Abstract
Background:
Abnormalities in circadian rhythms may be causal factors in development of major depressive disorder. The biology underlying a causal relationship between circadian rhythm disturbances and depression is slowly being unraveled. Although there is no direct evidence of dysregulation of clock gene expression in depressive patients, many studies have reported single-nucleotide polymorphisms in clock genes in these patients.
Methods:
In the present study we investigated whether a depression-like state in rats is associated with alternations of the diurnal expression of clock genes. The validated chronic mild stress (CMS) animal model of depression was used to investigate rhythmic expression of three clock genes: period genes 1 and 2 (Per1 and Per2) and Bmal1. Brain and liver tissue was collected from 96 animals after 3.5 weeks of CMS (48 control and 48 depression-like rats) at a 4h sampling interval within 24h. We quantified expression of clock genes on brain sections in the prefrontal cortex, nucleus accumbens, pineal gland, suprachiasmatic nucleus, substantia nigra, amygdala, ventral tegmental area, subfields of the hippocampus, and the lateral habenula using in situ hybridization histochemistry. Expression of clock genes in the liver was monitored by real-time quantitative polymerase chain reaction (PCR).
Results:
We found that the effect of CMS on clock gene expression was selective and region specific. Per1 exhibits a robust diurnal rhythm in most regions of interest, whereas Bmal1 and in particular Per2 were susceptible to CMS.
Conclusion:
The present results suggest that altered expression of investigated clock genes is likely associated with the induction of a depression-like state in the CMS model.
Keywords: animal model, circadian rhythm, clock gene expression, depression, stress
Introduction
Major depressive disorder is a condition with high prevalence in modern society. According to the World Health Organization, depression is currently ranked in the top five of the ten leading causes of the global burden of disease. The pathophysiology is complex and involves many brain regions. Furthermore, the symptomatology is heterogeneous and inconsistent. The core symptoms are anhedonia (lack of feeling pleasure) and depressed mood. Minor symptoms are irritability, difficulties in concentrating, feelings of worthlessness, guilt, agitation, and sleep disturbances (Kroenke et al., 2003).
Individuals that have an inherited abnormally-shifted or arrhythmic biological clock have a higher risk of becoming depressed (McClung, 2007). Jet lag and shift work conditions, disrupting the internal homeostasis, are also linked to major depressive disorder (Katz et al., 2002). Moreover, circadian manipulations, such as total sleep deprivation and bright light therapy, can reverse depressive symptoms within hours (Bunney and Potkin, 2008). Therefore, abnormalities in the circadian rhythm have been suggested to be a causal factor in the development of major depressive disorder.
Physiological and behavioral processes are governed by the master clock, the suprachiasmatic nucleus (SCN), located in the anterior part of the hypothalamus (Reppert and Weaver, 2002). Each cell in the SCN clock has a self-sustained molecular oscillator, which generates a rhythmic clock gene expression with a periodicity of about 24h (Reppert and Weaver, 2001). The individual neurons in the SCN are synchronized to a functional clock that drives circadian rhythms. Furthermore, the SCN clock receives daily information about environmental light and dark via a monosynaptic pathway from the retinal ganglion cells (Do and Yau, 2010).
Circadian oscillators are also present elsewhere in the central nervous system and in peripheral tissues, including the liver (Ramsey et al., 2007). Approximately 20 clock genes have been characterized, including the key genes such as period genes (Per) 1, 2, and 3, the cryptochrome genes 1 and 2, and brain and muscle arnt-like protein-1 (Bmal1/Aryl hydrocarbon receptor nuclear translocator-like [ARNTL1]).
The current study is based on the chronic mild stress (CMS) rat model of depression, which is a substantially validated model (Willner, 2005; Wiborg, 2013). It builds on the notion that exposure to unpredictable mild stress causes a reduction in the voluntary intake of a palatable solution of sucrose. Reduced sucrose intake is believed to be the consequence of a decrease in responsiveness to rewarding stimuli, mimicking the clinical core symptom of depression, anhedonia (Willner et al., 1987).
The diurnal rhythm of classical phase markers—melatonin, corticosterone, and core body temperature—has been shown to be abnormally regulated in anhedonic-like rats (Solberg et al., 2001; Ushijima et al., 2006; Christiansen et al., 2016). This finding may suggest underlying differences in the expression of core clock genes. To investigate this relationship further, we examined whether a stress-induced depression-like state was associate with changes in the rhythm of gene expression in sub-regions of the brain and in the liver. With this study we present for the first time a 24h expression profile analysis of three core clock genes in rats exposed to chronic mild stress.
Methods
Animals
Male Wistar rats were purchased from Taconic (6 weeks old, 120g). During the CMS protocol the animals were housed separately, except when grouping was applied as a stress parameter. Food and water was available ad libitum with an exception when food and/or water deprivation was applied as a stressor. Furthermore, the standard 12h light/dark cycle was only changed in course of the stress regime.
Sucrose Consumption Test
Sucrose consumption tests were performed as described previously (Christiansen et al., 2012). The rats were habituated to consume 1.5% sucrose solution during 1h weekly. Subsequently, the sucrose consumption test was performed once a week during the experiment. The first three measurements prior to CMS exposure established the baseline sucrose intake. Both the control and the CMS animals were food and water deprived for 14h prior to the sucrose consumption test. The sucrose index was used to characterize the individual hedonic status and was calculated for each measurement as a ratio between a current value of sucrose consumption test and the averaged baseline value. After 3.5 weeks of CMS exposure, anhedonic-like rats were selected based on a >30% decrease in sucrose consumption: 48 of them were used in the current study as well as 48 rats from the control group not challenged by stressors.
Chronic Mild Stress Model
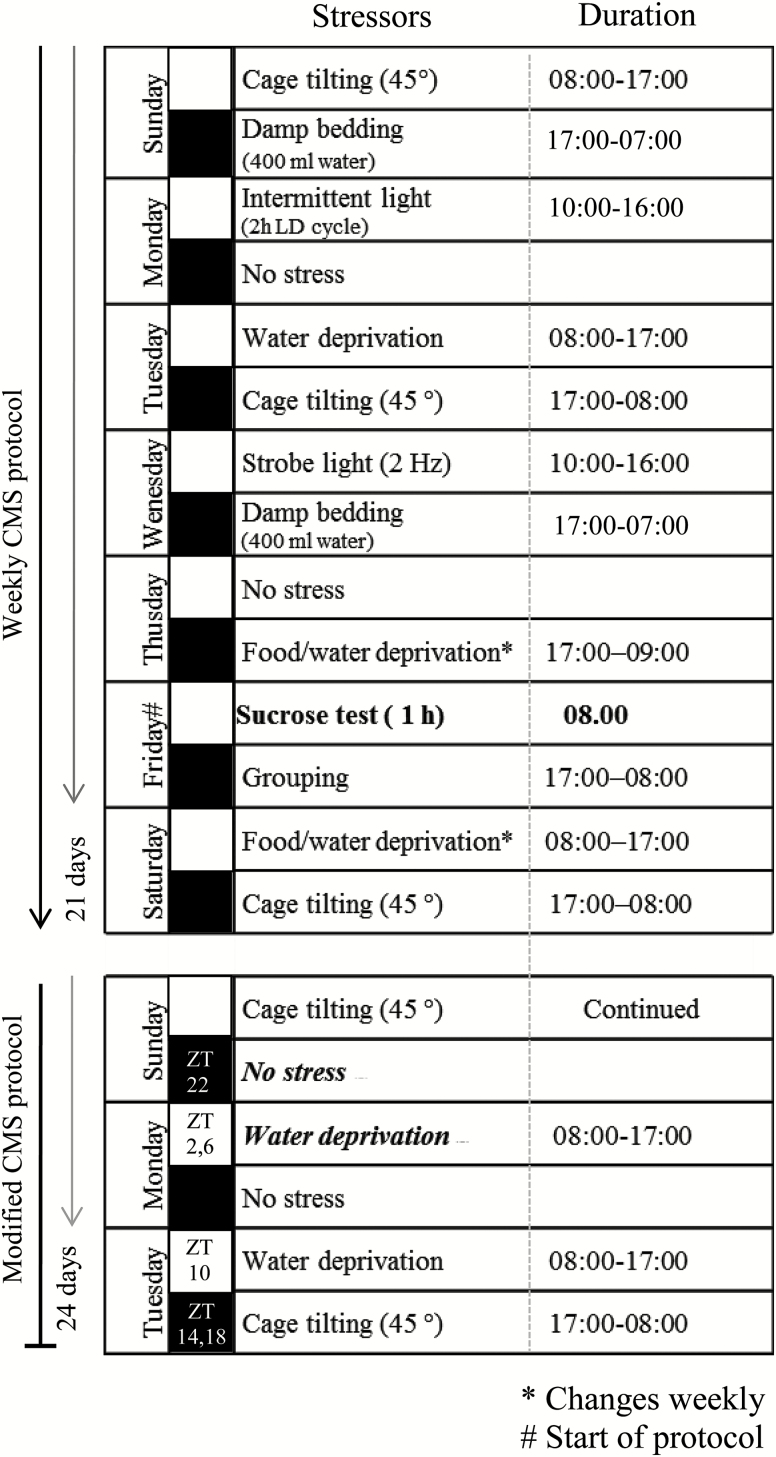
In short, the weekly stress protocol consisted of: one period of intermittent illumination, stroboscopic light, grouping, food or water deprivation, two periods of soiled cage and no stress, and three periods of box tilting (Figure 1, modified from Christiansen Hojgaard et al. [2016]). During grouping, rats were housed in pairs with different partners alternately serving as resident or intruder. All stressors lasted 10–14h. To minimize phase-shifting effects in the circadian system evoked by some of the individual stressors used in the original CMS protocol, changes were induced.
Figure 1.
Scheme of the stress protocol and the modifications applied during the diurnal study. Black boxes indicate the dark period of the 12h light/dark cycle. ZT, zeitgeber time. Figure is modified from Christiansen et al. (2016).
Experimental Design
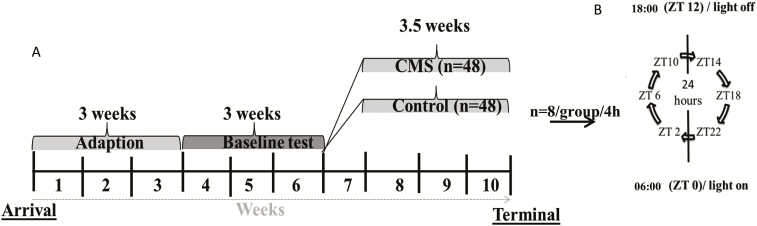
After adaptation to the animal facility and to sucrose consumption tests, the animals were randomly divided in two matched groups: stress unchallenged control and CMS. The animals used in the current study were selected based on their sucrose intake by the third week of CMS exposure (Figure 2A).
Figure 2.
(A) Experimental design and (B) a definition of zeitgeber times (ZT) during the daily 12h light/dark cycle.
After 3.5 weeks of CMS, eight animals from the control group and eight animals from the anhedonic-like group were killed by decapitation every 4h at the following zeitgeber times (ZTs): 2, 6, 10, 14, 18, and 22 (Figure 2B). Decapitation was initiated at ZT 22 (Figure 1).
Brain and liver tissues were removed, frozen on dry ice, and stored in -80◦C until further processing.
Handling and decapitation of the experimental animals during the dark phase was performed under dim red light. The total time of the procedure did not exceed 5min.
In Situ Hybridization Histochemistry
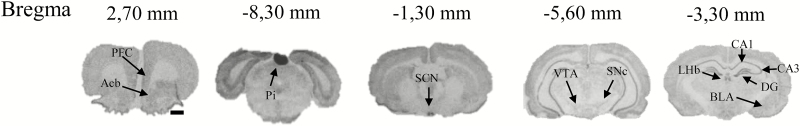
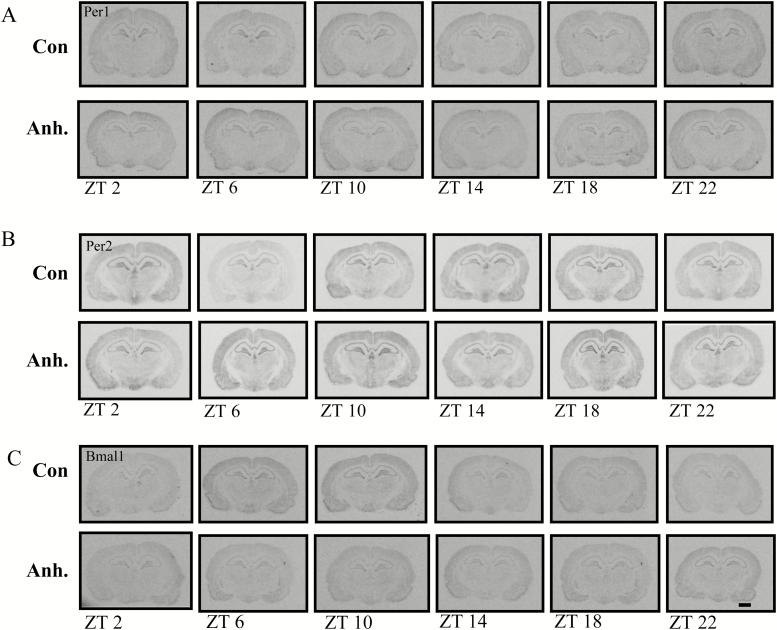
Frozen brains were cut in 12 µm thick coronal sections as consecutive series of ten on a cryostat (Leica, 3050 CS) at -20◦C. The following regions of interest were chosen: prefrontal cortex (infralimbic cortex) and shell of nucleus accumbens (bregma: 2.70mm); SCN (bregma:-1.30mm); hippocampus (CA1, CA3, and DG), lateral habenula, and the basolateral amygdala (bregma:-3.30mm); substantia nigra,pars compacta and the ventral tegmental (bregma:-5.60mm); and pineal gland (bregma:-8.30mm; Figure 3). The sections were mounted on superfrost plus glass slides immediately after sectioning. In situ hybridization histochemistry was performed using our previously described 33P-labelled sense and antisense c-RNA probes for vasoactive intestinal polypeptide, Per1, Per2 and Bmal1 (Nielsen et al., 1998; Fahrenkrug et al., 2008).
Figure 3.
Representative autoradiographic images of coronal brain sections with the corresponding bregma. Figure shows the in situ hybridization histochemistry of Per1 in the control group in the prefrontal cortex (PFC; the prelimbic part), nucleus accumbens (Acb), pineal gland (Pi), suprachiasmatic nucleus (SCN), substantia nigra (compacta part; SNc), ventral tegmental area (VTA), sub-regions of the hippocampus (CA1, CA3, and dentate gyrus [DG]), lateral habenula (LHb), and the basolateral part of the amygdala (BLA) at the time points indicated. Scale bar: applies to all images, 1mm.
Briefly, slides were adjusted to room temperature and fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) for 20min. Following PBS washing procedure, sections were permeabilised in 0.5 % triton X in PBS, and acetylated in 0.1 triethanol-amine for 10min. After consequent washing, the sections were incubated at 60◦C overnight with a hybridization buffer containing 1 x 107 c.p.m./ml of the (33P) UTP cRNA antisense probes. At the end, slides were rinsed in a gradient of sodium citrate saline, dehydrated in the ethanol gradient, and dried on air.
Finally, the sections were visualized on BioMax MR Film (Kodak) by 2 to 10 days of exposure. The intensity of the mRNA signal was evaluated by measurements of optical density for Per1, Per2, and Bmal1 expression using the ImageJ analyzing software (NIH). Measurements were performed on the left hemisphere, except for the SCN and the lateral habenula, where both hemispheres were used. Vasoactive intestinal polypeptide was used as a localization marker to evaluate expression in the SCN (data not shown).
Data are expressed as optical densities according to values calibrated with a 14C standard as references (Amersham). The level of mRNA in each brain was estimated by calculating the mean optical density from three sections. The calculated mean of these measurements was used to calculate the group mean and standard error of the mean (n = 8 in each group except for pineal gland where n = 5–8). Measurements obtained from the individual sections were corrected for nonspecific background by subtracting grey scale values from neighboring areas considered free of positive hybridization (Hannibal et al., 2001)
RNA Extraction, cDNA Synthesis, and Real-Time PCR Quantification
RNA extraction, cDNA synthesis, and the PCR reactions were performed as previously described (Fahrenkrug et al., 2006). Briefly, total liver RNA was extracted by the guanidinium thiocyanate-phenol-chloroform (TRIzol RNA isolation reagent) method (Chomczynski and Sacchi, 1987). RNA integrity was quantified by 28S/18S RNA ratio using Experion RNA StdSens Analysis Kit with the Experion Automated Electrophoreses System (Bio-Rad). RNA purity and concentration were determined by NanoDrop 1000 Spectrophotometer (Thermo Scientific). One µg of liver RNA was used for synthesis of cDNA using Taqman reverse transcription reagents (Applied Biosystems) in a total volume of 20 µl. The pooled cDNA was used to construct the standard curves.
The quantitative PCR was performed using a MX3000 Quantitative PCR System (Strategene) with Per1, Per2, Bmal1, and β2M primers and Taqman probes (Eurofin MWG Operon; Tables 1 and 2). It was verified that the amount of mRNA β2M, used as internal control, did not vary as function of time (data not shown). The multiplex principle was used to measure expression levels of gene of interest and the reference gene in the same well. All samples, standards, and the non-template-negative controls were made in duplicates in a final volume of 50 µl. The Taqman universal PCR master mix (Applied Biosystems) was used to run qPCRs.
Table 1.
Sequences of Primers Used for Real-Time Q-PCR
| Gene | Forward primers 5′ → 3′ | Reverse primers 5′ → 3′ |
|---|---|---|
| -Per1 | AGCTCTGCTGGAGACCACTGA | CACTCAGGAGACTATAGGCAATGGA |
| -Per2 | GCAGGCTCACTGCCAGAACT | CAAGATGATTCTATTCCAGAAGCATT |
| -Bmal1 | TGGACTGCAACCGCAAGAG | CCTTCCATGAGGGTCATCTTTG |
| -β 2 M | CGTGCTTGCCATTCAGAAAA | GAAGTTGGGCTTCCCATTCTC |
Table 2.
Sequences of TaqMan Probes and the Fluorescent Dye Used for Real-Time Q-PCR
| Gene | TaqMan probes and fluorescent dyes |
|---|---|
| -Per1 | FAM - CAGCAAGAGTACAAACTCACAGAGCCCATCC - TAMRA |
| -Per2 | FAM - AGCCCCAGCAAGTGATCGAGGACTAAG - TAMRA |
| -Bmal1 | FAM - CCATACTTTCTTGGTAGTTCAGTGGACTGC - TAMRA |
| -β 2 M | HEX - TCCCCAAATTCAAGTGTACTCTCGCCATC - TAMRA |
Statistical Analysis
All data were analyzed for normal distribution and presented as mean ± standard error of the mean (SEM). Data were analyzed using two-way ANOVA with CMS/stress factor (control vs. anhedonic-like) and ZT (zeitgeber 2, 6, 10, 14, 18, 22) as independent factors using statistical and data analysis package XLSTAT2010 (Addinsoft). Significance in differences by ZT as a main factor corresponds to the presence of diurnal rhythmic changes in the clock genes of interest. Bonferroni′s post hoc test was used for multiple comparisons in all tests. p < 0.05 was considered statistically significant.
Ethics
All the procedures involving animals are accepted by the Danish National Committee for Ethics in Animals Experimentation (Project ID: 2013-15-2934-00814).
Results
Sucrose Consumption Data
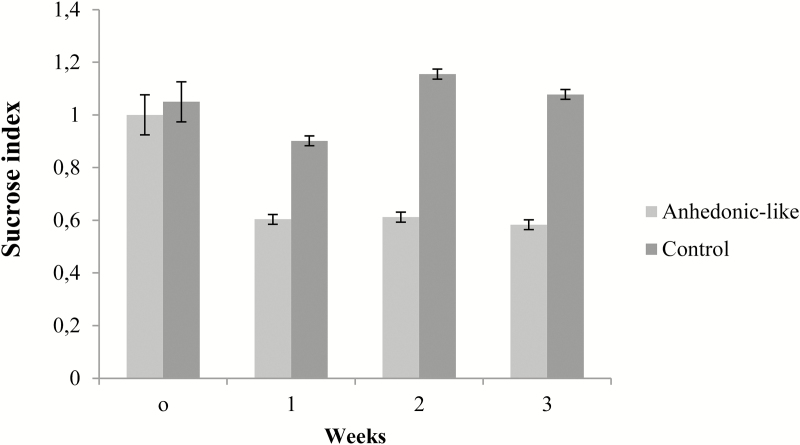
The stress-induced anhedonic-like state was assessed by the sucrose consumption test. Figure 4 shows the weekly sucrose intake for control and anhedonic-like rats during 3 weeks of CMS paradigm. Rats responding to the CMS paradigm by decreasing their sucrose intake more than 30% compared to baseline were designated as anhedonic-like. Therefore, all animals with an average sucrose index below 0.7 were classified as anhedonic-like.
Figure 4.
Effect of chronic mild stress (CMS) on sucrose intake.
Data from sucrose consumption tests expressed as a sucrose index for animals used in this study. SI was measured during 3 weeks of CMS for the anhedonic-like rats and the control rats. Data are presented as mean ± SEM.
Baseline sucrose intake in the control group was 14.80ml ± 0.45ml versus 15.97ml ± 0.39ml in the anhedonic-like group. The decrease in sucrose intake in the anhedonic-like group was consistent throughout the experiment.
Expression of Clock Genes in Sub Regions of the Brain
Using in situ hybridization histochemistry, we examined the diurnal rhythm of gene expression of the three key clock genes Per1, Per2, and Bmal1 in eleven sub-regions of the brain believed to be implicated in major depressive disorder.
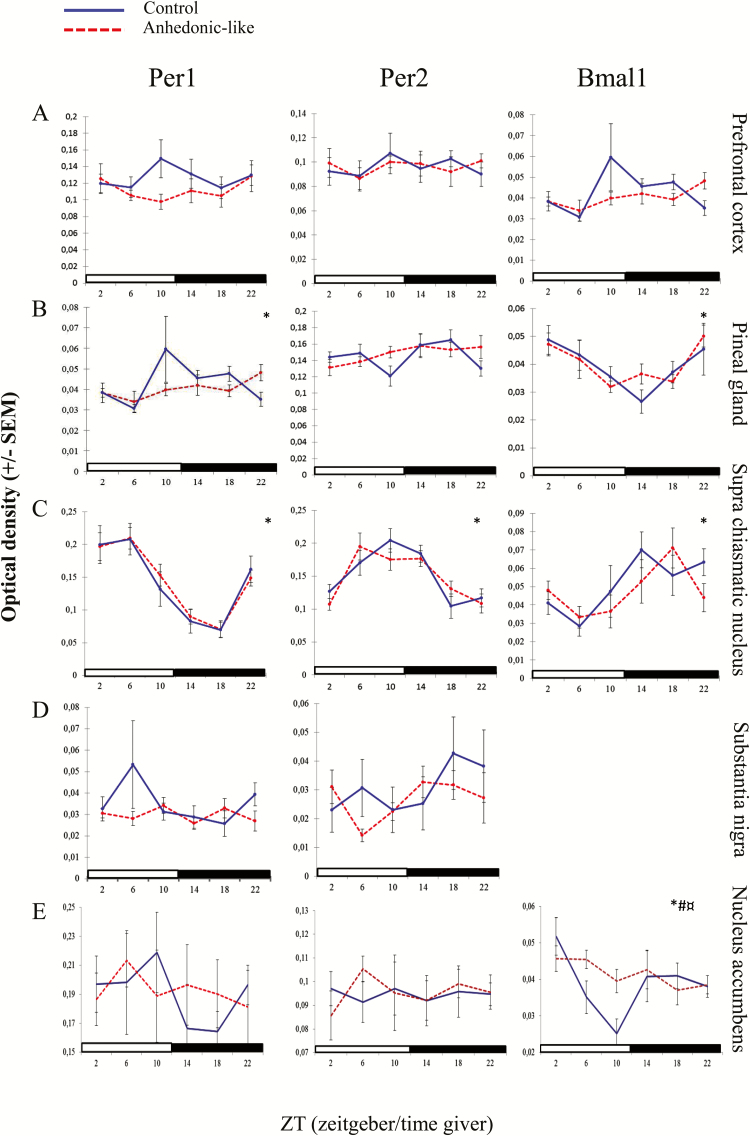
Expression of clock genes in the prefrontal cortex did not follow a diurnal rhythm (Figure 5A) and two-way ANOVA did not reveal any significant effect of CMS, time, or their interaction in any of the three genes in this region (Table 3).
Figure 5.
The effect of chronic mild stress (CMS) on clock gene expression in the brain.
The effect of CMS on the diurnal expression pattern of Per1, Per2, and Bmal1 in the (A) prefrontal cortex, (B) pineal gland, (C) suprachiasmatic nucleus, (D) substantia nigra, and (E) nucleus accumbens in anhedonic-like and control rats under 12h light/dark cycle conditions. The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as the time point when light in facilities switches on. Black bars indicate the dark period. The anhedonic-like group is indicated by a dashed line and the unchallenged control group by a solid line. Data are presented as mean ± SEM (n = 8 rats per time point per group, except for pineal gland where n = 5–8 in each group). Data for clock genes were subjected to two-way ANOVA to test for significant differences depending on effect of time (*), of CMS (#), and their interaction (¤).
Table 3.
A Statistical Overview of Results for Clock Gene Expression in Brain Regions
| Stress Factor | Time | Interaction | ||
|---|---|---|---|---|
| Prefrontal cortex | Per1 | F(1,95) = 2.89, | F(5,95) = 0.56, | F(5,95) = 0.94, |
| NS | NS | NS | ||
| Per2 | F(1,95) = 0.001, | F(5,95) = 0.47, | F(5,95) = 0.31, | |
| NS | NS | NS | ||
| Bmal1 | F(1,95) = 0.56, | F(5,95) = 1.98, | F(5,95) = 1.78, | |
| NS | NS | NS | ||
|
Pineal
gland |
Per1 | F(1,74) = 0.06, | F(5,74) = 5.75, | F(5,74) = 0.54, |
| NS | p < 0.0001 | NS | ||
| Per2 | F(1,79) = 0.16, | F(5,79) = 1.58, | F(5,79) = 1.44, | |
| NS | NS | NS | ||
| Bmal1 | F(1,78) = 0.40, | F(5,78) = 5.61, | F(5,78) = 0.70, | |
| NS | p < 0.0001 | NS | ||
|
Suprachias-matic nucleus
|
Per1 | F(1,93) = 0.03, | F(5,93) = 17.94, | F(5.93) = 0.17, |
| NS | p < 0.001 | NS | ||
| Per2 | F(1,93) = 0.001, | F(5,93) = 12.45, | F(5.93) = 1.16, | |
| NS | p < 0.001 | NS | ||
| Bmal1 | F(1,89) = 0.64, | F(5,89) = 4.04, | F(5.89) = 1.28, | |
| NS | p < 0.003 | NS | ||
|
Substantia nigra
|
Per1 | F(1,94) = 1.70, | F(5,94) = 0.79, | F(5,94) = 1.28, |
| NS | NS | NS | ||
| Per2 | F(1,94) = 0.70, | F(5.94) = 0.98, | F(5,94) = 0.81, | |
| NS | NS | NS | ||
| Bmal1 | - | - | - | |
| - | - | - | ||
|
Nucleus
accumbens |
Per1 | F(1.94) = 0.16, | F(5.95) = 1.02 | F(5.95) = 0.207 |
| NS | NS | NS | ||
| Per2 | F(1.94) = 0.57 | F(5.95) = 0.52 | F(0.49) = 0.78 | |
| NS | NS | NS | ||
| Bma1 | F(1,.5) = 11.78, | F(5.95) = 2.06 | F(5.95) = 4,081 | |
| p < 0.001 | p < 0.005 | p < 0.002 | ||
| CA1 | Per1 | F(1,95) = 4.98, | F(5,95) = 3.38, | F(5.95) = 0.088, |
| p < 0.03 | p < 0.008 | NS | ||
| Per2 | F(1,95) = 5.83, | F(5,95) = 0.68, | F(5.95) = 0.53, | |
| p < 0.02 | NS | NS | ||
| Bmal1 | F(1,95) = 0.05, | F(5,95) = 1.45, | F(5.95) = 0.87, | |
| NS | NS | NS | ||
| CA3 | Per1 | F(1,95) = 1.92, | F(5,95) = 2.92, | F(5.95) = 0.79, |
| NS | p < 0.002 | NS | ||
| Per2 | F(1,95) = 8.38, | F(5,95) = 1.74, | F(5.95) = 0.80, | |
| p < 0.005 | NS | NS | ||
| Bmal1 | F(1,95) = 0.46, | F(5,95) = 0.72, | F(5.95) = 0.67, | |
| NS | NS | NS | ||
|
Dentate
gyrus |
Per1 | F(1,95) = 0.5,8, | F(5,95) = 2.75, | F(5,95) = 1.20, |
| NS | p < 0.002 | NS | ||
| Per2 | F(1,95) =8.15, | F(5,95) = 1.07, | F(5,95) = 0.67, | |
| p < 0.005 | NS | NS | ||
| Bmal1 | F(1,95) = 0.28, | F(5,95) = 0.75, | F(5.95) = 0.46, | |
| NS | NS | NS | ||
|
Lateral habenula
|
Per1 | F(1,94) = 0.05, | F(5, 94) = 0.75, | F(5, 94) = 1.26, |
| NS | NS | NS | ||
| Per2 | F(1, 95)= 4.82, | F(5, 95) = 0.36, | F(5, 95) = 0.74, | |
| p < 0.05 | NS | NS | ||
| Bmal1 | F(1,74) = 0.49, | F(5, 94) = 0.72, | F(5, 94) = 0.47, | |
| NS | NS | NS |
Results of clock gene expression measurements subjected to two-way ANOVA to test for model differences with an effect of group, time, and their interaction. NS, not significant.
Expression of Per1 and Bmal1, but not Per2, in the pineal gland was found by two-way ANOVA to follow a diurnal rhythm (Table 3). Evidently a diurnal rhythmicity was observed in both groups of animals (Figure 6A, B, and C). Per1 expression was higher during the offset of light phase to onset of dark phase and in antiphase with Bmal1 that was higher during the offset of dark phase to the onset of light phase (Figure 5B). The time points for peak and nadir expressions in both groups were shown in Table 4.
Figure 6.
Representative images from the in situ hybridisation histochemistry for analysis of diurnal rhythm of Per1, Per2, and Bmal1 clock genes in coronal sections of the pineal gland in the rat brain. The experimental animals were decapitated after 3.5 weeks of chronic mild stress (CMS) every 4h. The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as the time point when light switches on. Decapitation was initiated at ZT 22. All animals were housed under a 12h light/dark cycle. Scale bar: applies to all images, 1mm.
X-ray images of the (A) Per1, (B) Per2, and (C) Bmal1 clock genes in the pineal gland in the control groups and the anhedonic-like groups at six time points.
Table 4.
Table of Peak and Nadir Values
| Control | Anh.-like | |||||
|---|---|---|---|---|---|---|
| Per1 | Per2 | Bmal1 | Per1 | Per2 | Bmal1 | |
| Pineal gland | ||||||
| Peak | 18 | - | 2 | 14 | - | 22 |
| Nadir | 10 | - | 14 | 6 | - | 10 |
| Suprachisamatic nucleus | ||||||
| Peak | 6 | 10 | 14 | 6 | 6 | 18 |
| Nadir | 18 | 18 | 6 | 18 | 2 | 6 |
| Hippocampus* | ||||||
| Peak | 22 | - | - | 18 | - | - |
| Nadir | 14 | - | - | 10 | - | - |
| Liver | ||||||
| Peak | 10 | 14 | 22 | 10 | 14 | 22 |
| Nadir | 2 | 2 | 10 | 2 | 2 | 10 |
* = (CA1, CA3, and DG)
Peak and nadir of the control and anhedonic-like (anh.-like) rats for Per1, Per2, and Bmal1 clock genes in the pineal gland, suprachiasmatic nucleus, hippocampus, and liver. Peak and nadir are presented as zeitgeber time (ZT), where ZT 0 is defined as a time point when light in facilities is on.
– = no diurnal rhythm measured by two-way ANOVA.
In the SCN, a pronounced diurnal rhythm of clock gene expression was observed in both groups for all three genes analyzed in the study (Figures 5C and 7A, B, and C). Two-way ANOVA revealed a significant effect of time with no difference between CMS and their interaction (Table 3). Peak level of Per1 in the anhedonic-like group was found at the same time point as in the control group (Table 4). Although the ANOVA-test did not show any significant interaction between stress factor and time, mean peak value for Per2 was 4 hours earlier in the anhedonic-like group compared to control animals. For Bmal1 peak value occurred 4 hours later compared to control rats (Table 4).
Figure 7.
Representative images from the in situ hybridisation histochemistry for analysis of diurnal rhythm of Per1, Per2, and Bmal1 clock genes in coronal sections of the suprachiasmatic nucleus (SCN) in the rat brain. The experimental animals were decapitated after 3.5 weeks of chronic mild stress every 4h. The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as the time point when light switches on. Decapitation was initiated at ZT 22. All animals were housed under a 12h light/dark cycle. Scale bar: applies to all images, 1mm.
X-ray images of the (A) Per1, (B) Per2, and (C) Bmal1 clock genes in the SCN in the control groups and the anhedonic-like groups at six time points.
In substantia nigra, the expression level of Per1 and Per2 was low and expression of Bmal1 was undetectable (Figure 5D). The two-way ANOVA did not reveal any significant differences between groups, time, or their interaction (Table 3).
Clock gene expression in the nucleus accumbens was calculated by two-way ANOVA and revealed a significant effect of groups, time, and their interaction for Bmal1, but not for Per1 and Per2 (Figure 5E; Table 3). Lack of rhythmicity in the expression of the Per1 gene may be explained by the large SEM.
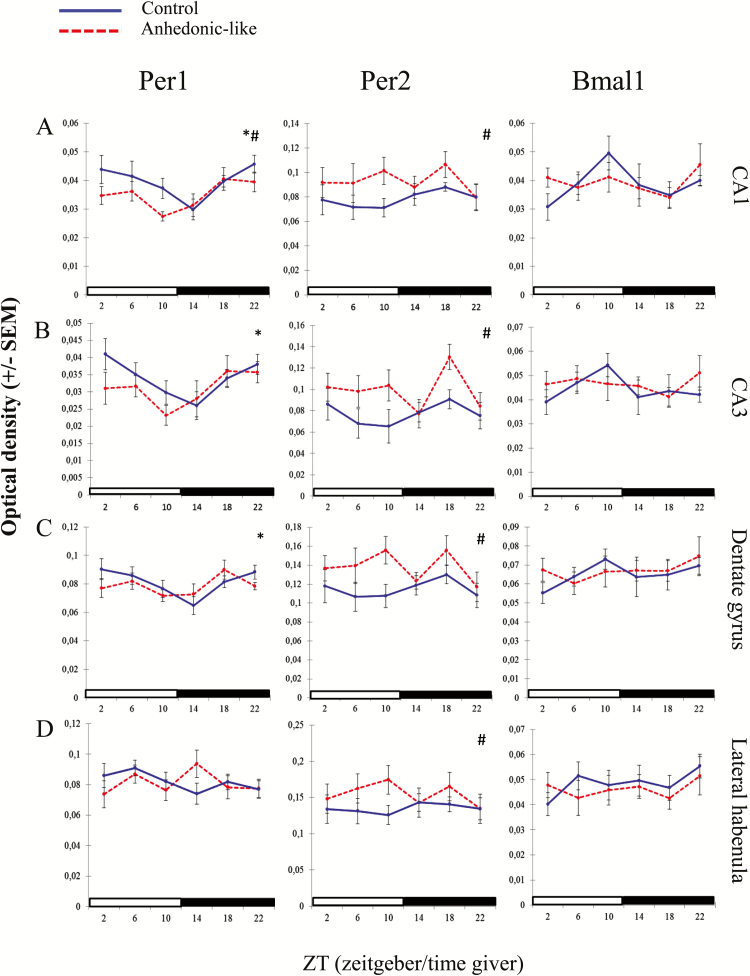
In the hippocampal CA1 area, differences between groups and time in Per1 expression were confirmed by the two-way ANOVA test with lower values of expression in the anhedonic-like group (Table 3). The two-way ANOVA indicated a difference between groups (Table 3) with increased expression of Per2 in the anhedonic-like group. For Bmal1 no significant differences were observed (Figures 8A and 9A, B, and C).
Figure 8.
The effect of chronic mild stress (CMS) on clock gene expression in the hippocampus and habenula.
The effect of CMS on the diurnal expression pattern of Per1, Per2, and Bmal1 in the (A) CA1, (B) CA3, (C) dentate gyrus, and (D) lateral habenula in anhedonic-like and control rats under 12h light/dark cycle conditions. The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as a time point when light in facilities switches on. Black bars indicate the dark period. The anhedonic-like group is indicated by a dashed line and the unchallenged control group by a solid line. Data are presented as mean ± SEM (n = 8 rats per time point per group, except for the pineal gland where n = 5–8 in each group). Data for clock genes were subjected to two-way ANOVA to test for significant differences depending on effect of time (*), of CMS (#), and their interaction.
Figure 9.
Representative images from the in situ hybridisation histochemistry for analysis of diurnal rhythm of Per1, Per2, and Bmal1 clock genes in coronal sections of the hippocampus in the rat brain. The experimental animals were decapitated after 3.5 weeks of chronic mild stress every 4h. The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as the time point when light switches on. Decapitation was initiated at ZT 22. All animals were housed under a 12h light/dark cycle. Scale bar: applies to all images, 1mm.
X-ray images of the (A) Per1, (B) Per2, and (C) Bmal1 clock genes in the hippocampus in the control groups and the anhedonic-like groups at six time points.
Figure 8 (B and C) demonstrates expression of clock genes in the CA3 and dentate gyrus regions of the hippocampus. Two-way ANOVA revealed differences with main effect of time for Per1 and main effect of groups for Per2 in both regions. Differences in expression of Bmal1 were insignificant (Table 3). ZTs for peak and nadir expression are shown in Table 4.
An effect of CMS on expression of clock genes in the lateral habenula was observed only as significant differences in the expression of Per2 between groups, thus anhedonic-like rats demonstrated an increased level of expression (Figure 8D; Table 3).
Optical density was very low to non-detectable in the ventral tegmental area and and in the basolateral amygdala (data not shown).
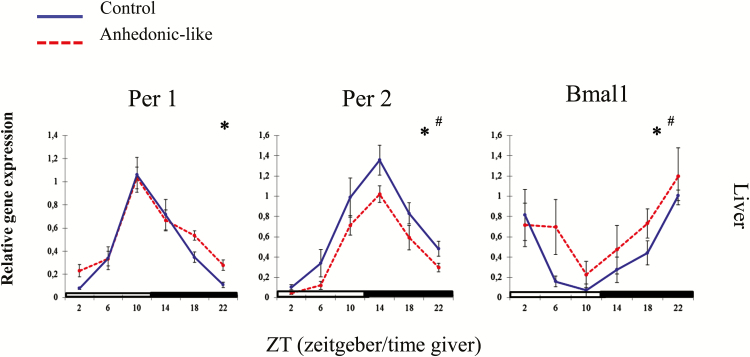
Hepatic Clock Gene Expression
Using q-PCR we measured the diurnal rhythm of gene expression of Per1, Per2, and Bmal1 in the liver of control and anhedonic-like rats (Figure 10). All clock genes were expressed in a rhythmic manner. Both groups displayed peak and nadir values at the same ZT for each of the three clock genes (Table 4). Expressions of Per1 and Per2 were in antiphase to Bmal1. Statistical analysis for Per1 expression in the liver revealed an effect of time, but no differences between groups or their interaction. Per2 and Bmal1 were also expressed in a rhythmic manner and significantly different between groups (Table 5).
Figure 10.
The effect of chronic mild stress on the 24h pattern of Per1, Per2, and Bmal1 in the liver in anhedonic-like and control rats under 12h light/dark cycle conditions.
The sampling time is indicated as zeitgeber time (ZT), where ZT 0 is defined as the time point when light in facilities switches on. Black bars indicate the dark period. The anhedonic-like group is indicated by a dashed line and the unchallenged control group by a solid line. Data are presented as mean ± SEM (n = 8 rats per time point per group, except for the pineal gland where n = 5–8 in each group). Data for clock genes were subjected to two-way ANOVA to test for significant differences depending on effect of time (*), of CMS (#), and their interaction.
Table 5.
A Statistical Overview of Results for Clock Gene Expression in the Liver
| Stress Factor | Time | Interaction | ||
|---|---|---|---|---|
| Liver | Per1 | F(1,90) = 2.52, NS |
F(5,90) = 36.154, p < 0.0001 |
F(5,90) = 0.95 NS |
| Per2 |
F(1,86) = 12.36, p < 0.001 |
F(5,86) = 30.99, p < 0.0001 |
F(5,86) = 0.38, NS |
|
| Bmal1 |
F(1,88) = 4.11, p < 0.05 |
F(5,88) = 6.93, p < 0.0001 |
F(5,88) = 0.61, NS |
Results of clock gene expression measurements subjected to two-way ANOVA to test for model differences with an effect of group, time, and their interaction. NS, not significant.
Discussion
In the present study we show the 24h rhythm (measured at 4h intervals) of three core clock genes in regions of the rat brain believed to be implicated in the depression-like state. Furthermore, we measured expression of the same clock genes in the liver. Our study found that clock genes in sub-regions of the hippocampal formation, the nucleus accumbens, and the liver were mostly affected by the stress paradigm. In general, the Per1 gene was mainly unaffected by the stress regime, whereas Bmal1 and Per2 were more stress susceptible.
An important study published in 2013 first demonstrated dysfunctioning clock genes in brains from depressed humans compared to controls (Li et al., 2013). Involvement of the clock genes in depression is also evident from several genetic studies. It has been reported that polymorphisms of clock genes is manifested in depressed patients (Partonen et al., 2007; Kripke et al., 2009; Wang et al., 2012; Kovanen et al., 2013; Hua et al., 2014; Shi et al., 2016).
From animal models, the clock genes have also been linked to depression-like behavior. Strong evidence is found in recent papers showing that SCN-specific Bmal1 knockdown mice exhibit depression-like behavior (Landgraf et al., 2016) and anhedonic-like behavior is observed in cryptochrome-deficient mice (Savalli et al., 2015). Further, studies have been published investigating the effect of stress on the expression of clock genes in rodents. However, different stressors were applied, such as post-traumatic stress disorder (Koresh et al., 2012), severe stressors (chronic unpredictable stress; Jiang et al., 2013), and systemic glucocorticoid administration and restraint stress (Al-Safadi et al., 2014). Furthermore, the use of different strains of animals (Erburu et al., 2015; Schaufler et al., 2016) and time points of day for decapitation makes it difficult to compare our results with these studies.
The CMS model is a well-established rat model of depression and has high face, predictive, and construct validity (Willner et al., 1992). However, when using this model to study chronobiology, it has to be noted that the CMS paradigm involves manipulating factors known as essential zeitgebers. In the current study, anhedonic-like rats were deprived of food twice a week. Since food is a known zeitgeber, this could pose a confounding factor on clock gene expression. Normally the feeding-fasting cycles are in phase with the rest-activity cycles. Therefore, rodents fed exclusively during the active period display a similar rhythmicity of liver gene expression compared to rodents fed ad libitum (Balsalobre et al., 2000; Le Minh et al., 2001). Further, it is only imposed meal times and temporally restricted feeding that are potent synchronizers for secondary clocks in peripheral organs such as the liver and in brain regions (for review see Challet and Mendoza [2010]). The CMS model relies on unpredictable stressors, therefore it is unlikely that nighttime food deprivation can take over as a zeitgeber.
A second zeitgeber potentially posing a problem for the present study was the light manipulations. However, to avoid this possible confounder, light manipulations were replaced by repeating other stressors in the final part of the study (see Figure 1). We argue that the measured disturbances in gene expression were induced by the combined stress load and not an effect of any individual stressor. In addition, light is known to shift the phase and we did not observe any shifting indicative for any influence of light manipulation. However, one cannot reject that the alternations of expression pattern could be due to acute stress. To exclude this possibility a separate study that examines circadian clock genes after acute stress influence is needed.
The SCN is considered the master clock of the body that hierarchically maintains stable phase relationships between endogenous rhythms and the cycle of light. One would therefore expect such a mechanism to be relatively stress insensitive. However, studies on the effect of stress on circadian oscillations are in disagreement. Some studies show that SCN is well protected against environmental disturbances (Meerlo et al., 2002; Liu et al., 2007) while other results indicate that stress is capable of perturbing the central circadian oscillator in the SCN (Jiang et al., 2011; Kerman et al., 2012).
The present study showed that the rhythm of clock gene expression was conserved in the SCN after stress exposure. However, changes in the mean time of peak were indicated for Per2 and Baml1. In agreement with other studies, a clear diurnal rhythm was observed for Per1, Per2, and Bmal1 expression with the peak and nadir of Per1 and Per2 in antiphase with Bmal1 (Nielsen et al., 2001; Girotti et al., 2009).
Comparing the expression levels between control and anhedonic-like rats showed that the expression pattern of Per1 in the SCN in both groups was almost identical, indicating Per1 is a very stable gene for maintaining the circadian rhythm. This is in line with a study showing that, following jet lag, the expression of Per1 re-entrains faster in the SCN than in any of the other examined tissues (Yamazaki et al., 2000). Moreover, Gilhoolet et al. (2011) showed that clamping the plasma corticosterone level did not alter expression of Per1 in the SCN, further supporting that Per1 is resistant to stress effects.
The prefrontal cortex is profoundly involved in sleep processes (Womack et al., 2013). Our findings demonstrate that the clock genes we studied are strongly expressed in the prefrontal cortex and, notably, they do not follow a diurnal rhythm (Figure 5A). This is in contrast to previously published data (Li et al., 2010; Edgar and McClung, 2013; Li et al., 2013; Erburu et al., 2015). However, the discrepancy probably relates to differences in the identity of the sub-regions being investigated. In our study, we analyzed the pre-limbic area of the prefrontal cortex, while related studies were referring to the entire prefrontal cortex.
A robust Per1 expression pattern was also demonstrated in specific subfields of the hippocampal formation, whereas Per2 was affected by CMS in all subfields (Figure 6A–C). Thus, a plausible mechanism whereby CMS alters the diurnal pattern of the classical phase markers, the extra SCN oscillators, and hepatics clock genes, without fundamentally altering the SCN rhythm, is by limbic regulation of the HPA axis function. Normally, the hippocampus has an inhibitory effect on the secretion of glucocorticoids, which is regulated by the HPA axis (Herman et al., 2005), and hyperactivity of the HPA axis in patients suffering from major depressive disorder is one of the most consistent findings in major depressive disorder (Pariante and Miller, 2001). Glucocorticoids are well known to affect the metabolic processes and to entrain the circadian rhythm in the peripheral organs such as the liver (Balsalobre et al., 2000). Our results are in agreement with this observation: in the liver Per2 and Bmal1 were significantly affected by the CMS regime, however, expression of Per1 was not significantly different from the control group.
Furthermore, our study indicates a main role of the hippocampus as one of the primary stress targets affecting the expression rhythmicity of clock-related genes. However, in the present study the rhythmical expression of clock genes in the hippocampus was only significant for Per1. Rhythmic expression of Per2 and Bmal1 was not statistically significant. Rhythmicity of clock gene expression in the hippocampus has, however, been stated by other groups: in rodents (Navigatore-Fonzo et al., 2014) and humans (Li et al., 2013).
Neurons localized in the SCN project to the pineal gland, the major site of melatonin synthesis (Kalsbeek et al., 2006). In major depressive disorder an abnormal rhythm of melatonin secretion is a prominent feature (Srinivasan et al., 2006). In our study the expression pattern of clock genes in the pineal gland was not affected by the CMS. The rhythm of expression of Per1 and Bmal1 in the pineal gland follows the rhythm of melatonin secretion. These results indicate that the clock genes do not affect the level of melatonin secretion. Thus it is likely that only light can affect the secretion of melatonin, further supporting melatonin as being a very stable proxy or marker of the circadian rhythm.
The pineal gland is connected to the lateral habenula, a structure that, according to anatomical and physiological studies, has been suggested to be implicated in the circadian timing system in rodents. (Tavakoli-Nezhad and Schwartz, 2006). The lateral habenula has therefore been suggested as a structure linking circadian rhythm abnormalities to mood alternations. In the present study, the analysis of clock gene expression in the lateral habenula confirms the stress-sensitive nature of Per2 expression, also reported in models manipulating the level of corticosterone (Segall et al., 2006; Segall and Amir, 2010; Logan et al., 2015).
The substantia nigra and the nucleus accumbens are key sub-regions involved in regulation of the dopaminergic system. Depression is frequently associated with a deficiency of dopamine (Dailly et al., 2004). Dopamine-producing neurons residing in these areas (Ungerstedt, 1971) are an important component of the reward system (Baik, 2013). We therefore investigated whether the anhedonic-like state affects the level of clock gene expression in the substantia nigra and the nucleus accumbens. In the substantia nigra, Per1 and Per2 are expressed in the two areas, but not in a statistically significant rhythmic manner. The level of Bmal1 is not detectable in the substantia nigra; however, in the nucleus accumbens the expression of Bmal1 is strongly affected by the CMS paradigm in the anhedonic-like rats. The expression pattern of the Bmal1 gene in the nucleus accumbens is in agreement with the literature (Masubuchi et al., 2000) and further indicates the involvement of the reward system in the depression-like state of CMS animals
Lastly, signal intensity of the clock genes is not equally strong in the different areas of the brain. In agreement with the literature, we measured strong signal intensities in the SCN and the hippocampus, while low to non-detectable signal intensities were measured in the ventral tegmental area and in the basolateral part of the amygdala. These data may further indicate the sub-oscillators that are most dominant for synchronizing the circadian rhythm.
In conclusion, our data provide evidence for altered expression pattern of clock genes, mainly Per2 and Bmal1, in sub-oscillators of the rat brain and in the liver. Since the development of an anhedonic-like condition is associated with alterations in clock gene expression, a normalization of this pattern is likely to be essential for the recovery from the pathological state.
Statement of Interest
None.
Acknowledgements
This work was supported by Danish Biotechnology Center for Cellular Communication. Stine Dhiin Hansen and Kim Henningsen are acknowledged for running the CMS model and their help during the 24h experimental work in the animal facilities.
References
- Al-Safadi S, Al-Safadi A, Branchaud M, Rutherford S, Dayanandan A, Robinson B, Amir S. (2014) Stress-induced changes in the expression of the clock protein PERIOD1 in the rat limbic forebrain and hypothalamus: role of stress type, time of day, and predictability. PLOS One 9(10):e111166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik JH. (2013) Dopamine signaling in reward-related behaviors. Front Neural Circuits 7:152. doi:10.3389/fncir.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289(5488):2344–2347. [DOI] [PubMed] [Google Scholar]
- Bunney JN, Potkin SG. (2008) Circadian abnormalities, molecular clock genes and chronobiological treatments in depression. Br Med Bull 86:23–32. [DOI] [PubMed] [Google Scholar]
- Challet E, Mendoza J. (2010) Metabolic and reward feeding synchronises the rhythmic brain. Cell Tissue Res 341(1):1–11. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162(1):156–159. [DOI] [PubMed] [Google Scholar]
- Christiansen S, Bouzinova EV, Palme R, Wiborg O. (2012) Circadian activity of the hypothalamic-pituitary-adrenal axis is differentially affected in the rat chronic mild stress model of depression. Stress 15(6):647–657. [DOI] [PubMed] [Google Scholar]
- Christiansen SL, Hojgaard K, Wiborg O, Bouzinova EV. (2016) Disturbed diurnal rhythm of three classical phase markers in the chronic mild stress rat model of depression. Neurosci Res. pii:S0168-0102(16)30001-3. doi:10.1016/j.neures.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Dailly E, Chenu F, Renard CE, Bourin M. (2004) Dopamine, depression and antidepressants. Fundam Clin Pharmacol 18(6):601–607. [DOI] [PubMed] [Google Scholar]
- Do MT, Yau KW. (2010) Intrinsically photosensitive retinal ganglion cells. Physiol Rev 90(4):1547–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar N, McClung CA. (2013) Major depressive disorder: a loss of circadian synchrony? Bioessays 35(11):940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erburu M, Cajaleon L, Guruceaga E, Venzala E, Munoz-Cobo I, Beltran E, Puerta E, Tordera RM. (2015) Chronic mild stress and imipramine treatment elicit opposite changes in behavior and in gene expression in the mouse prefrontal cortex. Pharmacol Biochem Behav 135:227–236. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug J, Georg B, Hannibal J, Hindersson P, Gras S. (2006) Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology 147(8):3769–3776. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug J, Hannibal J, Georg B. (2008) Diurnal rhythmicity of the canonical clock genes Per1, Per2 and Bmal1 in the rat adrenal gland is unaltered after hypophysectomy. J Neuroendocrinol 20(3):323–329. [DOI] [PubMed] [Google Scholar]
- Gilhooley MJ, Pinnock SB, Herbert J. (2011) Rhythmic expression of per1 in the dentate gyrus is suppressed by corticosterone: implications for neurogenesis. Neurosci Lett 489(3):177–181. [DOI] [PubMed] [Google Scholar]
- Girotti M, Weinberg MS, Spencer RL. (2009) Diurnal expression of functional and clock-related genes throughout the rat HPA axis: system-wide shifts in response to a restricted feeding schedule. Am J Physiol Endocrinol Metab 296(4): E888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal J, Jamen F, Nielsen HS, Journot L, Brabet P, Fahrenkrug J. (2001) Dissociation between light-induced phase shift of the circadian rhythm and clock gene expression in mice lacking the pituitary adenylate cyclase activating polypeptide type 1 receptor. J Neurosci 21(13):4883–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JP, Ostrander MM, Mueller NK, Figueiredo H. (2005) Limbic system mechanisms of stress regulation: hypothalamo-pituitary-adrenocortical axis. Prog Neuropsychopharmacol Biol Psychiatry 29(8):1201–1213. [DOI] [PubMed] [Google Scholar]
- Hua P, Liu W, Chen D, Zhao Y, Chen L, Zhang N, Wang C, Guo S, Wang L, Xiao H, Kuo SH. (2014) Cry1 and Tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J Affect Disord 157:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WG, Li SX, Zhou SJ, Sun Y, Shi J, Lu L. (2011) Chronic unpredictable stress induces a reversible change of PER2 rhythm in the suprachiasmatic nucleus. Brain Res 1399:25–32. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Li SX, Liu JF, Sun Y, Zhou SJ, Zhu WL, Shi J, Lu L. (2013) Hippocampal CLOCK protein participates in the persistence of depressive-like behavior induced by chronic unpredictable stress. Psychopharmacology (Berl) 227(1):79–92. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. (2006) SCN outputs and the hypothalamic balance of life. J Biol Rhythms 21(6):458–469. [DOI] [PubMed] [Google Scholar]
- Katz G, Knobler HY, Laibel Z, Strauss Z, Durst R. (2002) Time zone change and major psychiatric morbidity: the results of a 6-year study in Jerusalem. Compr Psychiatry 43(1):37–40. [DOI] [PubMed] [Google Scholar]
- Kerman IA, Clinton SM, Simpson DN, Bedrosian TA, Bernard R, Akil H, Watson SJ. (2012) Inborn differences in environmental reactivity predict divergent diurnal behavioral, endocrine, and gene expression rhythms. Psychoneuroendocrinology 37(2):256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koresh O, Kozlovsky N, Kaplan Z, Zohar J, Matar MA, Cohen H. (2012) The long-term abnormalities in circadian expression of Period 1 and Period 2 genes in response to stress is normalized by agomelatine administered immediately after exposure. Eur Neuropsychopharmacol 22(3):205–221. [DOI] [PubMed] [Google Scholar]
- Kovanen L, Kaunisto M, Donner K, Saarikoski ST, Partonen T. (2013) CRY2 genetic variants associate with dysthymia. PLOS One 8(8):e71450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. (2009) Circadian polymorphisms associated with affective disorders. J Circadian Rhythms 7:2. doi:10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke K, Spitzer RL, Williams JB. (2003) The patient health questionnaire-2: validity of a two-item depression screener. Med Care 41(11):1284–1292. [DOI] [PubMed] [Google Scholar]
- Landgraf D, Long JE, Proulx CD, Barandas R, Malinow R, Welsh DK. (2016) Genetic disruption of circadian rhythms in the suprachiasmatic nucleus causes helplessness, behavioral despair, and anxiety-like behavior in mice. Biol Psychiatry. PMID 27113500. doi:10.1016/j.biopsych.2016.03.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. (2001) Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J 20(24):7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ, Jr, Akil H, Bunney WE. (2013) Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci USA 110(24):9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SX, Liu LJ, Jiang WG, Sun LL, Zhou SJ, Le Foll B, Zhang XY, Kosten TR, Lu L. (2010) Circadian alteration in neurobiology during protracted opiate withdrawal in rats. J Neurochem 115(2):353–362. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129(3):605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Edgar N, Gillman AG, Hoffman D, Zhu X, McClung CA. (2015) Chronic stress induces brain region-specific alterations of molecular rhythms that correlate with depression-like behavior in mice. Biol Psychiatry 78(4):249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubuchi S, Honma S, Abe H, Ishizaki K, Namihira M, Ikeda M, Honma K. (2000) Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. Eur J Neurosci 12(12):4206–4214. [PubMed] [Google Scholar]
- McClung CA. (2007) Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther 114(2):222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerlo P, Sgoifo A, Turek FW. (2002) The effects of social defeat and other stressors on the expression of circadian rhythms. Stress 5(1):15–22. [DOI] [PubMed] [Google Scholar]
- Navigatore-Fonzo LS, Delgado SM, Golini RS, Anzulovich AC. (2014) Circadian rhythms of locomotor activity and hippocampal clock genes expression are dampened in vitamin A-deficient rats. Nutr Res 34(4):326–335. [DOI] [PubMed] [Google Scholar]
- Nielsen HS, Hannibal J, Fahrenkrug J. (1998) Embryonic expression of pituitary adenylate cyclase-activating polypeptide in sensory and autonomic ganglia and in spinal cord of the rat. J Comp Neurol 394(4):403–415. [DOI] [PubMed] [Google Scholar]
- Nielsen HS, Hannibal J, Knudsen SM, Fahrenkrug J. (2001) Pituitary adenylate cyclase-activating polypeptide induces period1 and period2 gene expression in the rat suprachiasmatic nucleus during late night. Neuroscience 103(2):433–441. [DOI] [PubMed] [Google Scholar]
- Pariante CM, Miller AH. (2001) Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry 49(5):391–404. [DOI] [PubMed] [Google Scholar]
- Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, Aron L, Rietschel M, Wellek S, Soronen P, Paunio T, Koch A, Chen P, Lathrop M, Adolfsson R, Persson ML, Kasper S, Schalling M, Peltonen L, Schumann G. (2007) Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med 39(3):229–238. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Marcheva B, Kohsaka A, Bass J. (2007) The clockwork of metabolism. Annu Rev Nutr 27:219–240. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. (2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63:647–676. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. (2002) Coordination of circadian timing in mammals. Nature 418(6901):935–941. [DOI] [PubMed] [Google Scholar]
- Savalli G, Diao W, Berger S, Ronovsky M, Partonen T, Pollak DD. (2015) Anhedonic behavior in cryptochrome 2-deficient mice is paralleled by altered diurnal patterns of amygdala gene expression. Amino Acids 47(7):1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufler J, Ronovsky M, Savalli G, Cabatic M, Sartori SB, Singewald N, Pollak DD. (2016) Fluoxetine normalizes disrupted light-induced entrainment, fragmented ultradian rhythms and altered hippocampal clock gene expression in an animal model of high trait anxiety- and depression-related behavior. Ann Med 48(1–2): 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall LA, Amir S. (2010) Exogenous corticosterone induces the expression of the clock protein, PERIOD2, in the oval nucleus of the bed nucleus of the stria terminalis and the central nucleus of the amygdala of adrenalectomized and intact rats. J Mol Neurosci 42(2):176–182. [DOI] [PubMed] [Google Scholar]
- Segall LA, Perrin JS, Walker SD, Stewart J, Amir S. (2006) Glucocorticoid rhythms control the rhythm of expression of the clock protein, Period2, in oval nucleus of the bed nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neuroscience 140(3):753–757. [DOI] [PubMed] [Google Scholar]
- Shi SQ, White MJ, Borsetti HM, Pendergast JS, Hida A, Ciarleglio CM, de Verteuil PA, Cadar AG, Cala C, McMahon DG, Shelton RC, Williams SM, Johnson CH. (2016) Molecular analyses of circadian gene variants reveal sex-dependent links between depression and clocks. Transl Psychiatry 6:e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg LC, Olson SL, Turek FW, Redei E. (2001) Altered hormone levels and circadian rhythm of activity in the WKY rat, a putative animal model of depression. Am J Physiol Regul Integr Comp Physiol 281(3):R786–794. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Smits M, Spence W, Lowe AD, Kayumov L, Pandi-Perumal SR, Parry B, Cardinali DP. (2006) Melatonin in mood disorders. World J Biol Psychiatry 7(3):138–151. [DOI] [PubMed] [Google Scholar]
- Tavakoli-Nezhad M, Schwartz WJ. (2006) Hamsters running on time: is the lateral habenula a part of the clock? Chronobiol Int 23(1–2): 217–224. [DOI] [PubMed] [Google Scholar]
- Ungerstedt U. (1971) Stereotaxic mapping of the monoamine pathways in the rat brain. Acta Physiol Scand Suppl 367:1–48. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Morikawa T, To H, Higuchi S, Ohdo S. (2006) Chronobiological disturbances with hyperthermia and hypercortisolism induced by chronic mild stress in rats. Behav Brain Res 173(2):326–330. [DOI] [PubMed] [Google Scholar]
- Wang J, Nuccio SR, Yang JY, Wu X, Bogoni A, Willner AE. (2012) High-speed addition/subtraction/complement/doubling of quaternary numbers using optical nonlinearities and DQPSK signals. Opt Lett 37(7):1139–1141. [DOI] [PubMed] [Google Scholar]
- Wiborg O. (2013) Chronic mild stress for modeling anhedonia. Cell Tissue Res 354(1):155–169. [DOI] [PubMed] [Google Scholar]
- Willner P. (2005) Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology 52(2):90–110. [DOI] [PubMed] [Google Scholar]
- Willner P, Towell A, Sampson D, Sophokleous S, Muscat R. (1987) Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology (Berl) 93(3):358–364. [DOI] [PubMed] [Google Scholar]
- Willner P, Muscat R, Papp M. (1992) Chronic mild stress-induced anhedonia: a realistic animal model of depression. Neurosci Biobehav Rev 16(4):525–534. [DOI] [PubMed] [Google Scholar]
- Womack SD, Hook JN, Reyna SH, Ramos M. (2013) Sleep loss and risk-taking behavior: a review of the literature. Behav Sleep Med 11(5):343–359. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288(5466):682–685. [DOI] [PubMed] [Google Scholar]