Abstract
Sorghum is a highly efficient C4 crop with potential to mitigate challenges associated with food, feed and fuel. TCP proteins are of particular interest for crop improvement programs due to their well-demonstrated roles in crop domestication and shaping plant architecture thereby, affecting agronomic traits. We identified 20 TCP genes from Sorghum. Except SbTCP8, all are either intronless or contain introns in the untranslated regions. Comparative phylogenetic analysis of Arabidopsis, rice, Brachypodium and Sorghum TCP proteins revealed two distinct classes categorized into ten sub-clades. Sub-clade F is dicot-specific, whereas A2, G1 and I1 groups only contained genes from grasses. Sub-clade B was missing in Sorghum, whereas group A1 was missing in rice indicating species-specific divergence of TCP proteins. TCP proteins of Sorghum are enriched in disorder promoting residues with class I containing higher percent disorder than class II proteins. Seven pairs of paralogous TCP genes were identified from Sorghum, five of which seem to predate Rice-Sorghum divergence. All of them have diverged in their expression. Based on the expression and orthology analysis, five Sorghum genes have been shortlisted for further investigation for their roles in regulating plant morphology. Whereas, three genes have been identified as candidates for engineering abiotic stress tolerance.
Manipulation of plant architecture has huge potential to improve the agronomic value of crop plants and has extensively been applied in domestication of grasses1. The molecular genetic characterization of the candidate genes, in the model systems rice and Arabidopsis, highlighted transcription factors as major players in shaping the plant morphology and architecture2,3.
With the discovery of the founding members, TB1 (Teosinte Branched 1) from maize, Cycloidea from snapdragon and PCFs (Proliferation Cell Factors) from rice, in the late 1990s, members of plant-specific TCP family of transcription factors, in particular, have been shown to play key roles in evolution of plant form and structure4. TB1 controls vegetative axillary meristem outgrowth and played major role in domestication of modern maize from the ancestral teosinte5. Cycloidea of snapdragon regulates floral asymmetry, whereas, rice PCFs have been shown to promote cell proliferation and organ growth6,7.
Till date, more than 30 TCP genes have been characterized in plants, using reverse genetics approaches, to play key roles in regulating leaf size and curvature, lateral organ development, internode elongation, circadian rhythms, floral transition, floral symmetry, pollen development and senescence8,9,10,11,12. Recent findings in Arabidopsis, rice and tomato unleashed the role of TCP genes in regulating plant immunity and abiotic stress tolerance as well13,14. However, bulk of the characterization of TCP gene functions has been carried out in the model plant species Arabidopsis11. Apparently, with the huge diversity in plant morphology and forms in monocots and dicots, the complete picture of TCP gene function and regulation cannot be obtained based on the studies done in Arabidopsis alone. Therefore, identification and functional characterization of these genes in the plants of agronomic importance is required for harnessing their full potential in crop improvement programs.
Sorghum has recently emerged as an excellent model system for C4 grasses, especially bioenergy crops15. The multipurpose varieties of Sorghum have the potential to mitigate some of the challenges associated with food, feed, forage and fuel security. Because of wide range of genotypic and phenotypic diversity; identification and characterization of key genes regulating plant morphology and architecture in Sorghum is required to gain insights into the genetic basis of morphological agronomic traits.
In this study, we have leveraged publicly available genomic and transcriptomic resources to interpret TCP gene functions in Sorghum. Analysis of predicted gene structures, protein attributes and gene expression in developmental stages and abiotic stress-treated samples revealed targets for functional genomic studies. Furthermore, comparative phylogenomic analysis of TCP family transcription factors in Arabidopsis and three poaceae species, including rice, Brachypodium and Sorghum, provided insights into evolution and diversification of TCP gene functions. The shortlisted TCP genes, in this study, are promising targets for engineering morphological agronomic traits and stress tolerance in Sorghum.
Results
Identification, phylogeny and classification of TCP proteins in Sorghum
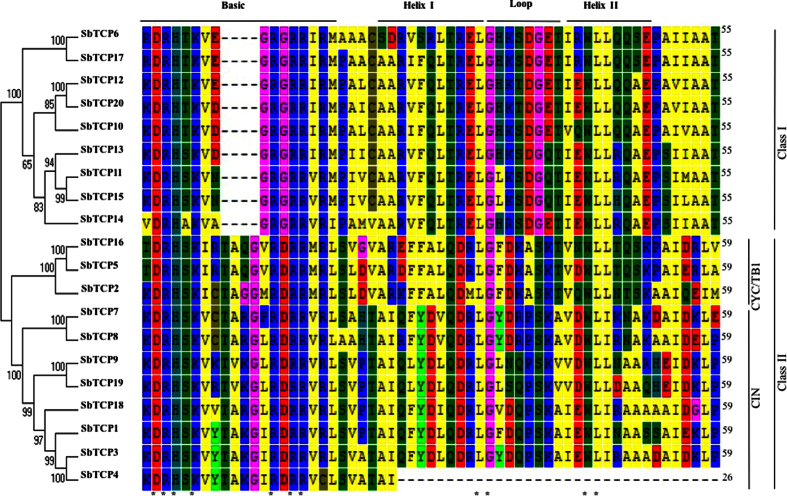
Using HMM profile of the TCP proteins, we retrieved 20 TCP proteins from Sorghum proteome (https://phytozome.jgi.doe.gov/pz/portal.html). The detailed information about their Phytozome gene IDs, number of transcripts, length of proteins, and isoelectric point is provided in Table 1. All TCP proteins are characterized by a conserved non-canonical bHLH domain, known as TCP domain. The presence of domain was confirmed using InterProScan and protein sequences, corresponding to TCP domain, were extracted. Multiple alignment of the TCP domain sequences followed by phylogenetic analysis revealed two distinct classes of TCP proteins. Class I comprised of nine proteins, whereas, eleven proteins clubbed into class II. Class II was further subdivided into CYCLOIDEA/TB1 (CYC/TB1) clade comprising of three proteins and CINCINNATA (CIN) clade with eight proteins (Fig. 1).
Table 1. TCP genes in Sorghum and their key features.
| S. No. | Accession IDs | Gene Name | Type | No. of Transcripts | Protein Length (aa) | Isoelectric Point (pI) | Predicted Subcellular Localization | Number of Predicted Disordered Regions | Percent Disordered Region |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Sobic.001G066100 | SbTCP1 | Class II | 2 | 389 | 9.12 | Nuclear | 8 | 46.27 |
| 389 | 9.12 | ||||||||
| 2 | Sobic.001G121600 | SbTCP2 | Class II | 1 | 380 | 8.5 | Nuclear | 5 | 70.79 |
| 3 | Sobic.002G035500 | SbTCP3 | Class II | 1 | 461 | 9.45 | Nuclear | 10 | 49.67 |
| 4 | Sobic.002G141450 | SbTCP4 | Class II | 1 | 80 | 10.03 | Chloroplast | 1 | 91.25 |
| 5 | Sobic.002G198400 | SbTCP5 | Class II | 1 | 268 | 5.82 | Nuclear | 5 | 65.67 |
| 6 | Sobic.002G268600 | SbTCP6 | Class I | 1 | 326 | 5.17 | — | 3 | 86.81 |
| 7 | Sobic.003G018700 | SbTCP7 | Class II | 3 | 462 | 6.46 | Nuclear | 13 | 48.48 |
| 462 | 6.46 | ||||||||
| 627 | 9.13 | 16 | 48.96 | ||||||
| 8 | Sobic.003G299700 | SbTCP8 | Class II | 1 | 451 | 6.62 | Nuclear | 10 | 52.33 |
| 9 | Sobic.003G305000 | SbTCP9 | Class II | 1 | 289 | 6.81 | — | 6 | 45.33 |
| 10 | Sobic.003G408400 | SbTCP10 | Class I | 1 | 325 | 6.34 | Cytoplasmic | 6 | 64 |
| 11 | Sobic.004G225400 | SbTCP11 | Class I | 1 | 222 | 9.9 | — | 4 | 59.46 |
| 12 | Sobic.004G237300 | SbTCP12 | Class I | 1 | 398 | 9.42 | Nuclear | 7 | 69.1 |
| 13 | Sobic.004G354700 | SbTCP13 | Class I | 1 | 243 | 10.09 | — | 5 | 62.14 |
| 14 | Sobic.006G025000 | SbTCP14 | Class I | 3 | 177 | 8.11 | Chloroplast | 3 | 87.01 |
| 177 | 8.11 | ||||||||
| 15 | Sobic.006G154000 | SbTCP15 | Class I | 1 | 207 | 9.77 | Nuclear/Chloroplast | 4 | 72.95 |
| 16 | Sobic.007G135700 | SbTCP16 | Class II | 1 | 257 | 6.23 | Nuclear | 6 | 66.15 |
| 17 | Sobic.007G182101 | SbTCP17 | Class I | 1 | 359 | 5.71 | Nuclear | 5 | 74.93 |
| 18 | Sobic.008G172200 | SbTCP18 | Class II | 1 | 302 | 6.59 | — | 7 | 50.66 |
| 19 | Sobic.009G195000 | SbTCP19 | Class II | 4 | 298 | 7.94 | — | 8 | 44.3 |
| 298 | 7.94 | ||||||||
| 268 | 7.9 | ||||||||
| 268 | 7.9 | ||||||||
| 20 | Sobic.010G092100 | SbTCP20 | Class I | 1 | 394 | 8.91 | Nuclear | 8 | 61.93 |
Figure 1. Multiple alignment of the TCP domains of Sorghum TCP proteins.
The domain sequences were extracted and aligned using clustal X. The dendrogram on the right shows phylogenetic grouping of TCP proteins based on the complete protein sequences with bootstrap values given on each branch. The colors indicate amino acids of different biochemical properties as obtained through MEGA 6.0. The basic region, helix I and II, and loop regions are marked on the top. The classes, defined on the basis of phylogenetic tree are marked on the left. The conserved residues are marked by black asterisks at the base of alignment. The numbers on the right indicate the length of TCP domains in each protein.
Except for SbTCP4 that contained only partial TCP domain, basic, helix I, loop and helix II regions could be demarcated in all of the Sorghum TCP proteins. TCP domain in class I proteins possess a deletion of four amino acids in the basic region and therefore, basic region in class I proteins only comprises of 16 amino acids as opposed to 20 amino acids in class II proteins. Seven highly conserved residues including Asp (D), His (H), Lys (K) and four residues of Arg (R) characterize the basic region of all TCP proteins (Fig. 1). The abundance of Lys and Arg in basic region of TCP domain likely contributes to the nuclear localization of these proteins4. The helix I, loop and helix II regions comprised of 11, 8 and 9 amino acids, respectively. Two residues each in helix I (Ala and Leu) and helix II (Trp and Leu) were also highly conserved whereas, only one amino acid (Gly) was conserved in the loop region (Fig. 1).
Genomic organization and duplication of Sorghum TCP proteins
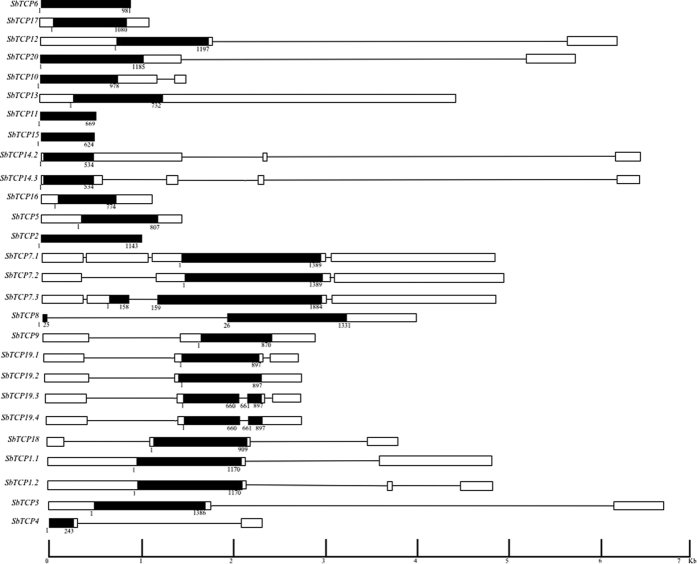
The information about the genomic location of TCP genes was obtained from Phytozome and used to map TCP genes on the Sorghum chromosomes. TCP genes were spotted on all chromosomes of Sorghum except chromosome five. Chromosome 2 and 3 contained four TCP genes each, whereas, chromosome 4 contained three genes. Similarly, chromosomes 1, 6 and 7 contained two TCP genes each, whereas chromosomes 8, 9 and 10 had single TCP gene each (Fig. 2). All the TCP genes are well spaced on the Sorghum chromosomes with no tandem duplicates inferred. Based on their location on respective chromosomes, the genes were named as SbTCP1-SbTCP20.
Figure 2. Chromosomal localization of duplicated TCP genes in Sorghum.
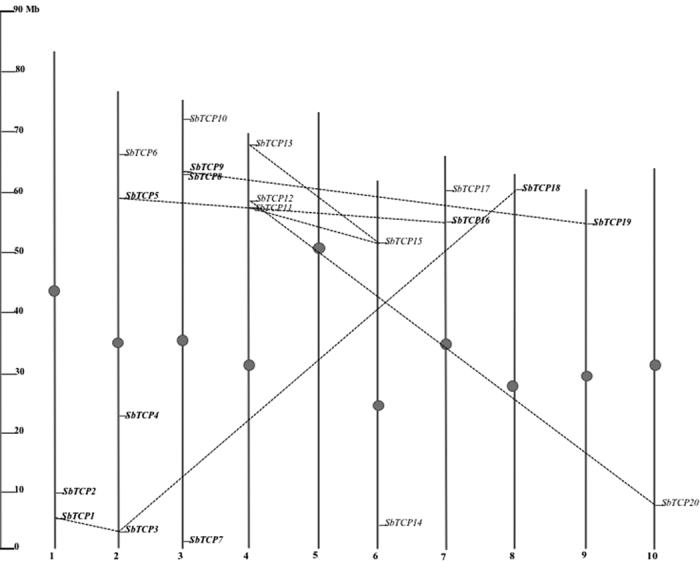
TCP genes were mapped on the Sorghum chromosomes using genomic information available in Phytozome. The vertical bars represent the chromosomes with numbers at the base of each bar representing chromosome number. The ruler on the left indicates the length of the chromosomes in Mb. Class II genes are shown in bold. The duplicated genes pairs in the segmental duplicated blocks are connected by dotted lines.
To understand the pattern of expansion and diversification of TCP genes in Sorghum, duplicated genes were identified using plant duplication database. Seven pairs of possible paralogs of TCP genes were identified on the duplicated blocks of Sorghum genome. Among class I genes, duplicated genes could be marked on the duplicated segments of chromosomes 1 and 2 (SbTCP1/SbTCP3); chromosomes 2 and 8 (SbTCP3/SbTCP18); chromosomes 2 and 7 (SbTCP5/SbTCP16); and, chromosome 3 and 9 (SbTCP9/SbTCP19). Notably, SbTCP3 is duplicated with both SbTCP1 and SbTCP18. Among class II genes, duplicated genes were found on chromosomes 4 and 6 (SbTCP13/SbTCP15 and SbTCP11/SbTCP15) and, chromosomes 4 and 10 (SbTCP12/SbTCP20). Again, SbTCP15 is duplicated with SbTCP11 as well as SbTCP13 indicating common ancestry of these genes. The percentage similarity among duplicated genes ranged from 49.5 (SbTCP1/SbTCP3) to 70.2% (SbTCP12/SbTCP20) (Supplementary Table 1).
The number of synonymous (Ks) and non-synonymous (Ka) substitutions per site of the duplicated TCP genes in Sorghum were determined using tools available with DNA Sequence Polymorphism (DnaSP) software and Plant Genome Duplication Database (PGDD). The Ka/Ks values for all pairs of duplicated genes were <1 implying that duplicated gene pairs are under purifying selection. The Ks values were not used to further predict the time of duplication events of duplicated genes as all the TCP genes of Sorghum have an average GC content higher than 75% in the third codon position (Supplementary Table 2). The average GC content for Sorghum TCP genes in the third codon position is estimated to be 85.7%.
Structural attributes of Sorghum TCP genes and proteins
The length of the coding regions of TCP genes in Sorghum ranged from 243 (SbTCP4) to 1884 bp (SbTCP8) with an average length of 1071 bp. Analysis of intron-exon distribution revealed eight TCP genes of Sorghum as intronless, whereas eight contained one intron, three genes contained two introns and one gene (SbTCP7) contained three introns (Fig. 3). In class II, all three genes belonging to CYC/TB1 clade were intronless, whereas those comprising CIN clade had at least one intron. Length of the introns in these genes ranged from 101 to 4341 bp. Out of 12 genes containing intron(s), 11 had introns in the untranslated regions (UTRs). Among these, one gene had intron in the 5′UTR and seven in the 3′UTR, whereas three genes had introns in both 5′ and 3′ UTRs (Fig. 3). Only one gene, SbTCP8 possessed intron in the coding sequence.
Figure 3. Structural organization of TCP genes in Sorghum.
The black boxes represent coding region, whereas, white boxes represent 5′ and 3′ Untranslated regions (UTRs). The introns in coding region or UTRs are shown by black lines. The scale bar at the base represents the length of the genes in Kb. Numbers provided with the black boxes mark exon boundaries and represent the actual size of the coding region.
Four TCP genes of Sorghum, one from class I (SbTCP14) and three from class II (SbTCP1, SbTCP7 and SbTCP19) were predicted to encode more than one transcript (gene models; Table 1) due to alternative splicing. However, except for one of the gene models of SbTCP7, all other gene models for the four genes encode for the same protein indicating that introns in the untranslated regions of these genes led to splicing variants (Fig. 3).
The length of the proteins ranged from 80 (SbTCP4) to 627 (SbTCP7) amino acids with an average of 325 amino acids. Earlier study in Arabidopsis reported that amino acid composition of TCP family proteins is typical of intrinsically disordered proteins with both class I and II proteins enriched in disorder promoting residues viz., Gln, Asn, Pro and Ser16. Analysis of TCP proteins of Sorghum with PONDR VL-XT predictor predicted degree of disorder ranging from 44.3 to 91.25% in these proteins (Table 1). The number of disordered regions in Sorghum proteins varied from 1 to 16. All the class I proteins showed high percent disorder (≥60%) compared to class II proteins where only four proteins had disorder percentage higher than 60%.
Predicted subcellular localization
Subcellular localization of TCP proteins was predicted using six different tools and an overall prediction was made by compiling the high confidence predictions from these tools (Table 1 and Supplementary Table 3). Ten of the Sorghum TCP proteins were predicted to localize in the nucleus. Two proteins, SbTCP4 and 14 were predicted to localize in the chloroplasts, whereas SbTCP10 was predicted to localize in the cytoplasm. The localization of SbTCP15 was predicted in both nucleus and chloroplast, suggesting that it may be required for coordinated expression in both nuclear and chloroplast compartments.
Expression analysis of Sorghum TCP genes
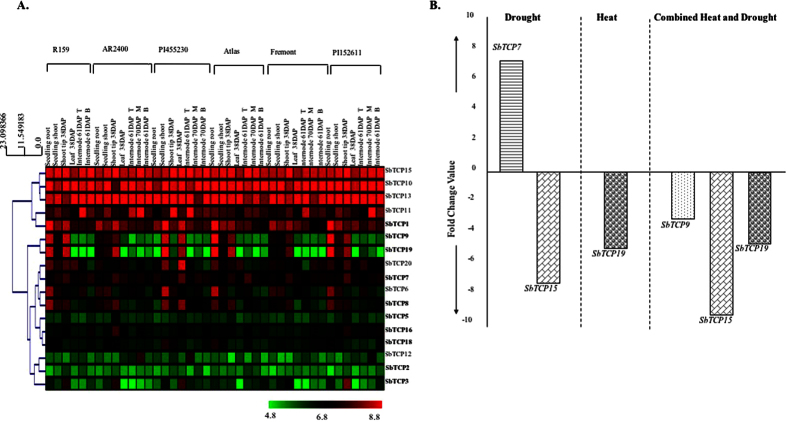
We used publicly available microarray-based (Affymetrix platform) expression data from six genotypes in five vegetative tissues including seedling root, seedling shoot, shoot tip, leaf and stem internodes for analyzing TCP gene expression. Out of 20 TCP genes, 17 are represented on the Affymetrix arrays (Fig. 4A). The expression data clearly revealed high-level ubiquitous expression of four class I genes, namely SbTCP5, 10, 11 and 13, in all the stages and genotypes. Whereas, SbTCP6 and 20 of class I exhibited dominant expression in seedling roots, shoots and shoot tips. Transcripts of SbTCP12 were specifically detected in limited portions of internodes of three bioenergy/high biomass lines, PI455230, PI152611 and AR2400. Genotype-specific variation in the expression pattern of TCP genes indicates differential activity of TCP genes in genotypes exhibiting different morphological features.
Figure 4. Expression analysis of TCP genes in vegetative tissues and in response to abiotic stress treatments.
(A) Expression analysis in vegetative tissues from diverse genotypes of Sorghum. Expression profile in six different genotypes of Sorghum in seedling root, seedling shoot, shoot tip, leaf and, stem internodes. The dendrogram on the left represents hierarchical clustering based on expression values. (B) Differential accumulation of TCP genes in response to heat, drought and combined stresses. The Y-axis represents the average fold changes between three biological replicates. Only the genes exhibiting more than two-fold change in all three replicates are shown.
Among class II genes, we noticed predominant expression of three genes including SbTCP8, 9 and 19 in tissues undergoing active cell division and elongation such as seedling shoots, roots and shoot tips. Whereas high expression of SbTCP1 and 7 was detected in most of the tissues analyzed. SbTCP5 expression was only detected in shoot tips, whereas SbTCP3 seemed to exhibit variable expression pattern in different genotypes mostly in shoot tip, seedling shoots and leaf tissue. We could not detect significant expression of SbTCP2 in any of the analyzed stages.
Further, the expression of all TCP genes was examined in microarray-based (Agilent platform) expression data in response to heat, drought and, combined heat and drought stress in Sorghum. Expression of only four TCP genes, SbTCP7, 9, 15 and 19, was affected by at least one of these stress treatments (Fig. 4B). SbTCP7 was the only gene upregulated in response to drought stress. SbTCP15 was downregulated in response to drought and combined stresses but remained unaffected by heat stress alone. Whereas SbTCP19 was downregulated by heat and combined stress but remained unaffected by drought stress; SbTCP9 was downregulated by combined stress but remained unaffected in response to individual stresses.
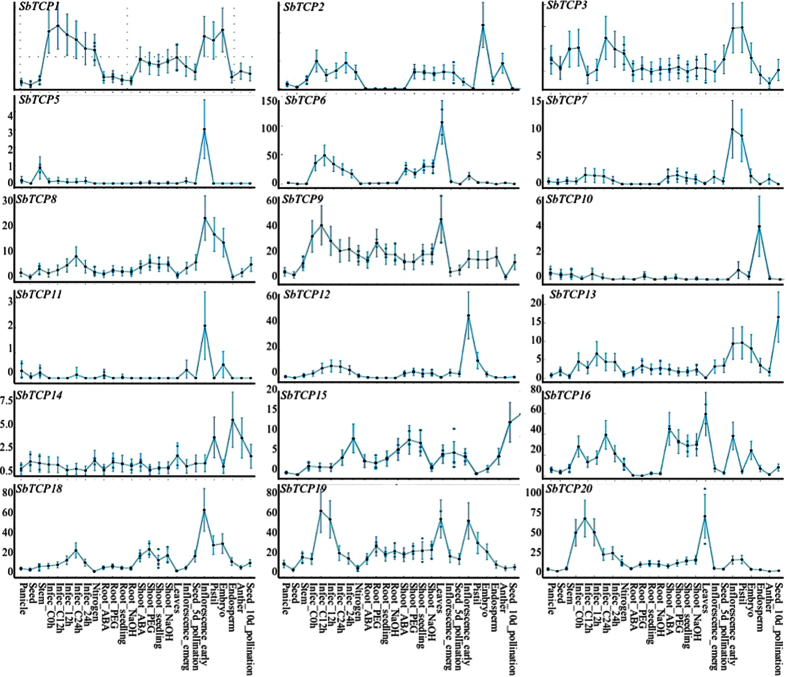
Furthermore, to analyze the expression of TCP genes in reproductive tissues and in response to biotic stress treatments, we queried RNA Sequencing (RNAseq) based expression data available in MOROKOSHI Sorghum Transcriptome database (http://sorghum.riken.jp/morokoshi/Home.html). Out of 20 TCP genes, 18 were present in MOROKOSHI database. Except for two class I genes, SbTCP10 and 20, which exhibited high accumulation in vegetative tissues, high-level expression of the rest 16 genes was detected in reproductive tissues. Overall, class I genes mostly exhibited high expression in vegetative tissues and early floral developmental stages, whereas class II genes were mainly detected in floral organs and seed tissues. In agreement with the previous observation, expression of SbTCP7 and 19 was affected by abiotic stress treatments. However, no obvious induction in response to infection with Bipolaris sorghicola was detected for any of the TCP genes (Fig. 5). Taken together, all of the Sorghum TCP genes exhibit high level expression in at least one of the analyzed stages except SbTCP4, 14 and 17 which were neither represented on chip nor detected in the RNAseq data.
Figure 5. RNAseq based expression patterns of Sorghum TCP genes.
Expression profiles for all the Sorghum TCP genes were downloaded from Morokoshi database. The X-axis represents the different stages of development or stress conditions and the Y-axis represents FPKM (fragments per Kb per million) values for TCP transcripts.
A comparison of expression patterns of duplicated genes SbTCP8 and 19 showed a highly similar expression pattern in vegetative tissues with correlation coefficient >0.9. However, both the paralogs exhibited different expression profiles in inflorescence stages indicating functional divergence in reproductive tissues between them (Fig. 5.). The correlation coefficient in expression patterns of the rest of the duplicated gene pairs range from −0.10 for SbTCP13/SbTCP15 to 0.72 for SbTCP5/SbTCP16 in vegetative tissues suggesting subfunctionalization or neofunctionalization as the most common fate of duplicated TCP genes in Sorghum. We blasted Sorghum orthologs in rice followed by reverse blast analysis to check if duplicated gene pairs predicted in Sorghum have arisen before Rice-Sorghum divergence. Five of the TCP gene duplications seem to predate Rice-Sorghum divergence with Sorghum paralogs SbTCP9/19 orthologous to rice paralogous genes OsTCP5/18; SbTCP5/16 paralogs of Sorghum orthologous to OsTCP22/24 of rice; SbTCP1/3 paralogs orthologous to rice PCF6/OsTCP21; SbTCP11/15 paralogs orthologous to rice OsTCP7/17 and SbTCP12/20 paralogs orthologous to rice OsTCP9/19 gene pairs (Supplementary Table 4).
Comparative phylogenetic and structural analysis of TCP proteins in Arabidopsis and grasses
In order to understand the evolutionary relationships among TCP proteins of Arabidopsis and grasses, we constructed an un-rooted neighbor joining phylogenetic trees using 88 complete protein sequences of TCP genes from Arabidopsis and three poaceae species (Fig. 6). These included 23 rice, 24 Arabidopsis, 21 Brachypodium and 20 Sorghum TCP proteins. The TCP proteins from these species could be categorized into two distinct classes, class I and class II. Class I comprised of 43 proteins including 10 rice, 9 Sorghum, 13 Arabidopsis and 11 Brachypodium proteins. Class I further could be subdivided into six sub-clades, named A, B, C, D, E and F. Sbu-clades A, B, D and E comprised of members from all four species. Sub-clade C however, was specific to monocots with no protein from Arabidopsis, whereas sub-clade F contained a single TCP protein AtTCP16 of Arabidopsis with no protein from grass genomes.
Figure 6. Phylogenetic relationship among TCP proteins in Arabidopsis, rice, Brachypodium and Sorghum.
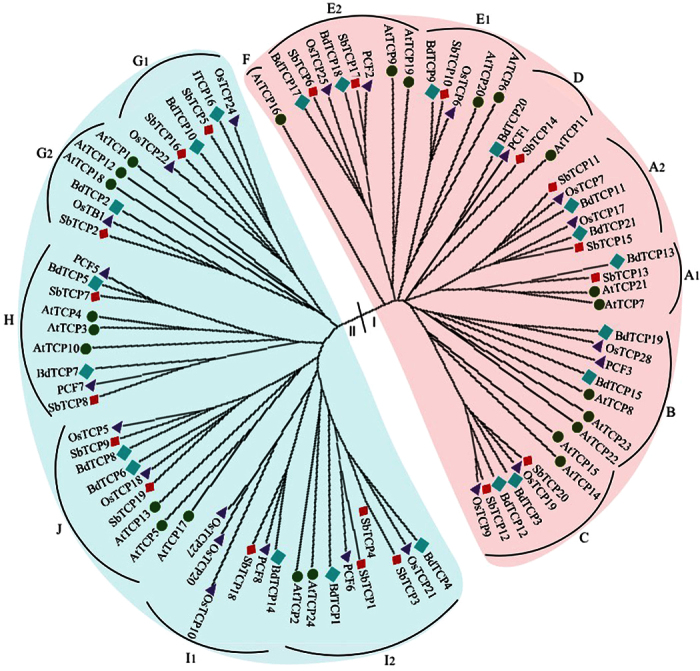
Full length amino acid sequences of TCP proteins of Arabidopsis, rice, Brachypodium and Sorghum were used to generate an unrooted Neighbor-joining radial tree constructed with 1000 bootstrap iterations. Different colors have been used to highlight class I and II proteins. Sub-clades have been marked in each class. Different symbols have been used to differentiate TCP prteins from different species.
Class II comprised of two major clades including CYC/TB1 and CIN clade. CYC/TB1 clade, containing single sub-clade G comprised of twelve genes with three genes each from all four species. Two distinct groups, named G1 and G2, were apparent in sub-clade G. The group G1 only comprised of monocot proteins including rice OsTCP22 and 24 (Retarded Palea 1); Brachypodium BdTCP10 and 16; and Sorghum SbTCP5 and 16. The Retarded Palea 1 (REP1) protein of rice in monocot-specific group G1 regulates palea development and floral zygomorphy17, characteristic to monocot species, indicating divergence of G1 group to regulate monocot-specific morphological features. Whereas, G2 comprised of three proteins of Arabidopsis (AtTCP1, 12 and 18) and one protein each from rice (OsTB1), Brachypodium (BdTCP2) and Sorghum (SbTCP2).
CIN clade of class II proteins comprised of 33 proteins with seven Brachypodium, ten rice, eight Sorghum and eight Arabidopsis proteins. These were organized into three distinct sub-clades named H, I and J, with each of them containing members from all four species. However, as expected, the TCP proteins from monocot species were more closely related to each other compared to the Arabidopsis orthologs.
Further, we used MEME motif search tool to identify the conserved motifs besides TCP domain in TCP proteins from all four species (Supplementary Fig. 1). A total of twenty motifs of lengths ranging from 6 to 57 amino acids were identified and their distribution in all TCP proteins was compared. The assignment of motifs reiterated the phylogenetic analysis and enabled a clear classification of class I and class II TCPs based on the motifs present in the two classes, indicating that similar motifs render similar functionalities to each class. Motifs 1, 2 and 3 represent TCP domains. All the class I proteins, except BdTCP13, possessed motif 1, whereas TCP domain in class II proteins, except SbTCP4, was represented by the motif 2. Motif 3 was present in all class I proteins and one class II protein namely, SbTCP4. Motif 9, representing arginine rich R domain, was specific to class II proteins and detected in thirteen proteins (BdTCP4, OsTCP21, SbTCP3, AtTCP24, AtTCP2, OsTCP10, OsTCP20, SbTCP2, OsTB1, BdTCP2, AtTCP18, AtTCP12 and AtTCP1). Whereas, motif 12, representing the target region of miR319, is specific to class II proteins and was detected in 15 TCP proteins (BdTCP4, OsTCP21, SbTCP3, SbTCP1, PCF6, BdTCP1, AtTCP24, AtTCP2, BdTCP14, PCF8, SbTCP18, AtTCP3, AtTCP4, BdTCP5 and PCF5) indicating the possibility of miRNA-mediated regulation of class II proteins. In addition to these motifs, we detected SP domain (rich in serine and proline residues), (PSVKHMFPFCDSSSPMDLPLYQQLQLSPPSPKPD), characteristic of TB1 like proteins in SbTCP2 and OsTB118 as well as coiled coil (CC) domain (AEPSIIAATGTGVTP) in eight class I (PCF1, AtTCP20, AtTCP11, AtTCP21, AtTCP7, BdTCP20, SbTCP13, SbTCP14) and six class II (PCF2, OsTCP28, AtTCP23, AtTCP9, SbTCP6, SbTCP17) proteins. The functional relevance of these domains is yet to be determined.
Comparative expression profiling of TCP genes in Sorghum, rice and Brachypodium
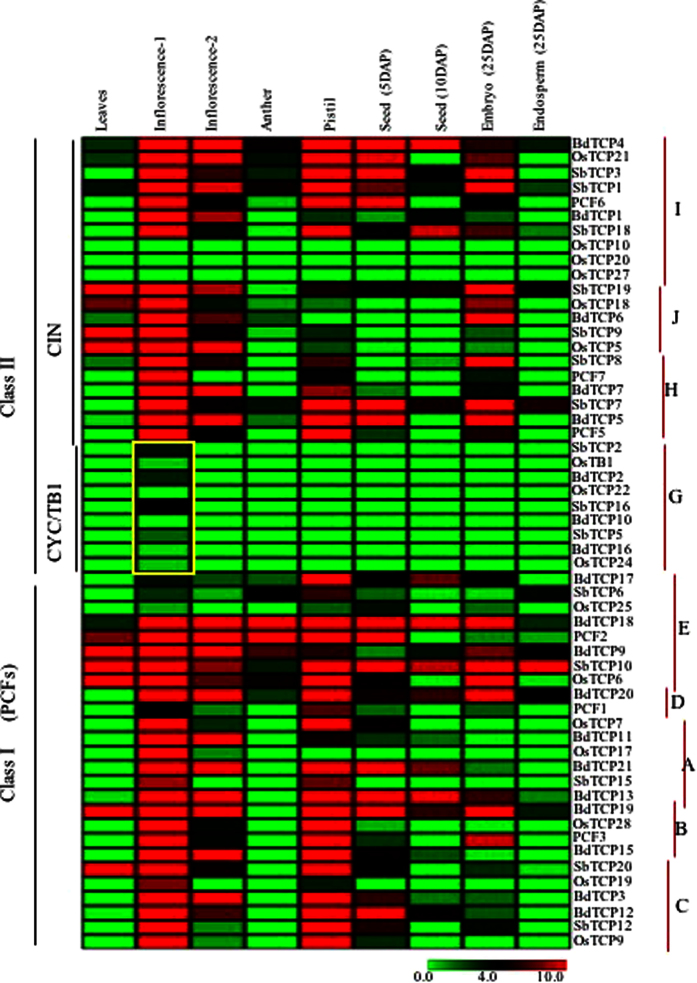
Earlier, we had compared the expression of TCP genes in rice and Arabidopsis by analyzing their expression patterns in similar stages of development19. To further investigate the conservation and/or diversification in the expression patterns of TCP genes in grasses, we leveraged the RNA sequencing based expression data available from physiologically similar tissues of three grass species representing three different lineages including the Pooideae (Brachypodium), Panicoideae (Sorghum) and Ehrhartoideae (rice) of family poaceae20. The comparative expression profiles of all the genes are illustrated in the order of their phylogenetic placement in the comparative phylogenetic tree (Fig. 7).
Figure 7. Comparative expression analysis of TCP genes in rice, Brachypodium and Sorghum.
Heat map was generated based on FPKM values obtained from the previously published transcriptomic data20. The scale bar at the base represents relative expression values.
In sub-clades A-E, we noticed that rice transcripts were mostly expressed in inflorescence and pistil tissues, whereas orthologous genes from Brachypodium and Sorghum were more broadly expressed with many of them expressing in seed tissues as well.
Sub-clade G includes CYC/TB1-like genes from all three species. Genes from all three species in this sub-clade exhibit low level but highly specific expression restricted to primordial inflorescence. Sub-clade H genes from all three species expressed in inflorescence, pistils and embryos. Transcripts of BdTCP4, OsTCP21, SbTCP1 and SbTCP3 of sub-clade I were detected in anthers, inflorescence, pistils and seed tissues. However, no expression was detected from OsTCP10, 20 and 27 genes of rice in any of the analyzed stages.
Sub-clade J genes were expressed in both leaves and reproductive tissues including inflorescence and embryos, whereas expression of sub-clade H genes was restricted to reproductive tissues. Overall, sub-clade G genes seemed to have highly conserved role in the primorida inflorescence in all three species, whereas, expression patterns of the genes in rest of the sub-clades seem to have diversified in all three species.
Discussion
Sorghum is a photosynthetically efficient C4 crop cultivated in wide range of agro-ecological conditions across 98 countries of the world21. It not only feeds over 750 million people in semi-arid tropical regions of Africa, Asia and Latin America22 but is also used as forage for dairy cattle and, has recently emerged as a promising feedstock for biofuels15. However, Sorghum varieties exhibit great deal of genotypic and phenotypic diversity in the field conditions. Based on the morphological and agronomic traits, Sorghum cultivars have been categorized into four ideotypes namely, grain, forage, energy and sweet sorghum. Interestingly, some of their morphological traits are directly associated with one or more agronomically important traits. For example, brown midrib phenotype is associated with the fodder quality in forage sorghum23, whereas green color of midrib and length of internodes are correlated with the sugar yield, juice volume and moisture in sweet sorghum24,25. Similarly, grain yield in grain Sorghum is affected by inflorescence exertion, flowering time, shape and size of the panicle15. Moreover, resistance to biotic and abiotic stresses may also be associated with plant morphology in Sorghum. For example, the genotypes with closed glumes have been shown to exhibit enhanced resistance to grain mold26. Therefore, directed modification of plant morphology in Sorghum can have profound effect on yield and varietal improvement.
Members of TCP gene family have been recruited to generate novel morphological traits during plant evolution and therefore, provide an important resource to generate desired phenotypes. TB1 of maize, a TCP family transcription factor, is a classic example demonstrating role of a single gene in crop domestication5. Sorghum ortholog of maize TB1, SbTB1 has also been implicated in regulating axillary bud outgrowth in response to light signals27. However, to realize the full potential of TCP genes in engineering Sorghum varieties and shortlist targets for engineering, a genome-wide analysis of TCP family in Sorghum was needed. Here, in this study, we provide a comprehensive overview of TCP gene family in Sorghum and based on the comparative phylogenomic analysis in Arabidopsis and three poaceae species short list candidates for experimental verification.
Arabidopsis and grass-diverged aspects of TCP family evolution
The number of TCP genes identified in Sorghum (20) is similar to that of Arabidopsis10 (24), rice19 (23) and Brachypodium (21). This result is consistent with the earlier observations that these species possess a similar number and sizes of gene families28,29. Domain alignment and phylogenetic analysis revealed similar organization with two distinct classes I and II of TCP proteins in Sorghum like that of rice and Arabidopsis. Among class II proteins, two distinctive clades, earlier named as CYC/TB1 and CIN clades in rice and Arabidopsis10, were identified. To infer the functions of the genes in these clades, we performed a comparative phylogenetic analysis of TCP genes in Arabidopsis and three poaceae species and employed classification scheme proposed earlier in cotton30 to classify them into sub-clades A to J. Members of all four species were identified from all the sub-clades except sub-clade F which seems to be specific to dicots and sub-clade B that seems to be missing in Sorghum.
On the other hand, in A1 group of sub-clade A, no rice protein was found. This group contained two Arabidopsis proteins (AtTCP7 and 21), one Sorghum protein (SbTCP13) and one Brachypodium protein (BdTCP13). AtTCP21, also known as CCA1 Hiking Expedition 1 (CHE1), is an important circadian regulator in Arabidopsis31. Orthology analysis using reverse blast analysis indicate that SbTCP13 and BdTCP13 in this group are orthologous to Arabidopsis AtTCP7, whereas CHE1 orthologs are missing in these species.
Conversely, three groups including group A2 of sub-clade A, group G1 of sub-clade G and I1 of sub-clade I only contain genes from grasses with no Arabidopsis ortholog. These genes seem to have arisen to cater monocot-specific traits. For example, Retarded Palea 1 (REP1) gene of rice controls palea development and floral zygomorphy in rice17. Floral zygomorphy is characteristic to all three poaceae species including rice, Sorghum and Brachypodium, whereas Arabidopsis has radially symmetrical flowers. Consistent with this observation, group G1 containing REP1 had no ortholog in Arabidopsis suggesting that this group has diverged to regulate floral asymmetry in grasses. Further characterization of monocot-diverged genes in these groups may provide candidates for improving agronomic potential of grasses.
Genome duplication seems to have played major role in the expansion of TCP family in higher plants. Seven pairs of paralogous genes were identified from Sorghum and an equal number of TCP gene pairs have been earlier reported from duplicated segments of the rice genome19. Most of the duplicated gene pairs detected in Sorghum seem to predate Rice-Sorghum divergence. However, these seem to have diverged in their structure and expression patterns indicating neo/subfunctionalization as major fate of duplicated genes in TCP family of Sorghum. Further, less than 1 Ka/Ks ratios between paralogous genes indicate purifying/stabilizing selection on the retained duplicates and their importance for the fitness of the plant.
Structure and expression of TCP genes indicate an inherent property as important transcriptional regulators during cell division
Structural analysis revealed presence of introns mainly in the UTR regions of TCP genes19 and a conserved motif representing target of miRNA-mediated posttranscriptional regulation11, indicating enhanced robustness to transcriptional or missplicing-related errors in these genes32. Moreover, all Sorghum TCP genes, except SbTCP7.3 and SbTCP8, lack introns in the coding regions which is generally associated with rapid regulation during cell division and cell differentiation33,34.
Furthermore, similar to reports in Arabidopsis, TCP proteins of Sorghum are also rich in disorder promoting residues. C-terminal intrinsically disordered region in Arabidopsis TCP8 has been shown to act as transactivation domain16. Presence of disordered regions and transactivation domains in TCP proteins indicate their activity as autonomous factors for activating transcriptional machinery and fine regulation during adaptive responses where gene expression would depend on tissue/stimulus-specific interactions.
The expression patterns of TCP genes also correlate well with their roles in regulating cell proliferation in meristematic tissues7. Expression analysis revealed high expression of Sorghum TCP genes in tissues undergoing active cell division such as seedlings shoots, roots, shoot tips and internodes, which is consistent to the reports in rice and Arabidopsis19. During reproductive development also, TCP genes usually express at high levels in early stages of inflorescence development.
Sorghum TCP candidates for engineering plant morphology and abiotic stress tolerance
TCP genes in model systems, rice and Arabidopsis have been shown to play key roles in integrating hormonal, environmental and developmental signals to regulate plant development and morphology11. Characterization of target genes in Sorghum will not only assist in modeling Sorghum varieties for desirable attributes but will also add another missing piece to the molecular genetic jigsaw puzzle of gene-environment interactions affecting morphological traits in crop plants.
SbTCP7 of Sorghum is orthologous to rice PCF5 and is the only gene upregulated in response to drought stress in our study. Negative regulation of PCF5 through miR319 confers increased tolerance against drought and salinity stress in rice35,36. MEME motif analysis revealed that SbTCP7 contained the motif 12 that corresponds to miR319 binding site, suggesting that SbTCP7 might also be involved in regulation of abiotic stresses in Sorghum through similar mechanism as in rice.
Another gene pair SbTCP9 and 19 of sub-clade J is orthologous to OsTCP5 and OsTCP18 of rice and were predominantly detected in meristematic tissues such as seedlings shoots, roots, shoot tips and leaves. Although nothing is known about OsTCP18 of rice, OsTCP5 has been shown to be regulated by strigalactones and cytokinins and is implicated in regulating mesocotyl length37. Arabidopsis orthologs (AtTCP5, 13 and 17) of these genes regulate cell proliferation and differentiation during leaf development38. When overexpressed, these conferred enhanced drought and salinity tolerance in Arabidopsis39. Both the Sorghum genes SbTCP9 and SbTCP19 were also downregulated by abiotic stress treatments indicating a possible involvement in abiotic stress response.
Furthermore, preferential location of introns in 5′ and 3′ UTR regions in all three genes, SbTCP7, 9 and 19, might not be circumstantial. Presence of introns in UTRs is a classic feature of substrates for non-sense mediated decay and posttranscriptional regulation40. SbTCP7 and 19 also exhibit alternative splicing which is an important component of stress response by regulating choice of splice sites41.
Based on the expression and orthology analysis, Sorghum genes of CYC/TB1 clade (SbTCP2, 5 and 16) in particular are good candidates for engineering shoot branching and panicle morphology. SbTCP2 is orthologous to rice TB1 that regulates tillering in rice18,42 and SbTCP5 is ortholog of rice REP1 involved in regulating palea development and floral zygomorphy17. Duplicated genes, SbTCP1 and 3 are orthologous to paralogous genes AtTCP2 and 24 of Arabidopsis and, PCF6 and OsTCP21 of rice. Both the genes in Arabidopsis and rice are regulated by miR319 and implicated in organ development and cold stress response, respectively. Both SbTCP1 and 3 contain target region of miR319 (motif 12) indicating conserved miRNA-mediated regulation of the orthologous genes in Sorghum as well.
Methods
Identification, chromosomal localization and duplication analysis
The Hidden Markov Model (HMM) profile for the TCP domain was generated from 1704 TCP proteins downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/)43 using HMMER V3.1b1 (http://hmmer.org/)44. Complete proteome of Sorghum was downloaded from Phytozome (http://www.phytozome.net/)45. Both in-house generated HMM profile and one (PF03634) downloaded from Pfam Database (http://pfam.xfam.org/)46 were independently used as a query to perform HMMER search against Sorghum proteome using default parameters with e-value cut off set to 0.01. The list of TCP proteins of rice and Arabidopsis was obtained from the previous studies19 and protein sequences were downloaded from rice genome annotation project database (http://rice.plantbiology.msu.edu/)47 and The Arabidopsis Information Resource (https://www.arabidopsis.org/)48 database, respectively. The TCP sequences for Brachypodium were downloaded from Grassius database (http://grassius.org/grasstfdb.html)49. To verify the presence of TCP domain, we scanned all the protein sequences using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search). The physical location of the Sorghum TCP genes was obtained from Phytozome (http://www.phytozome.net/) and used to localize TCP genes on respective chromosomes. Following the convention used in other species, the genes were named as SbTCP1 to SbTCP20, based on their location on the respective chromosomes. The information about duplicated genes was obtained from Plant Genome Duplication database (PGDD, http://chibba.agtec.uga.edu/duplication/). Both PGDD and DnaSp software were used to calculate rate of non-synonymous substitutions (Ka) and synonymous substitutions (Ks).
Phylogenetic analysis and identification of orthologs
Multiple sequence alignment of TCP proteins of Sorghum, rice, Arabidopsis and Brachypodium was performed using online Fast Fourier Transform tool (MAFFT) http://www.ebi.ac.uk/Tools/msa/mafft/)50 with local pair fast Fourier transformation and default parameters. We used MEGA 6.06 software (http://www.megasoftware.net/)51 to build the neighbor-joining phylogenetic tree from the sequence alignment using following parameters: p-distance model, pairwise gap deletion and 1000 bootstraps. A separate tree was constructed with only TCP proteins of Sorghum using same parameters. Further, to determine the homologous relationships among characterized TCP proteins of rice and Arabidopsis with that of Sorghum TCP proteins, we blasted Sorghum TCP proteins against rice and Arabidopsis proteomes downloaded from RGAP and TAIR databases, respectively. The first five hits with percent similarity higher than 30%, E-value 1e-5 and score cut off of 100 were reverse blasted against Sorghum proteome. If the same Sorghum protein that was queried in rice and Arabidopsis was obtained among the first three hits in Sorghum proteome, the genes were predicted to have orthologous relationship.
Gene structure analysis and identification of conserved motifs
The information about the gene length and distribution of exons, and introns in TCP genes was obtained from Phytozome. The Intron/Exon organization for TCP genes was determined by aligning the cDNA sequences to their corresponding genomic DNA sequences and using the result as the input for graphical display at the Gene Structure Display Server v2 (http://gsds.cbi.pku.edu.cn/). The intron/exon organization in UTR regions was checked manually.
Further, to identify the conserved motifs in TCP proteins, the online Multiple Expectation-Maximization for Motif Elicitation (MEME) (http://meme-suite.org/tools/meme)52 program was used using following parameters: length of motif minimum 6 and maximum 100. And maximum number of motifs set to 20.
Subcellular localization and disorder predictions
To predict the subcellular localization of TCP proteins, we used six independent prediction databases including Plant mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/)53, WoLFPSORT (http://www.genscript.com/wolf-psort.html)54, DISTILL (http://distill.ucd.ie/distill/)55, MultiLoc2 (http://abi.inf.uni-tuebingen.de/Services/MultiLoc2), SubLoc v1.0 (http://www.bioinfo.tsinghua.edu.cn/SubLoc/)56 and CELLO v2.5 (http://cello.life.nctu.edu.tw/)57. Except for Plant-mPloc, all other prediction programs provide confidence scores. The cutoff used to select high confidence predictions from WoLFPSORT, MultiLoc2, SubLoc v1.0 and CELLO v 2.5 were confidence score >12, confidence score >0.7, accuracy >95% and reliability score >4.0, respectively. For DISTILL predictions also, only high confidence results were considered. The final prediction was made after compiling only the high confidence predictions from these databases.
The disordered regions in TCP proteins were predicted using VLXT method in online Predictor of Natural Disordered Regions (PONDR®, http://www.pondr.com/). VLXT integrates all three feedforward neural networks including VL1 predictor, N-terminus predictor (XN) and the C-terminus predictor (XC) trained using disordered regions of varying length.
Expression analysis
For expression analysis of TCP genes, we used publicly available microarray and RNA Seq-based expression data for Sorghum. For expression analysis during vegetative stages of development, we used Affymetrix data submitted under accession number GSE49879 at NCBI-GEO58. Similarly, we used data generated in response to individual and combined heat and drought stresses using Agilent 28 K array59 submitted under accession number GSE48205 for expression analysis of TCP genes in response to abiotic stresses. We also downloaded expression profiles of all Sorghum TCP genes from MOROKOSHI database (http://sorghum.riken.jp/morokoshi/Home.html)60. For comparative transcriptomic analysis of TCP family genes in rice, Brachypodium and Sorghum, we used the already available RNAseq data generated in physiologically similar tissues of these species20.
Additional Information
How to cite this article: Francis, A. et al. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci. Rep. 6, 38488; doi: 10.1038/srep38488 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
R.S. and M.K.S. acknowledge the Ramalingaswami fellowship and grant from the Department of Biotechnology, Government of India.
The authors declare no competing financial interests.
Author Contributions R.S. conceived and supervised the complete study. A.F. performed the analysis and prepared the figures. N.D. and M.B. contributed to the bioinformatics analysis. R.S. and N.D. wrote the manuscript. M.K.S. and K.-H.J. participated in finalizing the manuscript. All authors read and approved the final manuscript.
04/06/2017
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
- Wang Y. & Li J. Molecular basis of plant architecture. Annu Rev Plant Biol 59, 253–279, doi: 10.1146/annurev.arplant.59.032607.092902 (2008). [DOI] [PubMed] [Google Scholar]
- Guo S. et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4, 1566, doi: 10.1038/ncomms2542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen G., Goossens A. & Pauwels L. Lessons from domestication: Targeting cis-regulatory elements for crop improvement. Trends Plant Sci 21, 506–515, doi: 10.1016/j.tplants.2016.01.014 (2016). [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J. & Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. The Plant Journal 18, 215–222 (1999). [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A. & Hubbard L. The evolution of apical dominance in maize. Nature 386, 485–488 (1997). [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., C. V., L. C. & Coen E. Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799 (1995). [DOI] [PubMed] [Google Scholar]
- S. K. & Ohashi Y. PCFI and PCF2 specifically bind to cis elements in the rice Proliferating Cell Nuclear Antigen gene. The Plant Cell 9, 1607–1619 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J., Sun Y., Blair P. & Mukhtar M. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci 20, 238–245, doi: 10.1016/j.tplants.2015.01.005 (2015). [DOI] [PubMed] [Google Scholar]
- Nicolas M. & Cubas P. TCP factors: new kids on the signaling block. Curr Opin Plant Biol 33, 33–41, doi: 10.1016/j.pbi.2016.05.006 (2016). [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M. & Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci 15, 31–39, doi: 10.1016/j.tplants.2009.11.003 (2010). [DOI] [PubMed] [Google Scholar]
- Li S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signaling & Behavior 10, e10441912–10441911, e10441912, doi: 10.1080/15592324.2015.1044192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K. et al. PvTB1, a Teosinte Branched1 gene homolog, negatively regulates tillering in switchgrass. Journal of Plant Growth Regulation 35, 44–53, doi: 10.1007/s00344-015-9505-x (2016). [DOI] [Google Scholar]
- Mukhopadhyay P. & Tyagi A. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci Rep 5, 9998, doi: 10.1038/srep09998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H. et al. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J 78, 978–989, doi: 10.1111/tpj.12527 (2014). [DOI] [PubMed] [Google Scholar]
- Mullet J. et al. Energy sorghum-a genetic model for the design of C4 grass bioenergy crops. J Exp Bot 65, 3479–3489, doi: 10.1093/jxb/eru229 (2014). [DOI] [PubMed] [Google Scholar]
- Valsecchi I. et al. The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol Biosyst 9, 2282–2295, doi: 10.1039/c3mb70128j (2013). [DOI] [PubMed] [Google Scholar]
- Yuan Z. et al. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149, 235–244, doi: 10.1104/pp.108.128231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. et al. The OsTB1 gene negatively regulates lateral branching in rice. The Plant Journal 33, 513–520 (2003). [DOI] [PubMed] [Google Scholar]
- Sharma R., Kapoor M., Tyagi A. & S. K. Comparative transcript profiling of TCP family genes provide insight into gene functions and diversification in rice and Arabidopsis. Journal of Plant Molecular Biology & Biotechnology 1, 24–38 (2010). [Google Scholar]
- Davidson R. M. et al. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. The Plant Journal 71, 492–502 (2012). [DOI] [PubMed] [Google Scholar]
- Pennisi E. How sorghum withstands heat and drought. Science 323, 573–573 (2009). [DOI] [PubMed] [Google Scholar]
- Ramatoulaye F., Mady C., Fallou S., Amadou K. & Cyril D. Production and use of sorghum: a literature review. Journal of Nutritional Health & Food Science 4, 1–4 (2016).28944285 [Google Scholar]
- Porter K., Axtell J., Lechtenberg V. & Colenbrander V. Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Science 18, 205–208 (1978). [Google Scholar]
- Burks P. S., Kaiser C. M., Hawkins E. M. & Brown P. J. Genome-wide association for sugar yield in sweet sorghum. Crop Science 55, 2138–2148 (2015). [Google Scholar]
- Gutjahr S. et al. Grain, sugar and biomass accumulation in tropical sorghums. I. Trade-offs and effects of phenological plasticity. Functional Plant Biology 40, 342–354 (2013). [DOI] [PubMed] [Google Scholar]
- Melake-Berhan A., Butler L. G., Ejeta G. & Menkir A. Grain mold resistance and polyphenol accumulation in sorghum. J. Agric. Food Chem. 44, 2428–2434 (1996). [Google Scholar]
- Kebrom T. H., Burson B. L. & Finlayson S. A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiology 140, 1109–1117 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- Vogel J. P. et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010). [DOI] [PubMed] [Google Scholar]
- Ma J. et al. Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci Rep 6, 21535, doi: 10.1038/srep21535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J., Breton G., Paser A. & Kay S. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack B. P., Arndt P. F., Duret L. & Crollius H. R. Preventing dangerous nonsense: selection for robustness to transcriptional error in human genes. PLoS Genet 7, e1002276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares D. C., Penkett C. J. & Bähler J. Rapidly regulated genes are intron poor. Trends in Genetics 24, 375–378 (2008). [DOI] [PubMed] [Google Scholar]
- Bicknell A. A., Cenik C., Chua H. N., Roth F. P. & Moore M. J. Introns in UTRs: why we should stop ignoring them. Bioessays 34, 1025–1034 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou M. et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161, 1375–1391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Li D., Zhou M. & Hu Q. Methods and compositions for enhanced resistance to abiotic stress in plants. U.S. Patent no. US 20130117882 A1 (May 9, 2013).
- Hu Z. et al. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol 55, 30–41, doi: 10.1093/pcp/pct150 (2014). [DOI] [PubMed] [Google Scholar]
- Baba K., Nakano T., Yamagishi K. & Yoshida S. Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol 125, 595–603 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K. & Shinozaki K. Plant resistant to environmental stress and method for producing the same. U.S. Patent no. US20130019340 A1 (Jan 17, 2013).
- Reddy A. S., Marquez Y., Kalyna M. & Barta A. Complexity of the alternative splicing landscape in plants. The Plant Cell 25, 3657–3683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S., Priest H. D., Megraw M. & Mockler T. C. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Current Opinion in Plant Biology 24, 125–135 (2015). [DOI] [PubMed] [Google Scholar]
- Choi M.-S. et al. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Reports 31, 57–65 (2012). [DOI] [PubMed] [Google Scholar]
- Guo A.-Y. et al. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Research 36, D966–D969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Research, W29–W37, doi: 10.1093/nar/gkr367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research 44, D279–D285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S. et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Research 35, D883–D887 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini T. Z. et al. The Arabidopsis Information Resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A. et al. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiology 149, 171–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. i. & Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- Bailey T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.-C. & Shen H.-B. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P. et al. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baú D. et al. Distill: a suite of web servers for the prediction of one-, two- and three-dimensional structural features of proteins. BMC Bioinformatics 7, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S. & Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17, 721–728 (2001). [DOI] [PubMed] [Google Scholar]
- Yu C. S., Chen Y. C., Lu C. H. & Hwang J. K. Prediction of protein subcellular localization. Proteins: Structure, Function, and Bioinformatics 64, 643–651 (2006). [DOI] [PubMed] [Google Scholar]
- Shakoor N. et al. A Sorghum bicolor expression atlas reveals dynamic genotype-specific expression profiles for vegetative tissues of grain, sweet and bioenergy sorghums. BMC Plant Biology 14, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. M. et al. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genomics 15, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita Y. et al. MOROKOSHI: Transcriptome database in Sorghum bicolor. Plant and Cell Physiology 56, e6–e6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.