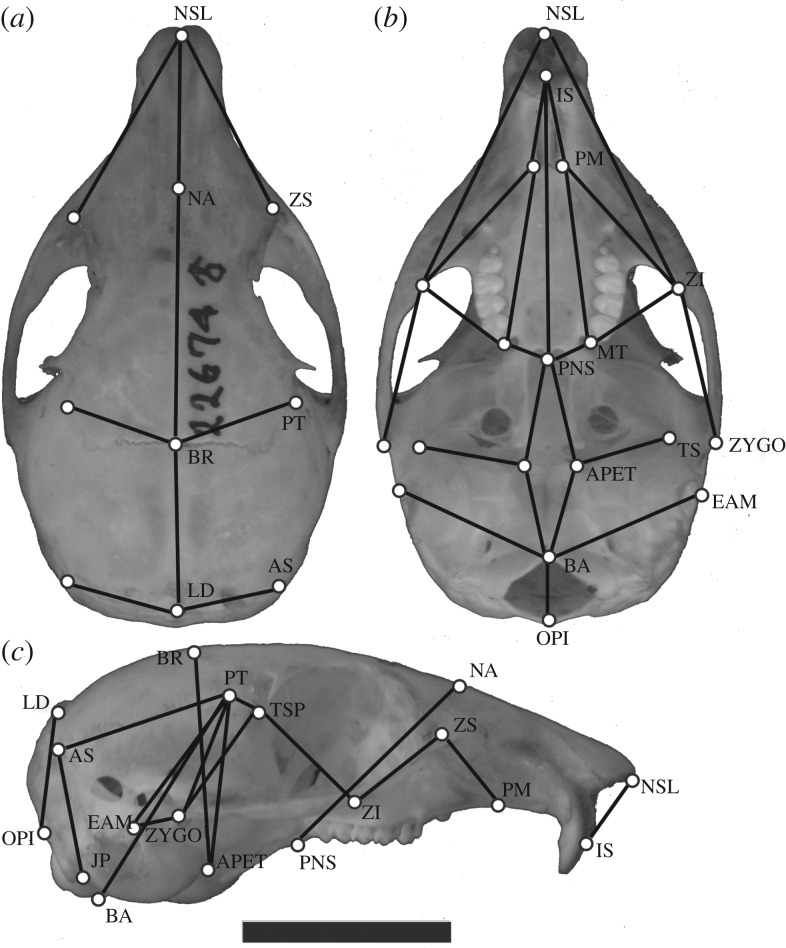
One of us (A.P.A.A.) recorded three-dimensional coordinates for 27 landmarks on 197 adult skulls of T. alpinus (Yosemite: 51 historical/38 modern; Southern Sierras: 75 historical/33 modern) and 481 adult skulls of T. speciosus (Yosemite: 77 historical/221 modern; Southern Sierras: 83 historical/100 modern). Adult specimens were defined by a fully erupted permanent premolar 4 and a completely fused basisphenoid–basioccipital suture. Based on these landmarks we calculated 35 linear distances, which were used in the subsequent analyses (figure 2). We chose to use linear measurements instead of landmark coordinates because we are primarily interested in differences in covariance structure. Proper estimation of the covariance structure in a geometric morphometrics approach would require much larger sample sizes than were available (at least three times the number of landmarks) [20].
Figure 2.
T. alpinus skull showing the 21 landmarks and 35 linear distances taken in each skull. Scale bar, 1 cm. (a) Dorsal view; (b) ventral view; (c) lateral view.
We used historical samples collected as part of a California-wide survey of terrestrial vertebrates conducted by Joseph Grinnell and colleagues from 1911 to 1915. Modern specimens were collected as part of the Grinnell Resurvey Project, an intensive resampling of Grinnell's historic sites that occurred from 2003 to 2013, and were collected under the auspices of the US National Park Service, specifically Yosemite and Sequoia-Kings Canyon National Parks, and under permit from the California Department of Fish and Wildlife, the Animal Care and Use Committee of the University of California at Berkeley, and followed the guidelines of the use of wild mammals in research developed by the American Society of Mammalogists [21]. All specimens are deposited in the Museum of Vertebrate Zoology at the University of California, Berkeley. Samples come from two independent transects, one through Yosemite National Park in the Sierra Nevada in central California and the other from the Southern Sierras, within and just east of Sequoia National Park approximately 180 km to the south. It has been previously shown [22] that the T. alpinus population from Yosemite has changed its skull morphology in a pattern compatible with directional selection when tested in a univariate context, with most changes being concentrated in the facial region for both transects. Also, while the Yosemite transect population has increased in size over the past century, the Southern Sierras population has decreased in size [22]. By contrast, comparable samples of T. speciosus had fewer traits that changed in a pattern compatible with directional selection in either transect, although in Yosemite most changes were also concentrated in the facial region.
(b). Matrix similarity
Here, we used the phenotypic matrix (P-matrix) as a proxy for its genetic counterpart, which is the evolutionary relevant parameter. Our decision to use P-matrices as a substitute for G-matrices is based on considerable evidence supporting their interchangeability, at least for morphological characters, especially in mammals [23–25]. Furthermore, to guarantee that our P-matrices were similar to other estimates of G, we compared our P-matrices with a Calomys expulsus G-matrix [26]. Also, we evaluated the overall similarity between the covariance matrices of all populations (historical and modern) using both the random skewers and the Krzanowski methods [27,28]. The random skewers method is based on the multivariate response to selection equation. This method consists of applying a set of random selection vectors (normalized to a length of one) to a pair of matrices and comparing the response vectors between the two matrices using a vector correlation procedure. The mean vector correlation between the matrices' evolutionary-response vectors is a measure of similarity. The more similar the response to selection, the higher the similarity between the matrices [28]. The Krzanowski method consists of comparing a pair of matrices by calculating the angles between the best-matched pairs of orthogonal axes (principal components) [27,29] (see the electronic supplementary material for further discussion). As matrices are estimated with error, we corrected matrix similarity values using matrix repeatability estimates [28] (electronic supplementary material, table S1).
(c). Directional selection versus genetic drift of skull traits
Before exploring whether directional selection had an impact on available phenotypic (co)variation, we tested whether natural selection was responsible for multivariate evolution between the historical and modern periods. To do so, we used the theoretical expectation that the amount of divergence expected for a population evolving under drift, in a multivariate framework, corresponds to a divergence matrix D. An estimation of this D-matrix can be made by D = Gt/Ne, where G represents the G-matrix of the ancestral population, t is the divergence time in generations, and Ne is the effective population size [1,30]. We estimated the D-matrix using the historical P-matrix as surrogate for the ancestral G-matrix (but see the electronic supplementary material for estimates using different matrices, table S5), Ne derived from upper and lower estimates for T. alpinus [31], and a generation time of 1 year [32]. Having estimated the D-matrix, we used it as the Σ parameter in a multivariate normal distribution, from which we randomly sampled 1000 vectors. Each of these vectors represents potential trait values for a population evolving under drift. The subtraction of each of those 1000 random vectors from the ancestral populations' trait means gave us estimations of morphological change expected under drift (Δz expected in a drift scenario). Finally, we compared the 95% probability distribution of the magnitude of morphological change expected under drift (which was estimated as the norm of the randomly sampled Δz vectors) with the empirical norm of Δz observed (i.e. estimation based on the historical and modern populations data, taking into account mean standard errors). We then concluded that directional selection was the main process responsible for observed divergence if the range of estimates from the magnitude of morphological divergence fell outside the 95% distribution expected under drift.
(d). Selection gradient estimate
In order to estimate the observed net selection gradient (β), which is the directional selection responsible for the observed morphological changes, we used the multivariate selection response equation [1]  where Δz is the response to selection estimated as difference between modern and historical observed trait means; G−1 is the inverse of the G-matrix (substituted by the historical P-matrix). Owing to the fact that inverted matrices are dominated by small eigenvalues usually estimated with a lot of noise, we controlled our P-matrices for noise using an eigenvalue extension method [33]. The calculated normalized β was then used as a benchmark to estimate changes in the pattern of (co)variation, as described below. Moreover, to compare the strength of selection between transects and species, we estimated the norm of the selection gradient standardized by trait means [34].
where Δz is the response to selection estimated as difference between modern and historical observed trait means; G−1 is the inverse of the G-matrix (substituted by the historical P-matrix). Owing to the fact that inverted matrices are dominated by small eigenvalues usually estimated with a lot of noise, we controlled our P-matrices for noise using an eigenvalue extension method [33]. The calculated normalized β was then used as a benchmark to estimate changes in the pattern of (co)variation, as described below. Moreover, to compare the strength of selection between transects and species, we estimated the norm of the selection gradient standardized by trait means [34].
(e). Effects of directional selection on skull P-matrices
As our main purpose was to compare changes in the pattern of (co)variation between time periods, we had to understand the possible distribution of those parameters in each species/area sample. To do so, we estimated P-matrices separately for the historical and modern samples, estimating a distribution of 1000 P-matrices by Monte Carlo resampling for each period, transect and species [35]. We then used these 1000 estimated matrices in subsequent analyses and considered a result significant whenever the observed modern P-matrix value fell outside the 95% distribution of the historical estimates [36]. To circumvent potential biases in our comparisons due to differences in sample sizes, the random populations sampled in the Monte Carlo procedure were set to the lowest sample size between the time periods compared.
To quantify the impact of natural selection on the P-matrices, we compared historical and modern P-matrices in relation to three different matrix features: (i) size, which can be described as the total amount of variation in the matrix or in a certain direction [34,37]; (ii) shape, which provides an indication of eccentricity, or how tight the correlations between traits are; and (iii) orientation in relation to the selection gradient.
In order to determine if the total amount of variance had changed from historical to modern samples, we estimated the trace of each covariance matrix from the Monte Carlo distribution. In addition, we determined if the amount of variance had changed in the direction of selection or in directions uncorrelated with the observed selection gradient. The amount of variance in any given direction was calculated as the evolvability in that direction [34], a metric which captures the ability of a population to evolve in the direction of a specified selection gradient. Evolvability can be measured as
where e(β) is the evolvability in the direction of a given selection gradient (β) and G is the G-matrix. To compare the effects of directional selection, we estimated the evolvability in the direction of the normalized observed selection gradient (βobs) for the distribution of P-matrices from the historical and modern periods. Moreover, we generated 1000 random selection gradient vectors uncorrelated with βobs. In order to obtain those sets of uncorrelated vectors, we first sampled 1000 normalized vectors from a normal distribution and applied the formula
where βr is the random selection gradient sampled from a normal distribution and βi is the uncorrelated resulting vector. We later normalized the random vectors to 1 and compared the evolvability potential of both the historical and modern P-matrices in those directions. Evolvability estimates were divided by the geometric means of all traits, thus accounting for scale differences between populations [34].
We also compared changes in the overall magnitude of integration between periods by estimating the coefficient of determination (r2) of the correlation matrices [38]. This coefficient is simply the average of the squared correlation coefficients and measures how tightly the correlations among traits are, with a higher estimate indicating higher correlations.
Lastly, we compared changes in the orientation of the P-matrix distributions between periods by estimating the correlation between the observed selection gradient and the first principal component of each of 1000 matrices from historical and modern times for both species and transects. All statistical analyses were done in the R environment for statistical computing [39] using the EvolQG package [35].
3. Results
(a). Matrix comparisons
Phenotypic matrices (P-matrices) were used as proxies for their underlying genetic matrices (G-matrices), which are the evolutionarily relevant parameter. Consequently, prior to any analyses we had to ensure that our estimated P-matrices were good estimates of the underlying G-matrices. To this end, we compared our P-matrices with a G-matrix derived from the distant related rodent Calomys expulsus [26] using both the random skewers and Krzanowski methods [8,28,29]. These comparisons showed a high degree of similarity between the P-matrices and the G-matrix, with comparisons from the random skewers method ranging from 0.61 to 0.81 (all significant, indicating that they are more similar than would be expected by chance) and those from the Krzanowski method ranging from 0.66 to 0.72 (electronic supplementary material, table S2). Although there is no consensus among researchers as to how similar a matrix has to be to be considered similar [23,40,41], these fairly high values indicated that our P-matrices were reasonable estimates of their genetic counterparts [24,41]. Moreover, comparisons of the P-matrices among populations showed that all were structurally similar, with random skewers estimates ranging from 0.81 to 0.95 and Krzanowski estimates ranging from 0.81 to 0.87 (electronic supplementary material, table S3).
(b). Directional selection versus genetic drift of skull traits
To determine whether the amount of divergence observed between historical and modern periods for each population was explained by genetic drift or by natural selection, we simulated the amount of morphological divergence expected by drift and compared it with the empirically measured magnitude of morphological change. For any effective population size adopted, the magnitude of empirically measured morphological change in the Southern Sierras was 5.7 and 10.95 times higher than that expected by drift for T. speciosus and T. alpinus, respectively. For the Yosemite transect, the same pattern was observed; the magnitude of empirically observed morphological change was 7.5 and 19.6 times higher for T. speciosus and T. alpinus, respectively, than that expected by drift. Therefore, data for both species support directional selection as the primary mode underlying the observed temporal changes (table 1).
Table 1.
Magnitude of morphological change (||Δz||) of skull traits of T. alpinus and T. speciosus observed (i.e. empirically measured) and expected under drift. Ne: effective population size based on [31]. ||Δz|| CI corresponds to the lower and upper estimates of the 95% confidence interval for the magnitude of morphological change, estimated as the norm of the Δz vector.
|
T. alpinus |
T. speciosus |
|||
|---|---|---|---|---|
| Yosemite | Southern Sierras | Yosemite | Southern Sierras | |
| ||Δz|| CI | ||Δz|| CI | ||Δz|| CI | ||Δz|| CI | |
| observed | 0.942–1.190 | 0.558–0.795 | 0.454–0.631 | 0.403–0.607 |
| Ne = 230019 | 0.020–0.051 | 0.020–0.051 | 0.026–0.062 | 0.025–0.071 |
| Ne = 430625 | 0.014–0.036 | 0.014–0.037 | 0.019–0.046 | 0.018–0.052 |
| Ne = 648513 | 0.011–0.030 | 0.011–0.030 | 0.015–0.037 | 0.014–0.042 |
Next, we estimated the standardized magnitude of morphological change (||Δzµ||) and selection gradient (||ßµ||), estimated as the norm of each vector, for both species to gauge the strength of selection. For T. speciosus from the Yosemite transect, we obtained ||Δzµ|| = 0.064 and ||ßµ|| = 15.457; comparable numbers for T. alpinus were ||Δzµ|| = 0.178 and ||ßµ|| = 39.388, which were 2.8 and 2.6 times higher, respectively, than those of T. speciosus. In the Southern Sierras, we obtained ||Δzµ|| = 0.063 and ||ßµ|| = 20.279 for T. speciosus, while the results for T. alpinus were ||Δzµ|| = 0.099 and ||ßµ|| = 27.062, values that were 1.57 and 1.33 times higher, respectively, than those of T. speciosus. In both cases, directional selection was stronger in T. alpinus than in T. speciosus.
(c). Effect of directional selection on morphological P-matrices
To determine the effect of directional selection on the P-matrices, we investigated the following changes between the historical and modern P-matrices: (i) the total amount of variation estimated by the matrices' traces; (ii) the amount of variation associated with the direction of selection and with directions uncorrelated with selection, calculated as evolvability divided by the geometric mean of the traits; (iii) the overall magnitude of correlation among traits estimated as the coefficient of determination of the correlation matrices (r2); and (iv) the orientation of the axis of greatest variation in relation to the selection gradient estimated by the correlations between PC1 and the selection gradients.
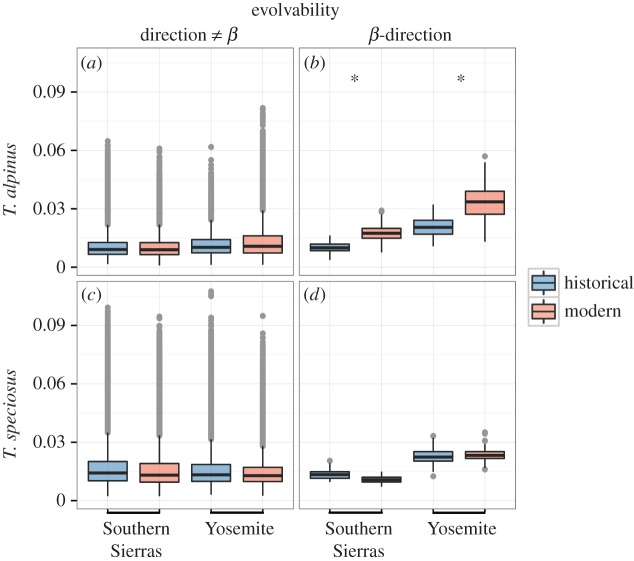
For T. alpinus from the Yosemite transect, the historical and modern matrix traces were 2.79 ± 0.22 s.d. and 3.33 ± 0.34 s.d., respectively, while for T. speciosus the corresponding values were 4.34 ± 0.28 s.d. and 4.12 ± 0.23 s.d., respectively. For the Southern Sierras transect, we obtained historical and modern matrix traces for T. alpinus of 2.63 ± 0.24 s.d. and 2.60 ± 0.25 s.d., respectively, while the corresponding values for T. speciosus were 4.68 ± 0.30 s.d. and 4.47 ± 0.31 s.d, respectively. Thus, the total amount of variation in each species-population comparison did not change temporally. However, the amount of variation in the direction of selection did increase for T. alpinus in both transects (figure 3b; Yosemite observed modern estimate = 0.033, 95% historical distribution = 0.0128–0.031; Southern Sierras observed modern estimate = 0.018, historical distribution = 0.006–0.015) but not for T. speciosus (figure 3d; Yosemite observed modern estimate = 0.024, 95% historical distribution = 0.015–0.029; Southern Sierras observed modern estimate = 0.011, historical distribution = 0.009–0.017). Importantly, there was no change in the amount of variation in either species or transect in directions uncorrelated with the selection gradient (figure 3a,c).
Figure 3.
Amount of variation in the direction of selection (β-direction) and in directions uncorrelated with selection (direction≠β) for T. alpinus and T. speciosus in historical and modern periods for both transects. The boxplots correspond to evolvability estimates from 1000 matrices through Monte Carlo sampling for the empirical selection gradient (b,d) and for random selection gradients uncorrelated with the empirical selection gradient (a,c). Asterisk denotes significant changes from historical to modern periods. (Online version in colour.)
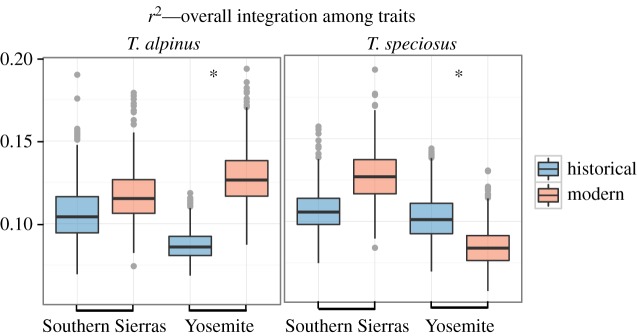
The overall magnitude of integration increased over time for T. alpinus in Yosemite, as the observed r2 index for the modern population (0.108) did not overlap with the historical distribution (0.072–0.106). For T. alpinus from the Southern Sierras, however, the overall magnitude of integration remained unaltered across time (observed modern r2 index = 0.094, 95% historical distribution = 0.079–0.140). We also observed idiosyncratic changes in T. speciosus, as the Yosemite population decreased its overall magnitude of integration (the observed modern value of 0.073 does not overlap the historical 95% distribution of 0.078–0.131), while the Southern Sierras population remained unaltered between the historical and modern periods (observed modern value = 0.118, 95% historical distribution = 0.085–0.133; figure 4).
Figure 4.
Overall magnitude of integration among traits for T. alpinus and T. speciosus in historical and modern periods for both transects. The boxplots correspond to the coefficient of determination of the correlation matrices (r2) estimated from 1000 matrices through Monte Carlo sampling. Asterisk denotes significant changes from historical to modern periods. (Online version in colour.)
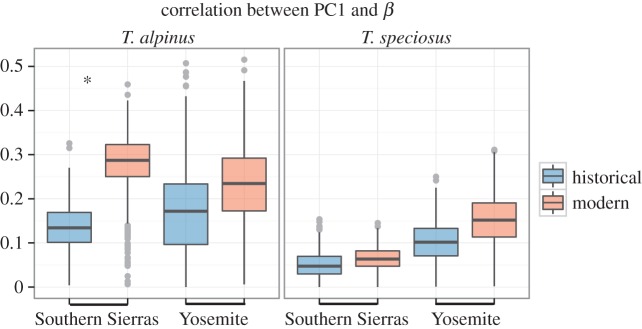
Lastly, the orientation of the axis of greatest variation in relation to the selection gradient, estimated by the correlations between PC1 and the selection gradients, did not change for most populations (figure 5). The only population where we observed an increase in the correlation between PC1 and the selection gradient was T. alpinus from the Southern Sierras, where the modern observed correlation of 0.31 was larger than the historical 95% distribution of 0.027–0.23.
Figure 5.
Orientation of the axis of greatest variation (PC1) in relation to the selection gradient (β) for T. alpinus and T. speciosus in historical and modern periods for both transects. The boxplots correspond to the correlation between both vectors (PC1 and β) estimated from 1000 matrices through Monte Carlo sampling. Asterisk denotes significant changes from historical to modern periods. (Online version in colour.)
4. Discussion
How species adapt to their environment is a fundamental question in biology, one dependent not only upon changes in species' environments (i.e. directional selection) but also on the amount of available phenotypic (co)variation. Our study investigated how these two interact over a period of approximately 100 generations in two co-distributed chipmunk species. We observed that some features of the available phenotypic (co)variation in cranial dimensions changed in response to directional selection, but in unpredictable ways.
Directional selection was a major component in skull evolution for both T. alpinus and T. speciosus, although the strength of selection, estimated as the selection gradient, for T. alpinus was stronger than for T. speciosus. The stronger selection gradient observed in T. alpinus populations supports the hypothesis that this species has been more sensitive to the environmental changes observed across its habitat range compared with T. speciosus [17,18,42]. As a species can become extinct when directional selection is too strong, one might think that T. alpinus is at a higher risk of extinction than is T. speciosus. Indeed, theoretical work has determined a threshold for the ratio of the amount of sustained environmental change, translated as the selection gradient, to the amount of variance available in a population above which the risk of extinction increases [43], and T. alpinus from both transects exhibited a ratio greater than this threshold [44]. However, as discussed below, available phenotypic (co)variation in T. alpinus has been redistributed between the historical and modern sampling periods to match the selection gradient, potentially enhancing the survival of this chipmunk.
The pattern of selection in each population is distinct. In T. alpinus from the Yosemite, most traits were selected to increase (electronic supplementary material, figure S2), which resulted in the observed size increase. On the other hand, T. alpinus from the Southern Sierras presented selection on most traits in the facial region to decrease, in congruence with the observed morphological changes in the population [22]. It is beyond the scope of our study to relate the selection gradients observed with possible selection agents. However, it is interesting to note that in all populations, one trait has been strongly selected to increase (IS–NSL; electronic supplementary material, figure S2). This trait (figure 2) represents the opening of the nasal cavity where the turbinate bones are located, which are responsible for water and heat regulation in mammals [45]. A possible explanation for this convergent change in all populations could involve selective pressure to increase the capacity to better regulate water and heat loss as climate has warmed. In Yosemite, for example, there has been as much as 3.5°C increase in average temperature, especially in the high country occupied by T. alpinus, over the past century [46].
The difference in selection strength between populations of T. alpinus and T. speciosus allowed us to further narrow our predictions about how the available phenotypic (co)variation is expected to change in a directional selection scenario under a model that allows for the evolution of the genotype–phenotype map [12,13,47]. As T. alpinus faced a stronger directional selection regime than did T. speciosus, we hypothesized that any changes in the available phenotypic (co)variation would be more pronounced in T. alpinus.
Our first prediction proposes that the amount of phenotypic variation would increase in the direction of the selective regime but not necessarily in other directions. Indeed, T. alpinus showed both increased variance in the direction of selection but not in other directions and an increase in the total amount of phenotypic variation for both populations examined. Furthermore, this increase was unique to each population because the direction of the net selection gradient is not the same on both T. alpinus populations (electronic supplementary material, figure S2). This was not the case for both populations of T. speciosus. In principle, increased (co)variation in the direction of selection is compatible with a hypothetical increase in the frequency of rare alleles, in which case it could be explained solely by additive genetic variance [10]. However, an increase in variance caused by rare alleles is thought to be transient and likely to most impact traits determined by a small number of alleles [48], which is unlikely to be the case for skull traits, and thus the impact of rare alleles is probably limited. Alternatively, a model accounting for epistatic interactions among traits could lead to this increased variation in the direction of selection, as indicated in figure 1. Under this model, selection acting in a given direction will favour alleles influencing the degree of correlation favoured by selection, which in turn will lead to changes in the amount of variation in this direction.
Our second prediction was that co-selected traits would increase their correlation. Indeed, Yosemite T. alpinus did show an increase in overall phenotypic integration among traits, confirming this prediction. This pattern is also in accordance with an epistatic model, where coordinate selection across multiple traits will lead to tighter correlations among them [6]. On the other hand, T. alpinus from the Southern Sierras did not exhibit an increase in overall correlation among traits. One possible explanation for these different spatial responses has to do with the direction in which selection acted, because selection in the Yosemite population was already in a dimension of relatively high variance, meaning that most traits were involved in the response to selection, while in the Southern Sierra population selection was not along a high variance dimension, as can be appreciated by comparing the evolvability boxplot distributions (figure 3). Therefore, we would expect fewer traits in the Southern Sierra transect to be co-selected, leading to the stability that we observed in the overall phenotypic integration among traits. Furthermore, Yosemite T. speciosus showed the opposite response, decreasing its overall degree of correlation, a pattern that would be expected in a drift scenario [11].
Lastly, we predicted a re-alignment of (co)variation patterns with the selection gradient. Even though this was not the case for three of the populations analysed (figure 5), the Southern Sierra sample of T. alpinus did exhibit increased correlation between PC1 and the selection gradient. Once again, this outcome might be linked to the direction in which selection was operating, because in the latter population, selection was not in a dimension of relatively high variation. On the other hand, for T. alpinus from Yosemite, the selection gradient was in a direction of high (co)variation, and thus a reorientation of the patterns of (co)variation would not have been necessary as sufficient variation was available for selection to act upon. These results are even more interesting if we link them to the evolvability results, as both T. alpinus populations increased the amount of variation along the selection gradient, but only one also re-aligned the matrix structure. Macroevolutionary studies in mammals have shown that the overall phenotypic correlation among traits and the amount of variance are very labile between groups, whereas the orientation of (co)variation is more conserved on a macroevolutionary time scale [24,25,49–52]. As we showed that a re-orientation of phenotypic (co)variation can be easily achieved under a model of sustained directional selection, a possible explanation for the widespread stability of the orientation of (co)variation on a macroevolutionary time scale could be a concordance between the adaptive landscape and patterns of (co)variation. On the other hand, this stasis pattern could be due to a high level of fluctuating selection, leading to the cancellation of directional selection effects on covariance matrices in the long run. An interesting next step would be to investigate additional cases where selection has not acted along an axis of major variance to see if the pattern reported here is robust.
An interesting aspect raised by our analysis is the striking contrast observed between species. While T. alpinus exhibited changes in the three aspects of its phenotypic (co)variation analysed, T. speciosus has remained fairly stable, with change observed only in the overall phenotypic correlation among traits, and in a direction opposite to the one we predicted. It is possible that the observed differences between these two species are related to the discrepant selection strengths they were subject to. Although we did observe a pattern consistent with directional selection for all populations, the selection pressure experienced by T. speciosus might have been weaker than that necessary to produce changes in phenotypic structure.
There are some caveats to our study that should be acknowledged. First, we worked with phenotypic instead of genetic (co)variances because of the difficulties in estimating the latter [51,53]. Although evolutionary quantitative genetics theory is based on genetic (co)variation, we assumed that our phenotypic estimates were appropriate substitutes for their genetic counterparts based on a substantial body of evidence showing that they are structurally similar, at least for morphological traits and particularly for mammals [23–25,54–57]. Furthermore, comparisons among the phenotypic (co)variation in the historical and modern samples of both chipmunk species and the genetic (co)variation of a third distant related rodent species (Calomys expulsus) showed that they were structurally similar, a result that supports our assumptions (for details on the reasoning behind this analysis, see [24]).
Second, we were able to estimate the net selection gradient only between the endpoints of the approximately 100 generations between the historic and modern periods. Although not ideal [58], this is the best approximation we have for the level of directional selection operating on both species between the two sampling periods.
Our study has several strengths. First, we examined well-sampled natural populations separated by multiple generations. Second, the large measured effective population size of our samples allowed us to overcome some of the caveats expected when working in experimental settings, which are frequently hampered by small effective sizes. Indeed, small effective sizes will affect any study designed to analyse the effects of directional selection because of the likelihood of substantial genetic drift. Moreover, the populations we examined were subject to differing selective regimes, in both direction and strength, over time. This allowed us to narrow the predictions and match our expectations to the different populations.
Our study examined a largely neglected aspect of evolutionary dynamics: the interaction between selective regimes and available phenotypic (co)variation. We suggest that the available multidimensional phenotypic (co)variation of a species can evolve quickly in natural populations under relatively strong directional selection, a hypothesis supported by both theoretical and simulation studies [12,13,59]. As species under strong directional selection tend to be more prone to extinction, our study, coupled with previous theoretical, computational and experimental knowledge, highlights one mechanism by which species may enhance their survival in the face of environmental change, namely a rapid reorganization of their available phenotypic (co)variation. This is especially relevant in the light of the ever-growing environmental threats to species survival.
Supplementary Material
Acknowledgements
We thank Joseph Grinnell for his foresight in assembling the specimens and accompanying detailed data for the historical samples we had available, and members of the Grinnell Resurvey Project for the modern samples and data. We thank C. Patton, L. Chow, P. Moore and G. Lefèvre for fieldwork assistance; Miriam Zelditch, A. Kauffmann-Zeh, associate editor Rita Mehta and two anonymous reviewers for helpful comments on previous versions of the manuscript; G. Garcia for kindly providing the G-matrix; and D. Houle for helpful discussions.
Data accessibility
Observed and estimated P-matrices and mean phenotypes for all populations are available on Dryad: http://dx.doi.org/10.5061/dryad.f8q6b [60].
Authors' contributions
A.P.A.A., J.L.P. and G.M. designed the study. A.P.A.A. and G.M. delineated the analyses. A.P.A.A. measured the specimens and analysed the data. A.P.A.A., A.H., G.M. and J.L.P. wrote the manuscript. All authors read, commented on and approved the final version of the manuscript.
Competing interests
The authors declare no competing financial interests.
Funding
Fieldwork was financed by grants from the Yosemite Conservancy, The National Park Service Inventory and Monitoring Program, the National Geographic Society (Grant 8190-07) and the National Science Foundation (DEB 0640859). This work was supported by grants from the Fundação de Amparo à Pesquisa de São Paulo (FAPESP) to A.P.A.A. (2010/52369-0 and 2012/00852-4), A.H. (2012/24937-9) and G.M. (2011/14295-7).
References
- 1.Lande R. 1979. Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33, 402–416. ( 10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- 2.Walsh B, Blows MW. 2009. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 40, 41–59. ( 10.1146/annurev.ecolsys.110308.120232) [DOI] [Google Scholar]
- 3.Schluter D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766–1774. ( 10.2307/2410734) [DOI] [PubMed] [Google Scholar]
- 4.Marroig G, Cheverud JM. 2005. Size as a line of least evolutionary resistance: diet and adaptive morphological radiation in new world monkeys. Evolution 59, 1128–1142. ( 10.1111/j.0014-3820.2005.tb01049.x) [DOI] [PubMed] [Google Scholar]
- 5.Arnold SJ, Bürger R, Hohenlohe PA, Ajie BC, Jones AG. 2008. Understanding the evolution and stability of the G-matrix. Evolution 62, 2451–2461. ( 10.1111/j.1558-5646.2008.00472.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roff DA, Fairbairn DJ. 2012. A test of the hypothesis that correlational selection generates genetic correlations. Evolution 66, 2953–2960. ( 10.1111/j.1558-5646.2012.01656.x) [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson GS, Fowler K, Partridge L. 1990. Resistance of genetic correlation structure to directional selection in Drosophila melanogaster. Evolution 44, 1990–2003. ( 10.2307/2409609) [DOI] [PubMed] [Google Scholar]
- 8.Blows MW, Higgie M. 2003. Genetic constraints on the evolution of mate recognition under natural selection. Am. Nat. 161, 240–253. ( 10.1086/345783) [DOI] [PubMed] [Google Scholar]
- 9.Delph LF, Steven JC, Anderson IA, Herlihy CR, Brodie ED III. 2011. Elimination of a genetic correlation between the sexes via artificial correlational selection. Evolution 65, 2872–2880. ( 10.1111/j.1558-5646.2011.01350.x) [DOI] [PubMed] [Google Scholar]
- 10.Barton NH, Turelli M. 1989. Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 23, 337–370. ( 10.1146/annurev.ge.23.120189.002005) [DOI] [PubMed] [Google Scholar]
- 11.Jones AG, Arnold SJ, Bürger R. 2003. Stability of the G-matrix in a population experiencing pleiotropic mutation, stabilizing selection, and genetic drift. Evolution 57, 1747–1760. ( 10.1111/j.0014-3820.2003.tb00583.x) [DOI] [PubMed] [Google Scholar]
- 12.Jones AG, Bürger R, Arnold SJ. 2014. Epistasis and natural selection shape the mutational architecture of complex traits. Nat. Commun. 5, 3709 ( 10.11038/ncomms4709)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavlicev M, Cheverud JM, Wagner GP. 2010. Evolution of adaptive phenotypic variation patterns by direct selection for evolvability. Proc. R. Soc. B. 278, 1903–1912. ( 10.1098/rspb.2010.2113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlicev M, Norgard EA, Fawcett GL, Cheverud JM. 2011. Evolution of pleiotropy: epistatic interaction pattern supports a mechanistic model underlying variation in genotype–phenotype map. J. Exp. Zool. B 316, 371–385. ( 10.1002/jez.b.21410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TF. 2013. Why epistasis is important for selection and adaptation. Evolution 67, 3501–3511. ( 10.1111/evo.12214) [DOI] [PubMed] [Google Scholar]
- 16.Cheverud JM. 1984. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 110, 155–172. ( 10.1016/S0022-5193(84)80050-8) [DOI] [PubMed] [Google Scholar]
- 17.Moritz C, Patton JL, Conroy CJ, Parra JL, White GC, Beissinger SR. 2008. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science 322, 261–264. ( 10.1126/science.1163428) [DOI] [PubMed] [Google Scholar]
- 18.Rubidge EM, Patton JL, Lim M, Burton AC, Brashares JS, Moritz C. 2012. Climate-induced range contraction drives genetic erosion in an alpine mammal. Nat. Clim. Change 2, 285–288. ( 10.1038/nclimate1415) [DOI] [Google Scholar]
- 19.Rubidge EM, Monahan WB, Parra JL, Cameron SE, Brashares JS. 2011. The role of climate, habitat, and species co-occurrence as drivers of change in small mammal distributions over the past century. Glob. Change Biol. 17, 696–708. ( 10.1111/j.1365-2486.2010.02297.x) [DOI] [Google Scholar]
- 20.Zelditch ML, Swiderski DL, Sheets HD, Fink WL. 2004. Geometric morphometrics for biologists: a primer, 1st edn Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 21.Sikes RS, Gannon WL. 2011. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J. Mammal. 92, 235–253. ( 10.1644/10-MAMM-F-355.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh RE, Assis APA, Patton JL, Marroig G, Dawson TE, Lacey EA. 2016. Morphological and dietary responses of chipmunks to a century of climate change. Glob. Change Biol. 22, 3233–3252. ( 10.1111/gcb.13216) [DOI] [PubMed] [Google Scholar]
- 23.Cheverud JM. 1988. A comparison of genetic and phenotypic correlations. Evolution 42, 958–968. ( 10.2307/2408911) [DOI] [PubMed] [Google Scholar]
- 24.Marroig G, Cheverud JM. 2001. A comparison of phenotypic variation and covariation patterns and the role of phylogeny, ecology, and ontogeny during cranial evolution of new world monkeys. Evolution 55, 2576–2600. ( 10.1111/j.0014-3820.2001.tb00770.x) [DOI] [PubMed] [Google Scholar]
- 25.Porto A, Oliveira FB, Shirai LT, de Conto V, Marroig G. 2009. The evolution of modularity in the mammalian skull I: morphological integration patterns and magnitudes. Evol. Biol. 36, 118–135. ( 10.1007/s11692-008-9038-3) [DOI] [Google Scholar]
- 26.Garcia G, Hingst-Zaher E, Cerqueira R, Marroig G. 2014. Quantitative genetics and modularity in cranial and mandibular morphology of Calomys expulsus. Evol. Biol. 41, 619–636. ( 10.1007/s11692-014-9293-4) [DOI] [Google Scholar]
- 27.Blows MW, Chenoweth SF, Hine E. 2004. Orientation of the genetic variance-covariance matrix and the fitness surface for multiple male sexually selected traits. Am. Nat. 163, 329–340. ( 10.1086/381941) [DOI] [PubMed] [Google Scholar]
- 28.Cheverud JM, Marroig G. 2007. Comparing covariance matrices: random skewers method compared to the common principal components model. Genet. Mol. Biol. 30, 461–469. ( 10.1590/S1415-47572007000300027) [DOI] [Google Scholar]
- 29.Krzanowsky WJ. 1979. Between-groups comparison of principal components. J. Am. Stat. Assoc. 74, 703–707. ( 10.1080/01621459.1979.10481674) [DOI] [Google Scholar]
- 30.Lande R. 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30, 314–334. ( 10.2307/2407703) [DOI] [PubMed] [Google Scholar]
- 31.Rubidge EM, Patton JL, Moritz C. 2014. Diversification of the alpine chipmunk, Tamias alpinus, an alpine endemic of the Sierra Nevada, California. BMC Evol. Biol. 14, 34 ( 10.1186/1471-2148-14-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid F. 2006. Peterson field guide to mammals of North America, 4th edn Boston, MA: Houghton Mifflin Harcourt. [Google Scholar]
- 33.Marroig G, Melo DAR, Garcia G. 2012. Modularity, noise and natural selection. Evolution 66, 1506–1524. ( 10.1111/j.1558-5646.2011.01555.x) [DOI] [PubMed] [Google Scholar]
- 34.Hansen TF, Houle D. 2008. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 21, 1201–1219. ( 10.1111/j.1420-9101.2008.01573.x) [DOI] [PubMed] [Google Scholar]
- 35.Melo D, Garcia G, Hubbe A, Assis AP, Marroig G. 2015. EvolQG—An R package for evolutionary quantitative genetics. F1000Research [Internet]. [cited 2015 Oct 28];4(925). See http://f1000research.com/articles/4-925/v1. [DOI] [PMC free article] [PubMed]
- 36.Manly BFJ. 2006. Randomization, bootstrap and Monte Carlo methods in biology, 3rd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 37.Hohenlohe PA, Arnold SJ. 2008. MIPod: a hypothesis testing framework for microevolutionary inference from patterns of divergence. Am. Nat. 171, 366–385. ( 10.1086/527498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheverud JM, Wagner GP, Dow MM. 1989. Methods for the comparative analysis of variation patterns. Evolution 38, 201–213. ( 10.2307/2992282) [DOI] [Google Scholar]
- 39.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See; http://www.R-project.org/. [Google Scholar]
- 40.Phillips PC, Arnold SJ. 1999. Hierarchical comparison of genetic variance-covariance matrices. I. Using the Flury hierarchy. Evolution 53, 1506–1515. ( 10.2307/2640896) [DOI] [PubMed] [Google Scholar]
- 41.Prôa M, O'Higgins P, Monteiro LR. 2013. Type I error rates for testing genetic drift with phenotypic covariance matrices: a simulation study. Evolution 67, 185–195. ( 10.1111/j.1558-5646.2012.01746.x) [DOI] [PubMed] [Google Scholar]
- 42.Hammond TT, Palme R, Lacey EA. 2015. Contrasting stress responses of two co-occurring chipmunk species (Tamias alpinus and T. speciosus). Gen. Comp. Endocrinol. 211, 114–122. ( 10.1016/j.ygcen.2014.11.013) [DOI] [PubMed] [Google Scholar]
- 43.Burger R, Lynch M. 1995. Evolution and extinction in a changing environment: a quantitative-genetic analysis. Evolution 49, 151–163. ( 10.2307/2410301) [DOI] [PubMed] [Google Scholar]
- 44.Assis APA, Zelditch ML, Patton JL, Marroig G. In preparation. Phenotypic responses of the Sierra Nevada chipmunk community to a century of environmental changes.
- 45.Schmidt-Nielsen K, Hainsworth FR, Murrish DE. 1970. Counter-current heat exchange in the respiratory passages: effect on water and heat balance. Respir. Physiol. 9, 263–276. ( 10.1016/0034-5687(70)90075-7) [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez P. 2016. Climate change trends, vulnerabilities, and carbon in Yosemite National Park, California, USA. Washington, DC: National Park Service, US Department of the Interior. [Google Scholar]
- 47.Wolf JB, Leamy LJ, Routman EJ, Cheverud JM. 2005. Epistatic pleiotropy and the genetic architecture of covariation within early and late-developing skull trait complexes in mice. Genetics 171, 683–694. ( 10.1534/genetics.104.038885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain K, Stephan W. 2015. Response of polygenic traits under stabilizing selection and mutation when loci have unequal effects. GenesGenomesGenetics 5, 1065–1074. ( 10.1534/g3.115.017970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lofsvold D. 1986. Quantitative genetics of morphological differentiation in Peromyscus. I. Tests of the homogeneity of genetic covariance structure among species and subspecies. Evolution 40, 559–573. ( 10.2307/2408577) [DOI] [PubMed] [Google Scholar]
- 50.Steppan SJ. 1997. Phylogenetic analysis of phenotypic covariance structure. I. Contrasting results from matrix correlation and common principal component analysis. Evolution 51, 571–586. ( 10.2307/2411129) [DOI] [PubMed] [Google Scholar]
- 51.Steppan SJ, Phillips PC, Houle D. 2002. Comparative quantitative genetics: evolution of the G matrix. Trends Ecol. Evol. 17, 320–327. ( 10.1016/S0169-5347(02)02505-3) [DOI] [Google Scholar]
- 52.Oliveira FB, Porto A, Marroig G. 2009. Covariance structure in the skull of Catarrhini: a case of pattern stasis and magnitude evolution. J. Hum. Evol. 56, 417–430. ( 10.1016/j.jhevol.2009.01.010) [DOI] [PubMed] [Google Scholar]
- 53.McGuigan K. 2006. Studying phenotypic evolution using multivariate quantitative genetics. Mol. Ecol. 15, 883–896. ( 10.1111/j.1365-294X.2006.02809.x) [DOI] [PubMed] [Google Scholar]
- 54.Cheverud JM. 1996. Quantitative genetic analysis of cranial morphology in the cotton-top (Saguinus oedipus) and saddle-back (S. fuscicollis) tamarins. J. Evol. Biol. 9, 5–42. ( 10.1046/j.1420-9101.1996.9010005.x) [DOI] [Google Scholar]
- 55.Roff DA. 1995. The estimation of genetic correlations from phenotypic correlations: a test of Cheverud's conjecture. Heredity 74, 481–490. ( 10.1038/hdy.1995.68) [DOI] [Google Scholar]
- 56.Reusch T, Blanckenhorn WU. 1998. Quantitative genetics of the dung fly Sepsis cynipsea: Cheverud's conjecture revisited. Heredity 81, 111–119. ( 10.1046/j.1365-2540.1998.00368.x) [DOI] [Google Scholar]
- 57.House CM, Simmons LW. 2005. The evolution of male genitalia: patterns of genetic variation and covariation in the genital sclerites of the dung beetle Onthophagus taurus. J. Evol. Biol. 18, 1281–1292. ( 10.1111/j.1420-9101.2005.00926.x) [DOI] [PubMed] [Google Scholar]
- 58.Jones AG, Arnold SJ, Burger R. 2004. Evolution and stability of the G-matrix on a landscape with a moving optimum. Evolution 58, 1639–1654. ( 10.1111/j.0014-3820.2004.tb00450.x) [DOI] [PubMed] [Google Scholar]
- 59.Melo D, Marroig G. 2015. Directional selection can drive the evolution of modularity in complex traits. Proc. Natl Acad. Sci. USA 112, 470–475. ( 10.1073/pnas.1322632112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Assis APA, Patton JL, Hubbe A, Marroig G. 2016. Data from: Directional selection effects on patterns of phenotypic (co)variation in wild populations. Dryad Digital Repository. ( 10.5061/dryad.f8q6b) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Observed and estimated P-matrices and mean phenotypes for all populations are available on Dryad: http://dx.doi.org/10.5061/dryad.f8q6b [60].