Abstract
RATIONALE
Bacteriophage P22 is believed to contain a total of 521 copies of 9 different proteins and a 41,724 base pair genome. Despite its enormous size and complexity, phage P22 can be electrosprayed, and it remains intact in ultra-high vacuum where its molar mass distribution has been measured.
METHODS
Phage P22 virions were generated by complementation in Salmonella enterica and purified. They were transferred into 100 mM ammonium acetate and then electrosprayed. The masses of individual virions were determined using charge detection mass spectrometry.
RESULTS
The stoichiometry of the protein components of phage P22 is sufficiently well-known that the theoretical molar mass can be determined to within a narrow range. The measured average molar mass of phage P22, 52,180±59 kDa, is consistent with the theoretical molar mass and supports the proposed stoichiometry of the components. The intrinsic width of the phage P22 mass distribution can be entirely accounted for by the distribution of DNA packaged by the headful mechanism.
CONCLUSIONS
At over 50 MDa, phage P22 is the largest object with a well-defined molar mass to be analyzed by mass spectrometry. The narrow measured mass distribution indicates that the virions survive the transition into the gas phase intact.
Keywords: charge detection mass spectrometry, bacteriophage P22, virus, DNA, induced charge
Introduction
A characteristic of a molecule is that it has a well-defined molar mass. This is true for large molecules like proteins and even some large protein complexes.[1,2,3,4,5,6,7,8] A striking example of the latter is provided by the recent measurement of the molar mass of the Prohead-1gp5 of the virus HK97 by mass spectrometry.[9] In this system, the capsid protein (gene product 5, or gp5) was expressed from a plasmid in the absence of the viral protease and as such, the empty immature virus proheads consist of exactly 420 copies of the capsid protein. Each HK97 capsid protein has a molar mass of 42,154 Da, leading to an overall theoretical molar mass for the Prohead-1gp5 of 17,704,680 Da.
With increasing size comes the possibility of heterogeneity. While there is no intrinsic heterogeneity in the HK97 Prohead-1gp5 mentioned above, in general heterogeneity in biological assemblies can arise from many sources, including variations in the number and composition of lipids, sugars, proteins and nucleic acids. Measuring the masses of heterogeneous objects by mass spectrometry is challenging. However, the measured mass distributions yield not only the average molar mass but also the dispersion, which can provide important information about compositional variability. Such information cannot be obtained from ensemble methods which provide only average values. A number of recent studies have demonstrated the value of measuring the masses of large objects, such as intact viruses.[7,8,9,10,11,12,13,14] For example, mass spectrometry has been used to resolve heterogeneity in recombinant adeno-associated virus (AAV) capsids,[14] and examine genome packaging in AAV gene therapy vectors.[12]
Here, we use charge detection mass spectrometry[15,16,17,18] (CDMS) to accurately measure the mass of infectious bacteriophage P22, a somewhat heterogeneous virus with a theoretical average molar mass of 51,613,585 Da. At >50 MDa and with 521 proteins, it is a remarkably massive and complex species. While masses have been measured for larger objects than phage P22 (including droplets, polymers, and nanoparticles) using specialized methods,[19,20,21,22,23,24,25] in those cases the molar mass is not well defined and a theoretical molar mass cannot be calculated; the measured mass distributions are very broad and depend on the conditions used to prepare the sample. On the other hand, biological assemblies generally have a defined composition and mass spectrometry can give important insight into their stoichiometry and heterogeneity. Phage P22 is by far the largest object with a well-defined molar mass to be analyzed by mass spectrometry.
P22 is a Salmonella-infecting bacteriophage which is widely studied as a model for tailed, double-stranded DNA (dsDNA) phages, the most common class of all viruses.[26] The structure and assembly of P22 is fairly well understood.[27] Figure 1 shows the components of the mature P22 virion.[28] P22 first assembles into a T=7, icosahedral procapsid containing 415 coat proteins (gp5), a 12-portal protein (gp1) complex which takes the place of one of the twelve vertices of the icosahedral capsid, ~100 copies of the assembly chaperone scaffolding protein (gp8),[29] and multiple copies each of the ejection proteins gp7, gp16, and gp20. The DNA is packaged into the capsid in an ATP-dependent fashion through a channel in the portal complex by the terminase complex (gp2 and gp3), which is not incorporated into the mature phage. DNA is actively packaged by the headful mechanism[28,30] from the P22 DNA that is a long concatemer in the cell. It is spooled into the capsid until the capsid is full, and then the DNA is cleaved. Slightly more than one genome is packaged per phage, so the ends of the DNA are redundant.[30] During DNA packaging, the capsid undergoes an expansion, and gp8 leaves through pores in the capsid. Plug proteins (12 copies of gp4 and 6 copies of gp10) and tail needle proteins (3 copies of gp26) are added to hold the DNA inside the capsid. Finally, tailspike proteins (18 copies of gp9) are added for cell recognition and attachment. The mature phage P22 therefore contains different numbers of 9 proteins, along with a distribution of DNA. We show below that its theoretical mass can be calculated to within a fairly narrow range.
Figure 1.
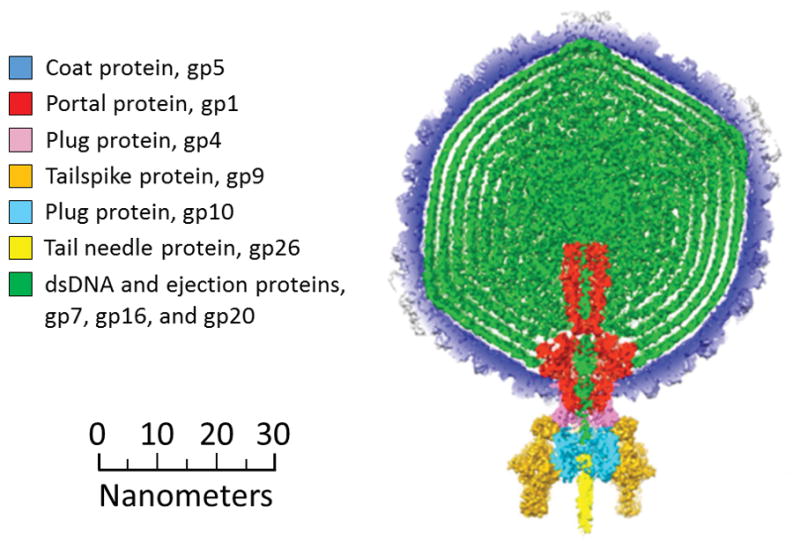
A diagram showing the components of infectious phage P22. Adapted from Ref. 28. The masses of the components are given in Table 1.
Experimental
Phage P22 Generation and Electron Microscopy
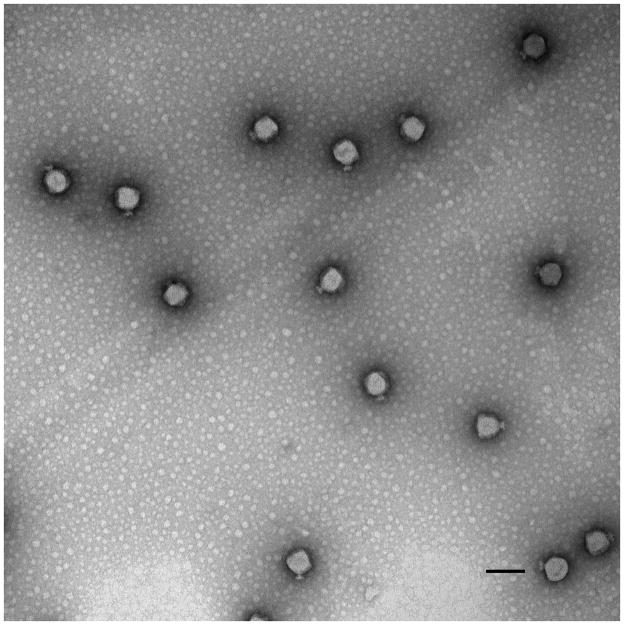
The phage P22 was generated by complementation in vivo. See supplemental information for details. CDMS measurements show that authentic phage P22 and phage P22 generated by complementation have the same masses. Samples for electron microscopy were prepared by applying phage onto 300-mesh carbon-coated copper grids (Electron Microscopy Sciences). The grids were stained with 1% uranyl acetate and visualized by an FEI Tecnai G2 Spirit BioTwin Transmission Electron Microscope as described elsewhere.[31] Figure 2 shows a TEM micrograph of infectious phage P22. The brightness of the particles indicates that they are filled, in this case with DNA and some protein. The phage diameter is ~65 nm, as expected.[32] The claw-like tail machinery is visible on most particles.
Figure 2.
Negative-stain, transmission electron micrograph of phage P22. The scale bar is 100 nm.
Charge Detection Mass Spectrometry
CDMS is a single-molecule technique where the m/z ratio and charge of individual ions are measured simultaneously, and then multiplied to give the mass. This process is repeated for thousands of ions to obtain a mass spectrum. The CDMS instrument[33,34,35] and data analysis methods[36,37] used in this work have been described in detail. Briefly, positive ions are generated by nano-electrospray. They are transmitted through several differentially-pumped regions containing RF ion guides, where the pressures are adjusted to optimize the signal. The ions are then accelerated through a 100 V potential difference, and focused into a dual hemispherical deflection analyzer (HDA). The HDA only transmits ions with a narrow band of kinetic energies centered on 100 eV/z. Finally, the ions pass into an ultra-high vacuum region containing an electrostatic ion trap with a charge detector tube embedded in it. When an ion enters the detector tube, it induces a charge that is picked up by a charge-sensitive preamplifier. The ion oscillates back and forth in the trap and passes through the detector tube many times which greatly improves the charge precision.[17] A trapping time of 91 ms was employed. The output from the preamplifier is digitized and analyzed with a Fortran program using fast Fourier transforms (FFTs). The signals for ions that are not trapped for the full period are discarded. The m/z is inversely proportional to the square of the ion’s fundamental frequency in the trap, and the charge is proportional to the sum of the fundamental and second harmonic peak magnitudes in the FFT. Thus, the FFT provides both the m/z and the charge measurement of each ion, which are multiplied to obtain mass. For CDMS, the phage P22 was transferred into 100 mM ammonium acetate by spin-column SEC (Bio-Rad Laboratories, Inc.).
Results and Discussion
Before presenting the measured mass spectrum of the phage, we tabulate the expected mass. The masses and stoichiometry of the components are given in Table 1. The P22 genome is linear dsDNA[38] that is 41,724 base pairs long with a known sequence.[39] The number of redundant base pairs as a consequence of headful packaging described above has been reported to range from 850 to 2350.[30] We use an average value of 1600 additional base pairs to give the mass of the dsDNA in Table 1. The stoichiometry of most of the proteins is precisely known;[32,40] the main source of uncertainty is the ejection proteins (gp7, gp16, and gp20). A variety of estimates have recently been given: 6–20 copies of each ejection protein,[41] and 12–20 copies.[26] The lower end of these ranges is incompatible with the measured mass of the intact phage. The most recent estimate of 11, 12, and 32 copies of gp7, gp16, and gp20, respectively,[42] is used in Table 1. Adding the masses of all the components together, we calculate the average total mass of the intact P22 virion as 51,613 kDa (Table 1).
Table 1.
Components of phage P22 and their masses.[28,32,39,42] The total phage mass was calculated by multiplying the mass of each component by its stoichiometry and summing all those masses together. For the genome the average DNA mass for the headful mechanism was used.30
| Component | Mass of one copy (Da) | Stoichiometry | Total Mass (Da) | |
|---|---|---|---|---|
| dsDNA | Genome + 1600 bp | 26,768,698 | 1 | 26,768,698 |
| Capsid | Coat protein, gp5 | 46,621 | 415 | 19,347,715 |
| Tail machine | Portal protein, gp1 | 82,612 | 12 | 2,889,621 |
| Plug protein, gp4 | 18,025 | 12 | ||
| Tailspike protein, gp9 | 71,857 | 18 | ||
| Plug protein, gp10 | 52,457 | 6 | ||
| Tail needle protein, gp26 | 24,603 | 3 | ||
| Ejection proteins | gp7 | 21,093 | (11) | 2,607,551 |
| gp16 | 64,358 | (12) | ||
| gp20 | 50,101 | (32) | ||
| Total Mass of Intact Virion (Da) | 51,613,585 | |||
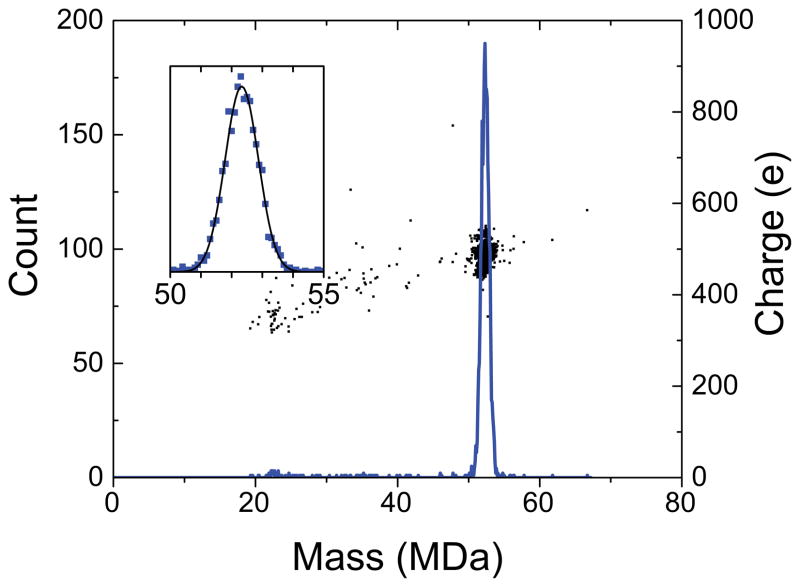
Figure 3 shows a typical CDMS mass spectrum of the intact phage generated by complementation in vivo. The spectrum was obtained by binning the measured masses for 2563 ions into 100 kDa bins. The spectrum shows a single peak centered around 52 MDa. There are no ions below 20 MDa because they are cut-off by the instrumental settings. The inset in Figure 3 shows an expanded view of the main peak. The experimental spectrum is represented by the blue points and the black line shows a Gaussian fit to the points. The Gaussian is a good fit to the measured peak. The Gaussian fit was used to determine the mean and the width of the mass distribution. The average mass from four independent measurements of the mean on two different samples is 52,180±59 kDa where the uncertainty is the standard deviation of the mean. The average full width at half maximum (FWHM) is 1,330±36 kDa. The width results from a combination of instrumental resolution, salt and solvent adducts, and intrinsic heterogeneity, as described in more detail below. The center of the mass distribution (the average mass) can be defined with an uncertainty which is much less than the width of the distribution.
Figure 3.
Mass spectrum of phage P22 measured with CDMS. The spectrum is a histogram constructed with 100 kDa bins. The inset shows an expanded view of the peak at around 52 MDa. In the inset, the blue points are the measured values and the black line is a Gaussian fit to the experimental data. The black points overlaying the main spectrum are a charge vs mass scatter plot showing that the average charge of the phage is about 490 e.
The points that overlay the mass spectrum in Figure 3 are a scatter plot of charge versus mass. There is a single cluster of points centered at around 490 elementary charges (e). Thus, the average m/z of the phage P22 ions is over 100 kDa. We have previously reported the mass distribution for immature, P22 procapsid-like particles.[43] The procapsid-like particles consisted of 420 copies of coat protein (gp5) and a distribution of scaffolding protein (gp8), as those were the only proteins expressed from the plasmid. The average charge determined here for the phage P22 (490 e) is larger than for the procapsid-like particles (425 e), which is consistent with the larger diameter of the phage according to the charge residue model of electrospray.[44] However, instrumental conditions and settings can have a significant effect on the charge of electrosprayed ions, so it is difficult to compare absolute charges measured in different experiments.
The measured average mass (52,180±59 kDa) is 567 kDa higher than the theoretical mass (51,613 kDa). Electrospray from non-denaturing solvents[45,46] (native electrospray), while enabling the analysis of complex and massive biological analytes by mass spectrometry, is known to lead to masses that are slightly larger than the theoretical mass due to adduction by salts, counter ions, and solvent.[47] In the recent measurement of the mass of the Prohead-1gp5 of HK97,[9] the measured mass was 1.3% larger than the theoretical mass. Here the percentage deviation is 1.1%. The reason that the percentage deviation is smaller here may be that the encapsidated DNA prevents adduction to the capsid interior, while the Prohead-1gp5 of HK97 is an empty shell.
The average FWHM of the peak due to phage P22 is 1,330±36 kDa. There are three main contributions to the width of peak: the mass resolution (which is known), the mass distribution due to adduct formation (which can be estimated), and the intrinsic heterogeneity in the mass of the phage P22 (which we would like to obtain). Assuming that all distributions are Gaussian, the intrinsic heterogeneity of the phage P22 is given by
| (1) |
where WIH is the FWHM of the intrinsic mass distribution, WMEAS is the FWHM of the measured peak, WRES is the FWHM from the mass resolution, and WADD is the FWHM due to adduct formation. The mass resolution depends on the uncertainties in the m/z and charge measurements. These uncertainties are well characterized[37] and lead to a combined relative root mean square deviation (RMSD) (Δm/m) of 0.0087 for ions with the trapping time, oscillation frequency and charge measured here. An estimate of the mass distribution due to adduct formation can be obtained from the effective resolving power (194) reported by Heck and collaborators for the Prohead-1gp5 of HK97.[9] We use the same value to estimate the width of the distribution for phage P22. With these two quantities and the average width of the measured peak, the intrinsic width of the phage P22 mass distribution is 747±66 kDa. If this distribution is entirely due to the distribution of packaged DNA, then the distribution has a FWHM of 1,213±107 base pairs. This value compares favorably with an earlier estimate of the width of 1,500 base pairs.[30] Thus, all the heterogeneity in the mass of the phage P22 can be attributed to the distribution of packaged DNA. Surprisingly then, these data indicate that there is little or no heterogeneity in the protein stoichiometries. While the proteins associated with the capsid and with the tail machine are forced into specific stoichiometries by geometry and their interactions within the phage, this does not appear to be true for the ejection proteins, so there could conceivably be a distribution for these proteins. However, the CDMS results show that if there is a distribution, it is narrow. The narrow intrinsic mass distribution measured for phage P22 also shows that almost all the ions in the spectrum are due to intact phage. A close inspection of Figure 3 shows that there are only a few ions in the spectrum that do not fall within the main peak. These may be a small fraction of procapsids which did not package DNA, or they may be misassembled particles. Thus, despite the size and complexity of phage P22, it survives the transition into the gas phase and remains intact in ultra-high vacuum.
The mass for the dsDNA given in Table 1 was calculated assuming that the DNA is packaged in an un-ionized state. In solution, however, the phosphates are thought to ionize, with neutrality retained through counter ions. The phosphate groups in the DNA backbone are known to have a strong affinity for Na+. If all the backbone phosphates were ionized and had Na+ counter ions the mass of the genome would increase by 1,906 kDa. This is incompatible with the measured mass, which indicates that the DNA is packaged predominantly in the un-ionized state. There is strong support for this conclusion. In recent CDMS measurements for adeno-associated virus (AAV), we were able to determine the mass of the empty capsid and the capsid with a full genome, for both single-stranded and self-complementary genomes.[12] The mass difference between the empty and full particles was in good agreement with the difference expected for unionized DNA. Even for electrosprayed DNA it appears that the number of counter ions is relatively small. Schultz and coworkers have measured the masses of electrosprayed, MDa-sized DNA and found that the fraction of counter ions decreased as the length of the DNA increased, and the mass approached the un-ionized mass for DNA masses of around 5 MDa.[48]
In conventional mass spectrometry where only the m/z is measured, the charge must be deduced from resolved peaks in the ensemble m/z spectrum in order to deduce the mass. The intrinsic mass heterogeneity for phage P22 is relatively small (747 kDa FWHM) compared to the average mass (52,180 kDa). However, with an average charge of 490 e it is impossible to resolve charge states in the m/z spectrum. Thus, it would be impossible to determine the mass distribution for phage P22 from the m/z spectrum alone. In this work we used CDMS where the m/z and charge of each ion are measured simultaneously, yielding the mass of each ion directly. CDMS is one of several single-molecule mass spectrometry methods that can be used to analyze large and heterogeneous objects.[49]
Conclusions
We have measured the molar mass distribution for infectious phage P22. Phage P22 contains a total of 521 copies of 9 different proteins along with dsDNA. Despite its enormous size and complexity, P22 phage survives the transition into the gas phase and remains intact in ultra-high vacuum. The average measured molar mass (52,180±59 kDa) is consistent with the theoretical molar mass and supports the proposed stoichiometry. The intrinsic width of the molar mass distribution (747±66 kDa) is entirely attributable to the distribution of DNA packaged by the headful mechanism, indicating that there is little or no heterogeneity in the protein components. Moreover, the average mass measurement shows that the DNA packaged within phage P22 is un-ionized rather than neutralized by counter ions. From a broader perspective, these results show that valuable information can be obtained from single-molecule mass measurements on objects with masses well beyond those usually studied by conventional mass spectrometry, providing important information on their composition and heterogeneity.
Supplementary Material
Acknowledgments
We thank Dr. Marie Cantino and Dr. Xuanhao Sun of University of Connecticut Bioscience Electron Microscopy Laboratory for assistance with transmission electron microscopy. We gratefully acknowledge the support of the NSF through award number CHE-1531823 to MFJ and the NIH for grant GM076661 to CMT.
References
- 1.Ganem B, Li YT, Henion JD. Observation of noncovalent enzyme-substrate and enzyme-product complexes by ion-spray mass spectrometry. J Am Chem Soc. 1991;113:7818. [Google Scholar]
- 2.Light-Wahl KJ, Schwartz BL, Smith RD. Observation of the noncovalent quaternary associations of proteins by electrospray ionization. J Am Chem Soc. 1994;116:5271. doi: 10.1016/1044-0305(94)85034-8. [DOI] [PubMed] [Google Scholar]
- 3.Loo JA. Observation of large subunit protein complexes by electrospray ionization mass spectrometry. J Mass Spectrom. 1995;30:180. [Google Scholar]
- 4.Rostom AA, Robinson CV. Detection of the intact GroEL chaperonin assembly by mass spectrometry. J Am Chem Soc. 1999;121:4718. [Google Scholar]
- 5.Wang Y, Schubert M, Ingendoh A, Franzen J. Analysis of non-covalent protein complexes up to 290 kDa using electrospray ionization and ion trap mass spectrometry. Rapid Commun Mass Spectrom. 2000;14:12. doi: 10.1002/(SICI)1097-0231(20000115)14:1<12::AID-RCM825>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Van Berkel WJH, Van Den Heuvel RHH, Versluis C, Heck AJR. Detection of intact megaDalton protein assemblies of vanillyl-alcohol oxidase by mass spectrometry. Prot Sci. 2000;9:453. doi: 10.1110/ps.9.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Nat Acad Sci USA. 2008;105:9216. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JCY, Zlotnick A, Jarrold MF. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc. 2014;136:3536. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa virus assemblies with native mass spectrometry. Angew Chem Int Ed. 2013;52:4020. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keifer DZ, Motwani T, Teschke CM, Jarrold MF. Acquiring structural information on virus particles with charge detection mass spectrometry. J Am Soc Mass Spectrom. 2016;27:1028. doi: 10.1007/s13361-016-1362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierson EE, Keifer DZ, Kukreja AA, Wang JC-Y, Zlotnick A, Jarrold MF. Charge detection mass spectrometry identifies preferred non-icosahedral polymorphs in the self-assembly of woodchuck hepatitis virus capsids. J Mol Biol. 2016;428:292. doi: 10.1016/j.jmb.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierson EE, Keifer DZ, Asokan A, Jarrold MF. Resolving Adeno-Associated Viral Particle Diversity With Charge Detection Mass Spectrometry. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Waterbeemd M, Snijder J, Tsvetkova IB, Dragnea BG, Cornelissen JJ, Heck AJR. Examining the heterogeneous genome content of multipartite viruses BMV and CCMV by native mass spectrometry. J Am Soc Mass Spectrom. 2016;27:1000. doi: 10.1007/s13361-016-1348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJR. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by orbitrap mass spectrometry. J Am Chem Soc. 2014;136:7295. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shelton H, Hendricks JCD, Wuerker RF. Electrostatic acceleration of microparticles to hypervelocities. J Appl Phys. 1960;31:1243. [Google Scholar]
- 16.Fuerstenau SD, Benner WH. Molecular weight determination of megadalton DNA electrospray ions using charge detection time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:1528. doi: 10.1002/rcm.1290091513. [DOI] [PubMed] [Google Scholar]
- 17.Benner WH. A gated electrostatic ion trap to repetitiously measure the charge and m/z of large electrospray ions. Anal Chem. 1997;69:4162. [Google Scholar]
- 18.Fuerstenau SD, Benner WH, Thomas JJ, Brugidou C, Bothner B, Siuzdak G. Mass spectrometry of an intact virus. Angew Chem Int Edit. 2001;113:559. doi: 10.1002/1521-3773(20010202)40:3<541::AID-ANIE541>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Nie Z, Cui F, Tzeng Y-K, Chang H-C, Chu M, Lin H-C, Chen C-H, Lin H-H, Yu AL. High-speed mass analysis of whole erythrocytes by charge-detection quadrupole ion trap mass spectrometry. Anal Chem. 2007;79:7401. doi: 10.1021/ac071207e. [DOI] [PubMed] [Google Scholar]
- 20.Peng W-P, Lin H-C, Lin H-H, Chu M, Yu AL, Chang H-C, Chen C-H. Charge-monitoring laser-induced acoustic desorption mass spectrometry for cell and microparticle mass distribution measurement. Angew Chem Int Ed. 2007;46:3865. doi: 10.1002/anie.200700309. [DOI] [PubMed] [Google Scholar]
- 21.Xiong C, Zhou X, Wang J, Zhang N, Peng W-P, Chang H-C, Nie Z. Ambient aerodynamic desorption/ionization method for microparticle mass measurement. Anal Chem. 2013;85:4370. doi: 10.1021/ac303565a. [DOI] [PubMed] [Google Scholar]
- 22.Mabbett SR, Zilch LW, Maze JT, Smith JW, Jarrold MF. Pulsed acceleration charge detection mass spectrometry: application to weighing electrosprayed droplets. Anal Chem. 2007;79:8431. doi: 10.1021/ac071513s. [DOI] [PubMed] [Google Scholar]
- 23.Doussineau T, Bao CY, Antoine R, Dugourd P, Zhang W, D’Agosto F, Charleux B. Direct molar mass determination of self-assembled amphiphilic block copolymer nanoobjects using electrospray-charge detection mass spectrometry. ACS Macro Lett. 2012;1:414. doi: 10.1021/mz300011b. [DOI] [PubMed] [Google Scholar]
- 24.Bell DM, Howder CR, Johnson RC, Anderson SL. Single CdSe/ZnS nanocrystals in an ion trap: charge and mass determination and photophysics evolution with changing mass, charge, and temperature. ACS Nano. 2014;8:2387. doi: 10.1021/nn405920k. [DOI] [PubMed] [Google Scholar]
- 25.Doussineau T, Désert A, Lambert O, Taveau J-C, Lansalot M, Dugourd P, Bourgeat-Lami E, Ravaine S, Duguet E, Antoine R. Charge detection mass spectrometry for the characterization of mass and surface area of composite nanoparticles. J Phys Chem C. 2015;119:10844. [Google Scholar]
- 26.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 Coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teschke CM, Parent KN. ‘Let the Phage Do the Work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J, Lander GC, Olia A, Li R, Casjens S, Prevelige PE, Jr, Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure. 2011;19:496. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prevelige PE, Jr, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J Mol Biol. 1988;202:743. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 30.Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J Mol Biol. 1988;199:467. doi: 10.1016/0022-2836(88)90618-3. [DOI] [PubMed] [Google Scholar]
- 31.Cortines JR, Motwani T, Vyas AA, Teschke CM. Highly specific salt bridges govern bacteriophage P22 icosahedral capsid assembly: identification of the site in coat protein responsible for interaction with scaffolding protein. J Virol. 2014;88:5287. doi: 10.1128/JVI.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Contino NC, Jarrold MF. Charge detection mass spectrometry for single ions with a limit of detection of 30 charges. Int J Mass Spectrom. 2013;345–347:153. [Google Scholar]
- 34.Contino NC, Pierson EE, Keifer DZ, Jarrold MF. Charge detection mass spectrometry with resolved charge states. J Am Soc Mass Spectrom. 2013;24:101. doi: 10.1007/s13361-012-0525-5. [DOI] [PubMed] [Google Scholar]
- 35.Pierson EE, Keifer DZ, Contino NC, Jarrold MF. Probing higher order multimers of pyruvate kinase with charge detection mass spectrometry. Int J Mass Spectrom. 2013;337:50. [Google Scholar]
- 36.Pierson EE, Contino NC, Keifer DZ, Jarrold MF. Charge detection mass spectrometry for single ions with an uncertainty in the charge measurement of 0.65 e. J Am Soc Mass Spectrom. 2015;26:1213. doi: 10.1007/s13361-015-1126-x. [DOI] [PubMed] [Google Scholar]
- 37.Keifer DZ, Shinholt DL, Jarrold MF. Charge detection mass spectrometry with almost perfect charge accuracy. Anal Chem. 2015;87:10330. doi: 10.1021/acs.analchem.5b02324. [DOI] [PubMed] [Google Scholar]
- 38.Byl CV, Kropinski AM. Sequence of the genome of salmonella bacteriophage P22. J Bacteriol. 2000;182:6472. doi: 10.1128/jb.182.22.6472-6481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedulla ML, Ford ME, Karthikeyan T, Houtz JM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. Corrected sequence of the bacteriophage P22 genome. J Bacteriol. 2003;185:1475. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJR. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J Mol Biol. 2008;379:385. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige PE, Jr, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 42.Casjens SR. Personal communication. University of Utah; Salt Lake City, UT: 2015. [Google Scholar]
- 43.Keifer DZ, Pierson EE, Hogan JA, Bedwell GJ, Prevelige PE, Jarrold MF. Charge detection mass spectrometry of bacteriophage P22 procapsid distributions above 20 MDa. Rapid Communications in Mass Spectrometry. 2014;28:483. doi: 10.1002/rcm.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez de la Mora J. Electrospray ionization of large multiply charged species proceeds via Dole’s charged residue mechanism. Anal Chim Acta. 2000;406:93. [Google Scholar]
- 45.Chowdhury SK, Katta V, Chait BT. Probing conformational changes in proteins by mass spectrometry. J Am Chem Soc. 1990;112:9012. [Google Scholar]
- 46.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.Lössl P, Snijder J, Heck AR. Boundaries of mass resolution in native mass spectrometry. J Am Soc Mass Spectrom. 2014;25:906. doi: 10.1007/s13361-014-0874-3. [DOI] [PubMed] [Google Scholar]
- 48.Schultz JC, Hack CA, Benner WH. Mass determination of megadalton-DNA electrospray ions using charge detection mass spectrometry. J Am Soc Mass Spectrom. 1998;9:305. doi: 10.1016/S1044-0305(97)00290-0. [DOI] [PubMed] [Google Scholar]
- 49.Keifer DZ, Jarrold MF. Single-molecule mass spectrometry. Mass Spectrom Rev. 2016 doi: 10.1002/mas.21495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.