Abstract
Background
Crohn's disease (CD) associated dysbiosis is characterized by a loss of Faecalibacterium prausnitzii, whose culture supernatant exerts an anti-inflammatory effect both in vitro and in vivo. However, the chemical nature of the anti-inflammatory compounds has not yet been determined.
Methods
Peptidomic analysis using mass spectrometry was applied to F. prausnitzii supernatant. Anti-inflammatory effects of identified peptides were tested in vitro directly on intestinal epithelial cell lines and on cell lines transfected with a plasmid construction coding for the candidate protein encompassing these peptides. In vivo, the cDNA of the candidate protein was delivered to the gut by recombinant Lactic Acid Bacteria to prevent DNBS-colitis in mice.
Results
The seven peptides, identified in the F. prausnitzii culture supernatants, derived from a single Microbial Anti-inflammatory Molecule (MAM), a protein of 15 kDa and comprising 53% of nonpolar residues. This last feature prevented the direct characterization of the putative anti-inflammatory activity of MAM-derived peptides. Transfection of MAM cDNA in epithelial cells led to a significant decrease in the activation of the NF-κB pathway with a dose-dependent effect. Finally, the use of a food-grade bacterium, Lactococcus lactis, delivering a plasmid encoding MAM was able to alleviate DNBS-induced colitis in mice.
Conclusion
A 15kDa protein with anti-inflammatory properties is produced by F. prausnitzii, a commensal bacterium involved in CD pathogenesis. This protein is able to inhibit the NF-κB pathway in intestinal epithelial cells and to prevent colitis in an animal model.
Keywords: microbiota, dysbiosis, Crohn’s disease, Faecalibacterium prausnitzii, anti-inflammatory protein
Background
Evidence from immunological, microbiological and genetic studies implicates abnormal host-microbial interactions in the pathogenesis of Crohn’s Disease (CD) [1] [2] [3] [4] [5]. Dysbiosis characterized by a reduction in bacterial biodiversity, lower bacterial population with anti-inflammatory properties and/or an increase in the proportion of bacteria with pro-inflammatory properties, has been observed in CD patients [6]. Several bacterial species have been associated with CD, including members of the Proteobacteria [7] such as adherent-invasive Escherichia coli [8], Campylobacter concisus [9] [10] and enterohepatic Helicobacter [11] [12]. So far, no definitive proof for any specific etiological agent has been highlighted. Additionally, several 16S rRNA sequencing-based studies have reported that members of the Firmicutes phylum were reduced in CD patients [13]. In previous work, we analyzed the composition of the ileal mucosa-associated microbiota of CD patients at the time of surgical resection for active disease, and six months later. Low proportions of Firmicutes, and particularly of Faecalibacterium prausnitzii, were consistently associated with an increased risk of post-operative recurrence of ileal CD [14]. Although controversial [15], we hypothesized that treatment with F. prausnitzii could be an effective strategy to counterbalance dysbiosis and reduce inflammation in CD patients. Following this, we and others demonstrated that F. prausnitzii exhibits anti-inflammatory effects both in vitro (cellular models) and in vivo (TNBS colitis model), associated with secreted metabolites that block NF-κB activation and IL-8 production by intestinal epithelial cells [14] [16] [17-18]. Moreover, gnotobiotic rodent models were previously used to show beneficial effects of F. prausnitzii on intestinal homeostasis and during an acute colitis [19] [20]. Identification of the active molecule(s) involved in this protective effect is of particular interest, since F. prausnitzii remains hardly cultivable due to its extreme oxygen sensitivity. Thus, finding the secreted molecule(s) responsible for this anti-inflammatory effect seems not only a cognitive issue but will also open the field of new therapeutic approach in CD.
The cellular and molecular effects of commensal and probiotic bacteria are now recognized. Some bacteria have been shown to reinforce the intestinal barrier through the production of a soluble factor either by normalizing intestinal permeability of inflamed tissues [21] or inducing the expression of defensins [22] and type 2 zona occludens proteins in tight junctions between epithelial cells [23]. In mice, some commensal bacteria such as segmented filamentous bacteria [24] [25], Bacteroides fragilis and Clostridia members are able to shape gut immune responses [26] [27] [28] [29]. Among the molecules secreted by non-pathogenic bacteria, which are responsible for cellular effects in the host, very few bioactive molecules have been identified [30] [31]. An example of previously identified bioactive molecules can be seen in Lactobacillus rhamnosus GG, which secretes two proteins, p75 and p40, which have been characterized as being able to inhibit epithelial cells apoptosis induced by pro-inflammatory cytokines [32]. Small molecules, less than 10 kDa, from Saccharomyces boulardii and Bacteroides thetaiotaomicron that interact with NF-κB pathway have been detected but remain unidentified [33] [34]. Searching for such bioactive molecules remains challenging as finding these molecules in a cultured supernatant containing thousands of molecules is comparable to searching for a needle in a haystack. Preliminary experiments indicated that various treatments of F. prausnitzii supernatant (heated > 70°c), enzyme digestion (trypsin, lipase, amylase) or filtration (MW<15kDa), do not suppress anti-inflammatory effects (Maubert M.A, unpublished data).
This prompted us to develop a peptidomic analysis of the supernatant in order to identify the presence of potential peptides derived from a unique and original protein from F. prausnitzii, which are able to interact with the NF-κB pathway in epithelial cells and are responsible for the anti-inflammatory effects.
Methods
Faecalibacterium prausnitzii bacterial strain, culture conditions and supernatant preparation
F. prausnitzii (strain A2-165) was grown overnight at 37°C in an anaerobic (90 % N2, 5% CO2 and 5% H2) workstation (Whitley A35 anaerobic workstation) in LyBHI broth, 37 g. L−1 of brain-heart infusion (Difco) and 5 g. L−1 of yeast extract (Conda) at pH 7. The culture supernatant of F. prausnitzii was obtained by centrifugation at 1700 g at 4°C for 20 min.
Faecalibacterium prausnitzii supernatant analysis
Fractionation of the F. prausnitzii culture medium
A solid/liquid extraction of culture medium using Waters Oasis HLB® SPE cartridges was carried out. Fractions were obtained by eluting with 20%, 40% and 80% of acetonitrile (F1, F2, F3 for F. prausnitzii supernatant and F1’, F2’, F3’ for LyBHI culture medium). After freeze-drying, these fractions were tested on epithelial cells through a cellular assay for anti-inflammatory effect (see below). For further investigations in mass spectrometry, F2/F2’ fractions were purified by preparative HPLC using a Waters Symmetry® C8 column (7.8 × 300 mm)
Comparative mass spectrometry analysis of F. prausnitzii culture medium
The bioactive anti-inflammatory fractions from the culture supernatant and LyBHI fractions eluted by SPE were analyzed by MALDI-TOF mass spectrometry (Voyager® DE Pro -AB Sciex).
Peptide identification and synthesis
Identification of peptides by FT-ICR mass spectrometry
The ions of interest were fragmented by a FT-ICR mass spectrometer, equipped with a 7T superconducting magnet (Apex Qe®, Bruker Daltonics). The peptides were then identified by complete de novo sequencing.
Peptides and functionalized peptides synthesis
All protected amino acids were commercially available from Iris Biotech GMBH or Bachem. Peptide syntheses were carried out on a 0.1-mmol scale using an ABI Model 431A peptide synthesizer (Applied BioSystems), starting from the appropriate Wang Tentagel resin, with 10 equiv. of the protected Fmoc-amino acid and HBTU/DIEA for the activation. The crude peptides were purified by HPLC to obtain purity over 97%.
Production of MAM protein in a bacterial heterologous system
The gene encoding the MAM protein was PCR amplified from genomic DNA. PCR product was digested, purified, and cloned in pSTABY plasmid and introduced in Escherichia coli. Protein extraction was carried out and verified by Coomassie blue staining and Western Blot.
Cellular assays for anti-inflammatory effect
Intestinal epithelial cell lines (Caco-2, HT29 and mucus-secreting HT29-MTX), from the European Collection of Cell Cultures (Wiltshire, United Kingdom) and INSERM U505 (Institut des Cordeliers, France), were cultured in supplemented DMEM (PAA) at 37°C in a 5% CO2 incubator. Cells were incubated with different F. prausnitzii supernatant fractions (1/100, 1/500 and 1/1000 dilutions in DMEM medium) or molecules of interest (2 to 10 μM) and, according to the cell lines used, stimulated with various pro-inflammatory cytokines (IL-1β 15 ng. mL−1 or TNFα 10 ng. mL−1). After 6h incubation, cell supernatants were removed for IL-8 analysis. Protein concentrations were determined in cell lysates using Bicinchoninic acid protein assay (Pierce, Rockford, IL) according to the manufacturers instructions. The IL-8 level was determined in duplicate in cell supernatants using ELISA kit DuoSet (R&D systems, Minneapolis, MN). Effect on NF-κB pathway (phosphorylated and total JNK, P38, ERK1/2, IκB, NF-κB) of F. prausnitzii supernatant was tested on Caco-2 cell line using multiplex assay (Bio-Plex®Bio-Rad).
Transfected cell lines and NFκB reporter assay
The cDNA encoding the protein ZP05614546.1 was cloned in 3xFlag (C-term) pCMV vector. Transfection was performed using transfectin™ Lipid reagent (Bio-Rad) in opti-MEM® medium (Gibco, Life Technologies) in different epithelial cells, namely HEK293T, HT29, and TLR4/MD2/CD14 stably-transfected HEK293T (Invivogen). Different concentrations of cDNA, depending on the considered plasmid, were used: 0.8 ng.μL−1 of the plasmid MAM, the plasmid containing cDNA of Carma1 and their empty equivalents, 0.08 ng.μL−1 of the NF-κB reporter plasmid and 0.2 pg.μL−1 of the Renilla luciferase control reporter vector. For HEK293T-TLR4/MD2/CD14, activation of the NF-κB pathway was performed by administration of LPS 100 ng.mL−1 (Sigma-Aldrich). Positive control of inhibition of NF-κB pathway was obtained with SN50 50μM (Enzo Life Sciences). NF-κB reporter assay was carried out, after 24h incubation, using Dual-Luciferase® Reporter Assay system (Promega). In the same manner, MAM activity on STAT3 pathway, activated by colivelin 0.1 nM (Santa Cruz Biotechnology), was evaluated.
Cellular imaging IF in Hela transfected cells
HeLa or HEK293T (both from American Type Culture Collection) cells were grown in DMEM (Invitrogen) with 10% FCS (HyClone) at 37°C and 5% CO2.
Twenty-four hours following transfection, cells were washed and fixed. Cells were stained using appropriate antibodies for 1h. Cells were imaged using a Leica SP5 confocal microscope.
In vivo assays for anti-inflammatory effect of MAM protein
Plasmid construction
MAM encoding plasmid (pILMAM) was created by a fusion between pIL253 [35] cut with PstI (Fermentas) and pCMV including DNA of MAM (see above) cut with Sbf1. Empty equivalent (pILEMPTY) was created using the same method, but with fusion of pIL253 empty pCMV. pILMAM and pILEMPTY were transformed in L. lactis MG1363 as described by Langella et al. (1993) [36]. L. lactis strains were thereafter grown on M17 medium.
DNBS-Induced Colitis
Mice were fed with L. lactis harboring pILMAM or pILEMPTY plasmid. After acclimatization, bacterial suspensions of L. lactis MG1363 (5.109 CFU in 200 μL), were administered to mice (C57BL/6, 6 weeks, male) daily by intragastric gavage from day 7 before until day 3 after induction of colitis. DNBS solution in 30% ethanol was administered intrarectally (at a dose of 100 mg. kg−1 body weight). Inflammation was monitored 72 h after DNBS administration. Mice were weighed before DNBS administration and at killing. Two groups of 8 mice were investigated, fed with L. lactis harboring pILMAM or pILEMPTY. Experiments were performed 3 times.
Inflammation Score Assessment of DNBS Colitis
The colon was removed, dissected free of fat and mesentery, carefully opened, and cleaned. Colon length was measured. Colonic damage and inflammation were assessed blindly according to the Wallace criteria. For histological assessment, a colon sample located in the most inflamed area was fixed in 4% paraformaldehyde acid (Sigma) and embedded in paraffin. Four micrometer sections were stained with hematoxylin/eosin and examined blindly according to the method described by Dieleman et al [37]. Histological scores were carried out according to Ameho criteria.
MAM detection from mice purified intestinal epithelial cells
Small and large intestine were dissected, washed, cut in 1cm pieces and incubated for 20 min at 37°C, 250 rpm in extraction buffer (EDTA 5 mM, DTT 0.145 mg. mL−1, 25 mM HEPES, 50 μM 2-mercaptoethanol (Sigma), RPMI (Lonza)). After incubation remaining solid tissues were withdrawn. Incubation mix containing epithelial cells was centrifuged and pellets lyzed by ultrasound in cold PBS (Lonza). After centrifugation supernatant was kept at −80°C in RNAlater (Qiagen) and RNA was extracted with RNeasy Mini Kit (Qiagen). Retro Transcription was performed using SuperScript® II Reverse Transcriptase (Sigma) with random primers. RT-PCR for MAM mRNA detection was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with the following primers: 5′-ACTCTGGTTGGCAACACCTT-3′ and 5′-CGATCGGGTTGCCCTTAACA-3′. Immunoblotting targeting the FLAG region of the MAM expressed protein (pILMAM construct) was used to detect MAM in purified epithelial cells from gut mucosa of sacrificed mice (on day 3).
Lymphocyte Isolation and Measurement of Cytokine Production
Lymphocyte suspensions were prepared from the Mesenteric Lymph Nodes (MLN) by pressing cells through a 70-mmol/L Falcon nylon cell strainer (BD Biosciences, San Jose, CA). Lymphocytes were counted by flow cytometry (Accuri C6), resuspended in culture medium (RPMI, Lonza with 100 Unit of Streptomicin Penicilin, PAA Laboratories and 10% SVF, Lonza) and activated with coated anti-mouse antibody CD3e and CD28 (eBioscience). Cytokine production was assessed in supernatant by ELISA (IL-17A and INF-γ, Mabtech) after 48h incubation.
MAM detection from dixenic mice
Caecum content of gnotobiotic mice harboring F. prausnitzii (A2-165) and Escherichia coli (K-12 JM105) subjected to 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced acute colitis were gifted by Sylvie Miquel and Muriel Thomas. To obtain dixenic E. coli/F. prausnitzii-diassociated mice, BALB/c germfree mice (obtained from the germfree rodent breeding facilities of Anaxem-Micalis; INRA, Jouy-en-Josas, France) were orally inoculated with a fresh culture of E. coli JM105 (108 to 109 CFU /ml). Following this, the E. coli monoassociated mice were inoculated with F. prausnitzii A2-165 as previously described [19]. One month after the stable implantation of the two strains, mice were intrarectally inoculated with 2,4,6-trinitrobenzenesulfonic acid (TNBS) (50 mg/kg body weight) to induce acute colitis, or with the vehicule (0.9% NaCl–ethanol) to obtain control mice [20]. Inflammation was monitored 48h after TNBS administration. 250 μL of phenol pH4,8 / chloroforme-isoamylalcohol (5 :1) (Sigma) was added to caecum contents and the mix was agitated strongly. After shaking, 12.5 μL of SDS 20% and 25 μL of sodium acetate at 3M pH8 was added. Samples were mixed with fast prep 40 s at power 5 and 20 s at power 5, followed by centrifugation for 15 min at 13000g. Superior phases were mixed with 250 μL of chloroforme-isoamylalcohol and centrifuged. Superior phases were treated with High Pure RNA Isolation Kit (Roche) according the manufacturers instructions. Retro Transcription was performed using SuperScript® II Reverse Transcriptase (Sigma) with the following primers: 5′-ACTCTGGTTGGCAACACCTT-3′ and 5′-CGATCGGGTTGCCCTTAACA-3′. RT-PCR for MAM mRNA detection was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with the following primers: 5′-ACTCTGGTTGGCAACACCTT-3′ and 5′-CGATCGGGTTGCCCTTAACA-3′.
Statistical analysis
Statistical analysis for significant differences was performed using Student’s t-test or Mann-Whitney test when appropriate using JMP® software (Abacus Concepts, Berkeley, CA). Animal experiments were performed three times. Statistical significance was considered when P < 0.05.
Results
Identification of MAM protein
Peptidomic analysis of F. prausnitzii supernatant
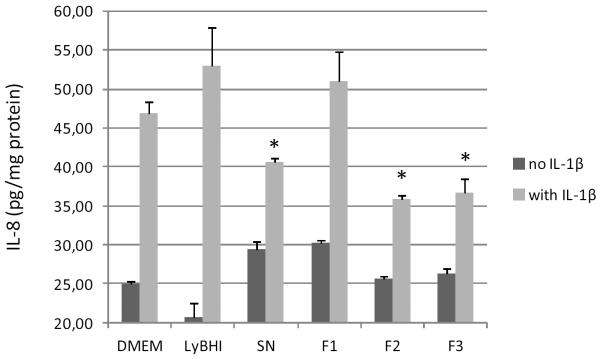
Two fractions (F2 and F3) from F. prausnitzii supernatant exerted inhibitory effects on IL1-β-induced IL-8 secretion in intestinal epithelial Caco2 cells (Figure 1). For comparison, the MALDI-TOF MS spectra generated from the culture medium fractions F2’ and F3’ and F. prausnitzii culture supernatant fractions F2 and F3 were also analysed.
Figure 1.
IL-8 response of human Caco2 cells to stimulation in DMEM medium with IL-1β, F. prausnitzii supernatant (SN) or LyBHI medium, and F. prausnitzii supernatant fractions F1 (20% of acetonitrile), F2 (40% acetonitrile) and F3 (80% acetonitrile). The values are expressed as the mean ± SEM in pg IL-8/mg protein. * p < 0.05 (compared to DMEM control with IL-1β in three experiments).
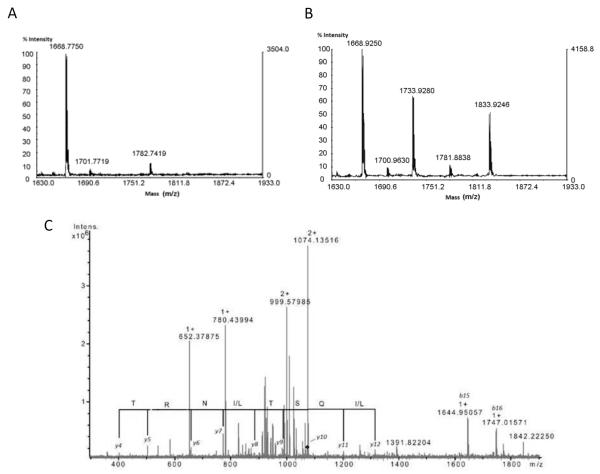
Analysis of F2’ and F3’ vs F2 and F3 using MALDI-TOF MS allowed us to detect seven ions ([M+H]+ m/z 1733.93, 1832.92, 1946.97, 2047.95, 2146.94, 2579.72 and 4601.06) in F2 and F3 fractions only (Figure 2 A-B and Supplementary files S1, S2, S3). The seven ions of interest were fragmented by an ESI FT-ICR mass spectrometer, and identified by de novo sequencing.
Figure 2.
(A) MALDI TOF MS spectra (zoom scan for the m/z range 1630–1933) generated from F2′ (A) and F2 fractions (B) showing two ions at m/z 1733.93, 1833.92, only in (B) MS spectrum. (C) FTICR CID spectrum of the [M + 2H]2+ m/z 1073.61 precursor ion (corresponding to the ion of interest [M + H]+ m/z 2146.94). De novo sequencing generated a probable partial amino acid sequence from singly charged ions. The accuracy of mass determination made it possible to attribute this series unambiguously and to differentiate between the isobaric amino acids K and Q.
To illustrate the identification of compounds, one ion of interest (here: m/z 2146.94) corresponding to [M+2H]2+ m/z 1073.61 was fragmented by collision induced dissociation (CID) (Figure 2 C). From various CID spectra (zoom, deconvoluted, product ion at [M+2H]2+ m/z 831.96), the complete sequence was finally obtained unambiguously: VT[I/L]VGNTF[I/L]QST[I/L]NRT[I/L]GV[I/L] (supplementary files S4, S5 and S6). The isomeric amino acids in square brackets [I or L] were undefined. The same procedure was used to determine the sequence of the 6 other peptides.
Identification of a protein and its derived peptides from F. prausnitzii
The in silico analysis of F. prausnitzii genome (Blast and ProteinInfo software with NCBInr) first allowed to complete the sequence of the peptide 4600.28 Da, removing any ambiguity between leucine and isoleucine, and then to demonstrate that the seven isolated peptides all derived from the F. prausnitzii protein ZP 05614546.1.
Sequences of protein ZP 05614546.1, called MAM (Microbial Anti-inflammatory Molecule or named after its discoverer Marie-Anne Maubert), and its derived peptides are presented in table 1.
Table 1.
Sequences of the 7 peptides identified in F. prausnitzii supernatant and sequence of the MAM protein.
| Pep1 | 1732.95 Da | GNTFLQSTINRTIGVL |
| Pep2 | 1832.02 Da | VGNTFLQSTINRTIGVL |
| Pep3 | 1945.10 Da | LVGNTFLQSTINRTIGVL |
| Pep4 | 2046.15 Da | TLVGNTFLQSTINRTIGVL |
| Pep5 | 2145.26 Da | VTLVGNTFLQSTINRTIGVL |
| Pep6 | 4600.28 Da | FSGNTTWKEVGNIGKNLFGTNVKGNPIEKNNFGDYAMNALGIA |
| Pep7 | 2578.40 Da | AAVYNLGVA PTKNTVKETE VKFTV |
|
Protein
ZP 05614546.1 |
MMMPANYSVIAENEMTYVNGGANFIDAIGAVTAPIWTLDNVKTFNTNIVTLVGNTFLQSTINRTIG VLFSGNTTWKEVGNIGKNLFGTNVKGNPIEKNNFGDYAMNALGIAAAVYNLGVA PTKNTVKETEVKFTV |
|
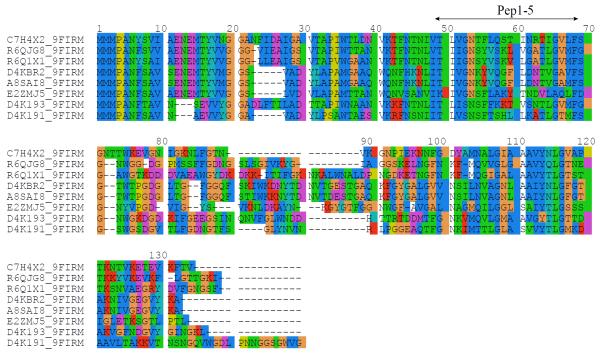
To investigate structure/function relationships of MAM protein, a sequence search was performed using the program BLAST. The BLAST search recovered several sequences that are homologous to MAM, all belonging to F. prausnitzii strains (Figure 3), with statistically significant, low e-values. The sequence alignment of MAM to its other F. prausnitzii counterparts (Figure 3) revealed high sequence identity (> 34%), with strong sequence conservation in the 1–71 N-terminal part and in the 105–118 segment (MAM numbering). The characterized peptides Pep1-5 corresponded to a region of the protein, which is conserved in F. prausnitzii orthologs.
Figure 3.
Sequence alignments between MAM protein and 7 other homologous proteins of F. prausnitzii. Sequences were identified using a BLAST search and aligned using ClustalW2 program. The protein identifiers correspond to the following F. prausnitzii strains: C7H4X2, A2-165; R6QJG8 and R6Q1X1, sp. CAG:82; D4KBR2, SL3/3; A8SAI8, M21/2; E2ZMJ5, KLE1255; D4K191 and D4K193, L2-6. The region corresponding to identified peptides Pep1-5 is shown by an arrow.
A sequence analysis based on ExPASy bioinformatics tools indicated that MAM does not contain regions of low sequence complexity and intrinsic disorder, and should therefore adopt a compact, globular fold. Although the sequence exhibited two highly hydrophobic regions around residues 23–38 and 103–121, MAM was not predicted as a transmembrane protein using DAS-TM, HMMTOP or PHDhtm programs.
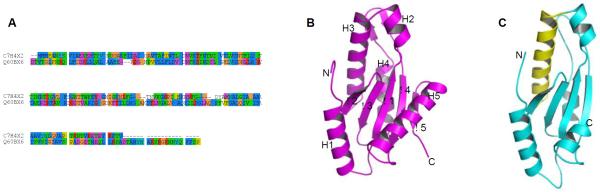
In order to get further structural information on MAM protein, the results of the BLAST search were examined using the hits displaying higher e-values. Some sequence similarity was observed with proteins belonging to other Firmicutes (Roseburia intestinalis) having a putative GGDEF domain. A sequence alignment of MAM with this GGDEF domain yielded 15% sequence identity and 38% sequence similarity over the whole sequence, both proteins having comparable sizes (Figure 4A). Based on these results, a homology model was tentatively built using Modeller program [38] (Figure 4C). The inspection of the model indicated that the spatial distribution of polar and hydrophobic residues was compatible with such a globular fold. GGDEF domains share a α/β topology typically composed of a five-stranded β-sheet core flanked by five α-helices (Figure 4B). The alignment showed that the short C-terminal β-strand was absent in the MAM model, as also observed in some GGDEF domains [39]. Interestingly, the region encompassing the characterized peptides Pep1-5 corresponds to helix H3 and is exposed at the surface of the model (Figure 4C).
Figure 4.
Homology model of MAM protein based on a GGDEF domain template. (A) Sequence alignment of MAM and GGDEF protein from M. capsulatus (Q60BX6); (B) X-ray structure of template protein (PDB entry 3ICL) showing the secondary structure elements; (C) three-dimensional model of MAM calculated with Modeller. The rms deviation on Cα positions of aligned residues is 1.1 Å. The region 49-68 corresponding to the identified peptides Pep1-5 is coloured in yellow.
Anti-inflammatory effect of the MAM protein
Direct anti-inflammatory effect of MAM protein and its derived peptides
MAM protein produced through heterologous expression system (e.g. E. coli) was systematically found in hydrophobic fractions, thus compromising any direct testing of anti-inflammatory effects. To circumvent this problem, MAM derived peptides were functionalized (pegylation, CPP coupling, modification of peptidic bond…), and/or mixed with amphiphilic partners to optimize their solubility. However, none of these compounds showed significant and reproducible anti-inflammatory effect in cellular assays. We thus decided to change our strategy. Previous experiments suggested that F. prausnitzii supernatant acts distally on NF-κB pathway in epithelial cells (data not shown). We reasoned that MAM or its derived peptides could act on intracellular targets. We thus assessed the effect of direct overexpression of MAM protein in epithelial cells.
Anti-inflammatory effect of MAM protein after transfection in eukaryotic epithelial cells
A full-length Flag-MAM cDNA was transfected in HEK293T and HT29 cells and the effect of the expressed protein was tested on NF-κB pathway, since preliminary experiments, using a multiplex assay (see Material and Methods), suggested an effect on the distal part of this pathway (data not shown).
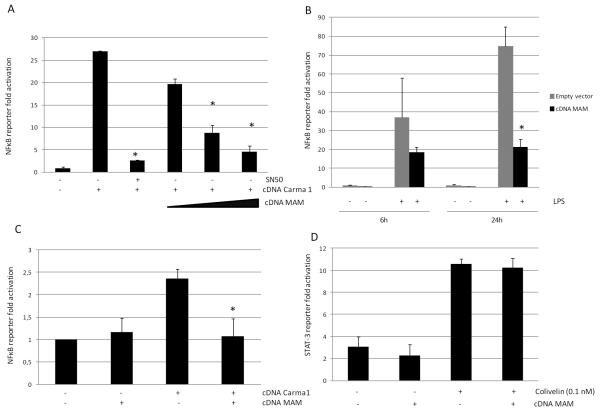
Expression of MAM was confirmed in HEK293T, MD2-TLR4-CD14 HEK293T and HT29 cells by Western blot analysis with anti-Flag antibody (supplementary file S7). Using a NF-κB reporter system in these human epithelial cells, we showed that expression of MAM protein was able to block NF-κB activation induced by Carma-1 or LPS in a dose-dependent manner (Figure 5 A-C). The specificity of the NF-κB activation in our system was validated by the inhibitor effect of SN50, a well known NF-κB inhibitor. Moreover, the specificity of the effect of MAM on the NF-κB pathway was highlighted by the lack of effect on STAT3 pathway in the same eukaryotic system (Figure 5 D).
Figure 5.
Decrease in activation of the NF-κB pathway after transfection of MAM protein in different epithelial cells: in HEK293T in a dose-dependent manner (A), in TLR4/MD2/CD14 stably transfected HEK293T stimulated by LPS 100 ng. mL−1 (B) and in intestinal cells HT29 (C). SN50 (50 μM) was used as positive control of NF-κB pathway inhibition in HEK293T. No activity of transfected MAM protein was observed on the STAT3 pathway activated by colivelin 0.1 nM (D). * p < 0.05 (compared to activation control).
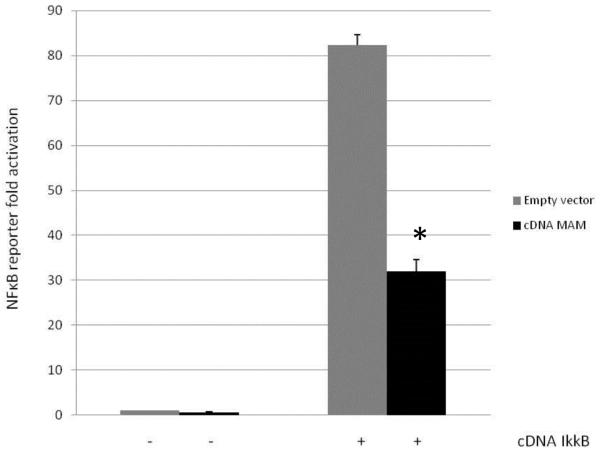
When NF-κB activation was performed using IκκB (distal part of the NF-κB pathway), MAM transfected cells still exhibited a reduced NF-κB activity (Figure 6).
Figure 6.
Decrease in activation of the NF-κB pathway after co-transfection of MAM protein and IκκB in HEK293T ( * p< 0.05)
Subcellular location of MAM protein in HeLa and in HEK293T cells
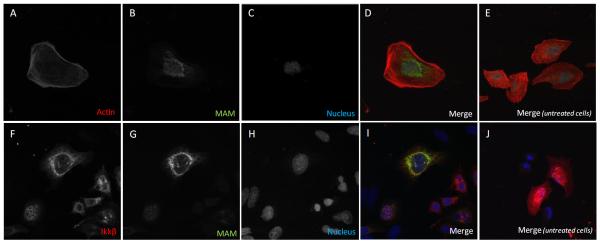
In agreement with this last result, MAM protein was localized around the cell nucleus in MAM-Flag transfected HeLa cells (Figure 7 A-D) and co-localized with IκκB in transfected HEK293T (Figure 7 E-H).
Figure 7.
Subcellular location of MAM protein in HeLa cells (A, B, C, D) and co-localisation of MAM protein with Iκκβ in HEK293T cells (F, G, H, I). E and J labels correspond to staining controls in untreated cells.
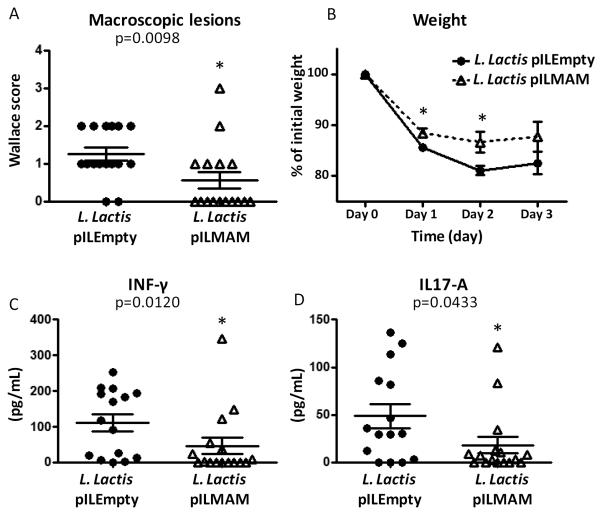
MAM effect in DNBS-induced colitis in mice
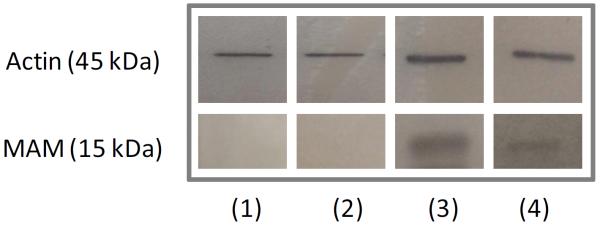
To mimic more adequately the in vivo situation, a food-grade bacterium, Lactoccocus lactis was modified to produce the cDNA coding for MAM, resulting in a kind of “in vivo transfection”. This allowed delivery of MAM cDNA directly by bacteria in mice subjected to DNBS treatment. Although no heterologous protein is produced by the bacterial strain, we verified that there is no difference in the growth between L. lactis pILEMPTY and pILMAM (supplementary file S8). To confirm that our strategy effectively leads to in vivo production of MAM, we performed western blot analysis on small bowel and colon epithelial cells of mice fed with L. lactis (Figure 8). Expression of MAM at the mRNA level was also confirmed (supplementary file S9). Weight loss was significantly lesser in mice fed with L. lactis pILMAM at day 1 and 2 (Figure 9B). After dissection of the gut, the Wallace score was also significantly lower in pILMAM (Figure 9A). We subsequently isolated lymphocytes from MLN of both mice groups and assessed their cytokine production following stimulation with anti-CD3 and anti-CD28 antibodies. The two pro-inflammatory cytokines IL-17A and INF-γ were produced in lower amounts in cells isolated from L. lactis pILMAM fed mice (Figure 9C-D). No histological differences were observed between mice fed with L. lactis pILMAM and L. lactis pILEMPTY (supplementary file S10). Taken together, these results show that L. lactis pILMAM exhibits significant protective effect in DNBS-induced colitis.
Figure 8.
Western blot with anti-flag antibody for MAM protein detection in small intestine enterocytes (1) and large intestine enterocytes (2) of mice fed with pILEmpty L. lactis and in small intestine enterocytes (3) and large intestine enterocytes (4) of mice fed with pILMAM L. lactis
Figure 9.
Effects of intragastric administration of L. lactis bacterial suspension (pILMAM or Empty equivalent) on TNBS-induced colitis in C57BL/6 mice considering Wallace score (A), weight after induction of colitis (B) and quantification using ELISA of IL-17A (C) and INF-γ (D) in colons obtained 48 h after DNBS colitis induction (in pg/mL of total proteins). The values are expressed as the mean ± SEM (*p < 0.05 compared to L. lactis pILEmpty controls).
MAM detection from dixenic mice
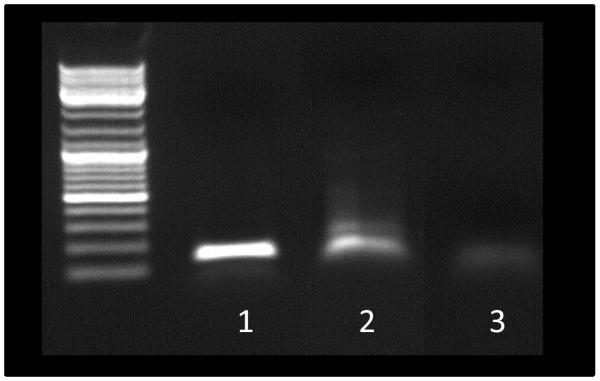
MAM mRNA was detected in vivo for the first time in our model of dixenic mice E. coli / F. prausnitzii (Figure 10). Sequencing confirmed that the detected mRNA was MAM encoding mRNA.
Figure 10.
PCR detection of MAM gene on RNA extract from feces of dixenic mice colonized with Faecalibacterium prausnitzii and Escherichia coli (1), on pILMAM plasmid (2, positive control) and on RNA extract of same sample (1) traited by RNAse before RT-PCR (3, negative control).
Discussion
Herein, we were able to identify a unique and original 15 kDa protein (ZP05614546.1), called MAM, with anti-inflammatory properties produced by F. prausnitzii. This protein and/or derived peptides involved in the anti-inflammatory effect are able to inhibit the NF-κB pathway in several intestinal epithelial cells lines. Interestingly, L. lactis delivering a MAM encoding plasmid was also able to prevent DNBS-colitis in mice.
This discovery is in line with a series of work performed by our group. Indeed, for many years, we and others have described CD-associated dysbiosis and observed a strong restriction in the biodiversity of the Firmicutes (one of the two major bacterial phyla in the normal gut microbiota [40]) and a quantitative decrease in bacteria belonging to Clostridium leptum group [6] [14] [41]. Faecalibacterium prausnitzii is a dominant species of this group. We have gone a step further, looking at the impact of these changes in gut microbiome on CD patient outcomes and inflammation pathways. In this setting, we were able to show that low level of F. prausnitzii in ileal mucosa of CD patients was predictive of postoperative recurrence [14] and again, more recently, we observed that low level in F. prausnitzii in faeces was predictive of CD relapse in patients in remission [42]. Moreover, this bacterium exerts anti-inflammatory properties in vitro and in vivo [14]. In this previous work the anti-inflammatory effects of F. prausnitzii were mediated by secreted molecules in the culture supernatant. These bioactive molecules were highly suspected to be protein-derived peptides from F. prausnitzii. Thus, it remained a major task to isolate these molecules, to characterize and to analyze their immunomodulatory effects.
We met several difficulties in achieving these goals. First, finding bioactive molecules in F. prausnitzii supernatant was challenging. We made it possible by using a bio-guided strategy generated from the culture medium fractions and differential MALDI-TOF MS analysis. Thanks to ultra-high resolution of the FT-ICR, the complete sequences of the peptides were finally obtained unambiguously by de novo sequencing. To note, the easy coupling of mass spectrometry with separation techniques, such as liquid chromatography, and its sensitivity makes it a method of choice for detection of various molecules present at trace levels in complex mixtures, such as a bacterial supernatant. Surprisingly, after in silico analysis, the seven peptides all originate from the same unknown protein ZP05614546.1 from F. prausnitzii. Some sequence similarities were observed with proteins belonging to other Firmicutes (Roseburia intestinalis) having a putative GGDEF domain. These proteins are described in the UnitProtKB database as putative since they have not yet been characterized and have unknown functions. Strikingly, MAM protein does not resemble any other known protein from Gram-positive or Gram-negative bacteria. These GGDEF domains are typically found in enzymes endowed with diguanylate cyclase activity and involved in the biosynthesis of cyclic di-GMP, a widespread signaling molecule in bacteria [43]. Accordingly, a BLAST search against the PDB database also revealed a hit with a protein from Methylococcus capsulatus containing a GGDEF domain. Although MAM protein might fold as a GGDEF domain, it lacks the critical catalytic residues (including the GGDEF sequence) that are involved in diguanylate cyclase activity. Furthermore, other residues involved in nucleotide binding such as the RXXD motif [39] are also missing. It is therefore very unlikely that MAM has any catalytic or regulatory functions involved in cyclic di-GMP signaling. Thus, no clear function could be assigned to this protein.
High prevalence of nonpolar residues prevented us to provide direct characterization of the putative anti-inflammatory activity. To overcome this technical lock, we applied a molecular approach by transfecting MAM cDNA in a series of epithelial cell lines and showing a significant decrease in the activation of the NF-κB pathway with a dose-dependent effect. Finally, a L. lactis strain delivering a MAM encoding plasmid was able to prevent DNBS colitis in mice. These results demonstrate that MAM protein supports, at least partly, the anti-inflammatory effect exerted by F. prausnitzii.
One can hypothesized that F. prausnitzii could exert its anti-inflammatory effect on host cells through many molecular patterns. In fact, when performing preliminary experiments on F. prausnitzii supernatant, we observed that anti-inflammatory effect was not abrogated by heat (above 70°c), enzyme digestion (trypsin, lipase, amylase) or MW filtration (below 15kDa) (Maubert M. A., unpublished data). This indicates that various metabolites other than MAM could contribute to this effect. Furthermore, it has been shown recently that F. prausnitzii is a major inducer of Clostridium-specific IL-10-secreting regulatory T cell subset present in the human colonic lamina propria and blood [44]. This indicates that several cell types are targeted through interactions between F. prausnitzii and the host to maintain and shape gut barrier immune function. In this setting, bacterial metabolites such as the abundant microbial-derived short-chain fatty acids (SCFA) have been identified to induce signaling effects regulating colonic regulatory T cell homeostasis [30]. To note human colonic butyrate producers are Gram-positive Firmicutes from which the two most abundant groups are related to Roseburia spp. and to F. prausnitzii. In fact, the intestinal tract of mammals is home to 1013 to 1014 commensal bacteria composed of hundreds of species of which certain are able to play specific roles in determining the immunological balance in host [27 45]. Atarashi and colleagues have recently demonstrated that Clostridial species induce regulatory T cells through SCFA pathway stimulating epithelial cells to produce TGFβ, contributing to regulatory T cells differentiation and expansion [46]. Other species-specific bacterial molecules, such as B. fragilis-derived polysaccharide A, have previously been demonstrated to have immunomodulatory functions [47] [48] [49]. Another study demonstrated that a mixture of probiotic strains, including Lactobacillus and Bifidobacterium, enhanced the production of TGF-β and IDO from dendritic cells and consequently induced Treg cells [50]. Thus, our work uncovering a new anti-inflammatory protein from F. prausnitzii, a major microbiota species of gut microbiota, reinforces the role of a metabolic interface of promiscuous bacterial molecules on gut mucosa physiology. However, a metabolomic approach based on gnotobiotic model permitted to propose other hypothesis concerning F. prausnitzii anti-inflammatory properties [20]. Authors identified various metabolites, particularly salicylic acid, specifically associated with the presence of F. prausnitzii. This study, associated to our results, confirm the complex action mechanisms of F. prausnitzii to limit the inflammatory process.
To conclude, our work opens new lines of evidence that the impact of CD-associated dysbiosis could, at term, change our practice and lead to novel strategies to prevent and treat inflammatory bowel diseases. Although discovering an anti-inflammatory molecule constitutes a first step towards new anti-inflammatory drugs in CD, MAM could represent a targeted biomarker for CD, regarding the value of F. prausnitzii for predicting CD relapse. In these perspectives, there is further need for deciphering the role of MAM in the gut ecosystem. In particular, emphasis should be placed on finding the function of MAM and the mechanism of its production within the bacteria before considering it a target for CD management.
Supplementary Material
Summary Box.
What is already known about this subject?
Crohn’s disease (CD) associated dysbiosis is characterized by low proportion of Faecalibacterium prausnitzii in fecal and mucosa-associated microbiome.
Loss of F. prausnitzii is predictive of CD relapse after surgery or in patients treated with immunosupressants.
F. prausnitzii exhibits anti-inflammatory effects in vitro and in vivo by secreted metabolites that block NF-κB activation.
What are the new findings?
F. prausnitzii produces bioactive peptides derived from a single 15 kDa protein (ZP05614546.1) of unknown function named Microbial Anti-inflammatory Molecule (MAM).
MAM expression in epithelial cells lines is able to block the NF-κB pathway.
Lactococcus lactis harboring a MAM-cDNA encoding plasmid is able to alleviate DNBS-colitis in mice
How might this impact on clinical practice in the foreseeable future?
Discovery of anti-inflammatory molecules from a commensal bacterium such as F. prausnitzii constitutes the first step towards new anti-inflammatory drugs in CD.
Knowing the predictive value of F. prausnitzii for CD relapse, MAM could represent a targeted biomarker for CD.
Acknowledgments
We thank Association François Aupetit (2009) and Agence Nationale de la Recherche (Mi2 2010) for funding this work. We also thank Lucette Groisard, Loïc Brot, Chantal Bridonneau and Isabelle Naas for technical assistance and Joëlle Masliah and Muriel Thomas for their scientific support. Marie-Anne Maubert thanks Jean Claude Tabet and the TGE High Field FT-ICR (CNRS) for providing the access to the FT-ICR mass spectrometer. Caecum content of Gnotobiotic mice harboring F. prausnitzii (A2-165) and Escherichia coli (K-12 JM105) subjected to 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced acute colitis have been gently given by Sylvie Miquel and Muriel Thomas. This last study was a part of the FPARIS collaborative project selected and supported by the Vitagora Competitive Cluster and funded by the French FUI (Fond Unique Interministériel; FUI no. F1010012D), the FEDER (Fonds Européen de Développement Régional; Bourgogne no. 34606), the Burgundy Region, the Conseil Général 21, and the Grand Dijon. This work was also supported by Merck Médication Familiale (Dijon, France) and Biovitis (Saint Étienne de Chomeil, France).
Abbreviations
- IBD
inflammatory bowel disease
- CD
Crohn’s disease
- MALDI-TOF
Matrix-Assisted Laser Desorption/Ionisation - Time Of Flight
- FT-ICR
Fourier Transform - Ion Cyclotron Resonance
- HPLC
High-performance liquid chromatography
- DNBS
Dinitrobenzene Sulfonic Acid
- TNBS
Trinitrobenzene Sulfonic acid
Footnotes
Philippe Seksik declares consulting fees from Abbvie, MSD and Biocodex. Harry Sokol received consulting fee from Danone and Enterome and lecture fee from Abbvie and Biocodex. For the other authors, they declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.MacDermott RP. Alterations of the mucosal immune system in inflammatory bowel disease. J Gastroenterol. 1996;31:907–16. doi: 10.1007/BF02358624. [DOI] [PubMed] [Google Scholar]
- 2.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 3.Allez M, Mayer L. Regulatory T cells: peace keepers in the gut. Inflamm Bowel Dis. 2004;10:666–76. doi: 10.1097/00054725-200409000-00027. [DOI] [PubMed] [Google Scholar]
- 4.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, Seksik P, Rigottier-Gois L, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–11. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhya I, Hansen R, El-Omar EM, et al. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–30. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 8.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 9.Man SM, Zhang L, Day AS, et al. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn's disease. Inflamm Bowel Dis. 2010;16:1008–16. doi: 10.1002/ibd.21157. [DOI] [PubMed] [Google Scholar]
- 10.Kaakoush NO, Mitchell HM. Campylobacter concisus - A new player in intestinal disease. Front Cell Infect Microbiol. 2012;2:4. doi: 10.3389/fcimb.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohr UR, Glasbrenner B, Primus A, et al. Identification of enterohepatic Helicobacter species in patients suffering from inflammatory bowel disease. J Clin Microbiol. 2004;42:2766–8. doi: 10.1128/JCM.42.6.2766-2768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Man SM, Zhang L, Day AS, et al. Detection of enterohepatic and gastric helicobacter species in fecal specimens of children with Crohn's disease. Helicobacter. 2008;13:234–8. doi: 10.1111/j.1523-5378.2008.00607.x. [DOI] [PubMed] [Google Scholar]
- 13.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 14.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen R, Russell RK, Reiff C, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol. 2012;107:1913–22. doi: 10.1038/ajg.2012.335. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Qiu X, Zhang H, et al. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS One. 2014;9:e109146. doi: 10.1371/journal.pone.0109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miquel S, Martin R, Rossi O, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–61. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Martin R, Chain F, Miquel S, et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis. 2014;20:417–30. doi: 10.1097/01.MIB.0000440815.76627.64. [DOI] [PubMed] [Google Scholar]
- 19.Wrzosek L, Miquel S, Noordine ML, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miquel S, Leclerc M, Martin R, et al. Identification of Metabolic Signatures Linked to Anti-Inflammatory Effects of Faecalibacterium prausnitzii. MBio. 2015;6 doi: 10.1128/mBio.00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mennigen R, Nolte K, Rijcken E, et al. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am J Physiol-Gastr L. 2009;296:G1140–G49. doi: 10.1152/ajpgi.90534.2008. [DOI] [PubMed] [Google Scholar]
- 22.Vora P, Youdim A, Thomas LS, et al. beta-Defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. Journal of Immunology. 2004;173:5398–405. doi: 10.4049/jimmunol.173.9.5398. [DOI] [PubMed] [Google Scholar]
- 23.Zyrek AA, Cichon C, Helms S, et al. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 2007;9:804–16. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- 24.Lecuyer E, Rakotobe S, Lengline-Garnier H, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–20. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Host interactions with Segmented Filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin Immunol. 2013;25:342–51. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouskra D, Brezillon C, Berard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–10. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 32.Yan F, Cao H, Cover TL, et al. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–75. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sougioultzis S, Simeonidis S, Bhaskar KR, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-kappaB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006;343:69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 34.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 35.Holo H, Nes IF. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989;55:3119–23. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langella P, Le Loir Y, Ehrlich SD, et al. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J Bacteriol. 1993;175:5806–13. doi: 10.1128/jb.175.18.5806-5813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieleman LA, Palmen MJHJ, Akol H, et al. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clinical and Experimental Immunology. 1998;114:385–91. doi: 10.1046/j.1365-2249.1998.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eswar N, Webb B, Marti-Renom MA, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. Chapter 2:Unit 2 9. [DOI] [PubMed] [Google Scholar]
- 39.Whitney JC, Colvin KM, Marmont LS, et al. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem. 2012;287:23582–93. doi: 10.1074/jbc.M112.375378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajca S, Grondin V, Louis E, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn's disease. Inflamm Bowel Dis. 2014;20:978–86. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 43.Ryan RP. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology. 2013;159:1286–97. doi: 10.1099/mic.0.068189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarrabayrouse G, Bossard C, Chauvin JM, et al. CD4CD8alphaalpha Lymphocytes, A Novel Human Regulatory T Cell Subset Induced by Colonic Bacteria and Deficient in Patients with Inflammatory Bowel Disease. PLoS Biol. 2014;12:e1001833. doi: 10.1371/journal.pbio.1001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 47.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 48.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 50.Kwon HK, Lee CG, So JS, et al. Generation of regulatory dendritic cells and CD4(+)Foxp3(+) T cells by probiotics administration suppresses immune disorders. P Natl Acad Sci USA. 2010;107:2159–64. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.