Abstract
Isomeric molecules present a challenge for analytical resolution and quantification, even with MS-based detection. The eight-aminobutyric acid (ABA) isomers are of interest for their various biological activities, particularly γ-aminobutyric acid (GABA) and the d- and l-isomers of β-aminoisobutyric acid (β-AIBA; BAIBA). This study aimed to investigate LC-MS/MS-based resolution of these ABA isomers as their Marfey's (Mar) reagent derivatives. HPLC was able to separate three Mar-ABA isomers l-β-ABA (l-BABA), and l- and d-α-ABA (AABA) completely, with three isomers (GABA, and d/l-BAIBA) in one chromatographic cluster, and two isomers (α-AIBA (AAIBA) and d-BABA) in a second cluster. Partially separated cluster components were deconvoluted using Gaussian peak fitting except for GABA and d-BAIBA. MS/MS detection of Marfey's derivatized ABA isomers provided six MS/MS fragments, with substantially different intensity profiles between structural isomers. This allowed linear deconvolution of ABA isomer peaks. Combining HPLC separation with linear and Gaussian deconvolution allowed resolution of all eight ABA isomers. Application to human serum found a substantial level of l-AABA (13 μM), an intermediate level of l-BAIBA (0.8 μM), and low but detectable levels (<0.2 μM) of GABA, l-BABA, AAIBA, d-BAIBA, and d-AABA. This approach should be useful for LC-MS/MS deconvolution of other challenging groups of isomeric molecules.
Keywords: Aminobutyric acid isomers, LC-MS/MS, Chromatographic deconvolution, Marfey's derivatization
1. INTRODUCTION
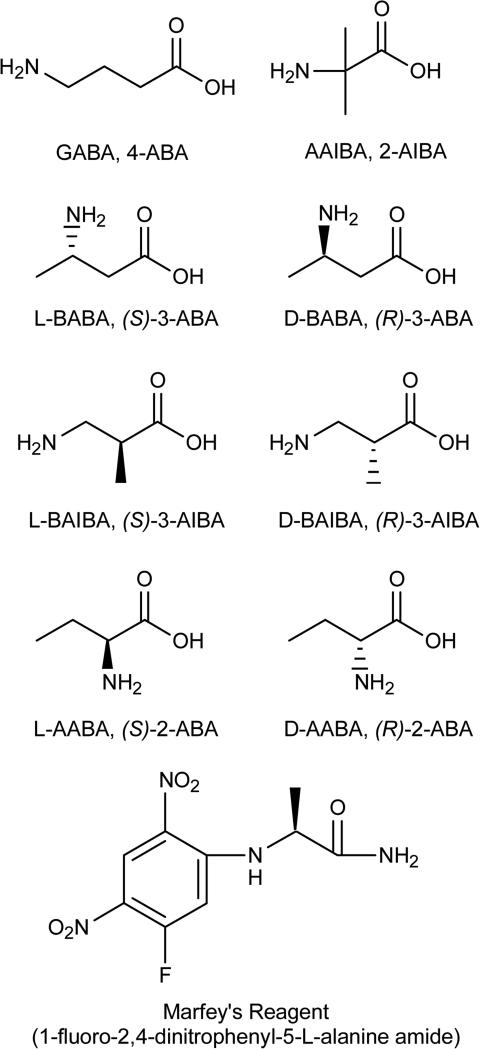
Isomeric aminobutyric acids comprise a set of eight different isomers; 4-aminobutyric acid (γ-ABA, GABA), (R)- and (S)-3-aminobutyric acid (d- and l-β-ABA; BABA), (R)- and (S)-2-aminobutyric acid (d- and l-α-ABA; AABA; Abu), (R)- and (S)-3-aminoisobutyric acid (d- and l-β-AIBA; BAIBA), and 2-aminoisobutyric acid (α-AIBA; AAIBA; Aib) (Fig. 1). GABA is a well-known and an important inhibitory neurotransmitter in the brain [1]. GABA levels are increased in pathological conditions such as Alzheimer's [2], pancreatic cancer [2], and immunogenic/inflammatory conditions [3]. BABA is a rarely occurring β-amino acid identified in tomato root extracts [4] and as an intermediate in Incednine biosynthesis in Streptomyces [5]. It has demonstrated protective action against plant pathogens when applied exogenously [4, 6-8], and has partial agonist activity against the ionotropic glycine neurotransmitter receptor in Xenopus oocytes (GlyR) [9]. AABA is a naturally occurring amino acid found in both its free form and as a constituent of ophthalmic acid (γ-l-Glu-l-AABA-Gly) [10]. It is also a component of many natural products including cyclosporine (l-AABA) and quinupristin (d-AABA). Both AABA and ophthalmic acid have been investigated as biomarkers for liver toxicity and alcohol abuse [11-14]. Abnormally high (>41 μM) serum AABA levels are found in septic patients, with high levels indicative of severe life threatening infections [15]. AAIBA is a natural amino acid found in some natural products such as Alamethicin [16, 17]. It is used in the design of short peptides with defined secondary structures [18], and in cell penetrating peptides in oligonucleotide delivery [19].
Figure 1.
Structures of the eight aminobutyric acid isomers, and Marfey's reagent.
Of particular recent interest is BAIBA [20]. BAIBA was first identified in 1951 as a new amino acid in human urine [21]. BAIBA can be generated by catabolism of valine to l-BAIBA [22-24] or catabolism of thymine to d-BAIBA [25]. Skeletal muscle is a major site for branched amino acid utilization [26], and during exercise, BAIBA levels are increased in the plasma of mice [27, 28]. BAIBA has been implicated in the beneficial effects of exercise on glucose home-ostasis and increased β-oxidation by hepatocytes via a PPAR-α mediated mechanism, and protection from metabolic diseases [27-29]. BAIBA has a number of observed biological activities such as influencing the development and progress of uremic toxemia [30, 31], increasing weight loss [32], and partial agonistic activity for the glycine receptor (GlyR) [9]. In obese patients, BAIBA improves glucose tolerance [32, 33], increases breakdown of lipids, and increases conversion of white fat to brown fat [34]. BAIBA is also a metabolic biomarker for breast cancer [35], liver and kidney malignancies [20, 36], acute and chronic myeloid leukemia [37], and hematological diseases [38].
Several early studies on the identification and characterization of BAIBA used GC-based stereospecific analyses to discriminate between d- and l-BAIBA [22-24, 39-44]. However, recent studies rarely use stereospecific detection methods, or pure BAIBA enantiomers. Separation of other ABA isomers have used GC- and HPLC-based methods [30, 31, 45-47]. A number of analytical approaches for ABA isomer resolution from extraterrestrial samples have also been reported ([48] and references therein). However, an approach capable of analytically resolving all the eight ABA isomers appears lacking, but would greatly facilitate studies on this interesting group of biological molecules. The purpose of this study was to investigate the potential for developing an LC-MS/MS method capable of resolving all eight ABA isomers for use in detailed studies of their biological activities. An effective method was developed by combining chromatographic resolution, and mathematical deconvolution of multichannel MS/MS date using a combination of Gaussian peak fitting and linear algebraic deconvolution. This study provides a template for similar efforts on other isobaric isomers.
2. EXPERIMENTAL
2.1. General
GABA, d- and l-BABA, d- and l-AABA, d- and l-BAIBA, AAIBA and d-Ala were purchased from Sigma-Aldrich (St. Louis, MO). C18-silica gel was obtained from Sep-Pak Cartridges from Waters (Milford, MA) and Marfey's reagent (1-fluoro-2,4-dinitrophenyl-l-5-alanine amide) was purchased from Novabiochem (a division of EMD Chemicals, Gibbstown, NJ). Solvents, HPLC water, and acetonitrile were obtained from Alfa Aesar (Ward Hill, MA); formic acid, acetone, and triethylamine (TEA) were purchased from Fisher Scientific (Pittsburgh, PA). Human sera was from Sigma-Aldrich (St. Louis, MO). Centrifuge operations were performed in a Sorvall RT6000 centrifuge, or a standard microcentrifuge. LC-MS/MS was performed on an AB Sciex 3200 QTrap mass spectrometer (Foster City, CA) coupled to a Shimadzu UFLC system (Columbia, MD) using electrospray ionization (ESI) and run with Analyst v. 1.4.2 software. Stock standard solutions (100 mM) of ABA isomers were made in HPLC grade water and stored at −20 °C. Working standard soluti ons of ABA isomers at 1 mM were made by dilution of stock standard solutions in HPLC water and stored at −20 °C.
2.2. Standard Marfey's Derivatization Reactions
A slightly modified version of the Marfey's derivatization technique followed previously [49, 50] was used. To 20 μL of the sample was added 20 μL of 20 mM Marfey's reagent in acetone, followed by 5 μL of 0.5 M TEA. The mixture was incubated overnight or for 4 days at 37 °C, depending on if AAIBA was of interest (since it reacts very slowly with Marfey's reagent). The reaction was quenched and diluted to 200 μL with 25/75/0.2% acetonitrile/water/formic acid.
2.3. Kinetic Characterization of Marfey's Derivatization Reactions
Different ABA isomers, especially the sterically constrained AAIBA isomer, were anticipated to have different reaction rates in the Marfey's derivatization reaction. To assess these differences, Marfey's derivatization reactions were set up as described above using well-resolved isomer mixtures: Mix 1 with 1 mM concentrations of l-BABA, GABA, and d-BABA; Mix 2 with 1 mM concentrations of d-BAIBA, l-AABA, and d-AABA; and Mix 3 with 1 mM concentrations of l-BAIBA and AAIBA. Reactions for kinetics characterization were performed by mixing 80 μL of a Mix (1, 2, and 3) with 80 μL of 20 mM Marfey's reagent, and 20 μL of 0.5 M TEA at 37 °C. Aliquots of 10 μL were removed at different times and the reaction was quenched by addition of 50 μL of 25/75/0.2% acetonitrile/water/formic acid, which acidified and stopped the reaction. Samples were run on the LC-MS/MS method described below, and UV 340 nm peak areas of Mar-ABA products used to determine reaction kinetics.
2.4. Chromatography Conditions
Chromatographic separations were performed on a Nucleodur 100-3 C8 125 × 2 mm column (Macherey–Nagel, Bethlehem, PA) at a flow rate of 0.3 mL/min. The mobile phases consisted of solvent A – water/0.1% formic acid, solvent B – 70/30/0.1% acetonitrile/water/formic acid, and solvent C – acetonitrile/0.1% formic acid. The gradient used in this study was 25% B for 5 min, followed by a ramp to 42.5% B over 35 min, and back to 25% B in 1 min, with a post run equilibration time of 6 min.
2.5. Quantitative Mass Spectral Tuning
Samples of Marfey's derivatized ABA isomers for MS/MS detection optimization were purified over C18 silica to remove salts as described previously [49, 50]. MS/MS detection optimization was performed on these purified samples using the automated quantitative optimization routine in Analyst. The optimized source parameters for detection of the parent ion (Q1, [M+H]+ = 356.2) were: declustering potential = 66 V, entrance potential = 5.5 V, source temperature = 450 °C, curtain gas = 20 psi, gas settings (GS1 and GS2) = 50 psi. The fragment ions used in this study and the optimized collision energies for their generation are summarized in Table 1.
Table 1.
Summary of Mar-ABA fragment ions (Q3) and collision energies (CE) used in this study
| Q3 | CE |
|---|---|
| 192.1 | 37 |
| 248.1 | 21 |
| 265.2 | 23 |
| 266.2 | 21 |
| 310.1 | 15 |
| 311.2 | 19 |
2.6. Preliminary Data Processing
Pre-processing of data was performed by translating Analyst WIFF format data files into NetCDF format files using the Analyst Translat.exe utility. Data in the NetCDF files was then imported into a Matlab data structure using Matlab's NetCDF file functions. MS channel data was collected discretely for 200 msec in each channel. Additionally, UV data was collected by the UV detector about 10 sec prior to the sample reaching the MS instrument. The Matlab cubic splines function (spline) was therefore used to align the data from each MS channel, as well as the UV data collected at 340 nm, to the same time vector as the first MS channel data. UV data was also baseline corrected. Gaussian peak fitting, as described further below, was used to confirm alignment, which demonstrated an RMSD of less than 0.05 sec between aligned MS and UV channel data.
2.7. Gaussian Peak Fitting and Deconvolution
Single component peaks in chromatograms were curve fit to the Gaussian peak equation (Eq. 1) using the Matlab fminsearch function.
| (1) |
For two overlapping peaks (the maximum number required in this study), curve fitting to the following sum of two Gaussian peaks equation was used to deconvolute the individual components:
| (2) |
(σ is the same in both terms, since it was observed to be essentially the same for all Mar-ABA isomer peaks.) In these expressions, Obs is the observed signal as a function of time, t is time, Base is the baseline of the curve, Hmax is the height of the peak, μ is the time at the center of the peak, and σ is the “standard deviation” of the peak. Note that for a Gaussian peak as defined above, the peak width at half-height (w½) = 2.355 σ, and the peak area (A) = H * σ * .
2.8. Multiple Channel MS (MRM) Linear Deconvolutio
The 6 MS fragments (channels) used in this study demonstrated different intensity ratios for the different ABA isomers. This potentially allows deconvolution of overlapping peaks into their components using a linear least squares approach. The expected multichannel chromatogram can be predicted given A – an n × m matrix with a column (m) for each component in the mixture each with n MS fragment channel intensities, × – an m × t matrix of the concentrations of each component as a function of time. The predicted n × t multichannel MS chromatogram b is then given by:
| (3) |
Since the intensity profile of the pure components (A) can be determined directly from LC-MS/MS chromatograms of the pure (well resolved) components, the observed multichannel chromatogram (b) of an overlapping mixture can be linearly deconvoluted in the least squares sense to give the component concentrations vs t profiles (x) using the Matlab backslash operator based formula:
| (4) |
During analysis of human serum samples, as described further below, substantial negative predicted component concentrations were obtained using Eq. 4, which are physically meaningless. The need for non-negative solutions in multivariate analyses is a common problem, and general approaches to non-negative least squares (NNLS) – where solutions to × in Eq. 4 are constrained to have values ≥ 0, are implemented in several analysis packages including Matlab. However, the Matlab implementation is for problems where both × and b are vectors, whereas the present case required a solution where × and b are both matricies. To address the need of the present analysis, the Matlab lsqnonneg function was used with iteration over the time dimension in the data matrix (b) to provide the deconvoluted (x) matrix:
| (5) |
where A is again the intensity vs species matrix (Eqs. 3 & 4) determined from LC-MS/MS chromatograms of pure (well resolved) Marfey's derivatized ABA standards.
2.9. Preparation and Analysis of Human Serum for ABA Isomer Composition
Human serum (2 mL) was treated with a three-fold excess of acetone (containing 10 μM d-Ala as internal standard) on ice for 5 min, and then centrifuged at 3300 g at 4 °C for 10 min. The supernatant was collected, dried under vacuum, and reconstituted in 300 μL of HPLC water/0.1% formic acid. A 10 μL aliquot of this sample was derivatized with 30 μL of 20 mM Marfey's reagent in acetone and 0.5 mM TEA in water for 4 days. The reaction was quenched by dilution to 200 μL with 25/75/0.2% acetonitrile/water/formic acid. A 30 μL aliquot of this Marfey's derivatized sample was injected for LC-MS/MS analysis. d-Ala was used as an internal standard for quantitative analyses. d-Ala was added to sample to provide 1 μM d-Ala in the sample prior to Marfey's derivatization, and quantified by LC-MS/MS as described previously [50].
2.10. Determination of linearity and absence of matrix effects
Serially diluted mixtures of 7 ABA isomers (excluding D-BAIBA) were prepared over the range of 0 to 400 pmol of injected analytes, and analyzed by LC-MS/MS and non-negative linear deconvolution as described above. Dilutions were also prepared in human serum to assess any effect of matrix and quantification.
3. RESULTS
3.1. Marfey's Derivatization Kinetics
It was anticipated, especially for the sterically hindered AAIBA isomer, that derivatization with Marfey's reagent might be slow. Reaction kinetics for Marfey's derivatization of individual ABA isomers were determined, and the results summarized in Table 2. The AAIBA isomer did indeed show a substantially slower reaction rate with Marfey's reagent than the other ABA isomers, such that complete (>95%) derivatization of this isomer requires a relatively long (54 hr) incubation.
Table 2.
Kinetics parameters for ABA isomer Marfey's derivatization reactions
| k (min−1) | t1/2 (min) | t95% (hrs) | |
|---|---|---|---|
| GABA | 0.035 | 20 | 1.4 |
| l-BABA | 0.012 | 58 | 4.2 |
| d-BABA | 0.012 | 57 | 4.1 |
| l-AABA | 0.026 | 27 | 1.9 |
| d-AABA | 0.053 | 13 | 0.93 |
| l-BAIBA | 0.080 | 8.7 | 0.62 |
| d-BAIBA | 0.081 | 8.6 | 0.62 |
| AAIBA | 0.00093 | 740 | 54 |
3.2. Chromatographic Separation and Resolution
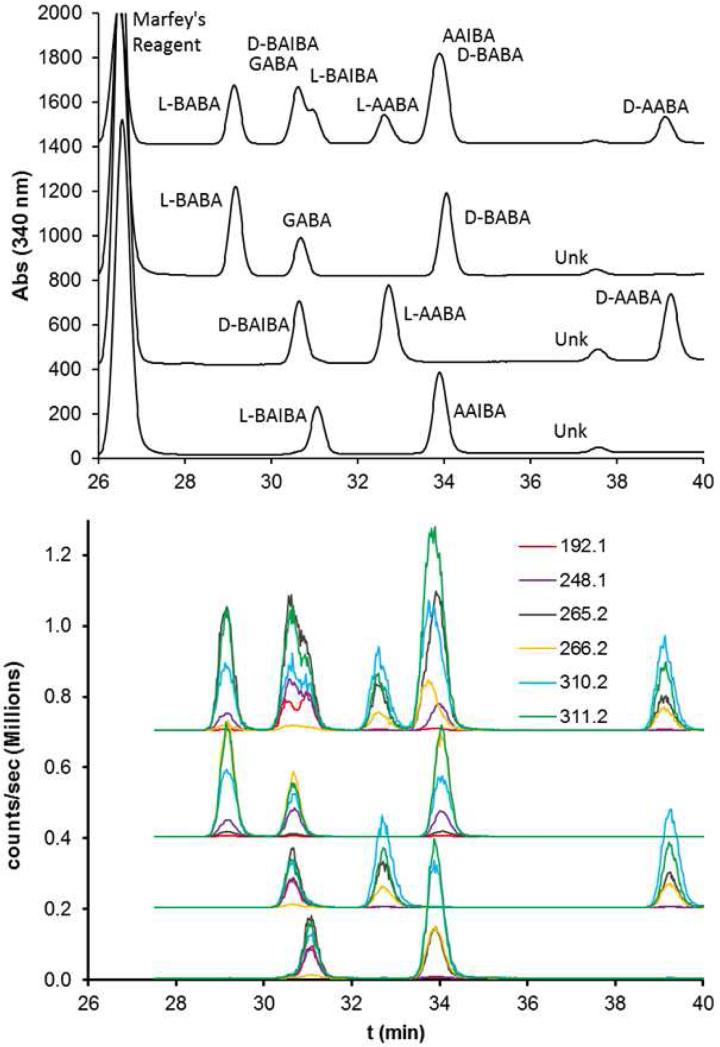
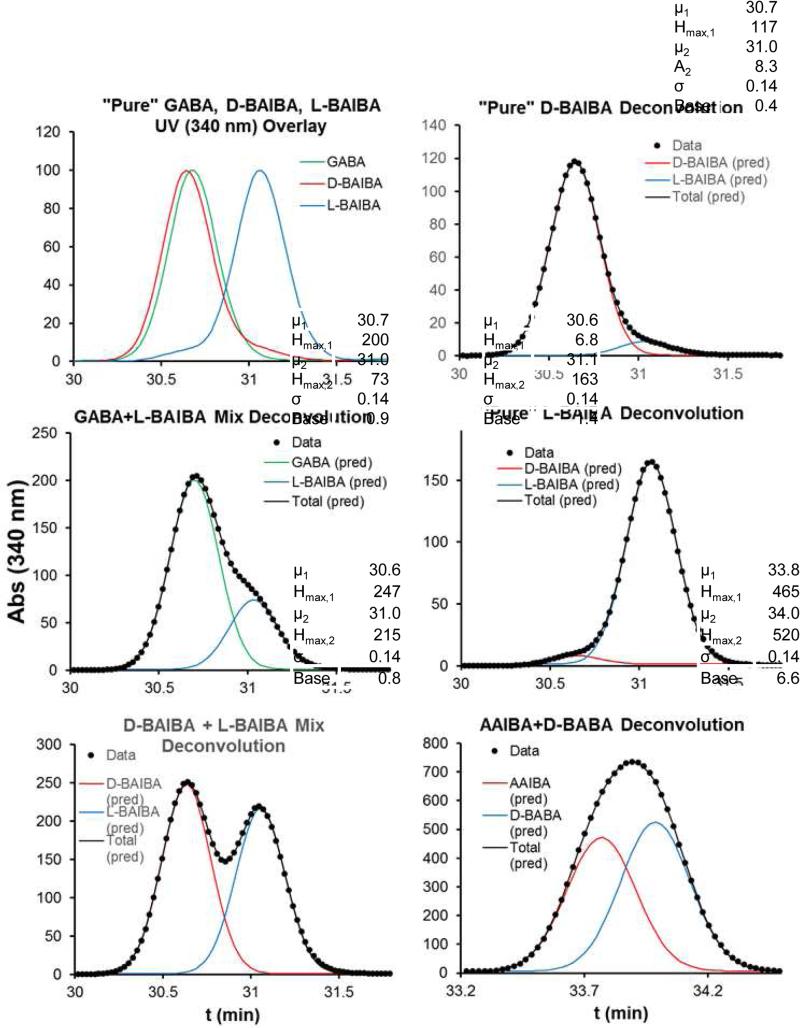
Significant effort was made to optimize resolution, with the initial goal of chromatographically resolving all eight Marfey's derivatized ABA isomers. Alternative gradients were investigated – including isocratic conditions, and inclusion of methanol in the solvent system – which we have previously found useful for differential shifting of overlapping Marfey's derivative peaks [51]. The best resolution obtained was with the gradient described above. Using this gradient, the Marfey's derivatives of ABA isomers showed good resolution for 3 of the 8 isomers (l-BABA, l-AABA, and d-AABA) and overlapping peaks in two separate clusters for the remaining 5 species (GABA+d-BAIBA+l-BAIBA, and AAIBA+d-BABA) (Fig. 2 top).
Figure 2.
Mar-ABA standard mix UV and MS/MS chromatograms. Upper panel: Mixture of all eight ABA isomers in top chromatogram, and well-resolved mixtures of component ABA isomers (Mix 1, 2, and 3, as defined in the text) in lower three chromatograms. Lower panel: Same chromatograms detected by MS/MS in positive mode. Q1 = 356.2 m/z with Q3 m/z as indicated in the legend.
3.3. MS/MS-Based Detection
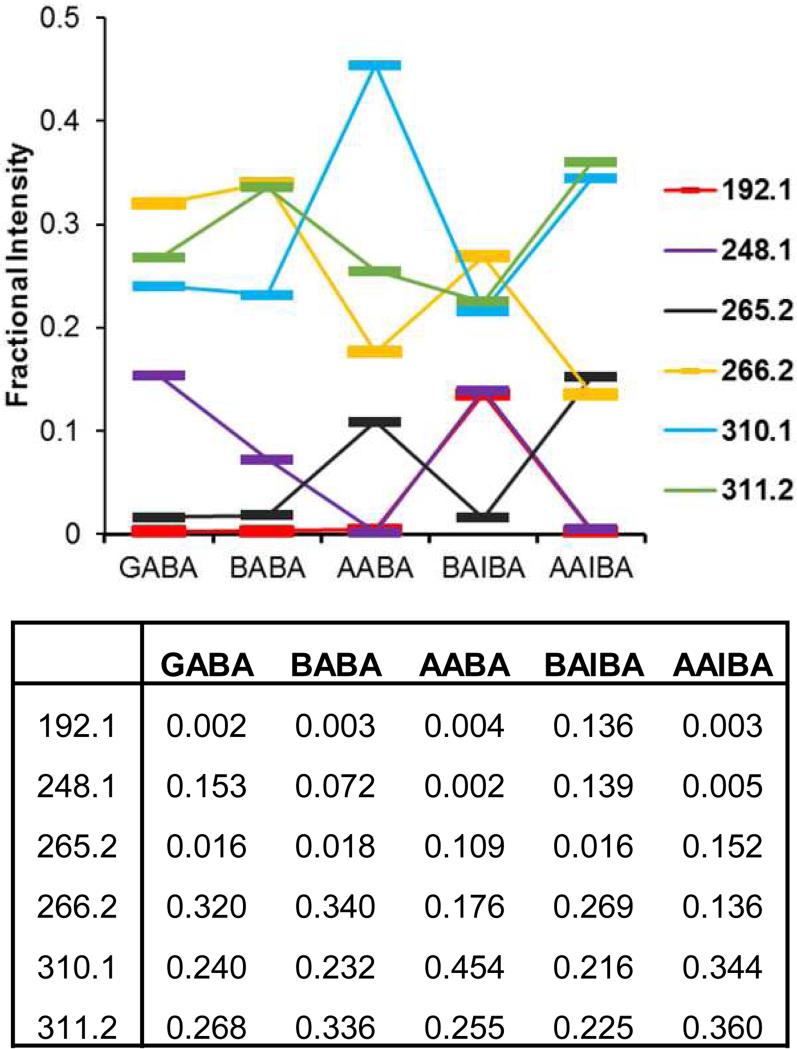
Standard mix chromatograms were also monitored by MS/MS using the settings and fragments summarized in Table 1 (Fig. 2 bottom). Fragment intensities profiles for Marfey's reagent derivatives of d- and l-stereoisomers (i.e. for Marfey's reagent derivatives of d- vs l-BABA, d- vs l-AABA, and d- vs l-BAIBA), were essentially indistinguishable (data not shown). However, distinguishable fragment intensity profiles were apparent between positional (α- vs β- vs γ-amino group) and structural (n-butyric vs isobutyric) ABA isomers (i.e. between GABA, BABA, AABA, BAIBA, and AAIBA) (Fig. 3). In the GABA/dl-BAIBA cluster, the 192.1 m/z fragment is 60-fold proportionately more intense for BAIBA than for GABA (or any other ABA isomer). In the AAIBA/d-BABA cluster, the 248.1 m/z fragment is 14-fold proportionately more sensitive for d-BABA, whereas the 265.2 m/z peak is 8.4-fold more sensitive for AAIBA.
Figure 3.
MS/MS fragment intensity profiles for Mar-ABA isomers normalized (to a sum of 1 for each entity). Connecting lines between adjacent entities are shown to highlight similarities and differences. Marfey's reagent derivatives of d- and l-isomers of BABA, AABA, and BAIBA gave indistinguishable intensity profiles (not shown).
3.4. Gaussian Peak Deconvolution of UV-Vis Chromatograms
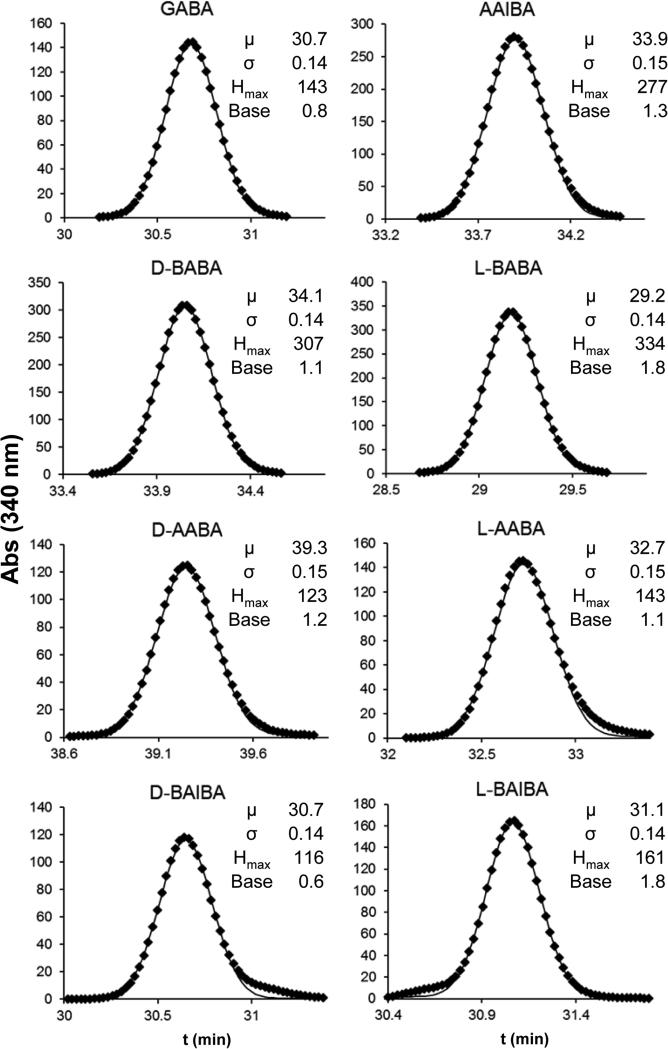
The partial separation of l-BAIBA from GABA and d-BAIBA (Fig 2) suggested that at least l-BAIBA could be quantified in a mixture using UV-vis monitored chromatograms. Given the high degree of peak overlap, Gaussian peak deconvolution was investigated as a means to quantify contributions from these overlapping components. Fitting of a Gaussian function to pure isomer UV-vis detected peaks demonstrated that the Marfey's derivative ABA isomer peaks eluted with a nearly perfect Gaussian peak shapes (Fig. 4). Overlays of UV monitored chromatograms of GABA, d-BAIBA, and l-BAIBA indicated that GABA and d-BAIBA were unresolved, but that l-BAIBA is sufficiently well resolved to be quantified in the presence of the other two (Fig. 5, top left panel). Gaussian peak deconvolution using the two component equation (Eq. 2) allowed quantification of even low relative levels of d- and l-BAIBA, and demonstrated low levels of opposite isomer impurity in commercial samples of d- and l-BAIBA (Fig 5, top two right panels). The overlapping peaks for AAIBA and d-BABA can be similarly deconvoluted using this approach (Fig. 5, bottom left panel).
Figure 4.
Gaussian peak fits to pure component chromatograms. Data (filled circles) and Gaussian fit curves (solid lines) for the “pure” component peaks for each Marfey's derivatized ABA isomer (data taken from the lower three UV 340 nm monitored chromatograms from the top panel in Figure 2). The small shoulders in the two BAIBA chromatograms (bottom panels) is due to traces of the opposing isomer, as illustrated in Fig 5 (top and center right panels).
Figure 5.
Gaussian peak fitting deconvolution of UV monitored chromatograms. Gaussian peak fitting deconvolution of overlapping UV 340 nm peaks in the GABA/d-BAIBA/l-BAIBA cluster (all panels except lower right) and in the AAIBA/d-BABA cluster (lower right panel).
3.5. Linear regression-based multichannel MS/MS signal deconvolution
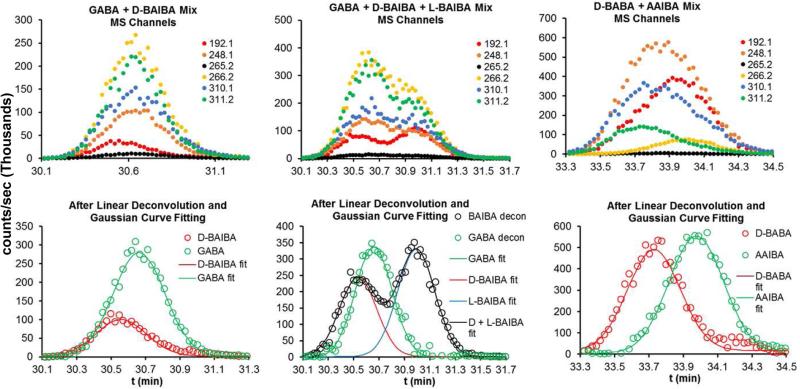
Each Mar-ABA derivative demonstrated a number of detectable fragments after collisionally induced dissociation, with substantial differences in the intensity profile for these fragments between ABA isomers (Fig. 3). Given these differences, the possibility of using a linear deconvolution approach was tested on a GABA+d-BAIBA MS/MS detected chromatogram (raw data shown in the top left panel of Fig 6). Linear deconvolution using the intensity profiles for GABA and BAIBA as matrix A in Eq. 4 (referred to as local deconvolution, since it uses only the component intensities expected in the narrow chromatography window under examination) gives the deconvoluted component data points shown in the bottom left panel of Fig. 6. These deconvoluted data points were then individually fit to the single component Gaussian curve equation (Eq. 1) (solid lines in the bottom left panel of Fig. 6). This approach was then tested for (local) deconvolution of a mixture of all three overlapping components in the GABA+d-BAIBA+l-BAIBA peak cluster (raw data in the top center panel of Fig. 6). Linear (local) deconvolution of this data gives the data points shown in the bottom center panel of Fig. 6. The resulting deconvoluted GABA data points were then fit to the single component Gaussian curve equation (Eq. 1) (solid green line in the bottom left panel of Fig. 6). The resulting deconvoluted BAIBA (d + l) data points (black circles) were then fit to the two component Gaussian curve equation (Eq. 2) to give the deconvoluted chromatogram for d-BAIBA (red line, lower center panel Fig. 6), l-BAIBA (blue line), and their sum (d + l, black line), demonstrating the combination of linear and Gaussian peak deconvolution for this three component mixture. The overlapping peaks in the multichannel MS chromatogram for AAIBA+d-BABA could also be deconvoluted similarly (Fig. 6, right panels).
Figure 6.
Combined Linear Regression and Gaussian Peak Fitting Deconvolution. Top left is the MS/MS detected chromatogram of a mixture of GABA+d-BAIBA, top center is of a mixture of GABA+d-BAIBA+l-BAIBA, and top right is of d-BABA and AAIBA. Open circles in the lower panels are the corresponding linearly deconvoluted data points. Lines in the lower panel represent Gaussian best-fit curves, as indicated in the captions.
3.6. Human Serum Analysis
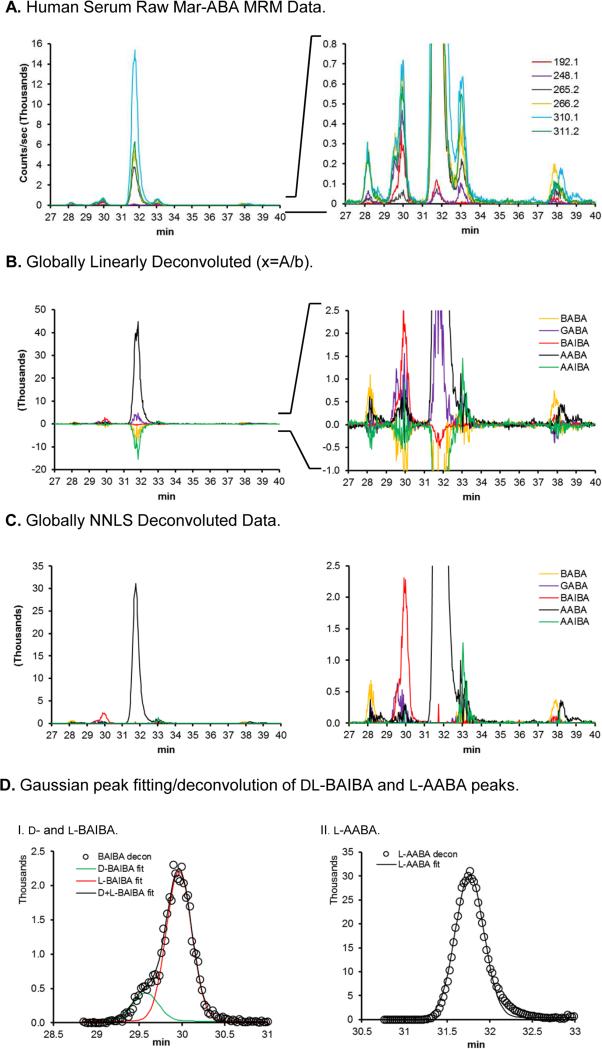
To demonstrate this approach on a relevant biological sample, it was then applied to commercially available human serum. Raw data is shown in Fig. 7A (full scale on the left, and expanded on the right). Global linear deconvolution (Eq. 4) (global referring to the use of the full intensity vs species matrix as A in Eq. 4) gave the globally deconvoluted chromatograms shown in Fig. 7B, in which substantial negative predicted concentrations were apparent. To avoid such negative predicted concentrations, which are physically meaningless, a non-negative least squares (NNLS) method was implemented and tested for use in global deconvolution (Eq. 5), which gave the deconvoluted chromatogram shown in Fig. 7C. The NNLS deconvoluted BAIBA chromatogram was further deconvoluted into its d- and l-BAIBA components by Gaussian peak fitting (Fig. 7D-I), with Gaussian peak fitting for the l-AABA peak also shown (Fig. 7D-II).
Figure 7.
Human serum Marfey's ABA derivative LC-MS/MS data and deconvolution.
3.7. Assessment of linearity and absence of matrix effects using NNLS deconvolution
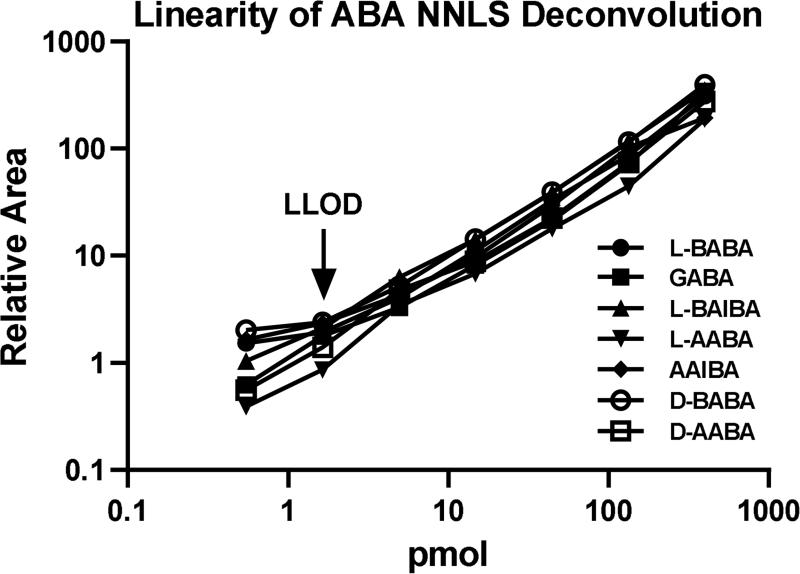
To assess linearity and matrix effects of the NNLS approach, serially diluted samples of 7 ABA isomers (lacking D-BAIBA, since it elutes very close to L-BAIBA and requires an additional Gaussian deconvolution step to fully resolve L- and D-BAIBA containing samples) were analyzed with this approach. Excellent linearity and sensitivity were observed with this approach from 0-400 pmol, with a LLOD of around 2 pmol of all tested analytes (Fig. 8). No matrix effects were apparent with human serum diluted samples (data not shown), consistent with our prior studies on LC-MS/MS quantification of other Marfey's reagent derivatized amino acid analytes [49-51].
Figure 8.
Semi-log vs. semi-log plot of NNLS deconvoluted peak areas vs pmol of injected analyte. A set of well-resolved ABA isomer mixes were used to determine the intensity matrix (A in eq. 3) for each analyte (Fig. 3). Each unknown multichannel chromatogram was then processed using the NNLS calculation given in eq. 5 to give deconvoluted pure component chromatograms for each unknown, and the appropriate peaks integrated to give corresponding areas. These areas are plotted on the y-axis (semi-log) vs the amount of the analyte injected (x-axis, also semi-log).
4. DISCUSSION
MS and MS/MS-based analytical methods allow the specific detection and quantification of analytes in complex mixtures. This approach is dependent on both chromatographic resolution and highly specific MS-based detection. However, isomeric/isobaric analytes with identical or similar chromatographic and MS characteristics, such as ABA isomers, remain challenging. ABA isomers are of current interest for their varied biological properties, which include the particularly interesting d- and l-BAIBA isomers. Chiral GC-MS approaches are capable of chromatographically resolving d- and l-BAIBA [23-26, 39, 42-45], and a two-dimension HPLC method with UV detection has been reported that is capable of resolving and quantifying ABA structural and stereoisomers from extraterrestrial samples [48], an alternative approach using generally available LC-MS/MS instrumentation would be highly desirable. The purpose of this study was to evaluate and demonstrate the potential of multichannel LC-MS/MS for the resolution of all eight ABA isomers as their Marfey's reagent derivatives, including l- and d-BAIBA.
Marfey's reagent is a chiral amine-derivatizing agent used to resolve and quantify d- and l-amino acids [52-54]. We have previously demonstrated the use of Marfey's reagent derivatization for the LC-MS/MS-based analysis of a number of amino acids and related compounds [49, 50], and this approach appeared applicable to the resolution and quantification of ABA isomers [55, 56]. Given their identical masses, it was expected that a significant degree or complete chromatographic resolution would be required for such an analysis. However, after investigating a number of different gradients and solvent mixtures it was found that complete resolution was not forthcoming (Fig. 2). Even when using a relatively long (shallow) gradient, complete resolution in UV monitored chromatograms could be obtained only for l-BABA, l-AABA, and d-AABA. GABA, d-BAIBA, and l-BAIBA eluted as closely overlapping peaks in one chromatographic cluster, and AAIBA and d-BABA in another cluster (Fig. 2).
Given the inability to develop a purely chromatography-based separation for the eight ABA isomers, the possibility that differences in MS/MS fragmentation patterns could provide a basis for resolving overlapping Mar-ABA isomer peaks was investigated. The six most intense fragments collectively among all the Mar-ABA isomers were used in this effort. MS/MS fragment intensity ratios were essentially identical for all D/L isomer pairs (Fig. 2 lower panel), meaning that d- and l- isomers cannot be distinguished using MS/MS fragmentation intensity data. However, substantial differences between different ABA positional (α- vs β- vs γ-amino group) and structural (n-butyric vs isobutyric) isomers were apparent (Fig. 3). These differences provide a possible means to deconvolute multichannel MS/MS fragment chromatograms into their component positional and structural isomers.
Another potentially useful feature of the observed chromatograms was that all pure compound peaks were found to have essentially perfect Gaussian (normal) peak shapes (Fig. 4), with the exception of l-BAIBA and d-BAIBA, both of which had significant shoulders apparent as discussed further below. This precise Gaussian behavior appeared to provide an additional means for peak deconvolution, especially for potential closely eluting ABA d/l isomer pairs given their indistinguishable MS/MS fragment intensity profiles (e.g. for d- and l-BAIBA). Gaussian peak deconvolution was demonstrated on UV 340 nm detected chromatograms of Marfey's derivatized ABA isomers in the two overlapping clusters (Fig. 5). This allowed resolution of all ABA isomers except GABA and d-BAIBA, which were nearly perfectly overlapping (Fig. 5 top left panel). This analysis also demonstrated that the commercially available d- and l-BAIBA samples were each significantly contaminated with the opposite isomer (Fig. 5 top 2 left panels).
Linear deconvolution of MS/MS detected chromatograms was then explored. The MS/MS chromatograms for GABA+d-BAIBA and GABA+d-BAIBA+l-BAIBA are shown in Fig. 6-top, and the linearly (local) deconvoluted data are shown as data points in Fig. 6-bottom. The d- + l-BAIBA linearly deconvoluted data points can be further deconvoluted by fitting to a two peak Gaussian equation (Eq. 2) to provide individual d-BAIBA and l-BAIBA component chromatograms (Fig. 6-bottom right). This approach thereby allows all three overlapping components in this cluster to be resolved using multiple fragment MS/MS chromatogram data and linear and Gaussian deconvolution. The overlapping multiple fragment MS/MS detected peaks in the AAIBA/d-BABA cluster could similarly be deconvoluted (data not shown), allowing all eight ABA isomers to be resolved and quantified by LC-MS/MS analysis.
This approach was then applied to the detection of ABA isomers in human serum. The raw data chromatogram is shown in Fig. 7A, demonstrating one pronounced MS/MS peak (l-AABA) and a number of weaker peaks. Application of global linear deconvolution (Fig. 7B) highlights the identity of the components of these peaks/clusters, which are as expected based on the retention of standards (the human serum data was collected on a different day than the Fig. 2-6 data, and shows somewhat different retention times). However, unconstrained (e.g. Eq. 3) global linear deconvolution was quite sensitive to noise in the MS/MS data, as indicated by the presence of negative intensity components in the linearly deconvoluted peaks clusters (Fig. 7B). While a local deconvolution approach could be applied, a global approach is both more straightforward and rigorous. Implementation of non-negative least squares deconvolution provided the deconvoluted chromatograms shown in Fig. 7C, which provided the desired all positive intensity deconvoluted chromatogram. This approach demonstrates good analytical linearity over a wide range (Fig. 8), and with no discernable human serum matrix effect. All of the peaks in the de-convoluted human serum chromatogram (Fig. 7C) occurred at the expected time except the late eluting (38 min) peak in the BABA channel. Both the large l-AABA peak at 31.6 min and the d- + l-BAIBA peaks at 29.5-29.9 min give well resolved deconvoluted chromatograms, as shown in Fig. 7C-I and 7C–II. The peak areas in the Fig. 7 correspond to a relatively high human serum levels of l-AABA (13 μM), an intermediate level of l-BAIBA (0.8 μM), and low but detectible levels (<0.2 μM) of GABA, l-BABA, AAIBA, d-BAIBA, and d-AABA. This l-AABA level determined in this preliminary analysis of commercial human serum is similar to levels determined in prior studies of 7-30 μM for healthy subjects [15]. The levels of d- and l-BAIBA of 0.8 μM (total) determined here is comparable to the value of 2 μM reported previously, as is the 1:4 d:l ratio [57].
A key feature of MS/MS based analytical methods is their selectivity for individual analytes in a complex mixture. However, in the case of mixtures isomeric/isobaric analytes this advantageous characteristic is lost when identical fragments are common among the analytes, as is the case for ABA isomers. Picking a single MS/MS channel specific to each analyte is simply not possible. As demonstrated in this study, linear deconvolution of multichannel MS/MS data analysis using non-negative least squares provides an elegant solution to this type of analytical problem by allowing multichannel MS/MS data to be transformed directly into individual component chromatograms. A related problem common to MS/MS based detection is the potential for interfering analytes to show up under the analyte peak of interest, and it is therefore common to select a second MS/MS channel to confirm selectivity. Linear deconvolution represents the logical extension of such an approach to the general case, since linear deconvolution utilizes all information on the multichannel MS/MS characteristics of an analyte/analytes. In the case of human serum analysis, an interfering component was observed in the BABA channel (Fig. 7C), which was however well removed chromatographically from the known L- and D-BABA retention times. This unknown interfering agent ran very close chromatographically to D-AABA. Single channel MS/MS data would likely (depending on which channel was selected) have confound these two peaks as one, and led to an erroneous results. The multichannel approach demonstrated provides a high information content (multichannel MS/MS) maximum likelihood (least squares) estimate of individual component chromatograms.
5. CONCLUSION
This study demonstrates the use of multivariate linear and Gaussian deconvolution for the LC-MS/MS-based resolution and quantification of all eight ABA isomers, and provides a valuable tool for studies of this interesting class of biological molecules. The methods developed in this study are also expected to be of general utility for the development of comprehensive MS/MS-based analytical approaches to other classes of isomeric molecules. Key findings of this study include: 1) Mar-ABA derivatives of d- and l-isomers give essentially identical fragment intensity profiles, but different ABA isomers give sufficiently different fragment intensity profiles to allow their linear deconvolution. 2) Both local and global deconvolution approaches are effective for linear deconvolution, but global deconvolution requires the use of a NNLS strategy to avoid physically unrealistic negative component concentrations. 3) All eight ABA isomers can be resolved and quantified using this approach. 4) BAIBA provides an intense 192.1 m/z fragment, which is uniquely characteristic of this biologically interesting isomer. 5) NNLS based linear deconvolution provides for a linear response curve over the range of 0-400 pmol. 6) Seven of the eight ABA isomers are detectible in human serum, with l-AABA predominating, followed by l-BAIBA.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health; NIAMS P01-AG039355 (LB) and NIAID R21-121903 (WG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bhat R, Axtell R, Mitra A, Miranda M, Lock C, Tsien RW, Steinman L. Inhibitory role for GABA in autoimmune inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2580–2585. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser E, Schoenknecht P, Kassner S, Hildebrandt W, Kinscherf R, Schroeder J. Cerebrospinal fluid concentrations of functionally important amino acids and metabolic compounds in patients with mild cognitive impairment and Alzheimer's disease. Neuro-degenerative diseases. 2010;7:251–259. doi: 10.1159/000287953. [DOI] [PubMed] [Google Scholar]
- 3.Barragan A, Weidner JM, Jin Z, Korpi ER, Birnir B. GABAergic signalling in the immune system. Acta physiologica. 2015;213:819–827. doi: 10.1111/apha.12467. [DOI] [PubMed] [Google Scholar]
- 4.Cohen Y. Local and systemic control of Phytophthora infestans in tomato plants by DL-3-amino-n-butanoic acids. Phytopathology. 1994;84:55–59. [Google Scholar]
- 5.Takaishi M, Kudo F, Eguchi T. A unique pathway for the 3-aminobutyrate starter unit from L-glutamate through beta-glutamate during biosynthesis of the 24-membered macrolactam antibiotic, incednine. Organic letters. 2012;14:4591–4593. doi: 10.1021/ol302052c. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerli L, Jakab G, Metraux JP, Mauch-Mani B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta -aminobutyric acid. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12920–12925. doi: 10.1073/pnas.230416897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao HH, Zhang M, Zhao H, Zhang Y, Wang XX, Guo SS, Zhang ZF, Liu TX. Deciphering the mechanism of beta-aminobutyric acid-induced resistance in wheat to the grain aphid, Sitobion avenae. PloS one. 2014;9:e91768. doi: 10.1371/journal.pone.0091768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luna E, van Hulten M, Zhang Y, Berkowitz O, Lopez A, Petriacq P, Sellwood MA, Chen B, Burrell M, van de Meene A, Pieterse CM, Flors V, Ton J. Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nature chemical biology. 2014;10:450–456. doi: 10.1038/nchembio.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmieden V, Betz H. Pharmacology of the inhibitory glycine receptor: agonist and antagonist actions of amino acids and piperidine carboxylic acid compounds. Molecular pharmacology. 1995;48:919–927. [PubMed] [Google Scholar]
- 10.Waley SG. Acidic peptides of the lens. The Biochemical journal. 1956;64:715–726. doi: 10.1042/bj0640715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medici V, Peerson JM, Stabler SP, French SW, Gregory JF, 3rd, Virata MC, Albanese A, Bowlus CL, Devaraj S, Panacek EA, Rahim N, Richards JR, Rossaro L, Halsted CH. Impaired homocysteine transsulfuration is an indicator of alcoholic liver disease. Journal of hepatology. 2010;53:551–557. doi: 10.1016/j.jhep.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur G, Leslie EM, Tillman H, Lee WM, Swanlund DP, Karvellas CJ, Group USALFS. Detection of Ophthalmic Acid in Serum from Acetaminophen-Induced Acute Liver Failure Patients Is More Frequent in Non-Survivors. PloS one. 2015;10:e0139299. doi: 10.1371/journal.pone.0139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dello SA, Neis EP, de Jong MC, van Eijk HM, Kicken CH, Olde Damink SW, Dejong CH. Systematic review of ophthalmate as a novel biomarker of hepatic glutathione depletion. Clinical nutrition. 2013;32:325–330. doi: 10.1016/j.clnu.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Waszkiewicz N, Poplawska R, Konarzewska B, Szajda SD, Galinska B, Rutkowski P, Lesniak R, Szulc A. Biomarkers of alcohol abuse. Part II. New biomarkers and their interpretation. Psychiatria polska. 2010;44:137–146. [PubMed] [Google Scholar]
- 15.Chiarla C, Giovannini I, Siegel JH. Characterization of alpha-amino-n-butyric acid correlations in sepsis. Translational research : the journal of laboratory and clinical medicine. 2011;158:328–333. doi: 10.1016/j.trsl.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Meyer CE, Reusser F. A polypeptide antibacterial agent isolated from Trichoderma viride. Experientia. 1967;23:85–86. doi: 10.1007/BF02135929. [DOI] [PubMed] [Google Scholar]
- 17.Payne JW, Jakes R, Hartley BS. The primary structure of alamethicin. The Biochemical journal. 1970;117:757–766. doi: 10.1042/bj1170757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahalakshmi R, Balaram P. Non-protein amino acids in the design of secondary structure scaffolds. Methods Mol Biol. 2006;340:71–94. doi: 10.1385/1-59745-116-9:71. [DOI] [PubMed] [Google Scholar]
- 19.Wada S, Urase T, Hasegawa Y, Ban K, Sudani A, Kawai Y, Hayashi J, Urata H. Aib-containing peptide analogs: cellular uptake and utilization in oligonucleotide delivery. Bioorganic & medicinal chemistry. 2014;22:6776–6780. doi: 10.1016/j.bmc.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 20.Griffith OW. Beta-amino acids: mammalian metabolism and utility as alpha-amino acid analogues. Annual review of biochemistry. 1986;55:855–878. doi: 10.1146/annurev.bi.55.070186.004231. [DOI] [PubMed] [Google Scholar]
- 21.Fink K, Henderson RB, Fink RM. Beta-aminoisobutyric acid, a possible factor in pyrimidine metabolism. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 1951;78:135–141. doi: 10.3181/00379727-78-19000. [DOI] [PubMed] [Google Scholar]
- 22.van Gennip AH, Kamerling JP, de Bree PK, Wadman SK. Linear relationship between the R- and S-enantiomers of a beta-aminoisobutyric acid in human urine. Clinica chimica acta; international journal of clinical chemistry. 1981;116:261–267. doi: 10.1016/0009-8981(81)90045-0. [DOI] [PubMed] [Google Scholar]
- 23.Landaas S, Solem E. High excretion of beta-aminoisobutyric acid in patients with ketoacidosis. Scand J Clin Lab Invest. 1983;43:95–97. [PubMed] [Google Scholar]
- 24.Roe CR, Struys E, Kok RM, Roe DS, Harris RA, Jakobs C. Methylmalonic semialdehyde dehydrogenase deficiency: psychomotor delay and methylmalonic aciduria without metabolic decompensation. Molecular genetics and metabolism. 1998;65:35–43. doi: 10.1006/mgme.1998.2737. [DOI] [PubMed] [Google Scholar]
- 25.Solem E, Jellum E, Eldjarn L. The absolute configuration of β-aminoisobutyric acid in human serum and urine. Clinica Chimica Acta. 1974;50:393–403. [Google Scholar]
- 26.Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annual review of nutrition. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 27.Roberts LD, Bostrom P, O'Sullivan JF, Schinzel RT, Lewis GD, Dejam A, Lee YK, Palma MJ, Calhoun S, Georgiadi A, Chen MH, Ramachandran VS, Larson MG, Bouchard C, Rankinen T, Souza AL, Clish CB, Wang TJ, Estall JL, Soukas AA, Cowan CA, Spiegelman BM, Gerszten RE. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell metabolism. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Delgado G, Martinez-Tellez B, Olza J, Aguilera CM, Gil A, Ruiz JR. Role of Exercise in the Activation of Brown Adipose Tissue. Annals of nutrition & metabolism. 2015;67:21–32. doi: 10.1159/000437173. [DOI] [PubMed] [Google Scholar]
- 29.Heath V. Metabolism: Feel the burn--muscle metabolite couples exercise to heat production. Nature reviews. Endocrinology. 2014;10:188. doi: 10.1038/nrendo.2014.4. [DOI] [PubMed] [Google Scholar]
- 30.Gejyo F, Kinoshita Y, Ikenaka T. Elevation of serum levels of beta-aminoisobutyric acid in uremic patients and the toxicity of the amino acid. Clinical nephrology. 1977;8:520–525. [PubMed] [Google Scholar]
- 31.Nyholm KK, Sjolin KE, Hammer M, Knudsen J, Stahl D, Nielsen HR. A study on the clinical significance of urinary beta-aminoisobutyric acid in patients with urothelial tumours. Biomedicine / [publiee pour l'A.A.I.C.I.G.] 1975;22:509–516. [PubMed] [Google Scholar]
- 32.Maisonneuve C, Igoudjil A, Begriche K, Letteron P, Guimont MC, Bastin J, Laigneau JP, Pessayre D, Fromenty B. Effects of zidovudine, stavudine and beta-aminoisobutyric acid on lipid homeostasis in mice: possible role in human fat wasting. Antiviral therapy. 2004;9:801–810. doi: 10.1177/135965350400900513. [DOI] [PubMed] [Google Scholar]
- 33.Begriche K, Massart J, Abbey-Toby A, Igoudjil A, Letteron P, Fromenty B. Beta-aminoisobutyric acid prevents diet-induced obesity in mice with partial leptin deficiency. Obesity. 2008;16:2053–2067. doi: 10.1038/oby.2008.337. [DOI] [PubMed] [Google Scholar]
- 34.Ginter E, Simko V. Recent data on obesity research: beta-aminoisobutyric acid. Bratislavske lekarske listy. 2014;115:492–493. doi: 10.4149/bll_2014_095. [DOI] [PubMed] [Google Scholar]
- 35.Corona G, Polesel J, Fratino L, Miolo G, Rizzolio F, Crivellari D, Addobbati R, Cervo S, Toffoli G. Metabolomics biomarkers of frailty in elderly breast cancer patients. Journal of cellular physiology. 2014;229:898–902. doi: 10.1002/jcp.24520. [DOI] [PubMed] [Google Scholar]
- 36.Tew KD, Taylor DM. The relationship of thymidine metabolism to the use of fractional incorporation as a measure of DNA synthesis and tissue proliferation. European journal of cancer. 1978;14:153–168. doi: 10.1016/0014-2964(78)90174-3. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen HR, Killmann SA. Urinary excretion of beta-aminoisobutyrate and pseudouridine in acute and chronic myeloid leukemia. Journal of the National Cancer Institute. 1983;71:887–891. [PubMed] [Google Scholar]
- 38.Enkhjargal T, Tserennadmid C. Urinary excretion of beta-aminoisobutyric acid in hematological diseases. Rinsho byori. The Japanese journal of clinical pathology. 2004;52:17–21. [PubMed] [Google Scholar]
- 39.Pollock G. The preparation of R(−−) beta-aminoisobutyric acid using Saccharomyces cerevisiae: an unexpected result. Analytical biochemistry. 1974;57:82–88. doi: 10.1016/0003-2697(74)90053-0. [DOI] [PubMed] [Google Scholar]
- 40.Solem E. The absolute configuration of beta-aminoisobutyric acid formed by degradation of thymine in man. Clinica chimica acta; international journal of clinical chemistry. 1974;53:183–190. doi: 10.1016/0009-8981(74)90097-7. [DOI] [PubMed] [Google Scholar]
- 41.Manning NJ, Pollitt RJ. Tracer studies of the interconversion of R- and S-methylmalonic semialdehydes in man. The Biochemical journal. 1985;231:481–484. doi: 10.1042/bj2310481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollitt RJ, Green A, Smith R. Excessive excretion of beta-alanine and of 3-hydroxypropionic, R- and S-3-aminoisobutyric, R- and S-3-hydroxyisobutyric and S-2-(hydroxymethyl)butyric acids probably due to a defect in the metabolism of the corresponding malonic semialdehydes. Journal of inherited metabolic disease. 1985;8:75–79. doi: 10.1007/BF01801669. [DOI] [PubMed] [Google Scholar]
- 43.van Gennip AH, van Bree-Blom EJ, Abeling NG, van Erven AJ, Voute PA. beta-Aminoisobutyric acid as a marker of thymine catabolism in malignancy. Clinica chimica acta; international journal of clinical chemistry. 1987;165:365–377. doi: 10.1016/0009-8981(87)90182-3. [DOI] [PubMed] [Google Scholar]
- 44.Podebrad F, Heil M, Beck T, Mosandl A, Sewell AC, Bohles H. Stereodifferentiation of 3-hydroxyisobutyric- and 3-aminoisobutyric acid in human urine by enantioselective multidimensional capillary gas chromatography-mass spectrometry. Clinica chimica acta; international journal of clinical chemistry. 2000;292:93–105. doi: 10.1016/s0009-8981(99)00210-7. [DOI] [PubMed] [Google Scholar]
- 45.Gamerith G, Brantner H. Thin-layer chromatographic identification and gas-liquid chromatographic separation of seven aminobutyric acids in the presence of protein and non-protein amino acids. Journal of Chromatography A. 1986;355:273–280. [Google Scholar]
- 46.Kittel A, Maas R, Konig J, Mieth M, Weiss N, Jarzebska N, Hohenstein B, Martens-Lobenhoffer J, Bode-Boger SM, Rodionov RN. In vivo evidence that Agxt2 can regulate plasma levels of dimethylarginines in mice. Biochemical and biophysical research communications. 2013;430:84–89. doi: 10.1016/j.bbrc.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Manier ML, Spraggins JM, Reyzer ML, Norris JL, Caprioli RM. A derivatization and validation strategy for determining the spatial localization of endogenous amine metabolites in tissues using MALDI imaging mass spectrometry. Journal of mass spectrometry : JMS. 2014;49:665–673. doi: 10.1002/jms.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamase K, Miyoshi Y, Mita M, Konno R. Chiral amino acid metabolomics in mammals - distribution and regulation of D-amino acids. J Pharmacol Sci. 2014;124:35p–35p. [Google Scholar]
- 49.Putty S, Vemula H, Bobba S, Gutheil WG. A liquid chromatography-tandem mass spectrometry assay for d-Ala-d-Lac: a key intermediate for vancomycin resistance in vancomycin-resistant enterococci. Analytical biochemistry. 2013;442:166–171. doi: 10.1016/j.ab.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 50.Jamindar D, Gutheil WG. A liquid chromatography-tandem mass spectrometry assay for Marfey's derivatives of L-Ala, D-Ala, and D-Ala-D-Ala: application to the in vivo confirmation of alanine racemase as the target of cycloserine in Escherichia coli. Analytical biochemistry. 2010;396:1–7. doi: 10.1016/j.ab.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Bobba S, Resch GE, Gutheil WG. A liquid chromatography-tandem mass spectrometry assay for detection and quantitation of the dipeptide Gly-Gln in rat brain. Analytical biochemistry. 2012;425:145–150. doi: 10.1016/j.ab.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adamson JG, Hoang T, Crivici A, Lajoie GA. Use of Marfey's reagent to quantitate racemization upon anchoring of amino acids to solid supports for peptide synthesis. Analytical biochemistry. 1992;202:210–214. doi: 10.1016/0003-2697(92)90229-z. [DOI] [PubMed] [Google Scholar]
- 53.Bhushan R, Bruckner H. Marfey's reagent for chiral amino acid analysis: A review. Amino Acids. 2004;27:231–247. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- 54.Marfey P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Calsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 55.Ilisz I, Berkecz R, Peter A. Application of chiral derivatizing agents in the high-performance liquid chromatographic separation of amino acid enantiomers: a review. Journal of pharmaceutical and biomedical analysis. 2008;47:1–15. doi: 10.1016/j.jpba.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 56.Peter A, Lazar L, Fulop F, Armstron DW. High-performance liquid chromatographic enantioseparation of beta-amino acids. Journal of chromatography. A. 2001;926:229–238. doi: 10.1016/s0021-9673(01)01078-0. [DOI] [PubMed] [Google Scholar]
- 57.Solem E, Agarwal DP, Goedde HW. The determination of beta-aminoisobutyric acid in human serum by ion-exchange chromatography. Clinica chimica acta; international journal of clinical chemistry. 1975;59:203–207. doi: 10.1016/0009-8981(75)90030-3. [DOI] [PubMed] [Google Scholar]