Abstract
Hepatocellular Carcinoma (HCC) is one of the most common malignant tumors with high incidence and mortality rate. Precision and effective biomarkers are therefore urgently needed for the early diagnosis and prognostic estimation. MicroRNAs (miRNAs) are important regulators which play functions in various cellular processes and biological activities. Accumulating evidence indicated that the abnormal expression of miRNAs are closely associated with HCC initiation and progression. Recently, many biomarker miRNAs for HCC have been identified from blood or tissues samples, however, the universality and specificity on clinicopathological features of them are less investigated. In this review, we comprehensively surveyed and compared the diagnostic, prognostic, and therapeutic roles of HCC biomarker miRNAs in blood and tissues based on the cancer hallmarks, etiological factors as well as ethnic groups, which will be helpful to the understanding of the pathogenesis of biomarker miRNAs in HCC development and further provide accurate clinical decisions for HCC diagnosis and treatment.
Hepatocellular Carcinoma (HCC) is the sixth most common cancer worldwide in terms of number of cases and the second major contributor to cancer mortality in man. The survival rates in the United States and developed countries are only 3% to 5%1,2. There are still no effective biomarkers for the early diagnosis and prognosis of HCC. Currently, only about 30% to 40% patients with HCC can get effective treatment at the right time3. It is extremely necessary to discover new biomarkers for precision diagnosis, prognosis and treatment of HCC.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs with 22–24 nucleotides in length. They play important roles in regulating human genes by inhibiting translation or cleavage. Recent studies showed that miRNAs were associated with a variety of important biological processes such as cell proliferation, development, and apoptosis4,5. Accumulating evidence indicated that miRNAs could be latent biomarkers in human cancers, including gastric cancer, lung cancer, prostate cancer, and breast cancer etc.6,7,8,9. Nowadays, extensive research efforts have demonstrated the biomarker role of miRNAs in HCC. For example, Jiang and his colleagues confirmed that miRNA panel assay (miR-10b, miR-106b and miR-181a) could be potential biomarkers for HCC preliminary screening10. He et al. focused on the applications of miRNAs from 13 studies and 21 sets of data and the association between the risk of HCC and miRNAs polymorphisms11. Another review summarized the function of circulating miRNAs12, and a meta-analysis included 14 studies involving 1,848 cases with HCC and 1187 controls concluded that the miRNA panels can be biomarkers for HCC with AUC = 0.99 (96% sensitivity and 96% specificity)13. Many comprehensive reviews recommend to pay attentions to the role and function of miRNAs in disease diagnosis, prognosis and therapy14,15,16,17,18,19,20,21,22. However, the differences in biological features of miRNAs between blood and tissues are still unclear, which limits the investigation on understanding clinical implications of miRNAs in different specimen.
In this review, we performed comprehensive functional analyses and comparisons of miRNA biomarkers in blood and tissues. The miRNA biomarkers in “tissues” were mainly extracted from liver tissues, adjacent noncancerous tissues or human HCC tissues whereas those in “blood” were collected from plasma, serum or whole blood samples. This review aims at comprehensively understanding the pathogenic mechanism and clinical value of HCC biomarker miRNAs, and providing insights into precision diagnosis and treatment of HCC.
Methods
Data collection
We systematically collected HCC biomarker miRNAs from citations in NCBI PubMed by retrieval formula “(liver cancer[tiab] OR intrahepatic bile duct[tiab] OR hepatocellular carcinoma[tiab] OR hepatoblastoma[tiab] OR cholangiocarcinoma[tiab]) AND (miRNA* OR microRNA*) AND (biomarker*[tiab] OR marker*[tiab] OR indicator*[tiab] OR predictor*[tiab])”. Here, studies in which miRNAs were exactly defined as markers or biomarkers were mainly considered, and those identified from body fluids such as saliva, urine and sweat were excluded as we only focused on miRNA biomarkers in blood and tissues. Besides, for further comparing the differentiation between HCC and cirrhosis and providing valuable strategies for the early detection of HCC, we also collected diagnostic miRNA biomarkers for liver cirrhosis using retrieval formula “cirrhosis[tiab] AND diagnos*[tiab] AND (miRNA* OR microRNA*) AND (biomarker*[tiab] OR marker*[tiab] OR indicator*[tiab] OR predictor*[tiab])”.
Target genes of miRNA biomarkers
The miRNA targets used in this study were integrated from both experimentally validated, i.e. miR2Disease23, TarBase (version 6.0)24, miRTarBase (version 4.5)25, miRecords (version 4.0)26 and computationally predicted, i.e. HOCTAR (version 2.0)27, ExprTargetDB28, and starBase (version 2.0)29 miRNA-target databases. To reduce false positives, we mainly selected miRNA-mRNA pairs validated by low-throughput experiments, i.e. real-time quantitative PCR, Western blot, etc. For computationally predicted pairs, they should reside in no fewer than two of the three prediction databases. Meanwhile, we unitized miRNA IDs according to the latest nomenclature in miRBase (release 21)30.
Functional survey of HCC biomarker miRNAs
The functions of HCC biomarker miRNAs are summarized based on the hallmarks of cancers31,32. Since some of the miRNAs are associated with liver injury and few of the miRNAs’ functions are unclear, we therefore grouped their functions into 12 categories as antigrowth signals, resisting cell death, avoiding immune destruction, tissue invasion and metastasis, tumor promotion inflammation, sustained angiogenesis, limitless replicative potential, genome instability and mutation, other clinicopathological features, liver injury, tumor suppressor/onco-miR, and unclear. Moreover, we compared the pathogenesis of HCC biomarker miRNAs based on etiological factors as well as ethnic groups, i.e. the effects of Hepatitis B Virus (HBV), Hepatitis C Virus (HCV) and ethnic variation on HCC development.
Pathway enrichment analyses
For better understanding the association between miRNAs and HCC pathogenesis, we mapped the targets of biomarker miRNAs onto signaling pathways using IPA (Ingenuity Pathway Analysis) program. The top 10 significantly enriched pathways (p-value < 0.01) were selected and further validated the correlation with HCC by PubMed literature exploration.
Results
Overview of the collected HCC biomarker miRNAs
After manually searching and checking in PubMed citations, a total of 50 and 18 diagnostic miRNA biomarkers in blood and tissues, respectively, were extracted from 44 articles (see Tables 1 and 2) and their clinicopathological features of HCC were further compared based on the hallmarks of cancer31, etiological factors and ethnic groups, respectively. As for prognostic and therapeutic biomarkers, respectively, 16 and 32 prognostic miRNAs in blood and tissues together with 8 therapeutic markers were collected according to records in 54 articles (see Tables 3, 4 and 5) and their clinicopathological features as well as functions were then explored.
Table 1. Diagnostic biomarkers in tissues for hepatocellular carcinoma.
| Reported ID | Offical ID | Sample | Ethnicity | Features | Expression | AUC | PMID | Validated Targets |
|---|---|---|---|---|---|---|---|---|
| miR-101 | miR-101-3p | 30 HC 67 CHB 61 HBV-LC 67 HBV-HCC | China | 1.inhibit HCC cell proliferation2.tumor suppressor3.promote apoptosis | down | CHB from HC 0.635 HBV-LC from HC 0.884 HBV-HCC from HC 0.788 | 2497195338 | Mcl-1, SOX9 |
| miR-126 | miR-126-3p | 19 HCV 6 HCC | Germany | tumor suppressor | down | NA | 2550007595 | NA |
| miR-127 | miR-127-3p | 33 HCC | China | tumor suppressor | down | NA | 2485484296 | NA |
| miR-130b | miR-130b-3p | 97 HCC | China | onco-miR | up | 0.914 | 2240334434 | RUNX3 |
| miR-139 | miR-139-5p | 31 CHB 31 HCC | China | 1.suppress metastasis and progression of cancer cells2.tumor suppressor | down | HCC from CH 0.761 (0.7701) | 2454928251 | Rho-kinase 2 |
| miR-148a | miR-148a-3p | 19 HCC | China | onco-miR | up | NA | 22496917106 | NA |
| miR-150 | miR-150-5p | 15 HC 15 ICC | China | tumor suppressor | up | 0.764 | 2548232097 | NA |
| miR-15b | miR-15b-5p | 96 HCC | China | preventing replicative stress in response to mitogenicsignalling | up | 0.982 | 2240334434 | NA |
| miR-182 | miR-182-5p | HCC | China | proliferation | up | NA | 2465362385 | IGF1R and GSK3B |
| miR-18b | miR-18b-5p | 110 HCC | Japan | 1.proliferation2.loss of cell adhesion ability | up | NA | 2349690152 | TNRC6B |
| miR-199a | miR-199a-5p | 17 CH 23 HCC | Egypt | NA | down | 0.856 | 2630275154 | Mitogen-activated protein kinase (MAPK) |
| miR-200a | miR-200a-3p | 29 HCC | Germany | suppress cancer cell migration | up | NA | 2489532653 | ZEB1/ZEB2 |
| miR-200b | miR-200b-3p | 29 HCC | Germany | suppress cancer cell migration | up | NA | 2489532653 | ZEB1/ZEB2 |
| miR-21 | miR-21-5p | 50 HC 30 LC 136 HCC | Japan | excessive secretion by primary cancer cells | up | CH from HC 0.773 HCC from HC 0.953 | 21749846110 | NA |
| miR-21 | miR-21-5p | 17 CH 23 HCC | Egypt | 1.cell growth2.migration3.invasion | up | 0.943 | 2630275154 | phosphatase and tensin homolog (PTEN) |
| miR-21 | miR-21-5p | 30 HC 97 HCC | China | 1.promote cell proliferation2.tumor invasion | up | NA | 2597303255 | PDCD4 and PTEN |
| miR-21 | miR-21-5p | 74 ICC | China | intrahepatic cholangiocarcinoma proliferation and growth | up | NA | 2580322956 | PTPN14 and PTEN |
| miR-214 | miR-214-3p | 9 HC 10 HCC | China | tumor suppressor | down | NA | 2478942039 | EZH2, CTNNB1 and CDH1 |
| miR-224 | miR-224-5p | 9 HC 10 HCC | China | 1.cell proliferation2.migration3.invasion4.anti-apoptosis | up | NA | 2478942039 | CD40 |
| miR-29a-5p | miR-29a-5p | 266 HCC | China | 1.tissue invasiveness and metastasis r2.tumor suppresso | up | 0.746 | 2328502257 | NA |
| miR-483-5p | miR-483-5p | 69 HC 69 HCC | America | anti-apoptotic oncogene | up | HCC from HC 0.827 | 2412741340 | NA |
Abbreviations and note: HC: healthy controls; CHB: patients with chronic type B hepatitis; CH: chronic hepatitis; HCV: hepatitis C virus; HCC: hepatocellular carcinoma; LC: liver cirrhosis; HBV: hepatitis B virus; ICC: Intrahepatic cholangiocarcinoma; NA: not available; 1: combination of plasma miRNA-139 with serum AFP; 2: combined miR-15b and miR-130b.
Table 2. Diagnostic biomarkers in blood for hepatocellular carcinoma.
| Reported ID | Offical ID | Sample | Source | Ethnicity | Features | Expression | AUC | PMID | Validated Targets |
|---|---|---|---|---|---|---|---|---|---|
| miR-199a-3p | miR-199a-3p | 156 HC 78 HCC | serum | China | invasion capability | down | 0.883 | 2561859958 | phosphorylated-S6 protein |
| miR-223 | miR-223-3p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | NA | down | 0.864(training set) 0.888(validation set) | 2210582259 | Stathmin1 |
| miR-101 | miR-101-3p | 30 HC 79 CHB 61 HBV-LC 67 HBV-HCC | serum | China | 1.inhibit HCC cell proliferation2.tumor suppressor3.promote apoptosis | down1 | CHB from HC 0.635 HBV-LC from HC 0.884 HBV-HCC from HC 0.788 | 2497195338 | Mcl-1, SOX9 |
| miR-106b | miR-106b-5p | 50 HC 31 CLD 27 HCC | blood | China | Proliferation | up | HCC from HC 0.89 HCC from CLD 0.81 CLD from HC 0.63 | 2576117910 | p21/E2F5 |
| miR-10b | miR-10b-5p | 50 HC 31 CLD 27 HCC | blood | China | 1.onco-miR2.liver injury | up | HCC from HC 0.85 HCC from CLD 0.73 CLD from HC 0.66 | 2576117910 | NA |
| miR-122 | miR-122-5p | 89 HC 48 CHB 101 HCC | blood | China | liver injury | up | HCC from HC 0.79 CHB from HC 0.93 | 2122961092 | NA |
| miR-122 | miR-122-5p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | 1.tumor size2.differentiation grade3.poor prognosis4.distance metastasis | down | 0.864(training set) 0.888(validation set) | 2210582259 | NA |
| miR-122 | miR-122-5p | 15 HC 30 DN 120 HCC | serum | China | 1.induce apoptosis2.suppress proliferation | up | 0.629 | 2626455344 | NA |
| miR-122 | miR-122-5p | 34 HC 70 HBV-HCC 48 CHB | serum | China | liver injury | up | HCC from HC 0.869 HBV-HCC from CHB 0.630 | 2217481893 | NA |
| miR-122-5p | miR-122-5p | 173 HC 233 LC 261 HCC | serum | China | 1.regulating hepatocyte development and differentiation2.apoptosis and suppress proliferation | down | 0.887(training sets) 0.879(validation sets) | 2523823886 | HepG2 and Hep3B cells |
| miR-1228-5p | miR-1228-5p | 173 HC 233 LC 261 HCC | serum | China | NA | up | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-122a | miR-122-5p | 85 volunteers matched | serum | China | tumor suppressor | down | 0.707(0.943)2 | 2372371398 | NA |
| miR-125b-5p | miR-125b-5p | 28 HC 24 CHB 22 HBV-LC 20 HBV-HCC | plasma | Turkey | suppress the cell growth | up | NA | 2459545033 | AKT |
| miR-130a | miR-130a-3p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | NA | up | HCV-HCC from HC 0.91 | 2635274042 | NA |
| miR-130b | miR-130b-3p | 97 HCC | serum | China | onco-miR | up | 0.914 | 22403344134 | RUNX3 |
| miR-139 | miR-139-5p | 31 CHB 31 HCC | plasma | China | 1.suppress metastasis and progression of cancer cells2.tumor suppressor | down | HCC from CH 0.761 (0.770)3 | 2454928251 | Rho-kinase 2 |
| miR-141-3p | miR-141-3p | 173 HC 233 LC 261 HCC | serum | China | NA | up | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-143 | miR-143-3p | 127 HC 118 CH 95 HCC | serum | China | differentiation | up | CH from HC 0.617 HCC from CH 0.795 | 2499365662 | FNDC3B |
| miR-146a | miR-146a-5p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | 1.suppresses HCC invasion2.exerted negative effects on anti-tumor immune response | up | HCV-HCC from HC 0.787 HCV-HCC from HCV-CLD 0.85 | 26352740142 | VEGF |
| miR-146a | miR-146a-5p | 313 HC 294 HCC | serum | China | onco-miR | NA | NA | 24816919107 | NA |
| miR-150 | miR-150-5p | 120 HC 110 CHB 120 HCC | serum | China | 1.tumor suppressor2.metastasis3.BCLC stage4.advanced TNM stages | down | 0.931 | 2621597060 | NA |
| miR-150 | miR-150-5p | 15 HC 15 ICC | plasma | China | tumor suppressor | up | 0.764 | 2548232097 | NA |
| miR-15b | miR-15b-5p | 96 HCC | serum | China | preventing replicative stress in response to mitogenicsignalling | up | 0.984 | 2240334434 | NA |
| miR-16 | miR-16-5p | 107 CLD 105 HCC | serum | America | 1.tumor suppressor2.apoptosis | down | NA | 2127858341 | BCL2, MCL1, CCND1, WNT3A |
| miR-17-5p | miR-17-5p | 28 HC 26 CHC 30 HCV-positive cirrhosis 8 HCC | blood | Turkey | NA | up | NA | 2539177181 | NA |
| miR-181a | miR-181a-5p | 50 HC 31 CLD 27 HCC | blood | China | tumor suppressor | down | HCC from HC 0.82 HCC from CLD 0.71 CLD from HC 0.64 | 2576117910 | NA |
| miR-182 | miR-182-5p | 40 HC 95 BLD 103 HCC | serum | China | 1.metastasis | up | 0.911 | 2590346661 | TP53INP1 |
| miR-18a | miR-18a-5p | 60 HC 30 HBV-CH 101 HBV-HCC | serum | China | 1.liver injury2.onco-miR | up | NA | 2286539994 | NA |
| miR-192 | miR-192-5p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | NA | up | 0.864(training set) 0.888(validation set) | 2210582259 | NA |
| miR-192 | miR-192-5p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | liver injury | up | HCV-HCC from HC 0.878 HCV-HCC from HCV-CLD 0.69 | 2635274042 | NA |
| miR-192-5p | miR-192-5p | 173 HC 233 LC 261 HCC | serum | China | NA | down | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-195 | miR-195-5p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | 1.onco-miR2.evading apoptosis3.tissue invasion and metastasis | down | HCV-HCC from HC 0.653 HCV-HCC from HCV-CLD 0.78 | 2635274042 | FGF7 and GHR |
| miR-196a | miR-196a-5p | 313 HC 294 HCC | serum | China | onco-miR | NA | NA | 24816919107 | NA |
| miR-199a-5p | miR-199a-5p | 173 HC 233 LC 261 HCC | serum | China | tumor suppressor | down | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-19a | miR-19a-3p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | 1.PV thrombosis2.invasion, satellite nodules and progression3.recurrence | down | HCV-HCC from HC 0.714 HCV-HCC from HCV-CLD 0.86 | 2635274042 | NA |
| miR-206 | miR-206 | 173 HC 233 LC 261 HCC | serum | China | NA | up | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-21 | miR-21-5p | 89 HC 48 CHB 101 HCC | blood | China | liver injury | up | HCC from HC 0.87 CHB from HC 0.91 | 2122961092 | NA |
| miR-21 | miR-21-5p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | tumor suppressor | up | 0.864(training set) 0.888(validation set) | 2210582259 | PTEN |
| miR-21 | miR-21-5p | 50 HC 30 LC 136 HCC | serum | Japan | excessive secretion by primary cancer cells | up | CH from HC 0.773 HCC from HC 0.953 | 21749846110 | NA |
| miR-21 | miR-21-5p | 30 HC 97 HCC | blood | China | 1.promote cell proliferation2.tumor invasion | up | NA | 2597303255 | PDCD4 and PTEN |
| miR-21 | miR-21-5p | 74 ICC | serum | China | intrahepatic cholangiocarcinoma proliferation and growth | up | NA | 2580322956 | PTPN14 and PTEN |
| miR-215 | miR-215 | 127 HC 118 CH 95 HCC | serum | China | metastasis | up | CH from HC 0.802 HCC from HC 0.816 | 2499365662 | NA |
| miR-221 | miR-221-3p | 10 HC 30 HCV 30 HCV-LC 30 HCV-HCC | serum | Egypt | anti-apoptotic | down | 0.655 | 2542932043 | NA |
| miR-223 | miR-223-3p | 89 HC 48 CHB 101 HCC | blood | China | liver injury | up | HCC from HC 0.86 CHB from HC 0.88 | 2122961092 | NA |
| miR-223-3p | miR-223-3p | 28 HC 26 CHC 30 HCV-LC 8 HCC | blood | Turkey | NA | down | NA | 2539177181 | NA |
| miR-223-3p | miR-223-3p | 28 HC 24 CHB 22 HBV-LC 20 HBV-HCC | plasma | Turkey | NA | down | NA | 2459545033 | NA |
| miR-24-3p | miR-24-3p | 46 HC 31 CLD 84 HCC | serum | China | 1.vascular invasion | up | HCC from CLD 0.636 (0.834)5 | 2512931263 | NA |
| miR-26a | miR-26a-5p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | lower miR-26a expression experienced worse survival but better response to interferon therapy | down | 0.864(training set) 0.888(validation set) | 2210582259 | NA |
| miR-26a-5p | miR-26a-5p | 173 HC 233 LC 261 HCC | serum | China | NA | down | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-27a | miR-27a-3p | 167 HC 169 CHB 141 LC 457 HCC | blood | China | onco-miR | down | 0.864(training set) 0.888(validation set) | 2210582259 | NA |
| miR-296 | miR-296-5p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | 1.metastasis2.tumor angiogenesis | up | HCV-HCC from HC 0.792 HCV-HCC from HCV-CLD 0.645 | 2635274042 | NA |
| miR-302c-3p | miR-302c-3p | 28 HC 26 CHC 30 HCV-positive cirrhosis 8 HCC | blood | Turkey | NA | up | NA | 2539177181 | NA |
| miR-30c-5p | miR-30c-5p | 28 HC 26 CHC 30 HCV-positive cirrhosis 8 HCC | blood | Turkey | 1.HCV-positive cirrhosis2.interferon-beta therapy | up | NA | 2539177181 | NA |
| miR-331-3p | miR-331-3p | 40 HC 95 BLD 103 HCC | serum | China | 1.proliferation2.metastasis | up | 0.89 | 2590346661 | PH |
| miR-34a | miR-34a-5p | 42 HC 125 HCV-CLD 112 HCV-HCC | blood | Egypt | child stage and BCLC score | up | HCV-HCC from HC 0.98 HCV-HCC from HCV-CLD 0.67 | 2635274042 | NA |
| miR-375 | miR-375 | 156 HC 78 HCC | serum | China | tumor suppressor | down | 0.637 | 2561859958 | NA |
| miR-375 | miR-375 | 210 HC 135 HBV 48 HCV 120 HCC | serum | China | NA | up | 0.96 | 21098710140 | NA |
| miR-433-3p | miR-433-3p | 173 HC 233 LC 261 HCC | serum | China | NA | up | 0.887(training sets) 0.879(validation sets) | 2523823886 | NA |
| miR-483-5p | miR-483-5p | 69 HC 69 HCC | serum | America | anti-apoptotic oncogene | up | HCC from HC 0.827 | 2412741340 | NA |
| miR-885-5p | miR-885-5p | 24 HC 23 CHB 26 LC 17 GC 9 ICC 6 FNH 46 HCC | serum | China | cholesterol reverse transport | up | 0.904 | 20815808111 | NA |
| let-7b | let-7b-5p | 15 HC 30 DN 120 HCC | serum | China | tumor suppressor | up | 0.645 | 2626455344 | NA |
| miR-203 | miR-203a-3p | 10 HC 30 non-cirrhotic HCV 25 HCV-related cirrhosis 23 HCV-HCC | serum | Egypt | 1.tumor-suppressive2.angiogenesis | down | HCC from non-HCC 0.76 | 27268654141 | NA |
| miR-885-5p | miR-885-5p | 192 HCC 96 LC 96 CHC 95 HC | serum | Egypt | 1.onco-miR2.liver injury | up | HCC from HC 0.63 HCC from LC 0.775 | 27271989120 | ISRE |
| miR-122 | miR-122-5p | 193 HCC 96 LC 96 CHC 95 HC | serum | Egypt | 1.tumor suppressor2.regulate lipid and cholesterol metabolism | up | HCC from HC 0.617 HCC from LC 0.617 | 27271989120 | ADAM17 |
| miR-29b | miR-29b-3p | 194 HCC 96 LC 96 CHC 95 HC | serum | Egypt | tumor suppressor | down | HCC from HC 0.766 | 27271989120 | NA |
| miR-221 | miR-221-3p | 195 HCC 96 LC 96 CHC 95 HC | serum | Egypt | 1.onco-miR2.apoptosis | up | HCC from LC 0.702 | 27271989120 | CDKN1B/p27CDKN1C/p57 |
| miR-181b | miR-181b-5p | 196 HCC 96 LC 96 CHC 95 HC | serum | Egypt | 1.onco-miR2.migration and invasion | up | HCC from LC 0.679 | 27271989120 | TIMP3 |
| miR-22 | miR-22-3p | 197 HCC 96 LC 96 CHC 95 HC | serum | Egypt | tumor suppressor | down | HCC from CHC 0.586 | 27271989120 | HDAC4 |
| miR-199a-3p | miR-199a-3p | 198 HCC 96 LC 96 CHC 95 HC | serum | Egypt | tumor suppressor | down | HCC from CHC 0.7 | 27271989120 | mTOR |
| miR-125b | miR-125b-5p | 56 HC 63 CHB 59 HBV-LC 64 HBV-HCC | plasma | China | 1.tumor suppressor2.migration and invasion3.cellular proliferation and cell cycle progression | down | HBV-HCC from HC 0.891 | 27152955121 | LIN28B |
| miR-96 | miR-96-5p | 104 HCC 100 CHB 90 LC 120 HC | serum | China | 1.onco-miR2.migration and invasion | up | HCC from CHB 0.803 | 26770453142 | NA |
| miR-126 | miR-126-3p | 28 HC 20 LC 59 HCC | plasma | India | NA | up | low AFP HCC from non-HCC 0.765 low AFP HCC from LC 0.643 | 26756996143 | APAF1, APC2, VEGFA, IRS1, CDKN2A |
| miR-224 | miR-224-5p | 26 HCC 22 LC 23 CHB 22 HC | serum | China | 1.migration and invasion2.suppress apoptosis | up | 0.88 | 26724963144 | NA |
Abbreviations and note: HC: healthy controls; CHB: patients with chronic type B hepatitis; CLD: chronic liver disease; HCV-CLD: non-malignant HCV-associated CLD patients; DN: chronic hepatitis B patients with pathologically proven DN; ICC: intrahepatic cholangiocellular carcinoma; LC: liver cirrhosis; HCV: hepatitis C virus HBV: hepatitis B virus; NA: not available; 1: upregulated in the HBV-LC group; 2: combined classifier (AFP and miRNA-122a); 3; combination of plasma miRNA-139 with serum AFP; 4: combined miR-15b and miR-130b; 5: Combined serum alpha-fetoprotein (AFP) and miR-24-3p.
Table 3. Prognostic biomarkers in tissues for hepatocellular carcinoma.
| Reported ID | Offical ID | Sample | Ethnicity | Features | Expression | PMID | Validated Targets |
|---|---|---|---|---|---|---|---|
| miR-101 | miR-101-3p | 20 HC 25 HBV-HCC | China | 1.HBsAg, HBV DNA level and tumor size | up | 24260081112 | NA |
| miR-101 | miR-101-3p | 130 HCC | China | tumor suppressor | down | 2317871399 | SOX9 |
| miR-101 | miR-101-3p | 30 HC 79 CHB 61 HBV-LC 67 HBV-HCC | China | 1.inhibit HCC cell proliferation2.tumor suppressor | up | 2497195338 | NA |
| miR-106b | miR-106b-5p | 104 HCC | China | 1.tumor size2. vascular invasion3. proliferation4. anchorage-independent growth of HCC cells5.metastasis | up | 2546644964 | NA |
| miR-122 | miR-122-5p | 60 HCC | China | 1.tumor suppressor2.maintenance of normal physiological metabolism | down | 26252254100 | PKM2 |
| miR-125b | miR-125b-5p | 49 HCC | China | tumor suppressor | down | 24811246101 | Eif5a2 |
| miR-1269 | miR-1269a | 95 HCC | China | 1.tumor nodes2.portal vein tumor embolus3.vaso-invasion4.tumor capsular infiltration5.expression of MTDH6.onco-miR7.carcinogenesis, metastasis and invasion of HCC | up | 2578504872 | AGAP1, AGK, BPTF, C16orf74, DACT1, LIX1L, RBMS3, ZNF706 and BMPER |
| miR-128-3p | miR-128-3p | 72 HCC | China | 1.suppress proliferation2.suppress metastasis | down | 2596236073 | PIK3R1 PI3K/AKT |
| miR-130a | miR-130a-3p | 102 HCC | China | 1.gender, HBsAgstatus, tumor size, and TNM stage2.tumor suppressor | down | 25218269102 | NA |
| miR-137 | miR-137 | 136 HCC | China | 1.vein invasion2.distant metastasis3.inhibition promotes HCC cell growth | down | 2497080835 | AKT2 |
| miR-146a | miR-146a-5p | 85 HCC | China | tumor suppressor | down | 24172202103 | ROCK1 |
| miR-155 | miR-155-5p | 100 HCC | China | 1.metastasis2.inhibits apoptosis | up | 2386366945 | NA |
| miR-155 | miR-155-5p | 216 HCC | China | onco-miR | up | 22629365108 | NA |
| miR-17-5p | miR-17-5p | 120 HCC | China | regulating proliferation and migration | up | 2258301165 | p38 MAPK-HSP27 |
| miR-182 | miR-182-5p | 81 HCC | China | 1.onco-miR2.motility and invasiveness | up | 2581340366 | FOXO1 |
| miR-182 | miR-182-5p | 86 HCC | China | intrahepatic metastasis | up | 2268171767 | MTSS1 |
| miR-183 | miR-183-5p | 81 HCC | China | 1.onco-miR2.motility and invasiveness | up | 2581340366 | FOXO1 |
| miR-185 | miR-185-5p | 41 NTR 54 TR | China | 1.suppress the tumor cell growth2.suppress invasive | down | 2364805436 | NA |
| miR-188-5p | miR-188-5p | 250 HCC | China | 1.suppress tumor cell proliferation2.suppress metastasis | down | 2599816374 | FGF5 |
| miR-18b | miR-18b-5p | 110 HCC | Japan | 1.proliferation2.loss of cell adhesion ability | up | 2349690152 | TNRC6B |
| miR-199a-5p | miR-199a-5p | 120 HCC | China | 1.Negatively Associated With Malignancies2.Regulates Glycolysis3.Lactate Production | down | 26054020145 | Hexokinase 2 |
| miR-206 | miR-206 | 147 HCC | China | 1.suppresses cell proliferation2.promotes apoptosis. | down | 2551308646 | NA |
| miR-21 | miR-21-5p | 50 HC 30 CH 136 HCC | Japan | NA | down | 21749846110 | NA |
| miR-21 | miR-21-5p | 112 HCC | China | 1.tumor differentiation2.TNM stage3.vein invasion | up | 2626162068 | NA |
| miR-21 | miR-21-5p | 119 HCC | China | 1.tumorinvasion, metastasis and prognosis2.promote cell proliferation and invasion3.inhibits cell apoptosis | up | 2515037347 | NA |
| miR-21 | miR-21-5p | 74 ICC | China | intrahepatic cholangiocarcinoma proliferation and growth | up | 2580322956 | PTPN14 and PTEN |
| miR-212 | miR-212-3p | 86 HCC | China | 1.inhibited cell proliferation2.induced apoptosis | down | 2634732148 | FOXA1 |
| miR-214 | miR-214-3p | 65 HCC | China | tumor suppressor | down | 23962428104 | FGFR-1 |
| miR-25 | miR-25-3p | 96 HCC | Iran | 1.TNM stage2.suppress proliferation3.suppress migration | up | 2620929669 | NA |
| miR-26a | miR-26a-5p | 120 HCC | China | 1.Cell Cycle2.angiogenesis | up | 2425942684 | CDK6, cyclin D1 |
| miR-26a | miR-26a-5p | 130 HCC | China | 1.suppress the tumor cell growth2.suppress invasive | down | 2338984837 | interleukin-6-Stat3 |
| miR-331-3p | miR-331-3p | 457 HCC | China | 1.Promotes Proliferation2.Metastasis | up | 2482530270 | Leucine-Rich Repeat Protein Phosphatase |
| miR-34a | miR-34a-5p | 120 HCC | China | 1.tumor size2.higher serum AFP level | down | 25596083113 | NA |
| miR-424 | miR-424-5p | 96 HCC | China | suppressed proliferation | down | 2631554187 | pRb-E2F pathway, Akt3 and E2F3 |
| miR-503 | miR-503-5p | 20 HCC | China | suppress metastasis | down | 2616326075 | PRMT1 |
| miR-744 | miR-744-5p | 96 HCC | China | 1.tumour suppressor2.tumor malignancy3.tumor cell proliferation4.invasion and migration5.HCC recurrence6.poor prognosis | down | 2554352176 | NA |
| miR-9 | miR-9-5p | 200 HCC | China | 1.tumour suppressor2.tumor stage3.venous infiltration | up | 2555220471 | NA |
| miR-96 | miR-96-5p | 81 HCC | China | 1.onco-miR2.motility and invasiveness | up | 2581340366 | FOXO1 |
| miR-125a | miR-125a-5p | 80 HCC | China | 1.Proliferation 2.Metastasis | down | 22768249146 | MMP11 and VEGF |
| miR-99a | miR-99a-5p | 142 HCC | China | tumor suppressor | down | 21878637147 | NA |
Abbreviations and note: HC: healthy controls; CHB: patients with chronic type B hepatitis; CLD: chronic liver disease; HCV-CLD: non-malignant HCV-associated CLD patients; DN: chronic hepatitis B patients with pathologically proven DN; ICC: intrahepatic cholangiocellular carcinoma; LC: liver cirrhosis; HCV: hepatitis C virus; HBV: hepatitis B virus; CH: chronic hepatitis; TR: treated recurrence group; NTR: none treated recurrence group; NA: not available.
Table 4. Prognostic biomarkers in blood for hepatocellular carcinoma.
| Reported ID | Offical ID | Sample | Source | Ethnicity | Features | Expression | PMID | Validated Targets |
|---|---|---|---|---|---|---|---|---|
| miR-1 | miR-1-3p | 54 LC 195 HCC | serum | Germany | 1.differentiation2.tumor suppressor | up | 2381024791 | NA |
| miR-101 | miR-101-3p | 20 HC 25 HBV-HCC | serum | China | 1.HBsAg, HBV DNA level and tumor size | up | 24260081112 | NA |
| miR-101 | miR-101-3p | 30 HC 79 CHB 61 HBV-LC 67 HBV-HCC | serum | China | 1.inhibit HCC cell proliferation2.tumor suppressor | up | 2497195338 | NA |
| miR-122 | miR-122-5p | 122 HCC | blood | China | 1.tumor suppressor2.proliferation3.differentiation4.regulation of cholesterol and lipid metabolisms5.stability and propagation of hepatitis C virus and hepatitis B infection | up | 2563644877 | NA |
| miR-122 | miR-122-5p | 120 HCC | plasma | South Korea | 1.hepatic necroinflammatory activity2.cell death3.tumor suppressor | up | 2612987849 | NA |
| miR-122 | miR-122-5p | 54 LC 195 HCC | serum | Germany | 1.liver transaminases2.MELD score | down | 2381024791 | NA |
| miR-128-2 | miR-128-2 | 20 HCC 20 HCC(PVTT) | serum | China | onco-miR | up | 25642945109 | NA |
| miR-150 | miR-150-5p | 120 HC 110 CHB 120 HCC | serum | China | 1.tumor suppressor2. metastasis3.BCLC stage4.advanced TNM stages | down | 2621597060 | NA |
| miR-16 | miR-16-5p | 60 HC 90 HCC | serum | China | 1.tumor size2.liver dysfunction and coagulation defect | down | 24697119114 | NA |
| miR-16 | miR-16-5p | 40 HCV 40 HCC | serum | Egypt | 1.apoptosis2.bilirubin | down | 2613372550 | NA |
| miR-17-5p | miR-17-5p | 96 HCC | blood | China | 1.metastasis2.TNM stage | up | 2310808678 | NA |
| miR-182 | miR-182-5p | 40 HC 95 BLD 103 HCC | serum | China | metastasis | up | 2590346661 | TP53INP1 |
| miR-199a | miR-199a-5p | 40 HCV 40 HCC | serum | Egypt | tumor size | down | 2613372550 | NA |
| miR-203a | miR-203a-3p | 90 HCV 152 HCV-HCC | serum | China | tumor suppressor | down | 26210453105 | Snal2 |
| miR-21 | miR-21-5p | 50 HC 30 CH 136 HCC | serum | Japan | NA | down | 21749846110 | NA |
| miR-21 | miR-21-5p | 74 ICC | serum | China | intrahepatic cholangiocarcinoma proliferation and growth | up | 2580322956 | PTPN14 and PTEN |
| miR-21 | miR-21-5p | 60 HC 90 HCC | serum | China | liver injury | down | 24697119114 | NA |
| miR-24-3p | miR-24-3p | 46 HC 31 CLD 84 HCC | serum | China | vascular invasion | up | 2512931263 | NA |
| miR-30c | miR-30c-5p | 90 HCV 152 HCV-HCC | serum | China | tumor suppressor | down | 26210453105 | EMT |
| miR-331-3p | miR-331-3p | 40 HC 95 BLD 103 HCC | serum | China | 1.proliferation2.metastasis | up | 2590346661 | PH |
| miR-335 | miR-335-5p | 125 HC 125 HCV/HBV 125 HCC | serum | China | response to TACE and clinical outcome | down | 26305026148 | NA |
| let-7f | let-7f-5p | 60 HC 90 HCC | serum | China | 1.tumor size2.early recurrence | down | 24697119114 | NA |
Abbreviations and note: PVTT: portal vein tumor thrombosis; LC: liver cirrhosis; HBV: Hepatitis B Virus; HCV: Hepatitis C Virus; HC: healthy controls; CHB: patients with chronic type B hepatitis; BLD: benign liver diseases; ICC: intrahepatic cholangiocellular carcinoma; CH: chronic hepatitis; NA: not available.
Table 5. Therapeutic biomarkers for hepatocellular carcinoma.
| Reported ID | Offical ID | Sample | Source | Ethnicity | Features | Expression | PMID | Validated Targets |
|---|---|---|---|---|---|---|---|---|
| miR-335 | miR-335-5p | 62 HCC | tissue | China | inhibit the proliferation and migration invasion | down | 25804796149 | ROCK1 |
| miR-192 | miR-192-5p | 59 HC 59 HCC | tissue | South Korea | increase tumor cell migration and invasion | down | 25065598150 | NA |
| miR-224 | miR-224-5p | 9 HC 10 HCC | tissue | China | 1.cell proliferation s2. migration3.invasion4.anti-apoptosi | up | 2478942039 | CD40 |
| miR-214 | miR-214-3p | 9 HC 10 HCC | tissue | China | tumor suppressor | down | 2478942039 | EZH2, CTNNB1 and CDH1 |
| miR-148a | miR-148a-3p | 19 HCC | tissue | China | onco-miR | up | 22496917106 | NA |
| miR-206 | miR-206 | 147 HCC | tissue | China | 1. suppress cell proliferation2.promote apoptosis. | down | 2551308646 | NA |
| miR-331-3p | miR-331-3p | 457 HCC | tissue | China | 1. promote proliferation2. metastasis | up | 2482530270 | Leucine-Rich Repeat Protein Phosphatase |
| miR-26a | miR-26a-5p | 120 HCC | tissue | China | 1. cell Cycle2. angiogenesis | up | 2425942684 | CDK6, cyclin D1 |
| miR-26a | miR-26a-5p | 130 HCC | tissue | China | 1. suppress the tumor cell growth2. suppress invasive | down | 2338984837 | interleukin-6-Stat3 |
Abbreviations and note: HC: healthy controls; NA: not available.
Functional characterization of HCC biomarker miRNAs based on cancer hallmarks
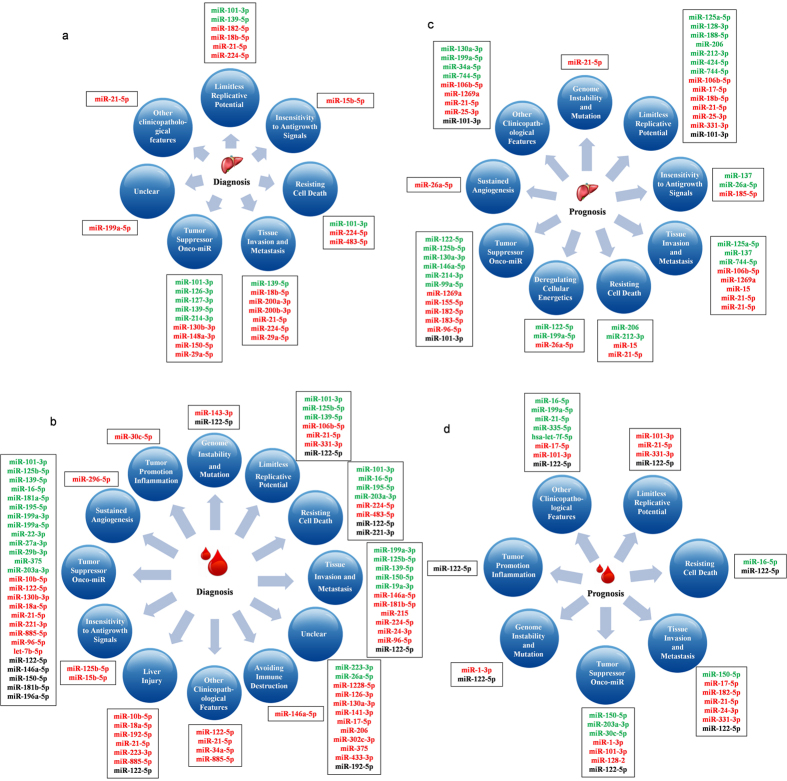
The functional characterization of HCC biomarker miRNAs are summarized from the primary references and classified into 12 categories as shown in Fig. 1. It indicates that the biomarker miRNAs are associated with all aspects of hallmarks of cancers and all the hallmarks lead to the cancer. Therefore, the personalized biomarkers are needed to precision diagnosis, prognosis and treatment of the complex HCC. The functions of the biomarker miRNAs are summarized as follows.
Figure 1. The correlation among clinicopathological features and reported HCC miRNA biomarkers.
Here, miRNAs in red and green, respectively, represent the up and down-regulated expression in tissues and blood. The miRNA in black means that its expression can be inconsistently up- or down- regulated in different reports. Sub-figure (a,b) represent clinicopathological features of diagnostic miRNA biomarkers in tissues and blood, respectively. Sub-figure (c,d) represent clinicopathological features of prognostic miRNA biomarkers in tissues and blood, respectively.
Insensitivity to Antigrowth Signals
Although it is unclear for the units and interconnections between the different kinds of antigrowth and differentiation-including signals and the core cell cycle machinery, an antigrowth signaling must be exist to circumvent developing HCC31. MiR-125b-5p and miR-15b-5p were the circulating diagnostic miRNA biomarkers associated with insensitivity to antigrowth signals and all of them were up-regulated and highly expressed in early-stage HCC cases33. Liu et al. combined miR-15b-5p and miR-130b-3p as a classifier for HCC detection, yielding a receiver operating characteristic curve area of 0.98 in their validation study, the same was found in tissue samples, miR-15-5p was also reported highly expressed34. As for prognostic biomarkers, three miRNAs related to insensitivity to antigrowth signals in the tissue samples were identified, including miR-137, miR-185-5p and miR-26a-5p. All of them were down-regulated in poor prognostic group which had a lower survival rate and shorter time to recurrence35,36,37.
Resisting Cell Death
Cancer cells evolve various ways to circumvent or restrict apoptosis. The diversity of apoptosis-avoiding machinery and program reflects the multiplicity of apoptosis-including signals that tumor cell populations experienced while their evolution to the malignant state32. In tissues, miR-101-3p, miR-224-5p and miR-483-5p were associated with resisting cell death. Among them, miR-101-3p was down-regulated whereas the remaining two were reported to be up-regulated38,39,40. Resisting cell death was significantly associated with lower expression of miR-101-3p, miR-16-5p, miR-195-5p, miR-203a-3p and miR-221-3p in blood samples38,41,42,43. Increased miR-221-3p, miR-224-5p, miR-483-5p and miR-122-5p expression were also detected in blood of HCC patients40,44. These above diagnostic biomarkers as classifiers for HCC detection, yielding a receiver operating characteristic curve area of 0.635 to 0.884 (see Tables 1 and 2). On the other hand, miR-155-5p, miR-206, miR-21-5p and miR-212-3p could be recognized as biomarkers for HCC prognosis in tissues. The expression levels of miR-155-5p and miR-21-5p were up-regulated whereas others were down-regulated45,46,47,48. Circulating miR-122-5p and miR-16-5p could be used as putative biomarkers for HCC. Among them, miR-122-5p and miR-16-5p were shown to be up and down-regulated, respectively49,50.
Avoiding Immune Destruction
According to the long-standing theory of immune surveillance proposes, most of solid tumors such as HCC appeared to have somehow controlled to avoid detection by the different kinds of arms of the immune system or could limit the extent of immunological killing, thus they could evade eradication by immune system32. Motawi and his colleagues overviewed that serum miR-146p-5p was up-regulated in HCC and showed the clinical value for HCV-related HCC diagnosis. This circulatory biomarker miRNA was reported to exerted negative effects on anti-tumor immune response42.
Tissue Invasion and Metastasis
Invasion and metastasis, complex and multi-step processes, are elementary factors that affects HCC patients survival rate and their genetic and biochemical mechanisms remain poorly understood31. In tissues, high expression of miR-18b-5p, miR-200a-3p, miR-200b-3p, miR-21-5p, miR-224-5p and miR-29-5p were most frequently to be detected in HCC, and miR-139-5p was down-regulated. Therefore, they were valuable for diagnosis of HCC39,51,52,53,54,55,56,57. Several circulating miRNA biomarkers also displayed signally correlation with tissue invasion and metastasis, including highly expressed miR-146a-5p, miR-181b-5p, miR-182-5p, miR-21-5p, miR-215, miR-24-3p, miR-224-5p, miR-296-5p, miR-331-3p and miR-96-5p and low expressed miR-125b-5p, miR-199a-3p, miR-122-5p, miR-139-5p, miR-150-5p, miR-195-5p and miR-19a-3p. The above diagnostic biomarkers could be used as classifiers for HCC detection, yielding a receiver operating characteristic curve area of 0.645 to 0.94342,51,55,56,58,59,60,61,62,63.
In tissues, with regard to up-regulated microRNAs in HCC tissues, highly expression of miR-106b-5p, miR-155-5p, miR-17-5p, miR-182-5p, miR-183-5p, miR-18b-5p, miR-21-5p, miR-25-3p, miR-331-3p, miR-9-5p and miR-96-5p were significantly correlated with invasion and metastasis45,47,52,56,64,65,66,67,68,69,70,71. The expression level of miR-1269a in HCC patients without portal vein tumor embolus was reduced72. In addition, the low expression of miR-125a-5p, miR-128-3p, miR-137, miR-185-5p, miR-188-5p, miR-26a-5p, miR-503-5p and miR-744-5p were detected in HCC tissues compared with their non-tumor livers and were involved in the multi-step processes35,36,37,73,74,75,76. There were six circulating prognostic biomarker miRNAs reported to be associated with tissue invasion and metastasis, including miR-122-5p, miR-17-5p, miR-182-5p, miR-21-5p, miR-24-3p and miR-331-3p, all of them were up-regulated in the group with low survival rate56,61,63,77,78. Meanwhile, the serum miR-150-5p was shown highly expressed in HCC patients after surgical operation and then low expressed after tumor relapsed60.
Tumor Promoted Inflammation
Inflammation has been proved to be existed at the earliest stage of tumor processes and to be capable of fostering the progression of incipient neoplasia into advanced tumors79. Besides chemicals, particularly reactive oxygen species were positively mutagenic for adjacent cancer cells, accelerating their genetic evolution towards the high malignant carcinoma80. In blood, the increased expression of miR-30c-5p could be used as a new classifier for HCV-positive HCC in early-stage81. In addition, hepatic necroinflammatory activity was associated with the high expression of miR-122-5p in plasma. The over expression of circulating miR-122-5p was a prognostic biomarker predicting the poor survival rate of patients underwent radio frequency ablation49.
Sustained Angiogenesis
Both oxygen and nutrients transported by vasculature are essential for cell survival and function. All cells in tissues obligate to live within 100 μm of a capillary blood vessel. The evidence showed that cells with aberrant proliferative lesions tended to lack angiogenic ability at first, and led to hinder the capability for expansion31. The development of angiogenic ability is vital for incipient neoplasia growth82,83. The over expression of circulating miR-296-5p was significantly associated with tumor angiogenesis42. In tissues, high expression of miR-26a-5p could suppress tumor angiogenesis in HCC by targeting HGF-cMet signaling, and it was a novel prognostic biomarker for HCC84.
Limitless Replicative Potential
There are three factors can lead to an uncoupling of the growth of a cell process from signals in their microenvironment, including insensitivity to antigrowth signals, resistance to apoptosis, and growth signal autonomy. Senescence, just like apoptosis, is as a protective system that could be activated by opposite growth signals or shortened telomeres that drives abnormal cells irreversibly into a G0-like state, and it could prevent further proliferation31. High expression of miR-182-5p, miR-18b-5p, miR-21-5p and miR-224-5p, together with the down-regulated expression of miR-101-3p and miR-139-5p not only played important roles in the regulation of cell proliferation and limitless replicative potential, but also were diagnostic signals for HCC38,39,51,52,54,55,56,85. High expression of miR-106b-5p, miR-21-5p, miR-331-3p and low expression of miR-101-3p, miR-125b-5p, miR-139-5p had great potential to be noninvasive and accurate circulating biomarkers for HCC preliminary screening10,38,51,55,56,61. Moreover, some opposite results about the expression levels of miR-122-5p were discussed44,86. In tissues, high expression of eight miRNAs (i.e. miR-101-3p, miR-106-5p, miR-17-5p, miR-18b-5p, miR-21-5p, miR-25-3p and miR-331-3p) and low expression of seven miRNAs (i.e. miR-125a-5p, miR-128-3p, miR-188-5p, miR-206, miR-212-3p, miR-424-5p and miR-744-5p) were outstandingly correlated with limitless replicative potential and could provide positive prognostic values for HCC38,46,47,48,52,56,64,65,69,70,73,74,76,87. Four prognostic circulating miRNAs associated with proliferation and limitless replicative potential, including miR-101-3p, miR-122-5p, miR-21-5p and miR-331-3p, were reported up-regulated in HCC patients38,56,61,77.
Genome Instability and Mutation
Multi-step cancer progression could be described as a series of genic clonal expansions. Acquiring the chance of an enabling mutant gene triggered these clonal expansions88,89,90. The widespread destabilization of genome is inherent to the vast majority of HCC cells32. The high expression of miR-122-5p and low expression of miR-143-3p in blood were prominently correlated with differentiation and genome instability. They could be used as noninvasive circulating biomarkers for diagnosis of HCC59,62,86. Up-regulated expression of miR-21-5p has been observed to be associated with genome instability and mutation, and it was a novel prognostic biomarker for HCC68. Patients with high serum concentrations of miR-1-3p and miR-122-5p showed a long overall survival time and these miRNAs could be used to assess the HCC staging scores77,91.
Liver injury
Biochemical molecules including miRNAs can be released into the circulation system due to the hypoxia and damage of liver cells. Accumulating reports indicated that serum miR-10b-5p, miR-122-5p, miR-18-5p, miR-192-5p, miR-21-5p, miR-223-3p and miR-885-5p were went up in patients with chronic hepatitis or HCC and they could serve as diagnostic biomarkers for liver injury but not specific for HCC10,42,92,93,94.
Tumor suppressor/onco-miR
Genetic suppressor and carcinogenicity interpreted the function of miRNAs from another perspective. In tissues, high expression of miR-150-5p and miR-29a-5p and low expression of miR-101-3p, miR-126-3p, miR-127-3p, miR-139-5p and miR-214-3p played tumor-suppressor roles and could be used as diagnostic biomarkers for HCC38,39,51,57,95,96,97. The circulating miR-101-3p, miR-122-5p, miR-125b-5p, miR-139-5p, miR-150-5p, miR-16-5p, miR-181a-5p, miR-199a-3p, miR-199a-5p, miR-203a-3p, miR-21-5p, miR-22-3p, miR-29b-3p, miR-375, let-7b-5p correlated with tumor suppressor and could be potential biomarkers to differentiate HCC from healthy controls10,38,41,44,51,58,59,60,86,97,98. On the other hand, miR-101-3p, miR-122-5p, miR-125b-5p, miR-130a-3p, miR-146a-5p, miR-214-3p and miR-99a-5p were considered as tumor suppressors in HCC and served as prognostic indicators for HCC38,99,100,101,102,103,104. Serum miR-1-3p, miR-101-3p, miR-122-5p, miR-150-5p, miR-203a-3p and miR-30c-5p were associated with suppressing tumorigenicity and new independent parameters of overall survival in HCC38,49,60,77,91,105.
The high expression of miR-130b-3p, miR-148a-3p, miR-181b-5p, miR-221-3p, miR-885-5p and miR-96-5p were functional in tumorigenicity and could be served as early diagnostic biomarkers for different tumor type34,106. Meanwhile, miR-10b-5p, miR-130b-3p, miR-146a-5p, miR-18-5p, miR-195-5p, miR-196a-5p and miR-27a-3p were related to carcinogenicity and played vital roles in HCC detection10,34,42,59,94,107. There were six miRNAs associated with oncogenicity and could be potential biomarkers for the overall survival of patients with HCC, including miR-1269a, miR-155-5p, miR-182-5p, miR-183-5p, miR-96-5p and miR-128-266,72,108,109.
Other clinicopathological features
Besides the above ten clinicopathological features and the hallmarks of cancer, biomarker miRNAs were also correlated with other clinicopathological features, such as secretion by primary cancer cells, child stage, cholesterol reverse transport, tumor size and recurrence, etc. Tomimaru et al. found that miR-21-5p was excessively secreted by primary cancer cells and could be a potential diagnostic biomarker for HCC110. Motawi and his colleagues identified that serum miR-34a-5p was correlated with child stage and BCLC score and could be used as an early biomarkers for HCC in high-risk group42. The miR-885-5p and miR-122-5p in serum was reported related to cholesterol reverse transport and assessment of liver pathologies111. In addition, miR-101-3p, miR-106b-5p, miR-130a-3p, miR-16-5p, miR-199a-5p, let-7f-5p and miR-34a-5p were found to have a significant correlation with tumor size in the tissue and serum of HCC patients50,64,102,112,113,114. The present literature also provided evidence that miR-130a-3p, miR-21-5p, miR-25-3p, miR-17-5p were independent prognostic factors and were associated with the TNM classification which is a universally accepted cancer staging system based on extension and size of the primary tumor (T), the adjacent lymph node (N), and the distant metastasis (M)68,69,78,102. The down-regulated expression of miR-774-5p and let-7f-5p can be considered as noninvasive biomarkers for predicting of the recurrence of HCC76,114.
Comparison of HCC biomarker miRNAs based on etiological factors and ethnic groups
Recently, accumulating evidence indicated that the occurrence and development of HCC are closely associated with etiological factors as well as ethnic groups. The differentiation between HCC and liver cirrhosis, for instance, is one of the main problems for the early detection of HCC. Moreover, different etiological factors such as HBV (Hepatitis B Virus) and HCV (Hepatitis C Virus) can also contribute to the HCC carcinogenesis. On the other hand, the incidence and mortality of HCC often showed different patterns among different ethnic groups. Hence it is necessary to compare HCC biomarker miRNAs based on etiological factors and ethnic groups.
Biomarker miRNAs for classifying of HCC and liver cirrhosis
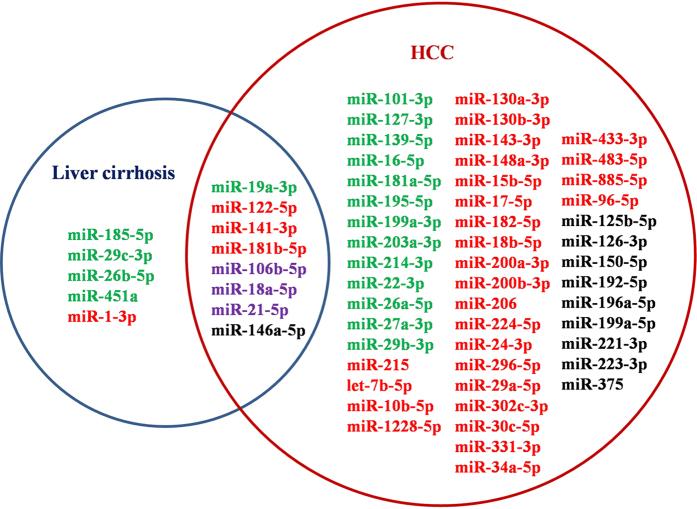
After manually searching for citations in PubMed, a total of 13 miRNA biomarkers for liver cirrhosis diagnosis were collected (see Table S1). We then compared them with HCC diagnostic miRNA biomarkers in order to screen key signatures for HCC early detection. As shown in Fig. 2, eight miRNAs, i.e. miR-106b-5p, miR-122-5p, miR-141-3p, miR-146a-5p, miR-181b-5p, miR-18a-5p, miR-19a-3p and miR-21-5p, were shared by cirrhosis and HCC. Interestingly, three of them (miR-106b-5p, miR-18a-5p and miR-21-5p) showed inverse expression patterns in cirrhosis and HCC groups. For example, the expression of miR-106b-5p (miR-106b) was down in cirrhosis samples115 whereas it turned out to be up-regulated in the blood of HCC patients10. In addition, miR-19a-3p (miR-19a) was reported as a useful molecular marker for monitoring the progression of liver fibrosis to cirrhosis and finally, to HCC42.
Figure 2. The Venn diagram of miRNA biomarkers for liver cirrhosis and HCC.
Here circles in blue and red, respectively, represent miRNAs for cirrhosis and HCC. The miRNAs in red and green represent the up- and down-regulated expression, respectively. The miRNAs in purple means they showed inverse expression patterns in cirrhosis and HCC samples and those in black means their expressions were inconsistently up- or down- regulated according to different literature reports.
The remaining 5 and 49 miRNAs, respectively, were specific to cirrhosis and HCC, which could be served as independent factors for classifying of cirrhosis and HCC. For example, miR-29c-3p showed significant positive correlations with the level of serum cholinesterase (CHE) and albumin (ALB) in liver cirrhosis patients, suggesting that the miRNA played functional roles in the establishment of liver cirrhosis116. Han et al. found that two miRNAs, i.e. miR-224 (miR-224-5p) and miR-214 (miR-214-3p), were significantly up- and down-regulated in HCC tissue samples respectively, which provided novel biomarker signatures for HCC diagnosis and treatment39.
It can be concluded that biomarker miRNAs revealed the pathogenesis of cirrhosis and HCC at the post-transcriptional level and could help deeply understand the differentiation between cirrhosis and HCC. From the perspective of precision medicine, HCC miRNA biomarkers, especially those specific to HCC, were indicators for capturing the early diagnostic signatures at the time of HCC initiation.
Biomarker miRNAs for monitoring the development of HBV/HCV-related HCC
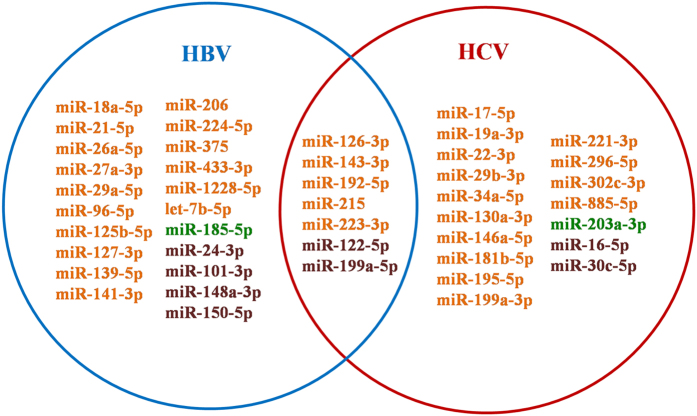
It has been widely acknowledged that the progression of HCC is closely affected by the infection of etiological factors, such as HBV, HCV, etc. On the other hand, miRNAs are reported to play crucial roles in HBV/HCV replication and pathogenesis117,118,119, i.e. they regulated HBV by directly binding to HBV transcripts or changing HBV gene expression at the transcriptional level118. For better investigating the influence of HBV/HCV on HCC development, miRNA biomarkers for HBV/HCV-related HCC were extracted from our collected dataset. As illustrated in Fig. 3, several miRNAs, i.e. miR-122-5p, miR-126-3p, miR-143-3p, miR-192-5p, etc., were functional in both HBV- and HCV-related HCC evolutionary progression. For example, Tan et al. found that serum miR-122-5p could be used as the diagnostic biomarker for detecting HBV-related HCC. Both the area under the receiver operating characteristic curve (AUC) and logistic regression model convinced the predictive power86. Meanwhile, the miRNA was also turned out to be effective for early detection of HCC on top HCV infection. Using the miRNA panel where miR-122-5p included, HCC patients could be classified from healthy controls and liver cirrhosis patients with high diagnostic accuracy120.
Figure 3. The Venn diagram of miRNA biomarkers for HBV/HCV-related HCC.
Here miRNA biomarkers for HBV/HCV-related HCC were extracted from our collected dataset. Circles in blue and red, respectively, represent miRNAs for HBV-related HCC and HCV-related HCC. The miRNAs in orange and dark green represent the diagnostic and prognostic markers, respectively. The miRNAs in brown means they had both diagnostic and prognostic role according to different literature reports.
There is still a large number of biomarker miRNAs that could be specifically used for monitoring the development of HBV/HCV-related HCC. Chen et al. analyzed the plasma samples from 242 individuals and uncovered that the expression of miR-125b-5p (miR-125b) was significantly down-regulated in HBV-induced HCC (HBV-HCC) patients compared to healthy controls as well as HBV groups without HCC121. Moreover, the low plasma level of miR-125b-5p also reflected the higher possibility of metastasis. Therefore, the miRNA held promise as a valuable diagnostic biomarker for HBV-HCC and HBV-infected patients with high HCC risks could be early detected by dynamically monitoring the changes of this miRNA. Liu et al. demonstrated that the expression levels of miR-30c-5p (miR-30c) and miR-203a-3p (miR-203a) were crucial indicators for predicting the poor prognosis of HCV-related HCC because the core protein of HCV could down-regulate the expression of miR-30c-5p and miR-203a-3p, resulting in the activation of epithelial-mesenchymal transition in normal hepatocytes as well as HCC tumor cells. As reported before, the activation process may contribute to the carcinogenesis of HCC105.
Understanding the pathogenesis of miRNA biomarkers in HBV/HCV-related HCC provided insights to evaluate the potential effects of HBV/HCV on HCC development, which will be helpful to the early and personalized detection of HCC.
HCC miRNA biomarkers within different ethnic groups
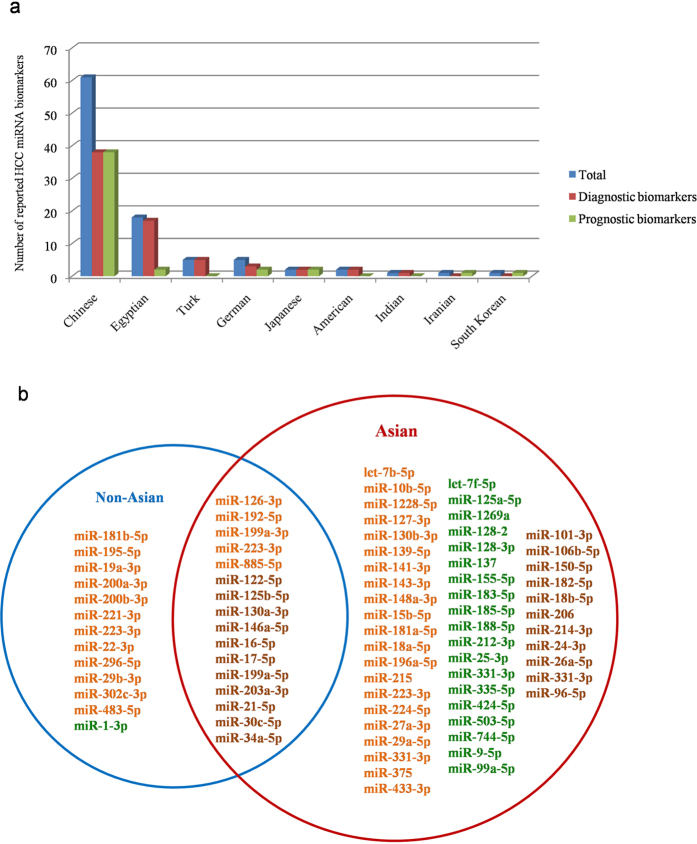
Genomic profiling of HCC tumors showed that HCC patients in different geographic regions tended to have specific recurrent molecular aberrations122. Asians, on the whole, achieved the highest HCC incidence according to the report by Wong et al.123. In terms of prognosis, the overall survival rate was also disparate among different ethnic groups124. Here we reorganized HCC miRNA biomarkers based on the ethnicity of patients described in each citation. As illustrated in Fig. 4a, most of the reported HCC miRNA biomarkers were related to Chinese population, which indirectly indicated the high risk or high incidence of HCC in China. For further exploring the ethnic specificity of HCC miRNA biomarkers, we then partitioned miRNAs into two categories based on the patient race, i.e. Asian-related (Chinese, Japanese, South Korean, Indian and Iranian) and non-Asian-related (Egyptian, American, Turk and German) HCC miRNA biomarkers. As shown in Fig. 4b, the number of Asian-specific HCC miRNA biomarkers is far more than that of non-Asian. We noticed that some miRNAs were reported to be functional in both Asian and non-Asian group. However, the expression pattern of them was sometimes quite different when they were involved in different pathogenic processes or belonged to different ethnic groups. For example, miR-125b-5p was associated with the biological behavior of HCC and had the diagnostic value of HCC for both Turks and Chinese. As in plasma samples of Chinese patients, it was found to be down-regulated121 whereas in Turks samples, its expression level was up33. For comparison of Egyptian and Chinese, the down-regulation of miR-146a-5p was correlated with HCC carcinogenesis and deterioration in Chinese population103, but in samples of Egyptian patients, it was inverse42.
Figure 4. HCC miRNA biomarkers in different ethnic groups.
Here miRNA biomarkers were classified based on the race/nation of patients described in each citation. Sub-figure (a) represents the distribution of reported HCC miRNA biomarkers in different national cohorts. Bars in blue, red and green mean the number of total, diagnostic and prognostic miRNA biomarkers, respectively. Sub-figure (b) is the Venn diagram of HCC miRNA biomarkers for Asian and non-Asian respectively. Circles in blue and red, respectively, represent Asian-related and non-Asian-related miRNA biomarkers. The miRNAs in orange and dark green represent the diagnostic and prognostic markers, respectively. The miRNAs in brown means they had both diagnostic and prognostic role according to different literature reports.
This ethnic difference may be caused by the heterogeneous pathogenesis, lifestyles and various factors including the diet, environmental exposures, etc. Moreover, the incidence of HBV/HCV infection in different countries is also inconsistent. Therefore, more in-depth researches on ethnically specific miRNA biomarkers is of clinical significance, which would provide personalized strategies for HCC diagnosis and treatment in the era of precision medicine.
Pathway enrichment analysis for targets of HCC miRNA biomarkers
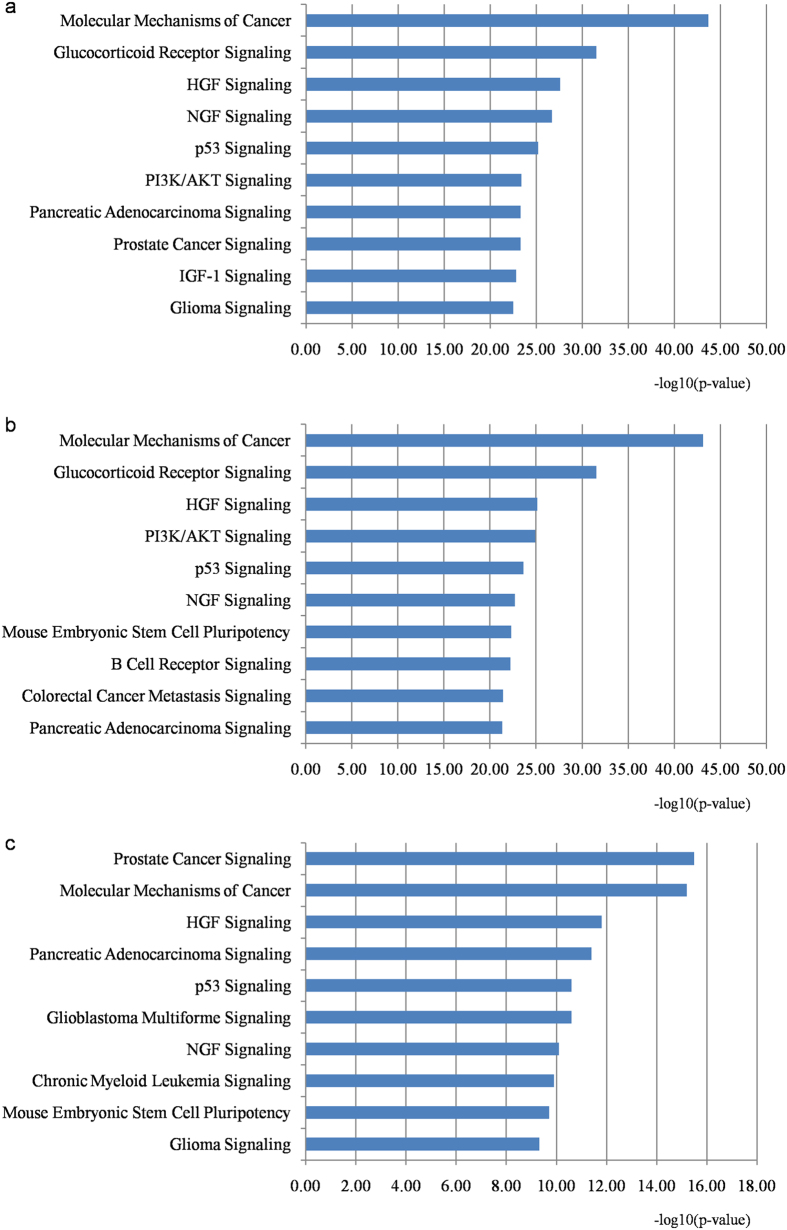
We performed the pathway enrichment analysis for targets of different types of reported miRNA biomarkers using IPA program. Here the targets of miRNA biomarkers originated from seven publicly available miRNA-target databases, including four experimentally validated databases and three computationally predicted databases (see Methods). For the three categories, i.e. the diagnostic, prognostic and therapeutic biomarker miRNAs, the top 10 significantly enriched pathways (p-value < 0.01) were chosen and shown in Fig. 5. The common enriched pathways among them were Molecular Mechanisms of Cancer, Glucocorticoid Receptor Signaling, HGF Signaling, NGF Signaling, p53 Signaling etc. Most of them are well-studied cancer associated pathways. Das et al. reported that the pathway Molecular Mechanisms of Cancer was potentially associated with recurrent HCC secondary to HCV following liver transplantation125. Glucocorticoids are involved in controlling many essential biological processes that are related to energy supply and growth control. The Glucocorticoid Receptor often functions as a cofactor of transcription factor STAT5 for growth hormone induced genes and Glucocorticoid Receptor Signaling has been turned out to be important in body growth, steatosis and metabolic liver cancer development126. The experimental result in mouse model demonstrated that the metabolic dysfunction and impairment of Glucocorticoid Receptor Signaling could cause steatosis and HCC in mice127. Wu et al. revealed that the HGF signaling could be activated by over expression of gene C1GALT1 in HCC via modulation of MET O-glycosylation and dimerization, which offered new insights into O-glycosylation and HCC pathogenesis128. Jin et al. indicated that p53 Signaling pathway was significantly dysregulated in HCC and it could reflect the development and progression of HCC129. Moreover, a number of genes participated in regulating human HCC by interacting with p53 Signaling pathway. For instance, the key gene RASSF10, which is located on chromosome 11p15.2, could suppress the growth of HCC via activating p53 Signaling pathway130. EGR1 is one of the key components in p53 Signaling, the re-expression of gene BCL6B in HCC cells could increase its expression and finally contribute to the activation of p53 Signaling131.
Figure 5. Top 10 pathways significantly enriched with targets of different biomarker miRNAs from HCC tissue and blood.
Sub-figure (a), (b), and (c) represent pathways enriched by targets of diagnostic, prognostic and therapeutic biomarker miRNAs, respectively. The statistical significance level (p-value) was negative 10-based log transformed.
Discussion
In this review, we made comprehensive functional survey and comparison of HCC diagnostic, prognostic and therapeutic miRNAs in blood and tissues. The number of diagnostic miRNA biomarkers in blood is approximately twice as much as those in tissues and meanwhile, the number of prognostic miRNA biomarkers in tissues is twice as much as those in blood. The reason for the statistical difference may be that many studies are inclined to investigate the noninvasive diagnostic miRNA biomarkers and researchers tend to use relatively stable hepatogenic biomarkers as prognostic indicators because miRNAs may be released into the blood selectively132,133. Most of the diagnostic, prognostic and therapeutic miRNA biomarkers are associated with one or two clinic pathological features in blood and tissues. A great number of prognostic biomarkers with high expression levels were detected in patients with shorter overall survival. Since the etiological factors as well as ethnic groups are closely associated with HCC carcinogenesis, we analyzed miRNA biomarkers by taking the HBV/HCV infection as well as regional variations into account in order to provide better clues for HCC pathogenic research. We mainly selected miRNAs which were explicitly reported as HCC markers/biomarkers in our current study. Besides, several miRNAs are still common and important during HCC development. For example, miR-142-3p was functional in HCC tumorigenesis and played a key role in regulating human RAC1 gene. The upregulation of miR-142-3p inhibited the expression level of RAC1 mRNA, suppressing the migration and invasion of HCC cells134. Interferon regulatory factor-1 (IRF-1) is a tumor-suppressor in HCC and its down-expression would help HCC tumors evade death. Yan et al. found that miR-23a was a negative regulator of IRF-1in HCC, which highlighted its importance in HCC initiation and progression135. Zhang et al. demonstrated that miR-99a could directly regulate AGO2 and control tumor growth in HCC, indicating the potential strategies for HCC treatment136.
HCC is a complex disease which is difficult for early diagnosis and treatment. The death rate of HCC remains high due to its poor prognosis. To some extent, miRNAs are effective biomarkers for HCC because of the noninvasive detection, good specificity and sensitivity. More systematic investigations and clinical experiments need to be done for better understanding the role and function of miRNA biomarkers in HCC pathogenesis137,138,139.
Additional Information
How to cite this article: Shen, S. et al. Biomarker MicroRNAs for Diagnosis, Prognosis and Treatment of Hepatocellular Carcinoma: A Functional Survey and Comparison. Sci. Rep. 6, 38311; doi: 10.1038/srep38311 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The work was supported by National Natural Science Foundation of China (NSFC) (grant Nos 91530320, 31670851, 31470821, 31400712).
Footnotes
Author Contributions S.S. and Y.L. contributed equally to the work. S.S. and Y.L. collected and analyzed the data; X.Y., L.S. and L.C. performed the computational analyses; S.S., Y.L., J.C. and B.S. wrote the manuscript; B.S. and L.Q. conceived and supervised the work jointly.
References
- Parkin D. M., Bray F., Ferlay J. & Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 55, 74–108 (2005). [DOI] [PubMed] [Google Scholar]
- Torre L. A. et al. Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108, doi: 10.3322/caac.21262 (2015). [DOI] [PubMed] [Google Scholar]
- Llovet J. M. et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst 100, 698–711, doi: 10.1093/jnci/djn134 (2008). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. Identification of candidate miRNA biomarkers from miRNA regulatory network with application to prostate cancer. J Transl Med 12, 66, doi: 10.1186/1479-5876-12-66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Qian L., Chen J., Chen W. & Shen B. Comparison of Prognostic MicroRNA Biomarkers in Blood and Tissues for Gastric Cancer. J Cancer 7, 95–106, doi: 10.7150/jca.13340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K. & Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med 5, doi: 10.3390/jcm5030036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanmi K., Ignacimuthu S. & Paulraj M. G. MicroRNA in prostate cancer. Clin Chim Acta 451, 154–160, doi: 10.1016/j.cca.2015.09.022 (2015). [DOI] [PubMed] [Google Scholar]
- Tan Z. et al. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/beta-catenin signaling in breast cancer. Oncotarget, doi: 10.18632/oncotarget.8119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Cheng Q., Zhang B. H. & Zhang M. Z. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: a validation set from China. Medicine 94, e603, doi: 10.1097/MD.0000000000000603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Zhang D. C. & Wei C. MicroRNAs as biomarkers for hepatocellular carcinoma diagnosis and prognosis. Clinics and research in hepatology and gastroenterology 39, 426–434, doi: 10.1016/j.clinre.2015.01.006 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. C., Xu Z., Zhang T. F. & Wang Y. L. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol 21, 9853–9862, doi: 10.3748/wjg.v21.i34.9853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. et al. MicroRNAs as a novel class of diagnostic biomarkers in detection of hepatocellular carcinoma: a meta-analysis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 35, 12317–12326, doi: 10.1007/s13277-014-2544-2 (2014). [DOI] [PubMed] [Google Scholar]
- George J. & Patel T. Noncoding RNA as therapeutic targets for hepatocellular carcinoma. Semin Liver Dis 35, 63–74, doi: 10.1055/s-0034-1397350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Ekanem N. R., Sakyi C. A. & Ray S. D. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev 81, 62–74, doi: 10.1016/j.addr.2014.10.029 (2015). [DOI] [PubMed] [Google Scholar]
- Hung C. H., Chiu Y. C., Chen C. H. & Hu T. H. MicroRNAs in hepatocellular carcinoma: carcinogenesis, progression, and therapeutic target. Biomed Res Int 2014, 486407, doi: 10.1155/2014/486407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gori M., Arciello M. & Balsano C. MicroRNAs in nonalcoholic fatty liver disease: novel biomarkers and prognostic tools during the transition from steatosis to hepatocarcinoma. Biomed Res Int 2014, 741465, doi: 10.1155/2014/741465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougelet A. & Colnot S. [microRNA: new diagnostic and therapeutic tools in liver disease?]. Med Sci (Paris) 29, 861–867, doi: 10.1051/medsci/20132910013 (2013). [DOI] [PubMed] [Google Scholar]
- Chai S. & Ma S. Clinical implications of microRNAs in liver cancer stem cells. Chin J Cancer 32, 419–426, doi: 10.5732/cjc.013.10038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wang J., Katayama H., Sen S. & Liu S. M. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma 60, 135–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano S. & Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 57, 840–847, doi: 10.1002/hep.26095 (2013). [DOI] [PubMed] [Google Scholar]
- Borel F., Konstantinova P. & Jansen P. L. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. Journal of hepatology 56, 1371–1383, doi: 10.1016/j.jhep.2011.11.026 (2012). [DOI] [PubMed] [Google Scholar]
- Jiang Q. et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic acids research 37, D98–104, doi: 10.1093/nar/gkn714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Corda B. & Hatzigeorgiou A. G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA 12, 192–197, doi: 10.1261/rna.2239606 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. D. et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research 39, D163–169, doi: 10.1093/nar/gkq1107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F. et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic acids research 37, D105–110, doi: 10.1093/nar/gkn851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino V. A. et al. HOCTAR database: a unique resource for microRNA target prediction. Gene 480, 51–58, doi: 10.1016/j.gene.2011.03.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E. R. et al. Exprtarget: an integrative approach to predicting human microRNA targets. PloS one 5, e13534, doi: 10.1371/journal.pone.0013534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. H., Liu S., Zhou H., Qu L. H. & Yang J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research 42, D92–97, doi: 10.1093/nar/gkt1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research 42, D68–73, doi: 10.1093/nar/gkt1181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- Giray B. G. et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Molecular biology reports 41, 4513–4519, doi: 10.1007/s11033-014-3322-3 (2014). [DOI] [PubMed] [Google Scholar]
- Liu A. M. et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2, e000825, doi: 10.1136/bmjopen-2012-000825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. L. et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget 5, 5113–5124, doi: 10.18632/oncotarget.2089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Q. et al. Metastasis-related miR-185 is a potential prognostic biomarker for hepatocellular carcinoma in early stage. Biomed Pharmacother 67, 393–398, doi: 10.1016/j.biopha.2013.03.022 (2013). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology 58, 158–170, doi: 10.1002/hep.26305 (2013). [DOI] [PubMed] [Google Scholar]
- Xie Y. et al. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther 15, 1248–1255, doi: 10.4161/cbt.29688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. et al. Identification of the typical miRNAs and target genes in hepatocellular carcinoma. Molecular medicine reports 10, 229–235, doi: 10.3892/mmr.2014.2194 (2014). [DOI] [PubMed] [Google Scholar]
- Shen J. et al. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol Biomarkers Prev 22, 2364–2373, doi: 10.1158/1055-9965.EPI-13-0237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K. Z., Zhang K., Li H., Afdhal N. H. & Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 45, 355–360, doi: 10.1097/MCG.0b013e3181f18ac2 (2011). [DOI] [PubMed] [Google Scholar]
- Motawi T. K., Shaker O. G., El-Maraghy S. A. & Senousy M. A. Serum MicroRNAs as Potential Biomarkers for Early Diagnosis of Hepatitis C Virus-Related Hepatocellular Carcinoma in Egyptian Patients. PloS one 10, e0137706, doi: 10.1371/journal.pone.0137706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Garem H. et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol 6, 818–824, doi: 10.4254/wjh.v6.i11.818 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. H. et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. International journal of cancer 138, 714–720, doi: 10.1002/ijc.29802 (2016). [DOI] [PubMed] [Google Scholar]
- Han Z. B. et al. [Expression and survival prediction of microRNA-155 in hepatocellular carcinoma after liver transplantation]. Zhonghua Yi Xue Za Zhi 93, 884–887 (2013). [PubMed] [Google Scholar]
- Yunqiao L., Vanke H., Jun X. & Tangmeng G. MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology 61, 1302–1307 (2014). [PubMed] [Google Scholar]
- Wang W. Y. et al. miR-21 expression predicts prognosis in hepatocellular carcinoma. Clinics and research in hepatology and gastroenterology 38, 715–719, doi: 10.1016/j.clinre.2014.07.001 (2014). [DOI] [PubMed] [Google Scholar]
- Tu H. et al. MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. Onco Targets Ther 8, 2227–2235, doi: 10.2147/OTT.S87976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cho H. J. et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem 48, 1073–1078, doi: 10.1016/j.clinbiochem.2015.06.019 (2015). [DOI] [PubMed] [Google Scholar]
- El-Abd N. E., Fawzy N. A., El-Sheikh S. M. & Soliman M. E. Circulating miRNA-122, miRNA-199a, and miRNA-16 as Biomarkers for Early Detection of Hepatocellular Carcinoma in Egyptian Patients with Chronic Hepatitis C Virus Infection. Mol Diagn Ther 19, 213–220, doi: 10.1007/s40291-015-0148-1 (2015). [DOI] [PubMed] [Google Scholar]
- Li T. et al. Downregulation of microRNA-139 is associated with hepatocellular carcinoma risk and short-term survival. Oncology reports 31, 1699–1706, doi: 10.3892/or.2014.3032 (2014). [DOI] [PubMed] [Google Scholar]
- Murakami Y. et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 13, 99, doi: 10.1186/1471-2407-13-99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayat S. A. et al. The microRNA-200 family–a potential diagnostic marker in hepatocellular carcinoma? J Surg Oncol 110, 430–438, doi: 10.1002/jso.23668 (2014). [DOI] [PubMed] [Google Scholar]
- Amr K. S. et al. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene 575, 66–70, doi: 10.1016/j.gene.2015.08.038 (2016). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): clinical analyses of patients and an HCC rat model. International journal of clinical and experimental pathology 8, 1466–1478 (2015). [PMC free article] [PubMed] [Google Scholar]
- Wang L. J. et al. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget 6, 5932–5946, doi: 10.18632/oncotarget.3465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. T. et al. MicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resection. PloS one 7, e52393, doi: 10.1371/journal.pone.0052393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Hou P., Wu Z., Wang T. & Nie Y. Circulating miR-375 and miR-199a-3p as potential biomarkers for the diagnosis of hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 36, 4501–4507, doi: 10.1007/s13277-015-3092-0 (2015). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 29, 4781–4788, doi: 10.1200/JCO.2011.38.2697 (2011). [DOI] [PubMed] [Google Scholar]
- Yu F., Lu Z., Chen B., Dong P. & Zheng J. microRNA-150: a promising novel biomarker for hepatitis B virus-related hepatocellular carcinoma. Diagn Pathol 10, 129, doi: 10.1186/s13000-015-0369-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chu F., Cao Y., Shao J. & Wang F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 36, 7439–7447, doi: 10.1007/s13277-015-3430-2 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Z. Q. et al. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol 9, 135, doi: 10.1186/1746-1596-9-135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F. L., Wang W. & Jia W. D. Diagnostic and prognostic significance of serum miR-24-3p in HBV-related hepatocellular carcinoma. Med Oncol 31, 177, doi: 10.1007/s12032-014-0177-3 (2014). [DOI] [PubMed] [Google Scholar]
- Li B. K. et al. Upregulation of microRNA-106b is associated with poor prognosis in hepatocellular carcinoma. Diagn Pathol 9, 226, doi: 10.1186/s13000-014-0226-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Jiang M., Yuan W. & Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg 25, 156–161, doi: 10.3109/08941939.2011.618523 (2012). [DOI] [PubMed] [Google Scholar]
- Leung W. K., He M., Chan A. W., Law P. T. & Wong N. Wnt/beta-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer letters 362, 97–105, doi: 10.1016/j.canlet.2015.03.023 (2015). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer 12, 227, doi: 10.1186/1471-2407-12-227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. S. et al. Increased expression of miR-21 predicts poor prognosis in patients with hepatocellular carcinoma. International journal of clinical and experimental pathology 8, 7234–7238 (2015). [PMC free article] [PubMed] [Google Scholar]
- Sadeghian Y. et al. Profiles of tissue microRNAs; miR-148b and miR-25 serve as potential prognostic biomarkers for hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine, doi: 10.1007/s13277-015-3799-y (2015). [DOI] [PubMed] [Google Scholar]
- Chang R. M., Yang H., Fang F., Xu J. F. & Yang L. Y. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology 60, 1251–1263, doi: 10.1002/hep.27221 (2014). [DOI] [PubMed] [Google Scholar]
- Cai L. & Cai X. Up-regulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in patients with hepatocellular carcinoma. Diagn Pathol 9, 1000, doi: 10.1186/s13000-014-0228-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan T. Q. et al. Upregulated MiR-1269 in hepatocellular carcinoma and its clinical significance. International journal of clinical and experimental medicine 8, 714–721 (2015). [PMC free article] [PubMed] [Google Scholar]
- Huang C. Y. et al. miR-128-3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients. Oncology reports 33, 2889–2898, doi: 10.3892/or.2015.3936 (2015). [DOI] [PubMed] [Google Scholar]
- Fang F. et al. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. Journal of hepatology 63, 874–885, doi: 10.1016/j.jhep.2015.05.008 (2015). [DOI] [PubMed] [Google Scholar]
- Li B., Liu L., Li X. & Wu L. miR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun 464, 982–987, doi: 10.1016/j.bbrc.2015.06.169 (2015). [DOI] [PubMed] [Google Scholar]
- Tan Y. L. et al. miR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clinics and research in hepatology and gastroenterology 39, 359–365, doi: 10.1016/j.clinre.2014.09.010 (2015). [DOI] [PubMed] [Google Scholar]
- Xu Y., Bu X., Dai C. & Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 36, 4773–4776, doi: 10.1007/s13277-015-3128-5 (2015). [DOI] [PubMed] [Google Scholar]
- Zheng J., Dong P., Gao S., Wang N. & Yu F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology 60, 549–552, doi: 10.5754/hge12754 (2013). [DOI] [PubMed] [Google Scholar]
- Qian B. Z. & Pollard J. W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51, doi: 10.1016/j.cell.2010.03.014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R. & Karin M. Immunity, inflammation, and cancer. Cell 140, 883–899, doi: 10.1016/j.cell.2010.01.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz Z. et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Molecular biology reports 42, 713–720, doi: 10.1007/s11033-014-3819-9 (2015). [DOI] [PubMed] [Google Scholar]
- Bouck N., Stellmach V. & Hsu S. C. How tumors become angiogenic. Adv Cancer Res 69, 135–174 (1996). [DOI] [PubMed] [Google Scholar]
- Hanahan D. & Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86, 353–364 (1996). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology 59, 1874–1885, doi: 10.1002/hep.26941 (2014). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. MiR-182 is up-regulated and targeting Cebpa in hepatocellular carcinoma. Chin J Cancer Res 26, 17–29, doi: 10.3978/j.issn.1000-9604.2014.01.01 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PloS one 9, e107986, doi: 10.1371/journal.pone.0107986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget 6, 27736–27750, doi: 10.18632/oncotarget.4811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M. & Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 19, 698–711, doi: 10.1016/j.devcel.2010.10.005 (2010). [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8, 286–298, doi: 10.1038/nrg2005 (2007). [DOI] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The epigenomics of cancer. Cell 128, 683–692, doi: 10.1016/j.cell.2007.01.029 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberle V. et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer 49, 3442–3449, doi: 10.1016/j.ejca.2013.06.002 (2013). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 50, 136–142, doi: 10.1002/mc.20712 (2011). [DOI] [PubMed] [Google Scholar]
- Qi P. et al. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PloS one 6, e28486, doi: 10.1371/journal.pone.0028486 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Guo Z., Wang J., Mao Y. & Gao Q. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci 57, 2910–2916, doi: 10.1007/s10620-012-2317-y (2012). [DOI] [PubMed] [Google Scholar]
- Peveling-Oberhag J. et al. MicroRNA Profiling of Laser-Microdissected Hepatocellular Carcinoma Reveals an Oncogenic Phenotype of the Tumor Capsule. Transl Oncol 7, 672–680, doi: 10.1016/j.tranon.2014.09.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. MicroRNA-127 post-transcriptionally downregulates Sept7 and suppresses cell growth in hepatocellular carcinoma cells. Cell Physiol Biochem 33, 1537–1546, doi: 10.1159/000358717 (2014). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncology reports 33, 819–825, doi: 10.3892/or.2014.3641 (2015). [DOI] [PubMed] [Google Scholar]
- Luo J. et al. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. Onco Targets Ther 6, 577–583, doi: 10.2147/OTT.S44215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS letters 586, 4362–4370, doi: 10.1016/j.febslet.2012.10.053 (2012). [DOI] [PubMed] [Google Scholar]
- Xu Q. et al. MicroRNA-122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncology reports 34, 2054–2064, doi: 10.3892/or.2015.4175 (2015). [DOI] [PubMed] [Google Scholar]
- Tsang F. H. et al. Prognostic marker microRNA-125b inhibits tumorigenic properties of hepatocellular carcinoma cells via suppressing tumorigenic molecule eIF5A2. Dig Dis Sci 59, 2477–2487, doi: 10.1007/s10620-014-3184-5 (2014). [DOI] [PubMed] [Google Scholar]
- Li B. et al. MicroRNA-130a is down-regulated in hepatocellular carcinoma and associates with poor prognosis. Med Oncol 31, 230, doi: 10.1007/s12032-014-0230-2 (2014). [DOI] [PubMed] [Google Scholar]
- Rong M., He R., Dang Y. & Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Upsala journal of medical sciences 119, 19–24, doi: 10.3109/03009734.2013.856970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li J., Wang X., Zheng C. & Ma W. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem Biophys Res Commun 439, 47–53, doi: 10.1016/j.bbrc.2013.08.032 (2013). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. Downregulation of miRNA-30c and miR-203a is associated with hepatitis C virus core protein-induced epithelial-mesenchymal transition in normal hepatocytes and hepatocellular carcinoma cells. Biochem Biophys Res Commun 464, 1215–1221, doi: 10.1016/j.bbrc.2015.07.107 (2015). [DOI] [PubMed] [Google Scholar]
- Yuan K. et al. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PloS one 7, e35331, doi: 10.1371/journal.pone.0035331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. et al. Association between miR-146aG >C and miR-196a2C >T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 35, 7775–7780, doi: 10.1007/s13277-014-2020-z (2014). [DOI] [PubMed] [Google Scholar]
- Huang Y. H. et al. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PloS one 7, e37188, doi: 10.1371/journal.pone.0037188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L., Xu L., Wang P. & Meng Z. Serum miR-128-2 serves as a prognostic marker for patients with hepatocellular carcinoma. PloS one 10, e0117274, doi: 10.1371/journal.pone.0117274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y. et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. Journal of hepatology 56, 167–175, doi: 10.1016/j.jhep.2011.04.026 (2012). [DOI] [PubMed] [Google Scholar]
- Gui J. et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 120, 183–193, doi: 10.1042/CS20100297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol Lett 6, 1811–1815, doi: 10.3892/ol.2013.1638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. et al. MicroRNA-34a expression is predictive of recurrence after radiofrequency ablation in early hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 36, 3887–3893, doi: 10.1007/s13277-014-3031-5 (2015). [DOI] [PubMed] [Google Scholar]
- Ge W. et al. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin Lab 60, 427–434 (2014). [DOI] [PubMed] [Google Scholar]
- Chen Y. J. et al. Circulating microRNAs as a Fingerprint for Liver Cirrhosis. PloS one 8, e66577, doi: 10.1371/journal.pone.0066577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B. X. et al. MicroRNA panels as disease biomarkers distinguishing hepatitis B virus infection caused hepatitis and liver cirrhosis. Scientific reports 5, 15026, doi: 10.1038/srep15026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. H., Yeh S. H. & Chen P. J. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochimica et biophysica acta 1809, 678–685, doi: 10.1016/j.bbagrm.2011.04.008 (2011). [DOI] [PubMed] [Google Scholar]
- Shrivastava S., Steele R., Ray R. & Ray R. B. MicroRNAs: Role in Hepatitis C Virus pathogenesis. Genes & diseases 2, 35–45, doi: 10.1016/j.gendis.2015.01.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Shi G. & Li N. The function of MicroRNA in hepatitis B virus-related liver diseases: from Dim to Bright. Annals of hepatology 14, 450–456 (2015). [PubMed] [Google Scholar]
- Zekri A. N. et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine, doi: 10.1007/s13277-016-5097-8 (2016). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Differential expression of plasma miR-125b in hepatitis B virus related liver diseases and diagnostic potential for hepatitis B virus induced hepatocellular carcinoma. Hepatology research: the official journal of the Japan Society of Hepatology, doi: 10.1111/hepr.12739 (2016). [DOI] [PubMed] [Google Scholar]
- Goossens N., Sun X. & Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepatic oncology 2, 371–379, doi: 10.2217/hep.15.26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R. & Corley D. A. Racial and ethnic variations in hepatocellular carcinoma incidence within the United States. The American journal of medicine 121, 525–531, doi: 10.1016/j.amjmed.2008.03.005 (2008). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Improved survival of patients with hepatocellular carcinoma and disparities by age, race, and socioeconomic status by decade, 1983-2012. Oncotarget, doi: 10.18632/oncotarget.10930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T. et al. Molecular Signatures of Recurrent Hepatocellular Carcinoma Secondary to Hepatitis C Virus following Liver Transplantation. Journal of transplantation 2013, 878297, doi: 10.1155/2013/878297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K. M. et al. Hepatic growth hormone and glucocorticoid receptor signaling in body growth, steatosis and metabolic liver cancer development. Molecular and cellular endocrinology 361, 1–11, doi: 10.1016/j.mce.2012.03.026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K. M. et al. Impairment of hepatic growth hormone and glucocorticoid receptor signaling causes steatosis and hepatocellular carcinoma in mice. Hepatology 54, 1398–1409, doi: 10.1002/hep.24509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. M. et al. C1GALT1 enhances proliferation of hepatocellular carcinoma cells via modulating MET glycosylation and dimerization. Cancer research 73, 5580–5590, doi: 10.1158/0008-5472.CAN-13-0869 (2013). [DOI] [PubMed] [Google Scholar]
- Jin B. et al. Identifying hub genes and dysregulated pathways in hepatocellular carcinoma. European review for medical and pharmacological sciences 19, 592–601 (2015). [PubMed] [Google Scholar]
- Jin Y. et al. RASSF10 suppresses hepatocellular carcinoma growth by activating P53 signaling and methylation of RASSF10 is a docetaxel resistant marker. Genes & cancer 6, 231–240, doi: 10.18632/genesandcancer.67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Epigenetic silencing of BCL6B inactivates p53 signaling and causes human hepatocellular carcinoma cell resist to 5-FU. Oncotarget 6, 11547–11560, doi: 10.18632/oncotarget.3413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigati L. et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PloS one 5, e13515, doi: 10.1371/journal.pone.0013515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. S. et al. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic acids research 38, 215–224, doi: 10.1093/nar/gkp857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. et al. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS letters 585, 1322–1330, doi: 10.1016/j.febslet.2011.03.067 (2011). [DOI] [PubMed] [Google Scholar]
- Yan Y., Liang Z., Du Q., Yang M. & Geller D. A. MicroRNA-23a downregulates the expression of interferon regulatory factor-1 in hepatocellular carcinoma cells. Oncology reports 36, 633–640, doi: 10.3892/or.2016.4864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis 3, e97, doi: 10.1038/oncsis.2014.11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang Y., Shen B. & Zhang D. Molecular signature of cancer at gene level or pathway level? Case studies of colorectal cancer and prostate cancer microarray data. Computational and mathematical methods in medicine 2013, 909525, doi: 10.1155/2013/909525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Identifying novel prostate cancer associated pathways based on integrative microarray data analysis. Computational biology and chemistry 35, 151–158, doi: 10.1016/j.compbiolchem.2011.04.003 (2011). [DOI] [PubMed] [Google Scholar]
- Chen J., Sun M. & Shen B. Deciphering oncogenic drivers: from single genes to integrated pathways. Briefings in bioinformatics 16, 413–428, doi: 10.1093/bib/bbu039 (2015). [DOI] [PubMed] [Google Scholar]
- Li L. M. et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer research 70, 9798–9807, doi: 10.1158/0008-5472.CAN-10-1001 (2010). [DOI] [PubMed] [Google Scholar]
- Khairy A., Hamza I., Shaker O. & Yosry A. Serum miRNA Panel in Egyptian Patients with Chronic Hepatitis C Related Hepatocellular Carcinoma. Asian Pacific journal of cancer prevention: APJCP 17, 2699–2703 (2016). [PubMed] [Google Scholar]
- Chen Y., Dong X., Yu D. & Wang X. Serum miR-96 is a promising biomarker for hepatocellular carcinoma in patients with chronic hepatitis B virus infection. International journal of clinical and experimental medicine 8, 18462–18468 (2015). [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. et al. Hepatic miR-126 is a potential plasma biomarker for detection of hepatitis B virus infected hepatocellular carcinoma. International journal of cancer 138, 2732–2744, doi: 10.1002/ijc.29999 (2016). [DOI] [PubMed] [Google Scholar]
- Lin L., Lu B., Yu J., Liu W. & Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clinics and research in hepatology and gastroenterology 40, 397–404, doi: 10.1016/j.clinre.2015.11.005 (2016). [DOI] [PubMed] [Google Scholar]
- Guo W. et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 62, 1132–1144, doi: 10.1002/hep.27929 (2015). [DOI] [PubMed] [Google Scholar]
- Bi Q. et al. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PloS one 7, e40169, doi: 10.1371/journal.pone.0040169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. The Journal of biological chemistry 286, 36677–36685, doi: 10.1074/jbc.M111.270561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Hu Y., Bai B. & Zhang S. Serum miR-335 Level is Associated with the Treatment Response to Trans-Arterial Chemoembolization and Prognosis in Patients with Hepatocellular Carcinoma. Cell Physiol Biochem 37, 276–283, doi: 10.1159/000430352 (2015). [DOI] [PubMed] [Google Scholar]
- Liu H., Li W., Chen C., Pei Y. & Long X. MiR-335 acts as a potential tumor suppressor miRNA via downregulating ROCK1 expression in hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 36, 6313–6319, doi: 10.1007/s13277-015-3317-2 (2015). [DOI] [PubMed] [Google Scholar]
- Yang Y. M. et al. Galpha12 gep oncogene deregulation of p53-responsive microRNAs promotes epithelial-mesenchymal transition of hepatocellular carcinoma. Oncogene 34, 2910–2921, doi: 10.1038/onc.2014.218 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.