Abstract
DNA lasers self-amplify optical signals from DNA analyte as well as thermodynamic differences between sequences, allowing quasi-digital DNA detection. However, these systems have drawbacks, such as relatively large sample consumption and complicated dye labelling. Moreover, although the lasing signal can detect the target DNA, it is superimposed on an unintended fluorescence background, which persists for non-target DNA samples as well. From an optical point of view, it is thus not truly digital detection and requires spectral analysis to identify the target. In this work, we propose and demonstrate an optofluidic laser that has a single layer of DNA molecules as gain material. A target DNA produces intensive laser emission comparable to existing DNA lasers, while any unnecessary fluorescence background is successfully suppressed. As a result, the target DNA can be detected with a single laser pulse, in a truly digital manner. Since the DNA molecules cover only a single layer on the surface of the laser microcavity, the DNA sample consumption is a few orders-of-magnitude lower than that of existing DNA lasers. Furthermore, the DNA molecules are stained by simply immersing the microcavity in the intercalating dye solution, and thus the proposed DNA laser is free of any complex dye-labelling process prior to analysis.
Introduction
A DNA sequence may have base changes induced by biological processes such as single-point mutation and cytosine methylation, and its analysis is of great interest in disease diagnosis and individualized medicine.1–10 Distinguishing a target DNA from the base-altered DNAs provides valuable information not only for clinical research, but also various genetics-related fields from molecular palaeontology to forensic science.11–16 Owing to the growing necessity of DNA detection, a number of DNA-analysing platforms, such as DNA microarrays and high resolution melting analysis, have been suggested over the past few decades.17–26 The majority of optical DNA detection systems rely on simple birefringence of DNA molecules27, 28 and fluorescence from dye-labelled DNA sequences, through changes in their intensity and/or color.21, 29–34 Meanwhile, a laser-based DNA analysis demonstrates an optical amplification of the signals inside the laser cavity, which results in several advantages over fluorescence-based methods, such as an improved discrimination ratio between target and non-target DNAs, and dramatically higher optical signal intensities.35–38 Moreover, thanks to the nonlinear nature of lasing phenomena, it is possible to distinguish the target DNA with its laser emission, which allows a quasi-digital detection scheme with only a single pulse of laser excitation.35–37
However, the existing laser-based DNA analysis systems, such as the molecular beacon laser,36 include dye labelling on the analyte DNA molecule, similar to traditional fluorescence based methods, and thus require complex sample preparation prior to analysis. Since the biomolecular sample is not abundant in most cases, a relatively large sample consumption is another drawback of the DNA lasers. The concentration of DNA/dye molecules in the gain medium of the DNA laser is a few orders-of-magnitude higher (a few tens to hundreds μM) than that of the traditional fluorescence-based methods (a few to sub-μM),21, 23–25, 35–40 and thus the DNA sample consumption of the laser-based DNA detection is significant. Even worse, the DNA/dye molecules that do not contribute to the laser are not simply wasted, but generate unintended fluorescence background superimposed on the laser signal. This background signal persists even for non-lasing non-target DNAs; thus, from an optical point of view, it is improper to argue that the existing laser-based DNA detection systems are truly digital.
In this article, we propose and demonstrate a novel optofluidic ring resonator (OFRR) laser system that can detect a target DNA sequence in a truly digital manner. Instead of utilizing bulk gain medium filling out the entire laser cavity, the proposed laser has a single layer of DNA molecules, which provides laser gain, on the OFRR surface. This laser system has successfully removed unintended fluorescence background from its optical signal, since all of the gain DNA molecules contribute to the laser phenomena. More importantly, when there is no laser from non-target DNAs, there is virtually no fluorescence from only a single layer of DNA molecules. Consequently, detection of the target DNA can be completed with only the presence of the optical signal, which is purely a laser emission, without any spectral analysis. Since only a small amount of DNA that can cover a single layer on the surface of the human-hair-thick fibre OFRR is required, the DNA sample consumption is dramatically lower than that of existing OFRR lasers in current DNA detecting platforms. The concentration of DNA molecules used in this laser is a few orders-of-magnitude lower, which is comparable to that of conventional fluorescence-based methods, and the corresponding DNA sample consumption in liquid volume is also extremely small, at only a few μL. Furthermore, the novel DNA laser is realized with DNA intercalating dye, and thus dye-labelling processes on the analyte DNA sample are not necessary. The OFRR with probe-target hybridized DNAs attached on the surface is stained with the intercalating dye molecules simply by immersing the fibre OFRR in the dye solution, which is followed by the DNA laser analysis.
Experimental
The preparation of an OFRR laser system is adapted from the previous work.41 A single mode optical fibre (SMF-28®, Corning®), with 125 μm diameter, is selected as a ring resonator microcavity of the laser system. The optical fibre OFRR is known to have an extremely high Q-factor that exceeds 106, which supports the whispering gallery mode (WGM) circulating along its outer surface.42–44
To prepare the fibre for surface functionalization, the polymer cladding is first removed from a 5-mm section at the tip of the fibre. The glass core of the fibre tip is then cleaned by multiple steps. It is sonicated in acetone, ethanol, and deionized (DI) water, for 15 min each, and then immersed in a hydrochloric acid / ethanol mixture (HCl:ethanol ratio of 1:1 by volume) for 30 min, followed by rinsing in DI water and drying. The clean fibre tip is then silanized by dipping into (3-aminopropyl)-trimethoxysilane (3-APTMS, 5 % in methanol) for 30 min. After rinsing with ethanol, the fibre is cured at 110 °C for 1 hr and cooled to room temperature.
To immobilize the DNA probes on the silanized fibre surface, the fibre tip is first activated with bis(sulfosuccinimidyl) suberate (BS3), which is a homofunctional amine-to-amine cross-linker. BS3 is dissolved in phosphate-buffered saline (PBS) with concentration of 0.1 mg/mL and the fibre tip is immersed in the solution for 30 min. After rinsing in pristine PBS, the activated fibre is incubated with streptavidin by dipping the fibre into 1 mg/mL streptavidin/PBS solution for 30 min. The concentration of streptavidin is set to be relatively high to ensure sufficient molecular coverage on the fibre surface after incubation. The streptavidin-incubated fibre is subsequently rinsed with PBS, DI water, and TAE (tris-acetate-EDTA)/12.5 mM MgCl2 buffer solution.
The probe single-stranded DNA (ssDNA) molecules are 40 bases long, and biotinylated at their 5’ ends. All of the 40-bases-long ssDNA samples (probe/target/single-base-mismatch) are purchased from Integrated DNA Technologies, and the base sequences are presented in Table S1 in the supplementary information. The probe DNA is dissolved in TAE/MgCl2 buffer solution with concentration 1 μM, and the fibre tip is dipped into the solution for 30 min. Since only a 2-mm length of the fibre tip is subject to the probe immobilization, only a few μL of the probe DNA solution is sufficient for this process. After the probe DNA molecules are attached to the fibre surface by well-known streptavidin-biotin binding,25, 45 the fibre tip with surface-immobilized probe DNAs can analyse any DNA molecule of interest in a liquid sample. Subsequently, the fibre tip DNA probe is dipped into unlabelled DNA analyte solution for hybridization, followed by DNA intercalating dye (SYTO®-13 Green Fluorescent Nucleic Acid Stain, Life-technologies) staining.
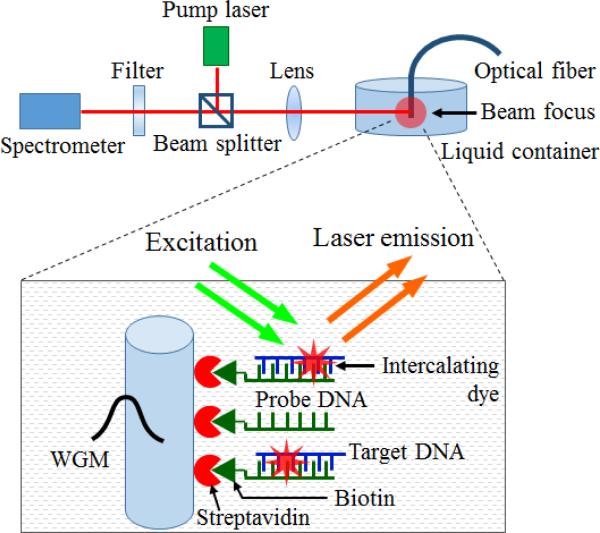
The optical setup utilized in our experiment is illustrated in Fig. 1. The fibre OFRR with surface-attached DNA molecules is immersed in TAE/MgCl2 buffer solution and pumped with an optical parametric oscillator (OPO) (Continuum, 5 ns pulsed laser, 20 Hz repetition rate) with 488 nm wavelength. The pump laser is focused on the 2-mm tip of the fibre through a confocal setup and the DNA laser emission is simultaneously collected by the spectrometer, (HR550i, Horiba Jobin Yvon).
Fig. 1.
Schematic of experimental setup: 2-mm tip of a fiber OFRR (SMF-28® optical fiber core) in liquid environment (tris-EDTA buffer solution) is pumped with OPO laser source through a confocal setup, and the laser emission is simultaneously collected by a spectrometer. The fiber OFRR supports WGM and its evanescent field interacts with surface-attached molecules, providing a feedback for lasing phenomena. Inset: Single molecular layer of DNA laser gain on the OFRR surface. The fiber OFRR is immersed in a series of solutions and finally functionalized with the probe ssDNA. The fiber DNA probe is hybridized with the target DNA in liquid sample, followed by the intercalating dye staining. The active DNA intercalating dye (SYTO®-13 Green Fluorescent Nucleic Acid Stain) combined with the hybridized DNA molecules contributes to the laser.
Results and discussion
The fibre OFRR with the probe DNAs immobilized on the surface is a DNA probe, which can be dipped into any liquid bio-sample for DNA analysis. The target ssDNA molecules that have complimentary base pairs with the probe ssDNA are prepared to demonstrate a target detection of our fibre DNA probe. To validate the specificity, the target ssDNA is dissolved in a TAE/MgCl2 buffer solution along with random ssDNA molecules (Sigma-Aldrich, average length of 2000 bases). The concentrations of the target DNA molecules and the random DNAs are 100 nM and 100 ng/μL, respectively. Considering the average molecular weight of the random DNAs (~1.1 MDa), the molar concentration of the random DNA sample is similar to that of the target DNA. The fibre DNA probe is immersed into the prepared analyte solution for 30 min at room temperature. The target DNA concentration in this experiment is more than 2 orders-of-magnitude lower than existing DNA lasers.35, 36 The lower target DNA concentration results in inferior lasing characteristics, as well as unreliable discrimination of the target and counterparts, due to an insufficient surface coverage, thus the analyte DNA concentration is kept no lower than 100 nM through the hybridization process. Since the fibre DNA probe is 125 μm in diameter and 2 mm long, only a few μL of the analyte solution is required to cover the entire surface of the probe and induce target DNA hybridization, similar to the probe DNA immobilization. After rinsing with the buffer solution, the fibre DNA probe with hybridized target DNA molecules is then stained with the DNA intercalating dye. The SYTO®-13 DNA intercalating dye, originally in dimethyl sulfoxide solution with 5 mM concentration, is five times diluted with the TAE/MgCl2 buffer solution. The fibre OFRR is dipped into the dye solution for 5 min at room temperature and we can expect the final molecular structure on the OFRR surface to be as illustrated in the inset of Fig. 1.
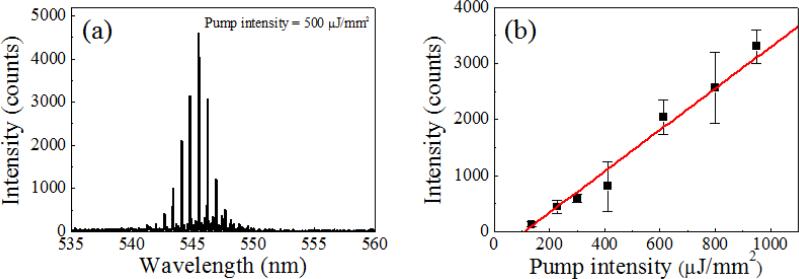
After intercalating dye staining, active fluorescent dyes combined with double stranded DNA (dsDNA) molecules are on the surface of the fibre OFRR, providing an optical feedback for lasing phenomena through interaction with the evanescent field of the WGM-confined light. The fibre OFRR is immersed in pristine TAE/MgCl2 buffer solution and optically pumped with OPO, as previously described. Fig. 2 depicts the laser emission spectrum when the pump intensity is 500 μJ/mm2, and the corresponding lasing characteristics of the DNA laser. Fig. 2(a) shows a typical lasing spectrum of a ring resonator laser. The center wavelength of the laser is 545.5 nm, and the free spectral range (FSR) is 0.6 nm. The FSR matches approximately well with calculations using the ring resonator diameter and effective refractive index. Meanwhile, when the DNA probe is immersed in a solution with the random DNA sample alone through the hybridization process, the fibre OFRR does not show laser emission, even at high pump intensity. Since only the intercalating dye combined to dsDNAs can contribute to the fibre OFRR laser, we can conclude that only the target DNA is hybridized with the probe DNA, as expected. This is direct proof that our DNA laser with the fibre OFRR and surface-immobilized DNA serves successfully as a target DNA molecular detector.
Fig. 2.
(a) Lasing spectrum from the optical fiber OFRR with hybridized target DNA. The fiber DNA probe is dipped into the liquid sample simultaneously containing the 40-bases-long target ssDNA and random ssDNA. The concentration of the target ssDNA is 100 nM and that of the random ssDNA sample is 100 ng/μL. Considering the average length of the random ssDNA, (~2000 bases, ~1.1 MDa) the molar concentration of the random ssDNA is approximately equal to that of the target. The OFRR with hybridized and stained DNA molecules is immersed in clean buffer solution, and optically analyzed. The pump intensity is 500 μJ/mm2. (b) Laser emission intensity as a function of the pump intensity. The estimated lasing threshold is 108 μJ/mm2. Error bars are obtained with five measurements.
Fig. 2(b) indicates the corresponding laser intensity as a function of OPO pump intensity. The presented laser intensity is spectrally integrated. The curve shows a clear lasing characteristic with a laser threshold of 108 μJ/mm2. Compared to the previous work with glass capillary OFRR and DNA/intercalating dye solution,35 the present lasing threshold is approximately two times lower, even though the DNA sample concentration in this work is more than 2 orders-of-magnitude lower, owing to DNA/dye molecules successfully attached and concentrated on the OFRR surface. However, the lasing threshold is higher than that of the recent OFRR laser with single molecular layer gain demonstrated with enhanced green fluorescent protein (eGFP) or Cy3/Cy5 dye labelled DNA.41 This is due mainly to the relatively low fluorescence quantum yield (QY) of the DNA intercalating dye, and imperfect coverage of the OFRR surface with DNA/dye molecules. Further evaluation of this issue follows later in this article.
To evaluate the specificity of our DNA detection system, the ssDNA molecule with the same molecular length as the target ssDNA but a single-base mismatch in the middle of the sequence is prepared (sequence is also provided in Table S1). The single-base mismatch DNA sample is dissolved in TAE/MgCl2 buffer solution with the same concentration as the target DNA solution (100 nM). The fibre DNA probe is dipped into both the target DNA solution and the single-base mismatch DNA solution for 30 min. This hybridization process is carried out at elevated temperature for improved discrimination between the target and the single-base-mismatched DNA. The hybridization temperature is limited to 55 °C, due to the instability of streptavidin-biotin binding at excessively high temperature.46 Due to this binding issue, it is impossible to denature dsDNA molecules with retained probe DNA immobilization, thus the DNA hybridized fiber is not reusable. After this hybridization process, both fibre OFRRs are rinsed with the buffer solution and stained with the DNA intercalating dye at room temperature, as in the previous experiments.
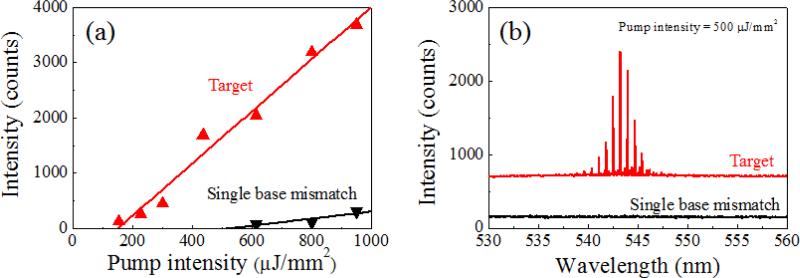
Fig. 3 shows a comparison between the lasing characteristics of the target DNA laser and the single-base-mismatched DNA laser. The two curves in Fig. 3(a) are the spectrally integrated laser intensities for the target DNA (red) and the single-base-mismatched DNA (black), as functions of the pump intensity. The target shows a clear lasing characteristic with a relatively low lasing threshold of 150 μJ/mm2 and high lasing efficiency, while the single-base mismatch has a higher lasing threshold of 530 μJ/mm2 with a lasing efficiency five times lower than that of the target. The thermodynamic difference between the target and the single-base-mismatched DNA is optically amplified through the laser phenomena,35, 37 resulting in this large difference in lasing characteristics, which enables digital target DNA detection. Fig. 3(b) presents lasing spectra from the target and the single-base mismatch when both fibre OFRRs are pumped at 500 μJ/mm2, where the curves have been vertically shifted for clarity. The target shows a laser emission spectrum nearly identical to that of the previous result in Fig. 2(a), while the single-base mismatch shows virtually no laser emission. Since the pump intensity is in between the lasing thresholds of the two, thanks to the nonlinear properties of the laser, we are able to distinguish the target DNA from the non-target counterpart only by the presence of the laser emission. For comparison, hollow-core glass capillary OFRR35, 47 is infiltrated with the target and single-base-mismatched DNA/intercalating dye solution and excited with an OPO optical pump using the same experimental setup at 55 °C. The resulting lasing spectra are presented in Fig. S1. Both cases show similar fluorescence backgrounds, although superimposed laser emissions differentiate the target DNA from single-base-mismatched DNA, and thus, spectral analysis is required to identify the target using these optical signals. Meanwhile, our DNA laser can distinguish the target DNA simply with the presence of an optical output, which is purely a laser signal. Therefore, we can claim that our DNA laser detection system is a truly digital and rapid method to distinguish the target DNA from others, with a single pulse of laser excitation, without any further spectral analysis. Furthermore, it is proven that our DNA detection platform is capable of discriminating even a single-base mismatch in the DNA sequence.
Fig. 3.
Comparison of lasing characteristics between target (red) and single-base-mismatched (black) DNA, 40 bases long: (a) Laser emission intensity as a function of the pump intensity. The lasing thresholds are 150 μJ/mm2 and 530 μJ/mm2 for the target and the single-base-mismatched DNA laser, respectively. The lasing efficiency also shows more than 5-fold decrease for the single-base-mismatched DNA sample. (b) Lasing spectra comparison when pump intensity is 500 μJ/mm2: Target DNA shows clear laser emission similar to Fig. 2(a), while the single-base-mismatch DNA shows zero signal. Through DNA hybridization, temperature is elevated to 55 °C to increase specificity. Note that the curves are vertically shifted for clarity.
As mentioned previously, the lasing characteristics of the proposed DNA laser are not superior to those in the earlier work41 with dye molecules non-specifically attached to the surface. Although relatively low fluorescence QY of the DNA intercalating dye utilized in this work (~0.40) is considered, the lasing threshold is higher than the expected. Since the surface density of the molecule might play a critical role in the lasing threshold, we can expect an improved lasing characteristic when the surface density of the DNA/dye single molecular layer is increased.
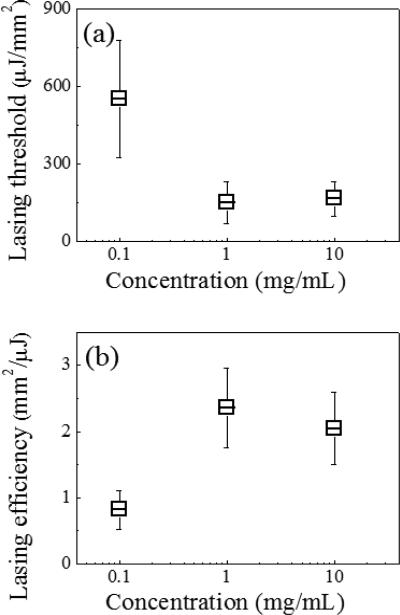
Fig. 4 illustrates the lasing thresholds (a) and efficiencies (b) of the DNA lasers with the hybridized target DNA, when the streptavidin concentration during the fibre DNA probe preparation is 10 times lower/higher than the proposed standard procedure. The fibre OFRR fabrication processes other than the streptavidin incubation, followed the standard recipe described above, including the DNA hybridization. When the BS3 treated fibre is incubated with 0.1 mg/ml streptavidin/PBS solution, (10 times lower than the standard) the corresponding laser has a lasing threshold of 550 μJ/mm2, approximately five times higher than the standard DNA laser with 1 mg/ml streptavidin incubation. The lasing efficiency is also three times lower. Both of these indicate that the surface density is approximately 60 % decreased, according to a previous theoretical analysis.41 However, a higher concentration of streptavidin does not result in an improved surface density or lasing characteristic. When the streptavidin concentration is 10 times higher, i.e., 10 mg/mL the corresponding lasing threshold is even slightly higher and the lasing efficiency is ~15 % lower than that of the standard DNA laser. This indicates that the surface density of the streptavidin cannot be increased further with higher concentration, owing to an issue of steric hindrance.45 We may have to also consider that the probe DNA suffers from the crowding effect when the surface density is excessively high,25, 48 which impairs the target DNA hybridization. It is certain that our standard DNA laser is in the optimal range for detecting small quantities of target DNA molecules and there is not much scope to improve its efficiency.
Fig. 4.
(a) Lasing thresholds and (b) lasing efficiencies as functions of streptavidin concentration during incubation. Preparation of the fiber OFRR other than the streptavidin incubation followed the standard procedure. Error bars are obtained with three measurements.
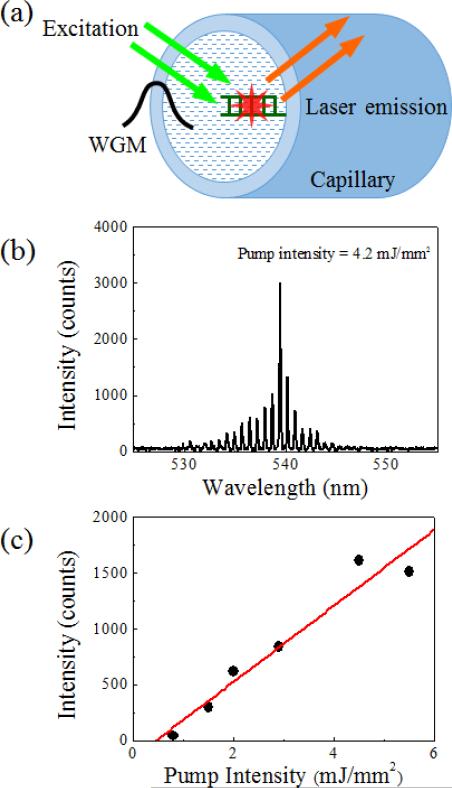
While the DNA laser proposed in this article is based on the optical fibre OFRR, it is possible to demonstrate the same DNA detection system with the traditional hollow-core glass capillary OFRR, which has advantages in integrating with microfluidic platforms over the fibre.35, 47, 49 Fig. 5(a) shows a schematic of a glass capillary OFRR laser with target-hybridized DNA and intercalated dye molecules attached on the inner surface. The glass capillary in this experiment has a diameter of 100 μm. A thinner wall gives improved lasing characteristics, owing to enhanced interaction between the evanescent field and the molecular gain; however, the wall thickness is maintained at 5 μm for mechanical reliability. Instead of immersing the entire OFRR into the buffer solution as required for the fibre, this capillary OFRR is rinsed and filled with the buffer solution after performing the same surface functionalization processes as with the fibre. Fig. 5(b) depicts a representative laser spectrum from the capillary OFRR DNA laser when it is pumped at 4.2 mJ/mm2. The curve shows a nearly identical spectrum to that of the fibre OFRR laser, while the FSR is slightly increased (0.72 nm), which corresponds to the smaller diameter. The lasing threshold estimated from the curve in Fig. 5(c) is approximately 500 μJ/mm2. The lasing threshold is increased by a factor of 5, compared to the fibre OFRR laser. This is caused by imperfect coverage of gain molecules on the inner surface of the capillary, as well as limited interaction between the WGM evanescent field and the molecular gain. Although the surface treatments after the cleaning processes are the same, the capillary cannot be sonicated during the cleaning process, owing to low mechanical reliability. This may be a main reason why the DNA/dye gain molecule coverage on the capillary inner surface is lower than the fibre surface, resulting in inferior lasing characteristics. However, this capillary OFRR allows an alternative method of introducing a DNA sample to the detection system, for example, connecting to up/downstream sample processing devices.
Fig. 5.
(a) Schematic of the traditional hollow-core glass capillary OFRR with stained DNA immobilized on the inner surface. The same optical setup is utilized to analyze the laser activity from this scheme. The wall thickness and the diameter of the glass capillary are 5 and 100 μm, respectively. (b) Lasing spectrum from the OFRR laser when the pump intensity is 4.2 mJ/mm2. (c) Laser emission intensity as a function of pump intensity. The estimated lasing threshold is 500 μJ/mm2. Note that the lasing threshold is increased by a factor of 5, owing to inefficient surface functionalization compared to the outer surface of the optical fiber. The wall thickness of the capillary OFRR also affects the performance of the laser. A thinner wall gives a lower lasing threshold as well as improved lasing efficiency, due to the limited penetration depth of the WGM evanescent field. However, the wall cannot be further thinned down from 5 μm, since excessively thin-walled glass capillary has very low mechanical reliability and can be broken by intensive external optical pumping.
Conclusions
In summary, we have demonstrated a novel optical DNA detection system based on the OFRR laser with a single layer of DNA molecules on its surface. The target DNA can be detected from its laser emission, while non-target DNA even with a single-base mismatch has virtually zero optical signal, since the intrinsic fluorescence background is successfully suppressed. This allows optical detection of DNA in a truly digital manner, with only the presence of an optical output. Therefore, our system dramatically facilitates and simplifies the target DNA detection. Moreover, the DNA sample consumption of our system is a few orders-of-magnitude smaller than that of existing laser-based DNA detection methods, owing to lower DNA molecular concentration as well as extremely small liquid sample volume. Since the DNA intercalating dye is utilized as laser dye gain, it is free from complex and expensive dye-labelling processes on DNA samples prior to analysis. With the compact and versatile geometry of the optical fibre/glass capillary OFRR laser system, we foresee numerous potential applications for our DNA detection platform to micro total analysis systems.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Research Foundation (NRF) and was funded by the Korean Government (MSIP) (2012M3A7B4049802). Chen and Fan: National Science Foundation (ECCS-1607250, DBI-1451127) and the National Institutes of Health (NIBIB-1R21EB016783).
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
References
- 1.Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 2.Christopoulos TK. Anal. Chem. 1999;71:425–438. [Google Scholar]
- 3.Robertson KD. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 4.Taylor RW, Turnbull DM. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson SP, Bartek J. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawkins RD, Hon GC, Ren B. Nat. Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupski JR. Trends Genet. 1998;14:417–426. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Shastry BS. Pharmacogenomics J. 2006;6:16–21. doi: 10.1038/sj.tpj.6500338. [DOI] [PubMed] [Google Scholar]
- 10.van't Veer LJ, Bernards R. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi R, Beroldingen C. H. v., Sensabaugh GF, Erlich HA. Nature. 1988;332:543–546. doi: 10.1038/332543a0. [DOI] [PubMed] [Google Scholar]
- 12.Wilson MR, Stoneking M, Holland MM, Dizinno JA, Budowle B. Crime Lab. Digest. 1993;20:68–77. [Google Scholar]
- 13.Kapitonov VV, Jurka J. Proc. Natl. Acad. Sci. USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckleton JS, Bright J-A, Taylor D. Forensic DNA Evidence Interpretation. CRC Press; 2004. [Google Scholar]
- 15.Shapiro B, Hofreiter M. Science. 2014;343:1236573. doi: 10.1126/science.1236573. [DOI] [PubMed] [Google Scholar]
- 16.Jablonski D, Shubin NH. Proc. Natl. Acad. Sci. USA. 2015;112:4852–4858. doi: 10.1073/pnas.1505146112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 18.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Berre V. Nucleic Acids Res. 2003;31:88e–88. doi: 10.1093/nar/gng088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Maier S, Nimmrich I, Yan PS, Caldwell CW, Olek A, Huang TH. J. Cell Biochem. 2003;88:138–143. doi: 10.1002/jcb.10313. [DOI] [PubMed] [Google Scholar]
- 21.Reed GH, Wittwer CT. Clin. Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 22.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery J, Wittwer CT, Palais R, Zhou L. Nat. Protoc. 2007;2:59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- 24.Sassolas A, Leca-Bouvier B. a. D., Blum L. ı. J. Chem. Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 25.Nimse SB, Song K, Sonawane MD, Sayyed DR, Kim T. Sensors. 2014;14:22208–22229. doi: 10.3390/s141222208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahrokhian S, Salimian R, Kalhor HR. RSC Adv. 2016;6:15592–15598. [Google Scholar]
- 27.Cha YJ, Gim MJ, Oh K, Yoon DK. ACS Appl. Mater. Interfaces. 2015;7:13627–13632. doi: 10.1021/acsami.5b03321. [DOI] [PubMed] [Google Scholar]
- 28.Cha YJ, Yoon DK. Adv. Mater. doi: 10.1002/adma.201604247. in press. [DOI] [PubMed] [Google Scholar]
- 29.Cox WG, Singer VL. Biotechniques. 2004;36:114–122. doi: 10.2144/04361RR02. [DOI] [PubMed] [Google Scholar]
- 30.Zeglis BM, Barton JK. Nat. Protoc. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Qian S, Chen X, Chen J, Lin Y, Liu J. RSC Adv. 2012;2:2562. [Google Scholar]
- 32.Valentini P, Pompa PP. RSC Adv. 2013;3:19181. [Google Scholar]
- 33.Zhang H, Li F, Dever B, Li XF, Le XC. Chem. Rev. 2013;113:2812–2841. doi: 10.1021/cr300340p. [DOI] [PubMed] [Google Scholar]
- 34.Nuhiji E, Mulvaney P. Small. 2007;3:1408–1414. doi: 10.1002/smll.200600676. [DOI] [PubMed] [Google Scholar]
- 35.Lee W, Fan X. Anal. Chem. 2012;84:9558–9563. doi: 10.1021/ac302416g. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Fan X. Angew. Chem. Int. Ed. 2012;51:1236–1239. doi: 10.1002/anie.201107381. [DOI] [PubMed] [Google Scholar]
- 37.Fan X, Yun SH. Nat. Methods. 2014;11:141–147. doi: 10.1038/nmeth.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Shopova SI, Wu CS, Arnold S, Fan X. Proc. Natl. Acad. Sci. USA. 2010;107:16039–16042. doi: 10.1073/pnas.1003581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tjong V, Tang L, Zauscher S, Chilkoti A. Chem. Soc. Rev. 2014;43:1612–1626. doi: 10.1039/c3cs60331h. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Liu H, Lee W, Sun Y, Zhu D, Pei H, Fan C, Fan X. Lab Chip. 2013;13:3351–3354. doi: 10.1039/c3lc50629k. [DOI] [PubMed] [Google Scholar]
- 41.Chen Q, Ritt M, Sivaramakrishnan S, Sun Y, Fan X. Lab Chip. 2014;14:4590–4595. doi: 10.1039/c4lc00872c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shopova SI, Zhu H, Fan X. Appl. Phys. Lett. 2007;90:221101. [Google Scholar]
- 43.Moon H-J, Chough Y-T, An K. Phys. Rev. Lett. 2000;85:3161–3164. doi: 10.1103/PhysRevLett.85.3161. [DOI] [PubMed] [Google Scholar]
- 44.Moon H-J, Chough Y-T, Kim JB, An K. Appl. Phys. Lett. 2000;76:3679–3681. [Google Scholar]
- 45.Wilchek M, Bayer EA, Livnah O. Immunol. Lett. 2006;103:27–32. doi: 10.1016/j.imlet.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 47.Lacey S, White IM, Sun Y, Shopova SI, Cupps JM, Zhang P, Fan X. Opt. Express. 2007;15:15523–15530. doi: 10.1364/oe.15.015523. [DOI] [PubMed] [Google Scholar]
- 48.Vainrub A, Pettitt BM. Biopolymers. 2003;68:265–270. doi: 10.1002/bip.10271. [DOI] [PubMed] [Google Scholar]
- 49.Suter JD, White IM, Zhu H, Shi H, Caldwell CW, Fan X. Biosens. Bioelectron. 2008;23:1003–1009. doi: 10.1016/j.bios.2007.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.