Abstract
Cerebral malaria (CM) is a severe complication of Plasmodium falciparum infection responsible for thousands of deaths in children in sub-Saharan Africa. CM pathogenesis remains incompletely understood but a number of effectors have been proposed, including plasma microparticles (MP). MP numbers are increased in CM patients’ circulation and, in the mouse model, they can be localised within inflamed vessels, suggesting their involvement in vascular damage. In the present work we define, for the first time, the protein cargo of MP during experimental cerebral malaria (ECM) with the overarching hypothesis that this characterisation could help understand CM pathogenesis. Using qualitative and quantitative high-throughput proteomics we compared MP proteins from non-infected and P. berghei ANKA-infected mice. More than 360 proteins were identified, 60 of which were differentially abundant, as determined by quantitative comparison using TMTTM isobaric labelling. Network analyses showed that ECM MP carry proteins implicated in molecular mechanisms relevant to CM pathogenesis, including endothelial activation. Among these proteins, the strict association of carbonic anhydrase I and S100A8 with ECM was verified by western blot on MP from DBA/1 and C57BL/6 mice. These results demonstrate that MP protein cargo represents a novel ECM pathogenic trait to consider in the understanding of CM pathogenesis.
Malaria infection caused by Plasmodium protozoan parasites still represents a major worldwide health problem affecting more than 200 million people and being responsible for the death of 600,000 of them, according to the latest WHO estimations1. Cerebral malaria (CM) is the most fatal malaria complication and affects mainly children under the age of 5 in sub-Saharan Africa2. CM prompt diagnosis remains difficult2 and despite available treatment, 15–20% of patients die, while 10–15% of cured patients will suffer from long-term neurological deficits3.
The pathological mechanisms of this complex neurological syndrome are still to be fully deciphered. The best described processes of CM pathogenesis include the sequestration of parasitized red blood cells (pRBC) in the brain microvasculature and an excessive activation of the immune response with production of pro-inflammatory cytokines4. An additional important feature is the increased number of microparticles (MP) in patients’ circulation. MP are submicron extracellular vesicles (100–1000 nm in size) released through a mechanism of outward blebbing of the plasma membrane by potentially all host cell types under physiological conditions or following stress and apoptosis5. Due to their process of formation, MP expose on their surface negatively charged phosphatidylserine residues and cellular markers specific to their cell of origin6. Importantly, they contain proteins, nucleic acids and lipids derived from the cytoplasm of the parent cell that they can convey to target cells, thus playing an important role in the intercellular communication and exchange of biological information7 as already shown in cancers, neurological disorders and cardiovascular diseases8,9,10. Interestingly, the proteomics characterisation of the cargo of plasma MP released under specific pathological conditions has already been found useful to identify new disease biomarkers and to propose new pathophysiological features11,12.
In CM patients, an increased number of MP originating from platelets, endothelial cells, monocytes and red blood cells has been shown in different clinical cohorts, where a significant correlation with the disease severity was proven13,14,15. Studies in the murine model of CM (experimental cerebral malaria – ECM), consisting of susceptible mouse strains (DBA/1, CBA and C57BL/6) infected with the Plasmodium berghei ANKA (PbA) parasite16, significantly contributed to further demonstrating that MP are not merely inert cellular products but active players in CM progression17,18. Indeed, similarly to human CM, increased numbers of cell-specific MP have been detected in ECM18 and mice showing a reduced release of MP (ABCA1−/− or pantethine-treated) are protected from the cerebral syndrome17,19. Additionally, adoptively transferred ECM plasma MP localised in inflamed vessels, elicited breakdown of the blood brain barrier and brain pathology similar to ECM18.
Although an important role of circulating MP in CM pathogenesis is now generally accepted20,21, the mechanisms through which these vesicles carry out their biological functions still need to be deciphered and their protein cargo is yet to be described. In this context, we hypothesized that the protein content of circulating host-derived MP produced during CM might be of central importance in the pathogenesis of this syndrome. To closely follow the development of the neurological syndrome, we took advantage of the well-established CM mouse model and we investigated the protein content of MP produced during ECM using high-throughput qualitative and quantitative proteomics and network analyses. Two selected ECM-MP proteins, carbonic anhydrase 1 (CA-I) and S100A8, were further verified on a larger number of samples and their abundance was proven to be increased within plasma MP specifically released during the infection.
Results
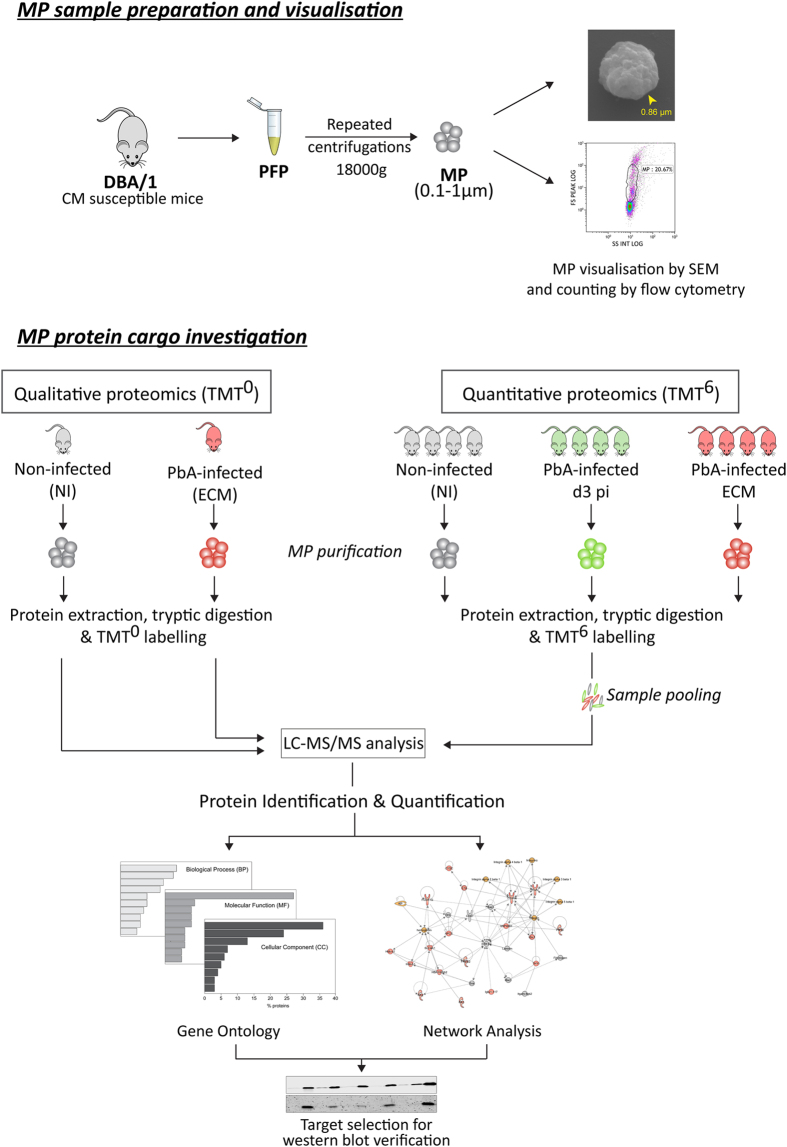
Experimental design
To obtain quantitative information on the protein cargo of MP released during different infection conditions we used the Tandem Mass TagTM (TMT) isobaric labelling technology (ThermoFisher Scientific).
To set up an efficient quantitative protocol to analyse plasma MP proteins obtained from individual mice, the TMTzero (TMT0) labelling approach was first applied. MP from one non-infected (NI) and one PbA-infected mouse (experimental cerebral malaria – ECM) were used. The lists of identified proteins were considered as “qualitative datasets”.
The protocol established with the TMT0 was then translated to the TMTsixplex (TMT6) format. To increase the number of tested samples, two parallel experiments were performed comparing plasma MP proteins from non-infected mice (NI, n = 4), PbA-infected mice at day 3 post-infection (d3 pi, n = 4) and PbA-infected mice at d8 post-infection when all the signs of cerebral malaria are detected (ECM, n = 4).
Qualitative and quantitative proteomics results were then compared and evaluated in the context of CM pathogenesis through gene ontology and network analyses, and further verified by western blot (WB) for two selected proteins, carbonic anhydrase I (CA-I) and S100A8 on two different mouse strains.
A graphical representation of the experimental design used in the present study is depicted in Fig. 1.
Figure 1. Experimental design. Graphical summary of the experimental design applied in the present study.
PFP = platelet free plasma; MP = microparticle; SEM = scanning electron microscopy; NI = non-infected; d3 pi = day 3 post-infection; ECM = experimental cerebral malaria (d8 post-infection). The mouse image was obtained at Pixabay.com.
MP visualisation by scanning electron microscopy (SEM) and counting by flow cytometry
To demonstrate the efficiency of our purification method, plasma MP from a NI mouse were imaged by SEM (Fig. 2). Figure 2A and C clearly show that the predominant population of purified vesicles has a size range compatible to that of MP (0.1–1 μm); only a minimal contamination of larger elements, which might correspond to cell debris or platelets, was visualised.
Figure 2. Murine plasma MP visualized by Scanning Electron Microscopy.
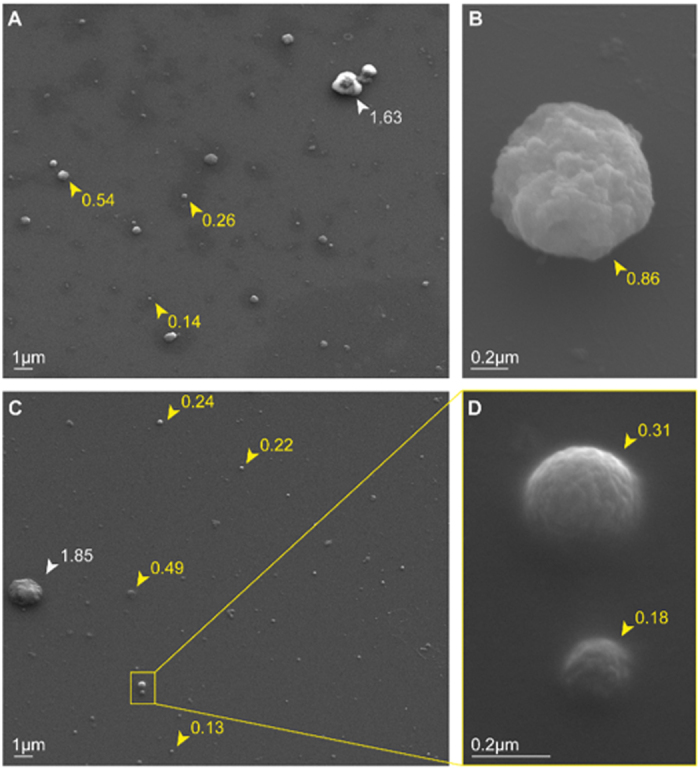
Plasma MP purified from a non-infected DBA/1 mouse have been imaged with a Zeiss Ultra FESEM. (A,C) Magnification x4000. The majority of the visualized vesicles have size corresponding to MP (0.1–1 μm - yellow arrowheads), while only one bigger element (white arrowheads) was visualised on each image probably corresponding to small aggregates of MP or microplatelets. (B,D) Visualisation of plasma MP at a higher magnification, ×23920 and 52160, respectively. Numbers beside arrowheads indicate the measured vesicle diameter expressed in μm.
When counted by flow cytometry in PFP samples, the number of Annexin V+ MP/μL in ECM PFP was 2.5 and 2.1 times higher than in NI and d3 pi PFP, respectively (Supplementary Figure S1).
Proteomics analyses
TMT0 qualitative experiment
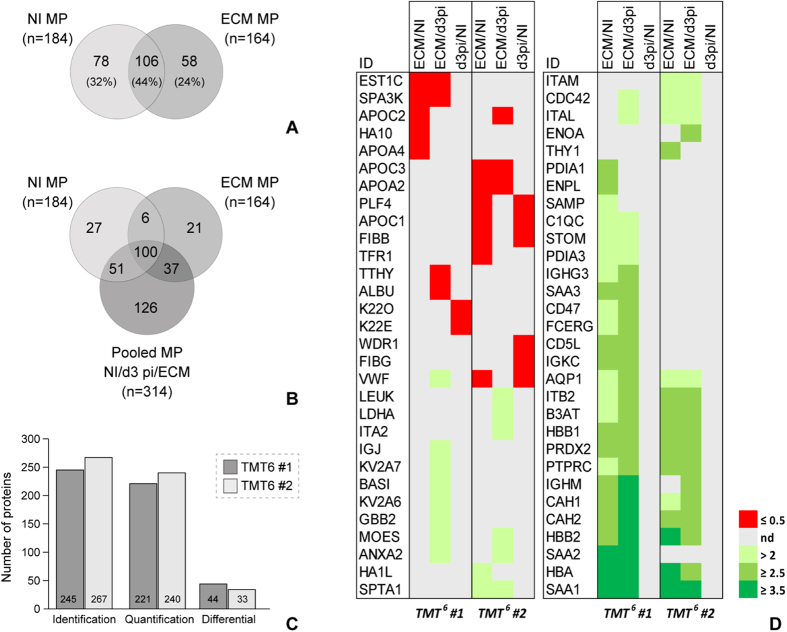
In the TMT0 experiment we identified 184 NI MP proteins and 164 ECM MP proteins (2 unique peptides, FDR ≤1%, Fig. 3A).
Figure 3. Identification and quantification results.
(A) Proteins identified in the plasma MP from a non-infected mouse (NI) and a PbA-infected mouse at the stage of experimental cerebral malaria (ECM) (TMT0 experiment). The two samples shared 44% of the identifications. Only proteins identified with minimum 2 peptides and FDR ≤1% have been considered. (B) Comparison of the proteins identified in the pooled NI – d3 pi – ECM samples (TMT6#1 and TMT6#2 experiments) and those identified in the individual NI and ECM samples (TMT0 experiment). Globally, 368 murine plasma microparticle proteins have been identified. (C) Quantitative results obtained from the two TMT6 experiments. Proteins differentially abundant in MP were defined as having a p-value < 0.001 and a ratio ≥2.1 or ≤0.5. (D) Heat map showing the level of expression of the proteins found to be significantly differentially abundant for the three computed ratios, i.e. ECM/d3 pi, ECM/NI and d3 pi/NI, in the two quantitative experiments. nd = non differential proteins.
The complete lists of identified proteins together with the identification details are reported in Supplementary MS Information-Identification. The percentage of TMT tagging – calculated as the proportion of peptides having a TMT0 at the N-terminus within the total identified peptides – was ≥97% in both samples, demonstrating the feasibility of the approach.
The comparison of the identified proteins between the two samples showed that 44% of the identifications were shared, 32% were only identified in the NI sample and 24% only in the ECM sample.
TMT6 quantitative experiments
In the two TMT6 quantitative experiments (TMT6-1 and TMT6-2), 314 proteins were overall identified with at least 2 unique peptides and FDR ≤1%. The good technical efficiency of the TMT6 experiments was evaluated through the percentage of labelled peptides (≥94%) and the distribution, across the 6 tags, of the mean relative intensity obtained for the spiked bovine beta-lactoglobulin (coefficient of variation – CV <20%).
Considering the two TMT0 and the two TMT6 experiments together, we globally identified 368 murine plasma MP proteins (2 unique peptides, FDR ≤1%) (Fig. 3B).
Quantitative information was obtained for 221 and 240 proteins in the TMT6-1 and TMT6-2 experiments, respectively and, of these, 67% were commonly quantified in the two experiments. Among all quantified proteins (TMT6-1 + TMT6-2), 60 were significantly different in abundance (Mann-Whitney U test, p ≤ 0.001) according to IsoQuant statistics and the calculated ratio threshold of 2.1 for increased proteins and 0.5 for decreased proteins (Fig. 3C,D and Supplementary MS information-Quantification). When the ECM/d3 pi ratio was considered, 38 proteins had significantly increased and 7 significantly decreased abundance. Thirty-one proteins were increased in ECM compared to NI MP, 26 of which had also higher abundance in ECM compared to d3 pi, and 12 proteins had an ECM/NI ratio ≤0.5. Finally, for the comparison d3 pi/NI, 8 proteins were significantly decreased and none were significantly increased, indicating that this is likely too early a time point in the infection to detect differences in MP cargo. This is in agreement with previous results on the number and phenotype of circulating MP in ECM18. The complete list of proteins whose abundance in MP was significantly affected during the infection is reported in Fig. 3D.
When Plasmodium berghei proteins were searched, two proteins - intra-erythrocytic P. berghei-induced structures protein 1 and merozoite surface protein-1 - were detected (2 unique peptides, FDR ≤1%) in the TMT0 ECM sample, after exclusion of all peptides also matching murine proteins (Supplementary MS information-Identification). In all other samples, no parasite protein could be identified with sufficient confidence.
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteome central.proteomexchange.org)22 via the PRIDE partner repository23 with the dataset identifier PXD003772.
Gene Ontology analysis
All murine plasma MP proteins identified in our proteomics experiments (n = 368) were subjected to Gene Ontology (GO) analysis. For each GO category (i.e., biological process – BP, molecular function – MF and cellular component – CC), the top-10 most represented GO terms are reported in Fig. 4.
Figure 4. Gene Ontology analysis.
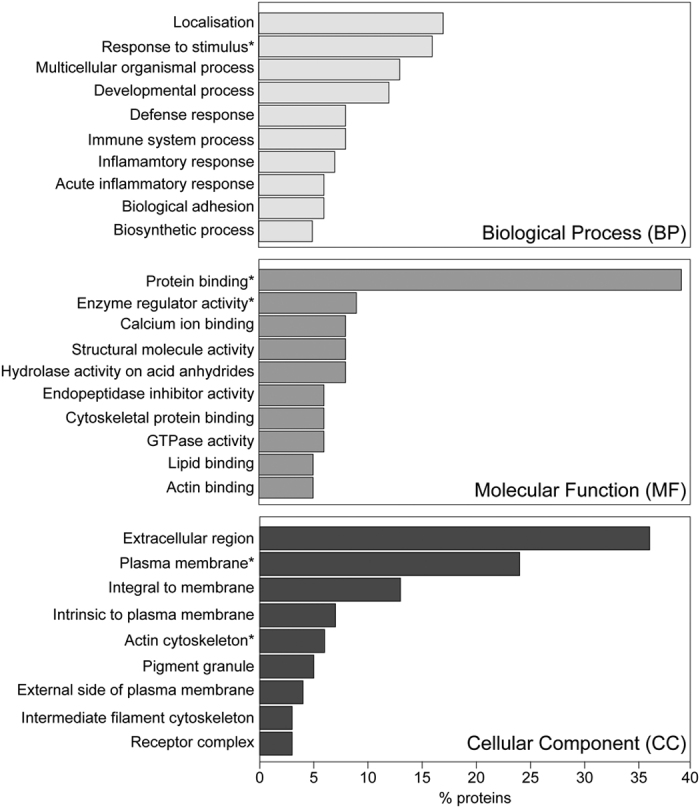
Top-10 GO terms represented across all 368 identified murine plasma MP proteins. (A) Biological Process GO terms; (B) Molecular Function GO terms; (C) Cellular Component GO terms. For each category only significantly represented GO terms have been considered (adjusted p-value < 0.0001) and the % of proteins belonging to each term is reported. *Indicates potentially relevant GO terms in the context of MP and ECM.
As shown, the majority of plasma MP proteins were involved in localisation processes and in response to stimulus, with the immune and inflammatory responses appearing among the top 10 BP, probably as a contribution of proteins whose expression was induced by the infection. Considering the CC terms, the identified proteins were highly significantly associated with the cytoskeleton, the plasma membrane and vesicles. Finally, the MF GO terms showed that the proteins identified from murine plasma MP have important active roles, being highly significantly associated with binding and regulatory activities.
Network and upstream regulator analyses
To highlight proteins associated with the ECM pathological state, a list comprising all those significantly overabundant in ECM MP compared either to d3 pi or to NI (n = 42) and all proteins uniquely identified in the TMT0 ECM MP sample (n = 21) was created (ECM-associated proteins, Table 1). Similarly, a list of NI-associated proteins (n = 43) comprised 16 proteins significantly overabundant in NI MP, compared either to d3 pi or to ECM MP, and 27 only identified in NI MP (Supplementary Table 1).
Table 1. ECM-associated proteins.
| # | AC | ID | Description | ECM/NI Ratio |
ECM/d3 pi Ratio |
||
|---|---|---|---|---|---|---|---|
| TMT6 #1 | TMT6 #2 | TMT6 #1 | TMT6 #2 | ||||
| 1 | P05366 | SAA1_MOUSE | Serum amyloid A-1 protein | 4.16 | 3.52 | 4.66 | 4.43 |
| 2 | P05367 | SAA2_MOUSE | Amyloid protein A | 3.81 | — | 4.71 | — |
| 3 | P01942 | HBA_MOUSE | Hemoglobin subunit alpha | 3.56 | 3.51 | 4.16 | 3.14 |
| 4 | P02089 | HBB2_MOUSE | Hemoglobin subunit beta-2 | 3.27 | 3.56 | 3.70 | 3.19 |
| 5 | P00920 | CAH2_MOUSE | Carbonic anhydrase 2 (CA-II) | 3.39 | 3.31 | 4.20 | 3.20 |
| 6 | P01872 | IGHM_MOUSE | Ig mu chain C region | 3.25 | — | 3.56 | 2.50 |
| 7 | Q61171 | PRDX2_MOUSE | Peroxiredoxin-2 (TSA) | 3.10 | 3.25 | 3.29 | 3.21 |
| 8 | P02088 | HBB1_MOUSE | Hemoglobin subunit beta-1 | 2.99 | 3.25 | 3.32 | 2.83 |
| 9 | P08113 | ENPL_MOUSE | Endoplasmin (GRP-94) (ERp99) | 3.02 | — | — | — |
| 10 | P04919 | B3AT_MOUSE | Band 3 anion transport protein, Isoform Kidney | 2.37 | 3.25 | 2.83 | 2.73 |
| 11 | Q9QWK4 | CD5L_MOUSE | CD5 antigen-like | 2.75 | — | 3.22 | — |
| 12 | P06800 | PTPRC_MOUSE | Receptor-type tyrosine-protein phosphatase C, Isoform 3 | 2.32 | 3.27 | 2.85 | 3.33 |
| 13 | P09103 | PDIA1_MOUSE | Protein disulfide-isomerase (PDI) (ER protein 59) | 2.75 | — | — | — |
| 14 | P01837 | IGKC_MOUSE | Ig kappa chain C region | 2.64 | — | 3.41 | — |
| 15 | P13634 | CAH1_MOUSE | Carbonic anhydrase 1 (CA-I) | 2.86 | 2.40 | 3.67 | 2.69 |
| 16 | P04918 | SAA3_MOUSE | Serum amyloid A-3 protein | 2.52 | — | 2.67 | — |
| 17 | P01831 | THY1_MOUSE | Thy-1 membrane glycoprotein | — | 2.51 | — | — |
| 18 | P11835 | ITB2_MOUSE | Integrin beta-2 | 2.17 | 2.70 | 2.46 | 2.70 |
| 19 | Q02013 | AQP1_MOUSE | Aquaporin-1 (AQP-1) (DER2) | 2.28 | 2.37 | 3.38 | 2.30 |
| 20 | P27773 | PDIA3_MOUSE | Protein disulfide-isomerase A3 (p58) (ERp57) (ERp60) | 2.28 | — | 2.42 | — |
| 21 | P01897 | HA1L_MOUSE | H-2 class I histocompatibility antigen, L-D alpha chain | — | 2.28 | — | — |
| 22 | P24063 | ITAL_MOUSE | Integrin alpha-L (LFA-1A) (Ly-15) | — | 2.18 | 2.36 | 2.23 |
| 23 | P05555 | ITAM_MOUSE | Integrin alpha-M, Isoform 2 | — | 2.14 | — | 2.28 |
| 24 | Q02105 | C1QC_MOUSE | Complement C1q subcomponent subunit C | 2.13 | — | 2.25 | — |
| 25 | P12246 | SAMP_MOUSE | Serum amyloid P-component (SAP) | 2.12 | — | — | — |
| 26 | P20491 | FCERG_MOUSE | High affinity Ig epsilon receptor subunit gamma | 2.12 | — | 3.15 | — |
| 27 | P60766 | CDC42_MOUSE | Cell division control protein 42 homolog | — | 2.12 | 2.19 | 2.17 |
| 28 | P08032 | SPTA1_MOUSE | Spectrin alpha chain, erythrocytic 1 | — | 2.10 | — | 2.06 |
| 29 | P03987 | IGHG3_MOUSE | Ig gamma-3 chain C region, Isoform 2 | 2.09 | — | 2.56 | — |
| 30 | Q61735 | CD47_MOUSE | Leukocyte surface antigen CD47 (IAP) | 2.08 | — | 2.76 | — |
| 31 | P54116 | STOM_MOUSE | Erythrocyte band 7 integral membrane protein | 2.08 | — | 2.30 | — |
| 32 | P17182 | ENOA_MOUSE | Alpha-enolase (NNE) | — | — | — | 2.51 |
| 33 | P07356 | ANXA2_MOUSE | Annexin A2 (PAP-IV) | — | — | 2.37 | 2.32 |
| 34 | P62880 | GBB2_MOUSE | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-2 | — | — | 2.28 | — |
| 35 | Q62469 | ITA2_MOUSE | Integrin alpha-2 (GPIa) | — | — | — | 2.27 |
| 36 | P01630 | KV2A6_MOUSE | Ig kappa chain V-II region 7S34.1 | — | — | 2.23 | — |
| 37 | P26041 | MOES_MOUSE | Moesin | — | — | 2.30 | 2.15 |
| 38 | P18572 | BASI_MOUSE | Basigin, Isoform 2 | — | — | 2.19 | — |
| 39 | P06151 | LDHA_MOUSE | L-lactate dehydrogenase A chain (LDH-A) (LDH-M) | — | — | — | 2.19 |
| 40 | P01631 | KV2A7_MOUSE | Ig kappa chain V-II region 26-10 | — | — | 2.13 | — |
| 41 | P15702 | LEUK_MOUSE | Leukosialin (Ly-48) | — | — | — | 2.12 |
| 42 | P01592 | IGJ_MOUSE | Immunoglobulin J chain | — | — | 2.07 | — |
| 43 | Q60963 | PAFA_MOUSE | Platelet-activating factor acetylhydrolase (PAF acetylhydrolase) | Only ID | |||
| 44 | P68254 | 1433T_MOUSE | 14-3-3 protein theta | Only ID | |||
| 45 | Q61646 | HPT_MOUSE | Haptoglobin beta | Only ID | |||
| 46 | P21981 | TGM2_MOUSE | Protein-glutamine gamma-glutamyltransferase 2 | Only ID | |||
| 47 | P17156 | HSP72_MOUSE | Heat shock-related 70 kDa protein 2 | Only ID | |||
| 48 | P84244 | H33_MOUSE | Histone H3.3 | Only ID | |||
| 49 | P31725 | S10A9_MOUSE | Protein S100-A9 (MRP-14) (p14) | Only ID | |||
| 51 | P27661 | H2AX_MOUSE | Histone H2AX (H2a/x) | Only ID | |||
| 52 | P01670 | KV3AI_MOUSE | Ig kappa chain V-III region PC 6684 | Only ID | |||
| 53 | P14426 | HA13_MOUSE | H-2 class I histocompatibility antigen, D-K alpha chain (H-2D(K) | Only ID | |||
| 54 | P0CW03 | LY6C2_MOUSE | Lymphocyte antigen 6C2 (Ly-6C2) | Only ID | |||
| 55 | Q8BFZ3 | ACTBL_MOUSE | Beta-actin-like protein 2 | Only ID | |||
| 56 | P01749 | HVM05_MOUSE | Ig heavy chain V region 3 | Only ID | |||
| 57 | P01646 | KV5AD_MOUSE | Ig kappa chain V-V region HP 123E6 | Only ID | |||
| 58 | P27005 | S10A8_MOUSE | Protein S100-A8 (MRP-8) | Only ID | |||
| 59 | P62806 | H4_MOUSE | Histone H4 | Only ID | |||
| 60 | P11672 | NGAL_MOUSE | Neutrophil gelatinase-associated lipocalin (NGAL) | Only ID | |||
| 61 | Q8CGP1 | H2B1K_MOUSE | Histone H2B type 1-K | Only ID | |||
| 62 | P50446 | K2C6A_MOUSE | Keratin, type II cytoskeletal 6 A (CK-6A) | Only ID | |||
| 63 | O08677 | KNG1_MOUSE | Kininogen-1 light chain, Isoform 3 | Only ID | |||
List of proteins significantly over-expressed in ECM either compared to d3 pi or to NI (TMT6 experiments #1 and #2) and proteins uniquely identified (Only ID) in the ECM sample (TMT0 experiment). —: non differential expression.
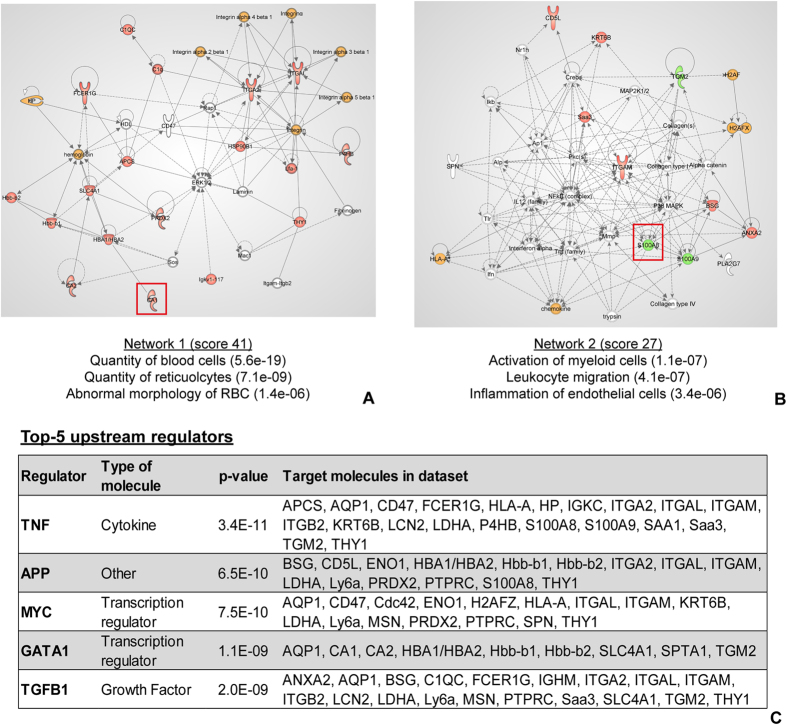
The 63 ECM-associated proteins were analysed with IPA Ingenuity to highlight significantly represented networks and predicted upstream regulators. The top two molecular networks are reported in Fig. 5A and B and for each the most interesting significant molecular functions were selected. The network with the highest score involved 18 molecules of the experimental dataset and was associated (p < 0.0001) with molecular functions affecting the quantity of blood cells and of reticulocytes, and RBC morphology. The second network, involving 13 molecules of the dataset, was significantly associated (p < 0.0001) with the activation of myeloid cells, leukocyte migration and inflammation of endothelial cells.
Figure 5. Network and upstream analyses of ECM-associated proteins.
(A,B) Most relevant networks showing the connectivity between MP proteins experimentally identified as ECM-associated (n = 63). Red: proteins identified as significantly over-expressed in ECM; Green: proteins uniquely identified in the ECM sample; Orange: multimeric proteins for which one or more chains have been identified as over-expressed in ECM. The most important biological functions, associated with each network, in the context of ECM, are reported at the bottom of the network. Protein targets selected for verification, CA-I and S100A8, are highlighted by a red square. (C) Top-5 molecules that are highly significantly likely to regulate the proteins experimentally found to be ECM-associated. For each regulator the list of target proteins in the experimental dataset is given. p-value: Fisher’s exact test, indication of the overlap between the proteins in the dataset and the genes known to be affected by the regulator.
The upstream regulator analysis predicted, with high significance, that tumour necrosis factor (TNF), amyloid precursor proteins, MYC, erythroid transcription factor (GATA 1) and transforming growth factor beta (TGFβ) were highly likely to regulate the expression of the proteins found to be associated with ECM MP. The detailed results of the upstream regulator analysis are reported in Fig. 5C.
The same analysis was performed on the list of NI-associated proteins (n = 43) and the top two networks were significantly associated with lipid metabolism and with haemostasis (Supplementary Figure S2).
CA-I and S100A8 verification by western blot
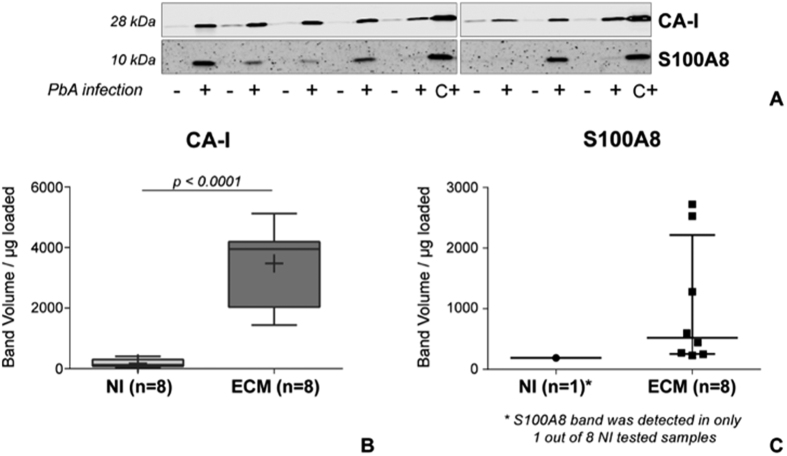
To verify our results, two proteins – carbonic anhydrase 1 (CA-I) and S100A8 - were selected, based on their biological functions and proteomics results, for further evaluation on a larger number of newly collected samples from DBA/1 mice (n = 16). CA-I (identified with 6 peptide spectral matches - PSM, 4 unique peptides and 17.31% sequence coverage) was quantified in both TMT6-1 and -2 experiments as overabundant in ECM MP compared to both NI (ECM/NI = 2.9 and 2.4, respectively) and d3 pi MP (ECM/d3 pi = 3.7 and 2.7, respectively). S100A8 was uniquely identified in ECM MP (3 PSM, 3 unique peptides and 54.55% sequence coverage).
As shown in Fig. 6, CA-I abundance was confirmed to be significantly higher in ECM MP (n = 8) compared to NI samples (n = 8) (t-test, p < 0.0001). A 10 kDa band corresponding to S100A8 protein was detected in all 8 ECM samples but in only 1 out of 8 NI MP, confirming proteomics results where it was only identified in PbA-infected MP. Unfortunately, to the best of our knowledge, a housekeeping protein for plasma MP has not been described yet. When the abundance of γ-actin was assessed on the same samples, a more intense signal was detected in NI samples indicating that actin is not an adequate loading control due to the high variability in protein composition of plasma MP, but also showing that our experiments were not biased towards a higher amount of proteins loaded in ECM samples (Supplementary Figure S3).
Figure 6. Western blot results for CA-I and S100A8 detection in murine MP.
(A) Detection of CA-I and S100A8 in MP samples (2.5 μg/lane left blot image, 1.8 μg/lane right blot image) from non-infected (−, NI, n = 8) and PbA-infected (+, ECM, n = 8) DBA/1 mice. A positive control (C+) consisting of murine spleen extract was included in each experiment. (B) Quantification of CA-I expression in murine plasma MP, showing its significantly higher expression in ECM samples (t-test). In each boxplot mean is reported as +. (C) S100A8 was detectable in only 1 out of 8 NI samples, while it was expressed in all ECM samples. The quantification of the detected bands is reported in the graph, where the horizontal line represents the median band volume and the error bars the interquartile range.
S100A8 and CA-I were also assessed in the platelet-free plasma (PFP) and MP-free plasma (MFP) of 10 out of the 16 samples (Supplementary Figure S4). CA-I was expressed in both PFP and MFP but no difference was observed between the 2 types of samples (PFP vs. MFP, Wilcoxon matched-pairs signed rank test) nor between groups (NI vs. ECM, Mann-Whitney U test). Interestingly, S100A8 was detected in only 2 out of 5 ECM PFP (and in the corresponding MFP) and in none of the NI samples.
To exclude the possibility of an association between the obtained results and the DBA/1 genotype, CA-I and S100A8 were assessed by WB in MP and PFP samples from C57BL/6 mice (n = 4 NI, n = 4 PbA-infected - ECM) (Supplementary Figure S5). CA-I was significantly more abundant in both MP and PFP samples from ECM mice compared to NI, while S100A8 was only detected in ECM MP, confirming the results obtained by both proteomics and WB in the DBA/1 model. S100A8 was not detected in the PFP of C57BL/6 mice, both NI and ECM.
Finally, the two proteins were also assessed in MP isolated from PFP obtained from children suffering either from CM or from asymptomatic malaria (AM), as well as from healthy community controls (CC) (n = 7 in each group) (Supplementary Figure S6). S100A8 was significantly (one-way ANOVA, p = 0.006) more abundant in MP from CM patients compared to both AM and CC (Dunnet’s multiple comparison post-test, p = 0.0069 and p = 0.0134, respectively). CA-I was not significantly different between the three groups, however its abundance in CM MP was almost double than in AM MP (mean CM/mean AM = 1.9).
Discussion
Host-derived plasma microparticles (MP) are well recognised as being involved in the pathogenesis of both human and experimental CM17,18,20,21, however the contribution of their protein cargo to the pathological mechanisms leading to this syndrome has not been deciphered yet. MP are known to contain proteins derived from the cell of origin, and this cargo doesn’t seem to be the consequence of a casual uptake from the parent cell’s cytoplasm, but rather the result of specific packaging mechanisms9,24 making these vesicles important players in intercellular communication11. To try to achieve new insights into the pathogenesis of CM and to better understand the role of MP in this syndrome, we have established and validated a qualitative and quantitative characterisation of the protein content of plasma MP released during ECM. To our knowledge, this is the first time that this strategy is applied to ECM and that the protein cargo of MP obtained from individual mice is successfully investigated by TMT quantitative proteomics. Only few other studies have used proteomics to investigate extracellular vesicles in malaria models, including P. yoeli-infected mice or in vitro cultured pRBC25,26.
Through our proteomics investigations we overall identified 368 plasma MP proteins, 290 of which were also quantified. These results are comparable with, or higher than, other proteomics studies on human circulating MP27,28 and murine plasma exosomes25. The list of identified proteins included some of the most abundant plasma proteins, such as albumin, which however were not significantly different between samples. Although a minimal contamination from plasma proteins can never be excluded, some of these proteins have already been reported to be present within MP even after 10 washes of the vesicle pellet or reported as part of the core proteome of human plasma MP27,29. When the immune-depletion of albumin and immunoglobulins was tested on our samples, the total number of identified proteins increased by 14% (data not shown); despite the increase in the number of identified proteins, such a sample preparation wouldn’t have been compatible with the downstream quantitative analysis of MP proteins derived from individual animals due to the limited amount of proteins available. For these reasons, we instead reduced the contamination of plasma proteins with two washes of the MP pellet, and we increased the number of identified peptides by using a long gradient at the LC-MS/MS level.
To comprehensively evaluate the MP protein cargo, we performed a gene ontology analysis and showed that these proteins reflect both MP mechanism of formation - with a significant representation of cytoskeletal and structural proteins - and their active biological functions, including binding, enzymatic and regulatory activities. This result further sustains that circulating MP released by the host in response to a certain stimulus, transport biologically active molecules that are likely to mediate physiological and pathological processes, as already proposed24,30,31,32. Interestingly, a number of proteins already proposed to be associated with parasite virulence and intra-erythrocytic life stage, including basigin, peroxiredoxin 2 and aquaporin 933,34,35, or associated to the phenomenon of adhesion to the brain microvasculature (integrins) have been identified within ECM MP.
A deeper analysis revealed that these ECM-associated proteins were interconnected in two distinct networks reflecting the pathological state of the sample from which they are derived. The first network involved molecules and pathways significantly associated with RBC number and morphology, indicating that RBC-MP represent an important proportion of circulating vesicles during ECM, as already reported18, and that their protein cargo reflects the known alteration of RBC occurring during the infection36. However, whether these proteins are specifically packaged into MP to propagate the infection or are just the result of RBC rupture following the maturation of the parasite is still to be determined. These proteomics results were further verified through the validation of the increased abundance in ECM MP of carbonic anhydrase I (CA-I), a protein participating in this network. CA-I, mainly expressed in RBC and in the gastro-intestinal tract37, belongs to a group of proteins involved in the maintenance of the acid-base homeostasis of blood and tissues through the reversible catalysis of carbon dioxide and bicarbonate conversion38. In severe malaria, the acid-base balance is often affected, as also indicated by the well-reported alteration of the lactate blood levels39 and, more recently, by the association of ECM with brain hypoxia and acidosis40. Interestingly, CA-I transcript was found to be up-regulated in the plasma of children experiencing severe malaria compared to mild malaria episodes occurring later in the same subjects41, further suggesting an interesting role for this protein in CM and raising the possibility that its increased plasma levels are due to MP. In our proteomics analyses we identified and quantified both CA-I and CA-II as significantly over-expressed in ECM MP compared to both NI and d3 pi MP. The muscle-derived CA isoenzyme, CA-III, had been previously identified by affinity proteomics as increased in CM children and as a discriminator between CM and both severe malaria anaemia (SMA) and uncomplicated malaria (UM)42.
In our study, CA-I expression was significantly increased in ECM MP compared to NI MP from DBA/1 mice when assessed by WB, however its plasma levels were not different between the two conditions. This might suggest that CA-I could be preferentially packaged in MP - probably derived from RBC - to be transported to other cells to counteract the cellular and tissue acidosis and hypoxia occurring during ECM. These results were confirmed in MP obtained from a different mouse strain (C57BL/6), showing that the increased CA-I expression in ECM MP is not genotype dependent. However, in contrast with what was obtained in DBA/1 mice, when assessed in plasma samples from C57BL/6 mice, CA-I concentration was higher in ECM compared to NI. More in-depth investigations are needed to better understand the contribution of the MP proteome within the plasma proteome.
The second network that was significantly represented among ECM-associated MP proteins comprised molecules implicated in myeloid cell migration, leukocyte activation and inflammatory response in endothelial cells. These functions are known to be important in the pathogenesis of both human and murine CM4,43,44 and, in recent years, particular attention has been paid to the role of the endothelium in CM pathogenesis45,46,47,48,49. The sequestration of different cell types at the brain microvasculature level - with predominant accumulation of pRBC in human CM and of leukocytes in ECM - is a typical trait of the syndrome44,47. This adhesion results in the activation of the endothelium and contributes to the endothelial damage associated with CM46,50. Interestingly, the association of markers of endothelial activation and of platelet adhesion with CM was also shown in patients’ plasma investigated by affinity proteomics42. The identification of these proteins further substantiates the active role of MP in ECM: by carrying important molecules between cells, MP could represent primary mediators in cell-to-cell interaction and activation occurring in CM.
Our hypothesis that MP contain proteins actively involved in CM pathogenesis, and in endothelial damage as pointed out by the network analysis, was further confirmed by the upstream regulation analysis, through which two of the most studied molecules in CM pathogenesis, TNF and TGFβ1, were predicted to regulate the MP proteins experimentally detected as ECM-associated. TNF has been reported to be involved, at different levels, in both human and murine CM, including in the interaction between pRBC and leukocytes with the endothelium, sequestration and the release of MP from endothelial cells20,21,51,52. Despite the role of TGFβ still being controversial, it has been shown that, on the focal brain obstruction, TGFβ released by adherent and activated platelets contributes to the cerebral endothelium damage53.
To further verify our proteomics and prediction analysis results, we assessed the abundance of one of the proteins involved in this second network - S100A8 - that was only identified in ECM-derived MP.
S100A8, also known as myeloid-related protein 8 (MRP-8), is a calcium-binding protein constitutively expressed mainly by neutrophils and monocytes54. In human cells and body fluids, it often exists as a heterodimer or tetramer with S100A9 (or MRP14)55, which we also identified as uniquely expressed in ECM MP. S100A8/A9 expression can be induced in monocytes, endothelial cells, keratinocytes or epithelial cells by mediators such as LPS and TNF56,57 and it is often increased in acute and chronic inflammatory conditions56, including rheumatoid arthritis, cancers and neurological disorders58,59 where they seem to play cytokine-like pro-inflammatory effects55.
Our verification data confirmed that S100A8 was detected within ECM MP but almost absent in NI MP, in both DBA/1 and C57BL/6 mice. Additionally, its expression was almost not-detectable in plasma samples from both mouse strains, suggesting that MP might be a preferential source of plasma S100A8.
In malaria, elevated serum or plasma levels of S100A8/A9 have been reported in two different studies. Bordmann et al. showed that in P. falciparum malaria-infected children, S100A8/A9 serum concentration correlated with both parasite load and temperature, even though these criteria are no longer sufficient to assess the disease severity60. More recently, in PbA-infected BALB/cA mice, an accumulation of S100A8+ and S100A9+ macrophages in the spleen was shown, as was an increased plasma concentration of the two proteins61. In our study, no difference was observed in S100A8 concentration in the spleen between NI and ECM DBA/1 mice (WB, data not shown), however it should be noted that different mouse strains are known to react differently to PbA-infection.
Based on their described biological properties, S100A8/A9 might be interesting players in CM. Indeed, S100A8/A9 are released by circulating neutrophils and monocytes upon contact with the inflamed endothelium57 in which they induce the transcription of genes encoding for pro-inflammatory chemokines and adhesion molecules such as ICAM-162. Additionally, S100A8/A9 have been implicated in leukocyte extravasation by increasing the binding capacity of leukocytes to endothelial ICAM-162.
The mechanism of secretion of these proteins in the extracellular environment is still not completely characterised. It has been proposed that their secretion could occur through a tubulin and calcium signalling dependent mechanism57,63, thus the identification of S100A8 in MP might support these pieces of evidence and represent a new mechanism contributing to S100A8/A9 plasma level.
In our study we focused on the characterisation of host proteins and the alteration of their abundance in MP during ECM infection; however, as reported in other works on the in vitro study of pRBC-MP26 or on the characterisation of exosomes from P. yoelii-infected mice25, parasite proteins can also be found in extracellular vesicles. Here, we identified only 2 parasite proteins (intra-erythrocytic P. berghei-induced structures protein 1 and merozoite surface protein-1) in ECM MP analysed using the qualitative approach, potentially as a result of a release of MP from pRBC. This number is lower than what was reported by others, however it should be considered that in order to avoid pooling of samples from multiple mice, our proteomics experiments were performed on a very small amount of protein (approximately 5 μg per sample). Assuming that, similar to exosomes, circulating MP can contain parasite proteins, these would be a minimal proportion of the total protein cargo requiring enrichment and fractionation steps for their detection. Such an investigation should be done in the future to decipher the in vivo interaction between the host and the parasite and the role of MP in this. Interestingly, one of the two PbA proteins that we identified within MP was already reported in P. falciparum-infected red blood cell MP produced in vitro26.
Our results indicate that the protein content of host-derived plasma MP is severely altered during ECM, compared to the early infection, and that some of the proteins appear to be selectively packaged into these vesicles during the infection. Nonetheless, complementary approaches, such as immune-staining of vesicles released under different infection conditions, should be used to confirm this specific packaging. To further evaluate the implication of MP-associated proteins, and in particular of CA-I and S100A8, in both human and murine CM, additional investigations are needed to establish their exact cellular source and role in disease pathogenesis, using in vitro and in vivo models. Despite the fact that the two proteins seem to be associated with ECM, as shown by our proteomics results on samples taken at day 3 post-infection, additional investigations should be done using non-CM models to prove the specificity of this association with the severe syndrome. Interestingly, when assessed in MP from human plasma samples from Ugandan children in a pilot study, the two proteins were increased in abundance in MP from children with CM compared to asymptomatic cases and this increase was statistically significant for S100A8. Asymptomatic malaria has recently been pointed out as a debilitating infection that should be considered seriously and treated64. These observations suggest that the two proteins might not only be associated to the severe syndrome in mice, but also in humans.
By describing for the first time the protein cargo of murine MP in general and more specifically those released during ECM, and by highlighting new potential molecular players, our results indicate that the cargo of plasma MP is affected during the severe infection and should be considered in the understanding of CM pathogenesis. Additionally, the differences in protein expression between MP and the corresponding PFP indicate that the qualitative and quantitative proteomics investigation of extracellular vesicles has great potential as a complementary strategy to the more common plasma proteomics in biomarker discovery research, since MP are likely to contain molecules specific to the infection or its stage.
The results here presented pave the way to future investigations in multiple directions including the further translation to human clinical samples to try and understand the contribution of plasma MP proteome to malaria severity and to identify new prognostic markers able to predict the short and long term outcome in CM patients.
Experimental Procedures
Mice and infection
DBA/1 and C57BL/6 CM-susceptible mice were infected with Plasmodium berghei ANKA (PbA) strain and developed a neurological pathology 6–8 days after infection17,65. Mice were infected and handled as reported elsewhere18. Blood was collected retro-orbitally under isoflurane-induced general anaesthesia either at day 3 post-infection (d3 pi) or at the stage of ECM (end-point) in 3.2% sodium citrate. Blood from NI mice was obtained following the same procedure. A total of n = 14 DBA/1 mice were used to perform proteomics experiments; while n = 16 DBA/1 and n = 8 C57BL/6 mice were used to perform verification experiments by WB. The detailed demographic description is reported in Supplementary Figure S1.
Ethical approval
All procedures performed in the present study have been approved by the Animal Ethics Committee of the University of Sydney (Projects n: K00/10-2010/3/5317 and K20/6-2011/3/5569) and adhered to the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Paediatric clinical samples
Platelet free plasma samples was obtained from children enrolled between 2008 and 2012 in Uganda in the context of a study whose details, inclusion and exclusion criteria are reported elsewhere66. Written informed consent was obtained from parents or guardians of study participants. The Institutional Review Boards for human studies at Makerere University School of Medicine (Study # 2008-033) and the University of Minnesota granted ethical approval for the study (Study # 0802M27022); all experiments were performed in accordance with relevant guidelines and regulations. The demographic description of the samples used in the present study is reported in Supplementary Figure S6.
MP preparation, counting by flow cytometry and visualisation by Scanning Electron Microscopy (SEM)
MP were prepared as previously reported67. Briefly, whole blood was centrifuged at 1,500 g for 15 min at room temperature to obtain platelet poor plasma (PPP). PPP was then centrifuged twice at 18,000 g for 3 min to remove possible platelet contaminations and/or cell debris and platelet free plasma (PFP) was obtained. MP were finally pelleted at 18,000 g for 45 min at 15 °C. To remove possible plasma protein contaminations (not specifically bound to MP), MP pellet was washed twice with PBS/3.2% sodium citrate and centrifuged at 18,000 g for 45 min after each washing step.
The number of MP in the PFP samples used for proteomics was counted by flow cytometry using Annexin V binding on a Beckman Coulter GalliosTM flow cytometer as previously reported13.
To confirm that the isolated population was consistent with plasma MP, vesicles purified from a NI mouse were also imaged by scanning electron microscopy. Samples were prepared as reported by Latham et al. and imaged with the Zeiss Ultra FESEM68. Imaging was carried out at the Australian Centre for Microscopy & Microanalysis (ACMM), The University of Sydney, Australia.
TMT0 experiment
A pilot experiment using the TMT0 approach was performed on MP proteins from one NI and one ECM mouse. For each sample, proteins were extracted from pelleted MP using repeated freeze/thaw cycles69, re-suspended in 0.1% Rapigest SF (Waters Corporation) in 0.1 M triethylammonium bicarbonate buffer (TEAB) pH 8.0, alkylated with 50 mM TCEP, reduced with 400 mM iodoacetamide and digested with 0.2 μg/μL trypsin overnight at 37 °C (Promega Corporation). Digested samples were then labelled with the TMT0 reagent (Thermo Fisher Scientific) following manufacturer’s instructions, except for the amount of tag used to label the samples. Due to the low amount of proteins in MP samples, one TMT vial (0.8 mg) was used to label two samples. After TMT labelling, samples were cleaned with C18 ultra-micro spin column (Harvard apparatus) and dried under vacuum prior to LC-MS/MS analyses.
TMT6 experiment
MP obtained from 400 μL of PFP from n = 4 NI, n = 4 d3 pi and n = 4 ECM mice were used to perform two parallel TMT6 experiments (TMT6-1 and TMT6-2). Protein extraction, peptide sample preparation and labelling were performed as reported for the TMT0. Additionally, each sample was spiked with 0.05 μg of bovine beta-lactoglobulin before protein reduction as an internal control.
Samples analysed in the first TMT6 experiment (TMT6-1) were labelled as follows: NI with the tags 126 and 127; d3 pi with the tags 128 and 129, and ECM with the tags 130 and 131. In the TMT6-2 experiment NI samples were labelled with the tags 128 and 129, d3 pi samples with the tags 130 and 131, and ECM samples with the tags 126 and 127.
After labelling, samples were combined, cleaned with C18 ultra-micro spin column and dried under vacuum prior to LC-MS/MS analyses.
Tandem mass spectrometry analyses, protein identification and quantification
Tandem mass spectrometry analyses were performed on a QExactive Orbitrap Plus (Thermo Electron) equipped with an Ultimate 3000 HPLC and autosampler system (Dionex). Peptide samples were concentrated and desalted onto a micro C18 pre-column (300 μm × 5 mm, Dionex) that was switched, after 4 min wash, into line with a fritless nano column (75 μm × 15 cm) containing C18 media (1.9 μm, 120 Å, Dr Maisch) manufactured according to Gatlin et al.70. For the TMT6 samples, the analytical peptide separation was run for 260 min at a flow rate of 200 nL/min; while for the TMT0 samples, a gradient of 140 min was applied. The detailed description of the MS/MS settings is reported as Supplementary information. Mass spectrometry analyses were carried out at the Bioanalytical Mass Spectrometry Facility, University of New South Wales, Australia.
MS/MS data were analysed using EasyProt platform v2.371. Raw data were converted into.mgf files and peak lists generated with MS convert (ProteoWizard 3.0.7529, http://proteowizard.sourceforge.net/tools.shtml). Protein identifications were obtained by searching peptide spectral matches against the SwissProt/UniProt database (version October 2014 - 546,790 entries) using the following settings: i) Mus musculus for the taxonomy; ii) trypsin as the digestion enzyme with only one missed-cleavage allowed; iii) carbamidomethylation of cysteines as fixed modification and oxidation of methionines as variable modification; iv) only peptides with at least 6 residues were considered for protein identification and v) the precursor ion tolerance was set to 10 ppm. Additionally, the TMT0 or the TMT6 modifications on lysines and peptide N-termini were added as fixed modifications in the respective experiments. Only proteins identified with at least 2 unique peptides and false discovery rate (FDR) ≤1%, calculated as reported in71, were considered as true identification matches.
For the quantitative experiment, protein relative quantification was obtained using IsoQuant embedded in the EasyProt platform. All peptides corresponding to one unique entry in the job, having FDR ≤1% and an intensity signal in each of the 6 TMT tags were considered for quantification.
For each TMT6 experiments, protein relative quantification was obtained computing the protein ratios ECM/d3 pi, ECM/NI and d3 pi/NI after isotopic purity correction according to the algorithm given by the manufacturer. To reduce biases due to potential differences in protein amount among the 6 TMT channels, the reporter ion intensities were normalized71 prior to ratio calculation and the distribution of the relative intensities of all peptides considered for protein quantification was checked for normality (Shapiro-Wilk normality test). The ratio threshold for considering a protein differentially abundant between the compared conditions was calculated72 and the significance level was set at 0.01 (Mann-Whitney U test). The technical performance of the two TMT6 experiments was evaluated through the rate of labelled peptides and the normalised relative intensity distribution of bovine beta-lactoglobulin across the 6 tags.
For all TMT0 and TMT6 experiments a second search against the UniProtKB/TrEMBL database (version December 2012 - 4,870,157 entries), choosing the Plasmodium berghei taxonomy (ID 5821) was done to search for parasite proteins.
Gene ontology, network and upstream regulator analyses
To highlight cellular mechanisms, biological processes and molecular functions associated with murine plasma MP proteins, the list of all identified proteins (TMT0 and TMT6) was submitted to gene ontology (GO) analysis using bioCompendium (http://biocompendium.embl.de/). Only GO terms significantly represented after correction for multiple comparisons were considered (adjusted p-value < 0.0001).
To evaluate the association of the identified MP proteins with CM pathology, two protein sub-datasets were created. A first dataset, called ECM-associated proteins, comprised all proteins found significantly over-abundant in ECM MP compared either to d3 pi or to NI MP in at least one of the two TMT6 quantitative experiments, and those exclusively identified in the ECM MP sample (TMT0 qualitative experiment). The second dataset, NI-associated proteins, included all proteins significantly increased in NI conditions in the TMT6 experiments and those only identified in the NI MP in the TMT0 experiment. Both datasets were analysed with Ingenuity® Pathway Analysis - IPA (Ingenuity® System, http://www.ingenuity.com). A network analysis was first performed to evaluate the molecular connectivity and interaction between the proteins in the datasets. This analysis is based on the assumption that highly connected molecules will have a higher influence on the network and that highly interconnected networks will have greater biological significance. Since a network is seen as a set of pathways, the different molecular functions determined to be significantly associated with the network can be highlighted73,74.
To evaluate ECM- or NI-associated proteins in a broader molecular and biological context, an upstream regulator analysis was also performed listing all genes, RNAs and proteins predicted to regulate the molecules in the experimental dataset.
Western Blot
Murine samples
The expression of two selected candidates, S100A8 and carbonic anhydrase 1 (CA-I), was assessed by WB in MP, PFP and MP-free plasma (MFP) samples.
MP protein concentration was determined after protein extraction using the Qubit protein assay (Life Technologies). For each MP sample, 1 to 2.5 μg of proteins were separated on a polyacrylamide gel and transferred onto a nitrocellulose membrane. For PFP and MFP 2.5 μL of sample were used.
CA-I was detected using a rabbit monoclonal anti-mouse CA-I antibody (0.63 μg/mL, Abcam) and a goat anti-rabbit IgG DyLightTM 800 conjugated secondary antibody (0.15 μg/mL). S100A8 was detected using a goat polyclonal anti-mouse S100A8 antibody (0.4 μg/mL, R&D Systems) and an anti-goat IgG IRDye® 800 conjugated, diluted 1/1000. Monoclonal mAb-2A3 against γ-actin, a kind gift from Christine Chaponnier (University of Geneva), was used at a 0.2 μg/mL and detected with an anti-mouse IgG DyLight 680 conjugated secondary antibody (0.15 μg/mL). The fluorescent signal was detected on an Odyssey Infrared Imaging System (LI-COR Biosciences) and images analysed with ImageQuant TL software (GE Healthcare Life Sciences). Protein expression in MP samples was determined as the measured band volume normalised to the amount of loaded protein.
To establish the appropriate number of mice to be used in WB experiments, a sample size calculation was done based on the variability of the normalised relative intensities of all the peptides used to quantify CA-I in both TMT6 experiments, after data logarithmic transformation to obtain a normal distribution. Considering an average standard deviation of 0.3 and 0.7 for ECM and NI samples, respectively, a sample size of n = 8 per group was estimated to be sufficient to detect a significant difference at 0.05 level and a fold-change of 2 with a power of 0.895 (two-sample t test for mean difference with unequal variances).
Human samples
MP were prepared from PFP as previously described13 and proteins extracted and measured as reported for the murine samples. For each sample, 2 to 3 μg of proteins were separated on 15% polyacrylamide gels. As these samples were obtained from young children the volume of plasma was minimal (160 μL), therefore due to the low amount of proteins obtained from CC MP, pools of two samples age and sex matched were analysed. S100A8 was detected using a rabbit polyclonal primary antibody (2.0 μg/mL, Abcam) and a goat anti-rabbit IgG DyLightTM 800 conjugated secondary antibody (0.15 μg/mL). CA-I was detected with the same antibody used for murine samples.
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, Inc.). Parametric or non-parametric statistical tests were applied according to data distribution. Comparisons between two unpaired groups were performed with the t-test or the Mann-Whitney U test, while comparisons between three unpaired groups were performed with the one-way ANOVA test followed by the Dunnet’s multiple comparison test. Paired groups were compared using the Wilcoxon matched-pairs signed rank test. All tests were two-tailed and the p-value significance was set at <0.05.
Additional Information
How to cite this article: Tiberti, N. et al. Exploring experimental cerebral malaria pathogenesis through the characterisation of host-derived plasma microparticle protein content. Sci. Rep. 6, 37871; doi: 10.1038/srep37871 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors thank Dr Ling Zhong and Prof Mark Raftery (Bioanalytical Mass Spectrometry Facility, University of New South Wales, Australia) for technical assistance for LC-MS/MS analyses; Alexandre Hainard and Trinidad Martin Campos (Proteomics Core Facility, University of Geneva, Switzerland) for EasyProt data analysis support and Dr Matthew Padula (School of Life Sciences, University of Technology Sydney) for critical reading of the manuscript. The authors acknowledge the Australian Microscopy & Microanalysis Research Facility at the University of Sydney. The authors also thank the PRIDE team for their support in MS data deposition to ProteomeXchange Consortium (dataset identifier PDX003307). This work was supported by the Australian National Health and Medical Research Council (#1028241 for VC and GG) and the National Institute of Health (1R01NS079873-01 for VC, R01 NS055349 for CJ and RO). N Tiberti was supported by the Swiss National Science Foundation (SNSF, grant n. P2GEP3_151822). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions N.T. and V.C. designed the study. N.T., A.J. and S.L.L. performed the experiments. N.T. performed data analyses. S.B. contributed to statistical analyses. C.J. and R.O. recruited the patients and provided clinical samples. G.E.G. and A.C. provided parts of reagents or analytical tools. N.T. and V.C. wrote the manuscript. All authors revised the manuscript.
References
- WHO. World malaria report (2014).
- Idro R., Marsh K., John C. C. & Newton C. R. Cerebral malaria: mechanisms of brain injury and strategies for improved neurocognitive outcome. Pediatr Res 68, 267–274, doi: 10.1203/00006450-201011001-00524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbeck G. L. et al. Blantyre Malaria Project Epilepsy Study (BMPES) of neurological outcomes in retinopathy-positive paediatric cerebral malaria survivors: a prospective cohort study. Lancet Neurol 9, 1173–1181, doi: 10.1016/S1474-4422(10)70270-2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm J. & Craig A. G. Pathogenesis of cerebral malaria–inflammation and cytoadherence. Front Cell Infect Microbiol 4, 100, doi: 10.3389/fcimb.2014.00100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J. H. et al. Microparticles as regulators of inflammation: novel players of cellular crosstalk in the rheumatic diseases. Arthritis Rheum 52, 3337–3348, doi: 10.1002/art.21350 (2005). [DOI] [PubMed] [Google Scholar]
- Burger D. et al. Microparticles: biomarkers and beyond. Clin Sci (Lond) 124, 423–441, doi: 10.1042/CS20120309 (2013). [DOI] [PubMed] [Google Scholar]
- Loyer X., Vion A. C., Tedgui A. & Boulanger C. M. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ Res 114, 345–353, doi: 10.1161/CIRCRESAHA.113.300858 (2014). [DOI] [PubMed] [Google Scholar]
- Gong J., Jaiswal R., Dalla P., Luk F. & Bebawy M. Microparticles in cancer: A review of recent developments and the potential for clinical application. Semin Cell Dev Biol 40, 35–40, doi: 10.1016/j.semcdb.2015.03.009 (2015). [DOI] [PubMed] [Google Scholar]
- Schindler S. M., Little J. P. & Klegeris A. Microparticles: a new perspective in central nervous system disorders. Biomed Res Int 2014, 756327, doi: 10.1155/2014/756327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barteneva N. S. et al. Circulating microparticles: square the circle. BMC Cell Biol 14, 23, doi: 10.1186/1471-2121-14-23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocsfalvi G. et al. Mass spectrometry of extracellular vesicles. Mass Spectrom Rev 35, 3–21, doi: 10.1002/mas.21457 (2016). [DOI] [PubMed] [Google Scholar]
- Katsuda T., Kosaka N. & Ochiya T. The roles of extracellular vesicles in cancer biology: toward the development of novel cancer biomarkers. Proteomics 14, 412–425, doi: 10.1002/pmic.201300389 (2014). [DOI] [PubMed] [Google Scholar]
- Pankoui Mfonkeu J. B. et al. Elevated cell-specific microparticles are a biological marker for cerebral dysfunctions in human severe malaria. PLoS One 5, e13415, doi: 10.1371/journal.pone.0013415 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes V. et al. Circulating endothelial microparticles in malawian children with severe falciparum malaria complicated with coma. JAMA 291, 2542–2544, doi: 10.1001/jama.291.21.2542-b (2004). [DOI] [PubMed] [Google Scholar]
- Nantakomol D. et al. Circulating red cell-derived microparticles in human malaria. J Infect Dis 203, 700–706, doi: 10.1093/infdis/jiq104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes V., de Souza J. B., Rénia L., Hunt N. H. & Grau G. E. Cerebral malaria: Which parasite? Which model? Drug Discovery Today: Disease Models 2, 141–147, doi: 10.1016/j.ddmod.2005.05.015 (2005). [DOI] [Google Scholar]
- Combes V. et al. ABCA1 gene deletion protects against cerebral malaria: potential pathogenic role of microparticles in neuropathology. Am J Pathol 166, 295–302, doi: 10.1016/S0002-9440(10)62253-5 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assaad F., Wheway J., Hunt N. H., Grau G. E. & Combes V. Production, fate and pathogenicity of plasma microparticles in murine cerebral malaria. PLoS Pathog 10, e1003839, doi: 10.1371/journal.ppat.1003839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penet M. F. et al. Protection against cerebral malaria by the low-molecular-weight thiol pantethine. Proc Natl Acad Sci USA 105, 1321–1326, doi: 10.1073/pnas.0706867105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes V., Coltel N., Faille D., Wassmer S. C. & Grau G. E. Cerebral malaria: role of microparticles and platelets in alterations of the blood-brain barrier. Int J Parasitol 36, 541–546, doi: 10.1016/j.ijpara.2006.02.005 (2006). [DOI] [PubMed] [Google Scholar]
- Combes V. et al. Microvesiculation and cell interactions at the brain-endothelial interface in cerebral malaria pathogenesis. Prog Neurobiol 91, 140–151, doi: 10.1016/j.pneurobio.2010.01.007 (2010). [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32, 223–226, doi: 10.1038/nbt.2839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41, D1063–1069, doi: 10.1093/nar/gks1262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mause S. F. & Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res 107, 1047–1057, doi: 10.1161/CIRCRESAHA.110.226456 (2010). [DOI] [PubMed] [Google Scholar]
- Martin-Jaular L., Nakayasu E. S., Ferrer M., Almeida I. C. & Del Portillo H. A. Exosomes from Plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PLoS One 6, e26588, doi: 10.1371/journal.pone.0026588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel P. Y. et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13, 521–534, doi: 10.1016/j.chom.2013.04.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little K. M., Smalley D. M., Harthun N. L. & Ley K. The plasma microparticle proteome. Semin Thromb Hemost 36, 845–856, doi: 10.1055/s-0030-1267038 (2010). [DOI] [PubMed] [Google Scholar]
- Ramacciotti E. et al. Proteomics of microparticles after deep venous thrombosis. Thromb Res 125, e269–274, doi: 10.1016/j.thromres.2010.01.019 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard O. et al. Quantitative proteome profiling of normal human circulating microparticles. J Proteome Res 11, 2154–2163, doi: 10.1021/pr200901p (2012). [DOI] [PubMed] [Google Scholar]
- Del Conde I., Shrimpton C. N., Thiagarajan P. & Lopez J. A. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 106, 1604–1611, doi: 10.1182/blood-2004-03-1095 (2005). [DOI] [PubMed] [Google Scholar]
- Hunter M. P. et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3, e3694, doi: 10.1371/journal.pone.0003694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling L. V. & Dalli J. Microparticles are novel effectors of immunity. Curr Opin Pharmacol 13, 570–575, doi: 10.1016/j.coph.2013.05.008 (2013). [DOI] [PubMed] [Google Scholar]
- Crosnier C. et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480, 534–537, doi: 10.1038/nature10606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela M. et al. Treatment of erythrocytes with the 2-cys peroxiredoxin inhibitor, Conoidin A, prevents the growth of Plasmodium falciparum and enhances parasite sensitivity to chloroquine. PLoS One 9, e92411, doi: 10.1371/journal.pone.0092411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Aquaporin 9 is the major pathway for glycerol uptake by mouse erythrocytes, with implications for malarial virulence. Proc Natl Acad Sci USA 104, 12560–12564, doi: 10.1073/pnas.0705313104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon C. A., Grau G. E. & Craig A. G. Malaria: modification of the red blood cell and consequences in the human host. Br J Haematol 154, 670–679, doi: 10.1111/j.1365-2141.2011.08755.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Cummings P. & Curtis P. The mouse carbonic anhydrase I gene contains two tissue-specific promoters. Mol Cell Biol 9, 3308–3313 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeva-Andany M. M., Fernandez-Fernandez C., Sanchez-Bello R., Donapetry-Garcia C. & Martinez-Rodriguez J. The role of carbonic anhydrase in the pathogenesis of vascular calcification in humans. Atherosclerosis 241, 183–191, doi: 10.1016/j.atherosclerosis.2015.05.012 (2015). [DOI] [PubMed] [Google Scholar]
- Newton C. R. et al. The prognostic value of measures of acid/base balance in pediatric falciparum malaria, compared with other clinical and laboratory parameters. Clin Infect Dis 41, 948–957, doi: 10.1086/432941 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrales P. et al. Cerebral tissue oxygenation impairment during experimental cerebral malaria. Virulence 4, 686–697, doi: 10.4161/viru.26348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupka M. et al. Mild Plasmodium falciparum malaria following an episode of severe malaria is associated with induction of the interferon pathway in Malawian children. Infect Immun 80, 1150–1155, doi: 10.1128/IAI.06008-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann J. et al. Affinity proteomics reveals elevated muscle proteins in plasma of children with cerebral malaria. PLoS Pathog 10, e1004038, doi: 10.1371/journal.ppat.1004038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer S. C. et al. Investigating the Pathogenesis of Severe Malaria: A Multidisciplinary and Cross-Geographical Approach. Am J Trop Med Hyg 93, 42–56, doi: 10.4269/ajtmh.14-0841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacer A. et al. Experimental cerebral malaria pathogenesis–hemodynamics at the blood brain barrier. PLoS Pathog 10, e1004528, doi: 10.1371/journal.ppat.1004528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Szestak T., Stins M. & Craig A. G. Amplification of P. falciparum Cytoadherence through induction of a pro-adhesive state in host endothelium. PLoS One 6, e24784, doi: 10.1371/journal.pone.0024784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai S. et al. Real-time imaging reveals the dynamics of leukocyte behaviour during experimental cerebral malaria pathogenesis. PLoS Pathog 10, e1004236, doi: 10.1371/journal.ppat.1004236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza M. C. et al. Lipoxin A4 attenuates endothelial dysfunction during experimental cerebral malaria. Int Immunopharmacol 24, 400–407, doi: 10.1016/j.intimp.2014.12.033 (2015). [DOI] [PubMed] [Google Scholar]
- Kim H., Higgins S., Liles W. C. & Kain K. C. Endothelial activation and dysregulation in malaria: a potential target for novel therapeutics. Curr Opin Hematol 18, 177–185, doi: 10.1097/MOH.0b013e328345a4cf (2011). [DOI] [PubMed] [Google Scholar]
- O’Regan N. et al. A novel role for von Willebrand factor in the pathogenesis of experimental cerebral malaria. Blood, doi: 10.1182/blood-2015-07-654921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon C. A. et al. Persistent endothelial activation and inflammation after Plasmodium falciparum Infection in Malawian children. J Infect Dis 209, 610–615, doi: 10.1093/infdis/jit419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez F., Barraud de Lagerie S., Fernandez C., Pino P. & Mazier D. Tumor necrosis factor alpha in the pathogenesis of cerebral malaria. Cell Mol Life Sci 60, 1623–1635, doi: 10.1007/s00018-003-2347-x (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer S. C. et al. Vascular endothelial cells cultured from patients with cerebral or uncomplicated malaria exhibit differential reactivity to TNF. Cell Microbiol 13, 198–209, doi: 10.1111/j.1462-5822.2010.01528.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer S. C. et al. TGF-beta1 released from activated platelets can induce TNF-stimulated human brain endothelium apoptosis: a new mechanism for microvascular lesion during cerebral malaria. J Immunol 176, 1180–1184 (2006). [DOI] [PubMed] [Google Scholar]
- Srikrishna G. S100A8 and S100A9: new insights into their roles in malignancy. J Innate Immun 4, 31–40, doi: 10.1159/000330095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann C. W. The multifunctional S100 protein family. Methods Mol Biol 172, 69–80, doi: 10.1385/1-59259-183-3:069 (2002). [DOI] [PubMed] [Google Scholar]
- Goyette J. & Geczy C. L. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids 41, 821–842, doi: 10.1007/s00726-010-0528-0 (2011). [DOI] [PubMed] [Google Scholar]
- Frosch M. et al. Myeloid-related proteins 8 and 14 are specifically secreted during interaction of phagocytes and activated endothelium and are useful markers for monitoring disease activity in pauciarticular-onset juvenile rheumatoid arthritis. Arthritis Rheum 43, 628–637, doi: 10.1002/1529-0131(200003)43:3<628::AID-ANR20>3.0.CO;2-X (2000). [DOI] [PubMed] [Google Scholar]
- Benoit S. et al. Elevated serum levels of calcium-binding S100 proteins A8 and A9 reflect disease activity and abnormal differentiation of keratinocytes in psoriasis. Br J Dermatol 155, 62–66, doi: 10.1111/j.1365-2133.2006.07198.x (2006). [DOI] [PubMed] [Google Scholar]
- Gebhardt C., Nemeth J., Angel P. & Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol 72, 1622–1631, doi: 10.1016/j.bcp.2006.05.017 (2006). [DOI] [PubMed] [Google Scholar]
- Bordmann G. et al. MRP 8/14 as marker for Plasmodium falciparum-induced malaria episodes in individuals in a holoendemic area. Clin Diagn Lab Immunol 4, 435–439 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi H. et al. The accumulation of macrophages expressing myeloid-related protein 8 (MRP8) and MRP14 in the spleen of BALB/cA mice during infection with Plasmodium berghei. Exp Parasitol 138, 1–8, doi: 10.1016/j.exppara.2014.01.003 (2014). [DOI] [PubMed] [Google Scholar]
- Viemann D. et al. Myeloid-related proteins 8 and 14 induce a specific inflammatory response in human microvascular endothelial cells. Blood 105, 2955–2962, doi: 10.1182/blood-2004-07-2520 (2005). [DOI] [PubMed] [Google Scholar]
- Rammes A. et al. Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the S100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway. J Biol Chem 272, 9496–9502 (1997). [DOI] [PubMed] [Google Scholar]
- Chen I. et al. “Asymptomatic” Malaria: A Chronic and Debilitating Infection That Should Be Treated. PLoS Med 13, e1001942, doi: 10.1371/journal.pmed.1001942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza J. B. & Riley E. M. Cerebral malaria: the contribution of studies in animal models to our understanding of immunopathogenesis. Microbes Infect 4, 291–300 (2002). [DOI] [PubMed] [Google Scholar]
- Shabani E. et al. High plasma erythropoietin levels are associated with prolonged coma duration and increased mortality in children with cerebral malaria. Clin Infect Dis 60, 27–35, doi: 10.1093/cid/ciu735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway J., Latham S. L., Combes V. & Grau G. E. Endothelial microparticles interact with and support the proliferation of T cells. J Immunol 193, 3378–3387, doi: 10.4049/jimmunol.1303431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham S. L. et al. Cooperation between beta- and gamma-cytoplasmic actins in the mechanical regulation of endothelial microparticle formation. FASEB J 27, 672–683, doi: 10.1096/fj.12-216531 (2013). [DOI] [PubMed] [Google Scholar]
- Comelli L. et al. Characterization of secreted vesicles from vascular smooth muscle cells. Mol Biosyst 10, 1146–1152, doi: 10.1039/c3mb70544g (2014). [DOI] [PubMed] [Google Scholar]
- Gatlin C. L., Kleemann G. R., Hays L. G., Link A. J. & Yates J. R. 3rd Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal Biochem 263, 93–101, doi: 10.1006/abio.1998.2809 (1998). [DOI] [PubMed] [Google Scholar]
- Gluck F. et al. EasyProt–an easy-to-use graphical platform for proteomics data analysis. J Proteomics 79, 146–160, doi: 10.1016/j.jprot.2012.12.012 (2013). [DOI] [PubMed] [Google Scholar]
- Tiberti N. et al. Discovery and verification of osteopontin and Beta-2-microglobulin as promising markers for staging human African trypanosomiasis. Mol Cell Proteomics 9, 2783–2795, doi: 10.1074/mcp.M110.001008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savli H., Szendroi A., Romics I. & Nagy B. Gene network and canonical pathway analysis in prostate cancer: a microarray study. Exp Mol Med 40, 176–185, doi: 10.3858/emm.2008.40.2.176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Gibby C. C., Yuan C., Wang J., Yeung S. C. & Shete S. Gene network analysis shows immune-signaling and ERK1/2 as novel genetic markers for multiple addiction phenotypes: alcohol, smoking and opioid addiction. BMC Syst Biol 9, 25, doi: 10.1186/s12918-015-0167-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.