Summary
In animals, double-stranded short interfering RNA (siRNA) and single-stranded microRNA (miRNA) regulate gene expression by targeting homologous mRNA for cleavage or by interfering with their translation, respectively [1–3]. siRNAs are processed from injected or transgene-derived, long, perfect double-stranded RNA (dsRNA), while miRNAs are processed from short, imperfect dsRNA precursors transcribed from endogenous intergenic regions [4–9]. In plants, both siRNAs and miRNAs activate cleavage of homologous RNA targets [10–12], but little is known about the genes controlling their production or action. The SGS2/SDE1 protein contributes to produce transgene siRNA [10], while DCL1 and HEN1 contribute to endogenous miRNA accumulation [8, 9]. Here, we show that: i) SGS2, SGS3 [13], AGO1 [14, 15], and HEN1 contribute to produce transgene siRNA involved in sense post-transcriptional gene silencing (S-PTGS); ii) HEN1, but not SGS2, SGS3, or AGO1, contributes to the accumulation of the endogenous miR171 miRNA and to the cleavage of Scarecrow target mRNA by miR171 [11]; iii) SGS2, SGS3, AGO1, and HEN1 contribute to resistance against cucumber mosaic virus [13, 15], but not to siRNA and IR-PTGS triggered by hairpin transgenes directly producing perfect dsRNA [16]; and iv) the actions of HEN1 in miRNA/development and siRNA/S-PTGS can be uncoupled by single-point mutations at different positions in the protein.
Results and Discussion
Through a forward genetic screen, we previously identified 30 Arabidopsis mutants deficient in S-PTGS triggered by the 35S-GUS sense transgene carried at the L1 locus [13–15, 17]. These 30 mutants were classified into 4 complementation groups: sgs1 (1 allele), sgs2 (18 alleles), sgs3 (5 alleles), and ago1 (6 alleles). SGS2 (also known as SDE1 [10]) is a putative RNA-dependent RNA polymerase (RdRp) [10, 13] sharing sequence similarity with QDE-1, which controls quelling in Neurospora [18], and RRF-1, which controls RNAi in C. elegans [19]. SGS3 is a coiled-coil protein of unknown function that has no obvious homolog in animals or fungi [13]. AGO1 belongs to the paz piwi domain (PPD) protein family of unknown function [14, 15] and shares sequence similarity with QDE-2, which controls quelling in Neurospora [18], AGO-2 and PIWI, which control RNAi in Drosophila [20, 21], and RDE-1, which controls RNAi in C. elegans [22]. Using a reverse genetics approach, we also reported that the ddm1 and met1 mutations in a SWI2/SNF2-like chromatin remodeling protein and a Dnmt1-like DNA methyltransferase controlling TGS also impaired S-PTGS at the L1 locus, although to a lower extent [23]. We pursued our forward genetic screen of the L1 EMS library and identified 14 additional mutants. These mutants were genetically classified by crossing with representative sgs1, sgs2, sgs3, ago1, ddm1, and met1 mutants. Together, the 44 EMS mutants deriving from line L1 belong to 6 complementation groups: sgs1 (1 allele), sgs2 (23 alleles), sgs3 (6 alleles), ago1 (12 alleles), met1 (2 alleles), plus a new mutant, called 23-2, defining a novel group that we describe in this paper.
Previous analysis of the sensitivity of sgs2, sgs3, and ago1 mutants to infection by different viruses revealed that mutants impaired in transgene S-PTGS are hypersensitive to infection by cucumber mosaic virus (CMV) due to a 5- to 6-fold overaccumulation of CMV RNA [13, 15]. This finding suggests that transgenes undergoing S-PTGS encode particular forms of RNA that share com mon features with viral RNAs targeted by the cellular PTGS machinery. Infection of 23-2 with CMV also revealed hypersensitivity and 5-fold overaccumulation of CMV RNA (Figure 1). This result therefore indicates that, with respect to CMV hypersensitivity, the 23-2 mutant behaves as the other mutants impaired in transgene S-PTGS that were identified through the same screen; this finding reinforces the mechanistic similarity between S-PTGS directed against sense transgenes and PTGS directed against CMV.
Figure 1. Phenotype and Virus Sensitivity of Line L1 and the 23-2 Mutant.

(A) Phenotype of line L1 and of the 23-2 mutant 2 weeks after mock infection or infection by CMV. All plants are the same age, indicating the delay in flowering of the 23-2 mutant.
(B) A close-up of (A) showing the leaf phenotype of the 23-2 mutant.
(C) CMV RNA accumulation in line L1 and in the 23-2 mutant, mock infected or infected by CMV. Standardization with a 25S rDNA probe is shown below.
GUS mRNA accumulation increased 14-fold in 23-2 plants compared with L1 plants (Figure 2) and leads to a 1000-fold increase in GUS activity (data available in the Supplemental Data available with this article online). However, the level of GUS mRNA accumulation was slightly lower in 23-2 compared with sgs2-1, sgs3-1, and ago1-27, which showed a 19-, 18-, and 17-fold increase in GUS mRNA accumulation, respectively (Figure 2), and a 3500-fold average increase in GUS activity compared with L1 plants (data available in the Supplemental Data). In addition, GUS activity in the 23-2 mutant was more variable from plant to plant and from one part of the plant to another than it was in the other mutants. The absence of full reactivation of the 35S-GUS transgene at the L1 locus and the variability of GUS expression could be attributed to epigenetic changes that have occurred at the L1 locus during the mutagenesis in addition to the impairment in PTGS that was demonstrated by CMV hypersensitivity and CMV RNA overaccumulation. However, the mutation in the 23-2 plants also partially impaired both S-PTGS of the 35S-GUS transgene carried by the L2 locus [17] and cosuppression of the endogenous NIA genes and the 35S-NIA2 transgene triggered by the 2a3 locus [17] (data available in the Supplemental Data). This finding suggests that the 23-2 mutation only partially impairs transgene-mediated S-PTGS and cosuppression. To confirm that there was residual PTGS activity in 23-2, we infected 23-2 plants with turnip mosaic virus (TuMV) or cucumber mosaic virus (CMV) expressing HC-Pro and 2b proteins that counteract PTGS, but not TGS, in Arabidopsis and tobacco [13, 16, 24–26]. Full transgene expression was observed in infected 23-2 plants (data available in the Supplemental Data), suggesting that the lack of full transgene expression in the 23-2 mutant is due to limited residual PTGS activity.
Figure 2. GUS mRNA and siRNA Accumulation in Wild-Type and Mutant Plants.
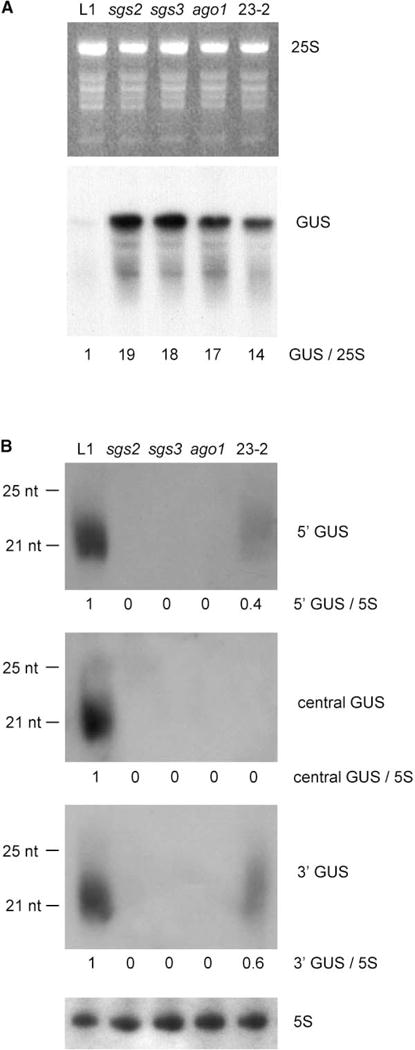
(A) mRNA extracted from leaves was hybridized with a GUS DNA probe. An ethidium bromide-stained gel is shown for standardization. The ratio between GUS and 25S signals is indicated below.
(B) siRNA extracted from leaves was hybridized with antisense RNA probes corresponding to the 5′ part (position 1–558), central part (position 558–789), or 3′ part (position 789–1865) of the GUS coding sequence. Similar results were obtained with sense probes. Hybridization with 5S RNA is shown for standardization. The ratio between GUS and 5S signals is indicated below.
S-PTGS in L1 plants correlated with the accumulation of GUS siRNA of both sense and antisense polarities, which correspond to various parts of the GUS coding sequence (Figure 2). As shown previously in the 6b5 tobacco line carrying the same 35S-GUS construct silenced by S-PTGS [27], ~21–22 nt long siRNAs were observed, but no ~25 nt siRNAs were visualized. No GUS siRNA signal could be detected in sgs2-1, sgs3-1, and ago1-27 mutants by using probes corresponding to the different parts of the transgene, even after long exposure. Furthermore, no sense or antisense siRNA could be detected in the 23-2 mutant by using a probe corresponding to the central part of the GUS coding sequence, but siRNAs of both polarities were still detectable, although at a level lower than in the L1 line, by using probes corresponding to the 5′ or 3′ region of the GUS coding sequence. This could reflect a differential effect of the protein impaired in the 23-2 mutant on the production or stabilization of different siRNA populations. Alternatively, some siRNAs could remain in the 23-2 mutant because they are not functional and therefore cannot act in the RISC complex.
Although they are impaired in S-PTGS and cosuppression triggered by various sense transgenes (L1, L2, 2a3), the sgs2, sgs3, and ago1 mutants are not affected in IR-PTGS triggered by hairpin constructs directly producing double-stranded RNA. This finding suggests that the SGS2, SGS3, and AGO1 proteins act upstream of dsRNA formation in transgenic plants carrying sense transgenes [16]. This hypothesis was confirmed by the complete disappearance of siRNA in sgs2-1, sgs3-1, and ago1-27 mutants (Figure 2), and this disappearance rules out the possibility that AGO1 could play a role similar to that of its homologs QDE-2 in Neuropora and AGO-2 in Drosophila, which are required for mRNA degradation by siRNA in the RISC complex [18–20]. Rather, AGO1 could play a role similar to that of its homolog PIWI in Drosophila and can participate in an early step of cosuppression [21]. Indeed, both ago1 and piwi mutants no longer accumulated siRNA, whereas qde-2 mutants still accumulated siRNA. Because GUS siRNAs are not totally absent in the 23-2 mutant, the corresponding protein could play a role downstream of dsRNA formation. To test this hypothesis, we introduced hairpin constructs directed against AP1, AG, or CLV3 endogenous genes in the 23-2 mutant. Transformants exhibiting a strong ap1, ag, or clv3 phenotype were obtained at a frequency similar (ca. 65%) to that observed in wild-type plants and in sgs2-1, sgs3-1, and ago1-27 mutants [16]. We also introduced the 306-0-1 locus carrying a 35S-ΔGUS-SUG hairpin construct directly producing dsRNA [16] by crossing into a L1-depleted 23-2 plant and found that siRNA accumulated at the same level in wild-type and mutant plants (data not shown). In addition, silencing of the target 35S-GUS transgene carried at the 6b4 locus [16] by the 306-0-1 locus was as efficient in wild-type plants and in 23-2 (data available in the Supplemental Data). These results indicate that, like SGS2, SGS3, and AGO1, the protein impaired in the 23-2 mutant does not contribute to siRNA and IR-PTGS triggered by hairpin constructs directly producing long, perfect dsRNA.
Whereas sgs2 and sgs3 mutants have no obvious phenotypes [13], ago1 mutants exhibited developmental abnormalities, ranging from leaf serration and reduced fertility in hypomorphic alleles to complete sterility and eventual death in null alleles [14, 15]. The 23-2 mutant also exhibited developmental abnormalities, including narrowing leaves, late flowering, and reduced fertility (Figure 1), but this phenotype did not resemble that of ago1 mutants. Segregation analyses of 750 F2 plants derived from a cross between 23-2 and L1 showed that this developmental phenotype cosegregates with the release of S-PTGS and suggests that both effects result from a single mutation. This mutation was mapped to a 60-kb interval between markers CER442391 and CER442404 on chromosome 4 (data available in the Supplemental Data). This region covered by BAC T13K14 contains the HEN1 gene previously identified through a screen for mutations that enhance the hua1-1 and hua2-1 mutations, which cause floral abnormalities [28], and was subsequently shown to control the accumulation of endogenous miRNA [8]. Crosses performed between 23-2 and hen1-1, hen1-2, or hen1-3 alleles ([28]; X.C., unpublished data) yielded F1 progenies exhibiting the mutant phenotype and undergoing S-PTGS of GUS (data not shown). This finding indicates that 23-2 is a hen1 allele, but that hen1-1, hen1-2, and hen1-3 alleles are not impaired in S-PTGS and act in a dominant manner over the 23-2 allele with regard to S-PTGS. Sequencing of the HEN1 gene in 23-2 revealed a G-to-A transition at position 25911 of BAC T13K14 (position 2412 of the predicted cDNA At4g20910), resulting in a Glu-to-Lys change in the C terminus of the protein. Introduction of the wild-type HEN1 gene in the 23-2 plants completely restored both a wild-type phenotype and S-PTGS (data available in the Supplemental Data), demonstrating the involvement of HEN1 in both processes. The 23-2 mutant was therefore renamed hen1-4.
Previous analyses revealed that the hen1-1 mutation affects the accumulation of endogenous miRNA, indicating that HEN1 acts in miRNA metabolism [8]. However, the effect on the accumulation of the corresponding mRNA target has not yet been investigated. Analysis of the accumulation of the endogenous miR171 miRNA revealed that it accumulates at similar levels in wild-type (L1) plants and in sgs2, sgs3, and ago1 mutants but cannot be detected in the 23-2/hen1-4 mutant (Figure 3A). Correspondingly, the Scarecrow SCL6-III (At3g60630) mRNA that is targeted for cleavage by miR171 [11] accumulated at similar levels in wild-type (L1) plants and in sgs2, sgs3, and ago1 mutants but was increased 4-fold in the 23-2/hen1-4 mutant (Figure 3B). These results therefore clearly demonstrate that the hen1-4 mutation affects both endogenous miRNA (miR171) or transgene siRNA (GUS) accumulation, resulting in reduced cleavage of the corresponding mRNA targets.
Figure 3. miR171 miRNA and SCL mRNA Accumulation in Wild-Type and Mutant Plants.
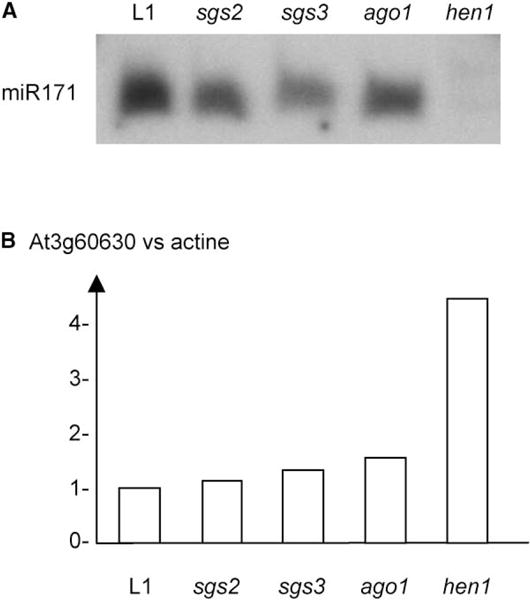
(A) Small RNA extracted from wild-type and mutant flowers was hybridized with a probe complementary to miR171.
(B) Total RNA extracted from wild-type and mutant flowers was quantified for SCL6-III mRNA relative accumulation by real-time PCR by using primers surrounding the cleavage site. Quantifications are normalized with actine2 transcript. The wild-type value is 1. AU, arbitrary unit.
The dual effect of the hen1-4 mutation on the accumulation of both siRNA and miRNA and on the cleavage of the corresponding mRNA target suggests common features between these two pathways in plants. This is the first report of a mutation affecting both pathways in plants. The dcr-1 mutation affecting the double-stranded specific RNase III DICER has been shown to affect siRNA and miRNA only in C. elegans [29], whereas the caf mutation in the Arabidopsis DCL1 gene affects miRNA, but not siRNA [30]. However, the actions of HEN1 in miRNA/development and siRNA/S-PTGS can be uncoupled by single-point mutations at different positions in the protein; this finding suggests that they may represent two partially overlapping activities or two distinct activities that have been combined on a single molecule. The fact that the hen1-4 mutant is impaired in miRNA/development and in siRNA/S-PTGS (triggered by sense transgenes), but not in siRNA/IR-PTGS (triggered by hairpin transgenes), also suggests the existence of additional steps in the two former pathways. In animals, it is known that miRNAs derive from short RNA precursors (ca. 70-nt long) that fold into partial dsRNA molecules [4–6]. Conversely, hairpin transgenes triggering IR-PTGS produce long, perfect dsRNA molecules with a loop. Thus, HEN1 could play a role in the stabilization or processing of imperfectly folded dsRNA molecules. How sense transgenes triggering S-PTGS actually produce dsRNA is still not known, but it requires the action of a putative RdRP encoded by the SGS2/SDE1 gene [10, 13]. This enzyme is assumed to synthesize antisense molecules complementary to the sense mRNA transcribed from the transgene. It is therefore reasonable to think that it could elongate partially folded dsRNA molecules to produce long, perfect dsRNA molecules. The HEN1 protein could participate in the initiation of this reaction by stabilizing partially folded dsRNA molecules corresponding to sense transgenes. If HEN1 actually contributes to the stabilization or processing of imperfect dsRNA, its requirement should not be all or nothing, but conversely should depend on the relative stability of the partially folded dsRNA. This hypothesis is consistent with the fact that some miRNAs are not totally eliminated in hen1 mutants [8]. Alternatively, or in addition to this latter role, HEN1 could also play a role in the stabilization of some siRNA or miRNA after processing by a DICER-like enzyme. Indeed, siRNAs produced by the central part of the GUS coding sequence are totally absent in the hen1-4 mutant, whereas siRNAs produced by the 5′ and 3′ parts are only reduced (Figure 2), suggesting that not all siRNA have the same action or stability. The residual siRNA and PTGS activity observed in the hen1-4 mutant could also result from the presence of an active (transcribed) gene besides HEN1 on BAC T13K14 that putatively encodes a protein (At4g20920) sharing 66% identity and 75% similarity with HEN1 (At4g20910). Because there is only 3 kb between HEN1 and this gene, it will be very difficult to obtain a double mutant to see if it is totally impaired in siRNA accumulation and S-PTGS.
Experimental Procedures
Plant Material
L1, L2, 6b4, 306-1, and 2a3 lines as well as sgs2-1, sgs3-1, and ago1-27 mutants used in this study were previously described [13, 15–17]. Details for all genetic analyses, loci introgression, and mutation mapping are available in the Supplemental Data.
Molecular Biology
Proteins were extracted and GUS activity was analyzed as described before [13]. All RNAs (mRNA, siRNA, and miRNA) were extracted and analyzed as described by Mallory et al. [27]. Northern blots were quantified by using a phosphorimager apparatus. Real-time PCR for quantification of the SCL6-III transcripts (At3g60630) was done as follows: PolydT cDNAs were made by using the Invitrogen cDNA first strand synthesis system, and 5 μg total RNA was extracted with Tri-reagent (MRC). Quantifications were performed on a Biorad IQcycler apparatus with the Quantitech SYBR green kit (Qiagen) upon recommendations of the manufacturer. PCR was carried out in 96-well optical reaction plates heated to 95°C for 10 min to activate hotstartTaq DNA polymerase, followed by 50 cycles of denaturation at 95°C for 30 s and annealing-extension at 60°C for 45 s. Target quantifications were performed with specific primer pairs designed for each side of the cleavage site by using Beacon Designer from Biosoft. The primers used for SCL6-III (At3g60630) are 5′-ACCAAGACCAGTCAGCGGTAATC-3′ and 5′-AGTGTCGTCGTTGTTGTTGTTAAGG-3′. Results are normalized with actine2 (At3g18780) by using 5′-GCACCCTGTTCTTCTTACCG-3′ and 5′-AACCCTCGTAGATTGGCACA-3′. Each quantification was made in triplicate. For each couple of primers, conditions were, as recommended, 1≥E≥0.85 and r2≥0.985, where E is the PCR efficiency and r2 corresponds to the correlation coefficient obtained with the standard curve.
Supplementary Material
Acknowledgments
We thank Allison Mallory and Vicki Vance for helpful advice on small RNA analyses and Jean-Marie Pollien and Hervé Ferry for taking care of the plants. This work was partly supported by a grant from the European Union Biotechnology Program (B104-CT96-0253) to H.V. and M.F., a grant from Rhobio to H.V. and J.-B.M., a PhD fellowship from the French Minister of Research to F.V., and a grant from the National Institutes of Health (1 R01 GM61146-01) to X.C.
Footnotes
Supplemental Data
Supplemental Data including S-PTGS and IR-PTGS activity in the hen1-4 mutant as well as the mapping and restoration of the hen1-4 mutation are available at http://images.cellpress.com/supmat/supmatin.htm.
References
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Plasterk RH. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 7.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 11.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 12.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 14.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 17.Elmayan T, Balzergue S, Beon F, Bourdon V, Daubremet J, Guenet Y, Mourrain P, Palauqui JC, Vernhettes S, Vialle T, et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 20.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 21.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:1–20. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 22.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 23.Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 24.Marathe R, Smith TH, Anandalakshmi R, Bowman LH, Fagard M, Mourrain P, Vaucheret H, Vance VB. Plant viral suppressors of post-transcriptional silencing do not suppress transcriptional silencing. Plant J. 2000;22:51–59. doi: 10.1046/j.1365-313x.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 25.Mette MF, Matzke AJ, Matzke MA. Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr Biol. 2001;11:1119–1123. doi: 10.1016/s0960-9822(01)00315-3. [DOI] [PubMed] [Google Scholar]
- 26.Beclin C, Berthome R, Palauqui JC, Tepfer M, Vaucheret H. Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology. 1998;252:313–317. doi: 10.1006/viro.1998.9457. [DOI] [PubMed] [Google Scholar]
- 27.Mallory AC, Reinhart BJ, Bartel DP, Bowman L, Vance VB. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA. 2002;99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finnegan EJ, Margis R, Waterhouse PM. Post-transcriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr Biol. 2003;13:236–240. doi: 10.1016/s0960-9822(03)00010-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


