SUMMARY
Four classes of floral homeotic MADS domain proteins specify the identities of the four organ types in an Arabidopsis flower. While the activities of the MADS domain proteins are essentially confined to the flower or to the inflorescence, several genes, such as APETALA2, HUA1 and HUA2, also act outside the flower in addition to their organ identity functions inside the flower. We identified a new gene, HUA ENHANCER 1 (HEN1) from a sensitized genetic screen in the hua1-1 hua2-1 background that is compromised in floral homeotic C function. We showed that HEN1, like the C function gene AGAMOUS, acts to specify reproductive organ identities and to repress A function. HEN1 also shares AG’s non-homeotic function in controlling floral determinacy. HEN1 may achieve these functions by regulating the expression of AG. hen1 single mutants exhibit pleiotropic phenotypes such as reduced organ size, altered rosette leaf shape and increased number of coflorescences, during most stages of development. Therefore, HEN1, like the A function gene AP2, plays multiple roles in plant development as well as acting in organ identity specification in the flower. HEN1 codes for a novel protein and is expressed throughout the plant.
Keywords: Flower development, Organ identity, C function, HUA1, HUA2, HEN1, AGAMOUS
INTRODUCTION
Three classes of floral homeotic genes, known as the A, B and C genes, act in overlapping domains in the floral meristem to specify the identities of the four floral organ types (Coen and Meyerowitz, 1991; Meyerowitz et al., 1991). While the A, B and C genes each act in two adjacent whorls, the SEPALLATA (SEP) 1, 2 and 3 genes (also referred to as the class E genes) act in whorls 2, 3 and 4 to specify petal, stamen and carpel identities together with the A, B and C genes (Pelaz et al., 2000; Theißen and Saedler, 2001).
With the exception of the A function gene APETALA2 (AP2), the A, B, C and E genes code for MADS domain DNA-binding proteins (Yanofsky et al., 1990; Ma et al., 1991; Jack et al., 1992; Mandel et al., 1992; Shiraishi et al., 1993; Huang et al., 1993; Goto and Meyerowitz, 1994; Jofuku et al., 1994; Riechmann et al., 1996; Mandrel and Yanofsky, 1998) that likely act as transcription factors in multimeric complexes (Gutierrez-Cortines and Davies, 2000; Honma and Goto, 2001). The expression of the floral homeotic MADS box genes is largely restricted to the flower and to the specific floral whorls where they function (Yanofsky et al., 1990; Drews et al., 1991; Ma et al., 1991; Jack et al., 1992; Mandrel et al., 1992; Goto and Meyerowitz, 1994; Mandrel and Yanofsky, 1998). In fact, ectopic expression of the floral homeotic MADS box genes in leaves can convert leaves to floral organs (Honma and Goto, 2001; Pelaz et al., 2001), suggesting that the A, B, C and E MADS box genes constitute most, if not all, of the flower-specific regulators of floral organ identities. However, genes that play pleiotropic roles in the development of the plant may also serve as regulators of floral organ identities. One such example is the A function gene AP2 (Jofuku et al., 1994; Okamuro et al., 1997).
AG is essential for stamen and carpel identities and floral determinacy. AG also antagonizes A function by restricting the activities of AP2 and the expression of APETALA1 (AP1) to the outer two floral whorls (Bowman et al., 1991b; Gustafson-Brown et al., 1994). AG controls floral determinacy by repressing the expression of WUSCHEL, a gene that specifies stem cell fates, in the center of the flower (Schoof et al., 2000; Lenhard et al., 2001; Lohmann et al., 2001). How AG specifies stamen and carpel identities at the molecular level is less well understood. Since AG likely functions in complexes involving the B and E proteins, the B, C and E MADS domain proteins probably act at the same developmental hierarchy in the floral homeotic pathways.
Several other genes that act in the specification of carpel and/or stamen identities have been identified. CRABS CLAW (CRC), encoding a protein with zinc finger and helix-loop-helix domains, and SPATULA (SPT), encoding a basic helix-loop-helix protein, control aspects of gynoecium development (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Heisler et al., 2001). Either crc-1 or spt-2 reduces the carpelloidy present in the first whorl organs of ap2-2 ag-1 flowers, suggesting that the two genes also function in carpel identity specification (Alvarez and Smyth, 1999). Two other genes, HUA1 and HUA2, were found to control stamen and carpel identities, antagonize A function, and specify floral determinacy (Chen and Meyerowitz, 1999). These functions of HUA1 and HUA2 were only recognized in genetic backgrounds in which AG activities are compromised, such as in the weak ag-4 allele (Sieburth et al., 1995) and in the ag-1/+ background. HUA2 codes for a novel protein (Chen and Meyerowitz, 1999) that has transcription activation activity (unpublished results), and HUA1 codes for a nuclear RNA-binding protein with CCCH zinc fingers (Li et al., 2001). The molecular relationship among HUA1, HUA2 and AG has yet to be determined.
In order to identify additional players that specify reproductive organ identities and/or control floral determinacy, we performed a genetic screen in the hua1-1 hua2-1 double mutant background. Two recessive mutations in the HUA ENHANCER 1 (HEN1) locus strongly enhance the hua1-1 hua2-1 homeotic phenotypes in the inner two whorls. The hen1 mutations also convert the determinate hua1-1 hua2-1 floral meristems to indeterminate meristems, suggesting that HEN1 also controls floral determinacy. The hen1 mutant plants have shorter stems and smaller leaves that differ in shape from those of wild type. Therefore, HEN1 is a gene that plays multiple roles in plant development.
MATERIALS AND METHODS
Plant strains and EMS mutagenesis
The strains used in this study are all in the Landsberg erecta (Ler) background, except that wild-type Columbia plants were used for mapping. ag-4 (Sieburth et al., 1995), ap1-1 (Irish and Sussex, 1990; Bowman et al., 1993), ap2-2 (Bowman et al., 1991b), hua1-1 (Chen and Meyerowitz, 1999) and hua2-1 (Chen and Meyerowitz, 1999) have been characterized previously. hen1-1 and hen1-2 were isolated in this study.
An EMS mutagenesis screen was carried out in the hua1-1 hua2-1 background. 10,000 hua1-1 hua2-1 seeds were treated with 0.2% EMS/0.01% Tween 20 for 12 hours, and then planted onto soil. M2 seeds were collected from single M1 plants. Among 1551 M2 families screened for floral homeotic phenotypes in the inner two whorls, two lines were isolated that were later found to contain mutant alleles at the HEN1 locus. hen1-1 was backcrossed to Ler three times before further genetic analysis while hen1-2 was backcrossed twice.
Complementation tests and construction of mutant combinations
The hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 plants show complete male sterility and extremely reduced female fertility. For complementation tests, we first crossed each triple mutant to hua1-1 hua2-1 to obtain hua1-1 hua2-1 hen1-1/+ and hua1-1 hua2-1 hen1-2/+ plants. Plants of these two genotypes were then crossed to each other. Among 29 F1 plants from this cross, 7 showed the triple mutant floral and vegetative phenotypes, suggesting that the two mutations are allelic.
The hua1-1 hua2-1 hen1-1 ap1-1 mutant was generated by crossing hua1-1 hua2-1 hen1-1/+ plants to hua1-1 hua2-1 ap1-1 plants. In the F2 population, plants that were homozygous for hen1-1 were first identified by their vegetative phenotype and examined for the presence of ap1-1-like floral phenotypes (the presence of flowers at the base of the first whorl organs and lack of second whorl petals) to identify the quadruple mutant. Similarly, ap2-2 was introduced into the hua1-1 hua2-1 hen1-1 background. In this case, a total of 336 F2 plants were screened to obtain three quadruple mutants because HEN1 and AP2 are linked by approximately 20 cM.
Map-based cloning of HEN1
hua1-1 hua2-1 hen1-1/+ plants were crossed to wild-type plants of the Columbia ecotype. F2 seeds were harvested from single F1 plants. The F2 families that segregated hen1-1 were identified by the presence of the hen1-1 vegetative and the hua1-1 hua2-1 hen1-1 floral phenotypes. In these F2 families, 796 F2 plants that were homozygous for hen1-1 were identified with the vegetative phenotypes. Genomic DNA was isolated from each of the 796 hen1-1 plants. Initial mapping with 40 such plants using simple sequence length polymorphism (SSLP) and cleaved amplified polymorphic sequences (CAPS) markers located HEN1 to chromosome IV. Additional CAPS markers based on polymorphisms between Ler and Col provided by Cereon allowed the mapping of HEN1 to a single BAC. Information on these markers can be obtained at the Chen lab web site (http://waksman.rutgers.edu/~xuemei).
HEN1 cDNA and genomic clones
The HEN1 full-length cDNA sequence from the WS ecotype was already deposited into GenBank under the name GENEY in a study that was focused on its neighboring gene, POLLENLESS3 (Sanders et al., 1999). We screened the Weigel floral cDNA library and obtained a partial HEN1 cDNA from the Ler ecotype. The 5′missing portion of the Ler cDNA was obtained by reverse transcription reactions followed by polymerase chain reactions (RT-PCR). The sequence of the HEN1 cDNA from Ler agrees with the published sequence of GENEY except for three nucleotides, which are likely due to ecotype polymorphisms. The HEN1 cDNA from Ler is in GenBank under the accession number AF411383.
For complementation tests, the HEN1 genomic region was amplified from the BAC T13K14 with Ex-Taq (Panvera) using primers HEN1p10 (5′ KpnI-tcatggattcgtggtatagcgttactt 3′) and HEN1p11 (5′ KpnI-gccctcgtgaaaaagatcaagaacgc 3′) and cloned into pPZP211 (Hajdukiewicz et al., 1994) to result in pPZP211-HEN1p10/p11. This clone contains the entire HEN1 genomic region with 1847 bp upstream of the initiation codon but excludes the neighboring genes on either side. This clone was used to transform hen1-1 and hua1-1 hen1-1 hua2-1/+ plants using vacuum infiltration (Bechtold et al., 1993).
Scanning electron microscopy
Tissues were fixed and critical point dried according to the method of Bowman et al. (Bowman et al., 1991b). Images were captured with an AMRAY-1830 I microscope.
RNA filter and in situ hybridization
Total and poly(A)+ RNA isolation was carried out as described previously (Chen and Meyerowitz, 1999). For RNA filter hybridization, approximately 1 µg of poly(A)+ RNA was used for each sample. A 978 bp region of the HEN1 cDNA was amplified by PCR with T13K14p21 (5′-TCA GGA TCC ACT GCC AAAGA-3′) and T13K14 p22 (5′-GTT TGG CAA AGC TTC CTGTG-3′) and used as the HEN1 probe.
In situ hybridization with radioactive probes was carried out as described previously (Chen and Meyerowitz, 1999). In situ hybridization with digoxigenin (DIG)-labeled probes was carried out according to the protocol described at www.wisc.edu/genetics/CATG/barton/protocols.html. The plasmids used for AG, AP1, AP3, PI, SEP1, SEP2 and SEP3 probes are pCIT565 (Yanofsky et al., 1990), pKY89 (Gustafson-Brown et al., 1994), pD793 (Jack et al., 1992), pcPINX (Goto and Meyerowitz, 1994), pCIT4221 (Ma et al., 1991), pSR12 (Savidge et al., 1995) and pAGL9 (Mandrel and Yanofsky, 1998), respectively. For HEN1, the 978 bp T13K14p21/p22 fragment was amplified with Pwo polymerase and cloned into pCR-BluntII-TOPO (Invitrogen) in two orientations to result in pTOPO-cHEN1S and pTOPO-cHEN1A. pTOPO-cHEN1S and pTOPO-cHEN1A were digested with BamHI and HindIII, respectively, and transcribed with T7 polymerase for generating the sense and antisense probes.
RESULTS
hen1 single mutant phenotypes
The hua1-1 hua2-1 double mutant exhibits vegetative as well as floral phenotypes: slightly smaller leaves, shorter stems and flowers with weak carpel-to-sepal transformation (Chen and Meyerowitz, 1999; see below). An EMS genetic screen was carried out to search for mutations that enhance the hua1-1 hua2-1 floral phenotypes. Among 1551 M2 families screened, two recessive hen1 alleles, hen1-1 and hen1-2, were isolated. While the hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers showed severe loss-of-C-function phenotypes (see below), the triple mutant plants were also much smaller than the hua1-1 hua2-1 double mutant (data not shown), suggesting that HEN1 functions in vegetative development. In fact, while hen1-1 and hen1-2 single mutants lack obvious floral homeotic phenotypes, several non-homeotic defects are clearly visible.
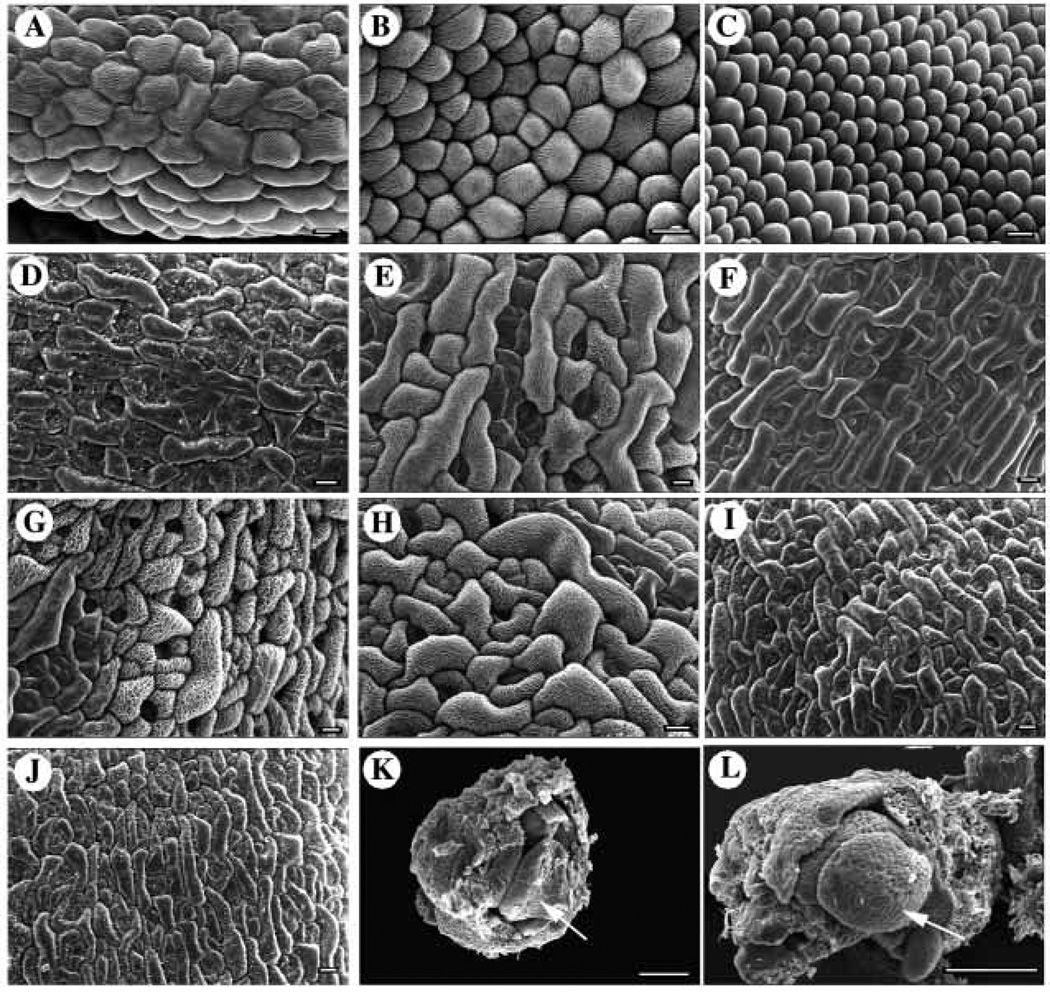
hen1-1 and hen1-2 plants are smaller in stature compared to wild-type plants. Most aerial organs in hen1 mutants are reduced in size: leaves and floral organs are smaller (Fig. 1A–C, E), and stems are shorter (data not shown). The hen1 (throughout the paper, both alleles are referred to when allele numbers are not specified) cotyledons, however, are similar in size to those of wild type (data not shown). In addition to reduced size, hen1 leaves are shaped differently: they are more pointed at the apical end (Fig. 1B,C) and the edges tend to curl upward (Fig. 1B). Flower density also appears to be much higher in hen1 inflorescences (Fig. 1F,G). However, this could be a secondary effect of reduced internode elongation in hen1 stems. Scanning electron microscopy (SEM) revealed that cells in mature hen1-1 leaves and petals are smaller than those in mature Ler leaves and petals (Fig. 1H–M).
Fig. 1.
hen1-1 and hen1-2 single mutant phenotypes. (A–C) Rosettes of (A) a Ler, (B) a hen1-1 and (C) a hen1-2 plant, around the time of bolting. Rosette leaves in hen1-1 and hen1-2 tend to be pointed (arrows), and the edges tend to curl up slightly (arrowhead). (D) A hen1-1 plant containing the HEN1p10/p11 transgene. (E) A Ler (left) and a hen1-1 (right) mature flower at the same magnification. (F) A Ler and (G) a hen1-1 inflorescence. (H–J) Scanning electron micrographs of (H) a Ler, (I) a hen1-1 and (J) a hen1-2 mature leaf. (K–M) Scanning electron micrographs of (K) a Ler, (L) a hen1-1 and (M) a hen1-2 mature petal. Scale bars, 100 µm in H–J, and 10 µm in K–M.
hen1-1 and hen1-2 single mutants exhibit delayed transition to producing flowers on the inflorescence stems (Table 1). Under our long-day conditions (16 hour light/8 hour darkness), both hen1-1 and hen1-2 plants produce more cauline leaves with axillary inflorescences before producing single flowers on the inflorescence stems than Ler plants, with hen1-1 being stronger than hen1-2. The two mutations, however, appear to affect rosette leaf number in opposite ways: hen1-1 plants having a slight increase and hen1-2 plants having a decrease in rosette leaf number (Table 1).
Table 1.
Rosette and cauline leaf numbers in various genotypes under long-day conditions
| Genotype | Rosette leaf | Cauline leaf | n |
|---|---|---|---|
| Ler | 7.0±0.7 | 2.8±0.4 | 40 |
| hen1-1 | 9.7±1.5 | 7.1±1.2 | 40 |
| hen1-2 | 4.8±0.7 | 3.5±0.7 | 80 |
| hen1-1 T2* | 10.2±0.9 | 3.9±0.6 | 20 |
Values are mean±standard deviation.
T2 transgenic plants that are homozygous for hen1-1 and either homozygous or hemizygous for the HEN1p10/p11 transgene.
hen1-1 and hen1-2 single mutants also exhibit reduced fertility. Male and female fertility both appear to be affected. Anthers from early-arising hen1-1 and hen1-2 flowers often fail to dehisce. Although late-arising flowers do produce pollen, the amount of pollen produced appears to be greatly reduced. In addition, when the stigma is pollinated with wild-type pollen, the siliques elongate to the same extent as they would after self-pollination: mature hen1 siliques reach approximately 1/4–1/2 of the length of a wild-type silique (data not shown). Seed set from hen1 siliques is accordingly reduced (data not shown).
hen1 mutations enhance the hua1-1 hua2-1 homeotic phenotypes
While hua1-1 hua2-1 flowers have stamens in the third whorl (Fig. 2A), all flowers of hua1-1 hua2-1 hen1-1 plants have primarily petals in the third whorl (Fig. 2B,F). Some of the medial third whorl organs occasionally exhibit stamen characteristics, i.e., structures that resemble anther thecae are present (data not shown). hua1-1 hua2-1 hen1-2 flowers have petals and frequently staminoid petals (organs with white pigmentation but containing anther thecae-like structures) in the third whorl (Fig. 2C), suggesting that hen1-2 is weaker than hen1-1. In most hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers, the fourth whorl gynoecia resemble those in hua1-1 hua2-1 flowers except that the stigmatic papillae usually consist of two or more lobes (Fig. 2B,C). In late-arising flowers, though, the fourth whorl gynoecia can have greatly enlarged ovaries on top of gynophores (Fig. 2D). Inside these ovaries, additional floral organs are present (Fig. 2E). Occasionally, late-arising hua1-1 hua2-1 hen1-1 flowers resemble those of ag-3 in that the identities of the two reproductive whorls are completely lost and internal flowers are found in the center (Fig. 2F).
Fig. 2.
Floral phenotypes. (A) A hua1-1 hua2-1, (B) a hua1-1 hua2-1 hen1-1, and (C) a hua1-1 hua2-1 hen1-2 flower. (D) A late-arising hua1-1 hua2-1 hen1-2 flower. The gynoecium is enlarged with a gynophore at the base (arrowhead). (E) A late-arising hua1-1 hua2-1 hen1-2 flower with additional floral organs (arrowhead) inside the carpels. (F) A late-arising hua1-1 hua2-1 hen1-1 flower with flowers (arrowhead) in place of carpels. (G) A hua1-1 hen1-1 and (H) a hua2-1 hen1-1 flower. (I) A flower of a hua1-1 hua2-1 hen1-1 plant containing the HEN1p10/p11 transgene. Arrows, third whorl organs. Scale bars, 1 mm.
The homeotic transformation in the third whorl and the defect in floral determinacy in late flowers are manifested only in hua1-1 hua2-1 hen1 triple mutant plants. Like hua1-1 hua2-1 flowers, hua1-1 hen1 and hua2-1 hen1 flowers have stamens and carpels in the inner two whorls (Fig. 2G,H, and data not shown). The hen1-1 or hen1-2 single mutant flowers also show normal identities in the four floral whorls (data not shown).
Homeotic transformation at the cellular level
The homeotic transformation in the third whorl of hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers was further confirmed by SEM. Cells that are shaped like pieces of a jigsaw puzzle are found on the surface of wild-type (Smyth et al., 1990) and hua1-1 hua2-1 third whorl stamens (Fig. 3A). The adaxial and abaxial surfaces of wild-type petals consist of small cone-shaped and cobble stone-like cells, respectively. The epidermal cells of hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 third whorl organs resemble petal cells (Fig. 3B,C). Furthermore, a study of ontogeny revealed that the homeotic transformation in the third whorl of hua1-1 hua2-1 hen1-1 flowers can be observed as early as stages 7–8, when the stamens assume the first signs of differentiation (Smyth et al., 1990). In stages 7–8 flowers of the hua1-1 hua2-1 genotype, the third whorl organs become spade-shaped, showing signs of differentiation into the anther and the filament (Fig. 3K). In hua1-1 hua2-1 hen1-1 flowers of similar stages, however, the third whorl organs are flat, resembling perianth organs in shape (Fig. 3L). In wild-type flowers, the third whorl stamen primordia are initiated earlier than the second whorl petal primordia (Smyth et al., 1990). The third whorl petals develops sooner than the second whorl petals in hua1-1 hua2-1 hen1-1 flowers (data not shown), suggesting that the third whorl petals still behave as third whorl organs in timing of organ initiation, despite the alteration in identity.
Fig. 3.
Scanning electron microscope images of Ler, hen1-1, hua1-1 hua2-1, hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers and/or floral organs. (A–C) The adaxial surface of the third whorl organs from (A) hua1-1 hua2-1, (B) hua1-1 hua2-1 hen1-1 and (C) hua1-1 hua2-1 hen1-2 flowers. Petal-type cells are found in the third whorl organs in the two triple mutants (B,C). (D) The top portion of a Ler ovary, showing valve cells that lack cuticular thickenings. (E,F) The tip (E) and the bottom (F) of a hua1-1 hua2-1 ovary. Some valve cells at the tip of the ovary exhibit cuticular striations that mimic sepal cells (E). The bottom of the hua1-1 hua2-1 ovary is similar to that in Ler in terms of cell surface characteristics. (G,H) The bottom portion of ovaries from (G) a hua1-1 hua2-1 hen1-1 and (H) a hua1-1 hua2-1 hen1-2 flower. The cells resemble sepal cells. (I,J) Top portions of the ovaries from (I) a hua1-1 hen1-1 and (J) a hua2-1 hen1-1 flower. The cells appear wild type. (K,L) Stage 7–8 flowers of (K) hua1-1 hua2-1 and (L) hua1-1 hua2-1 hen1-1 genotypes, with sepals dissected to reveal the third whorl organs (arrows) that differ in shapes. Scale bars, 10 µm except in K and L (100 µm).
Homeotic transformation in the fourth whorl of hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers was also observed with SEM. The gynoecium can be subdivided into the stigma, the style and the ovary along the longitudinal axis. Epidermal cells characteristic of the stigma and style are found at the expected positions along the gynoecia of hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 flowers (data not shown). Ovary epidermal cells in the two triple mutants, however, differ from those in wild-type and hua1-1 hua2-1 flowers. Epidermal cells along the wild-type ovary lack epicuticular striations (Fig. 3D and data not shown). While cells on the bottom half of the ovary or those along the medial edges of the top half of the ovary in the hua1-1 hua2-1 gynoecium are normal (Fig. 3F), cells in the lateral portions of the top half of the ovary (coincident with the bulge in the hua1-1 hua2-1 gynoecium) show epicuticular thickenings that resemble sepal cells (Fig. 3E). Large cells that are present in abaxial sepals are also found (Fig. 3E), suggesting a carpel-to-sepal transformation in the upper lateral positions in the hua1-1 hua2-1 ovary. This partial carpel-to-sepal transformation in hua1-1 hua2-1 ovaries is enhanced by hen1-1 and hen1-2. In hua1-1 hua2-1 hen1-1 and hua1-1 hua2-1 hen1-2 gynoecia, epidermal cells all over the ovaries exhibit epicuticular thickenings (Fig. 3G,H). The ovary cell types appear normal in hua1-1 hen1, hua2-1 hen1 and hen1 gynoecia (Fig. 3I,J, and data not shown).
Expression of floral homeotic genes in hua1-1 hua2-1 hen1-1 mutants
To begin to understand the molecular basis of the homeotic phenotypes in the triple mutants, we studied the expression of the A, B, C and E genes in the mutants. While the expression of classe B (APETALA3 and PISTILLATA) and class E genes (SEP1, 2, 3) was not detectably different in hua1-1 hua2-1 hen1, hua1-1 hua2-1 and wild-type flowers (data not shown) (Jack et al., 1992; Flanagan and Ma, 1994; Goto and Meyerowitz, 1994; Savidge et al., 1995; Mandrel and Yanofsky, 1998), significant difference was detected for classes A and C gene expression between hua1-1 hua2-1 hen1 and hua1-1 hua2-1.
Class A: AP1
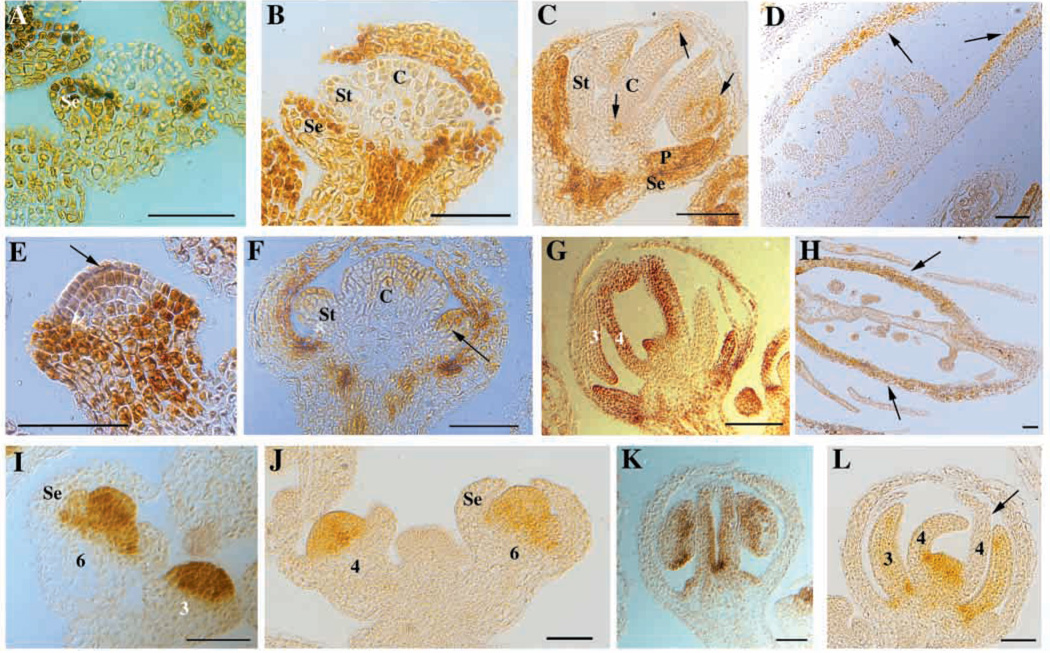
AP1 is expressed throughout stages 1–2 flower primordia and becomes restricted to the outer two whorls starting at stage 3, when AG is expressed in the inner two whorls and acts to repress AP1 expression (Mandrel et al., 1992; Gustafson-Brown et al., 1994). In hua1-1 hua2-1 flowers, AP1 expression patterns are similar to those in wild type in stages 1-6 flowers (Chen and Meyerowitz, 1999) (Fig. 4A,B). After stage 6, low levels of AP1 RNA were detected in a patchy pattern in stamens and carpels (Fig. 4C,D) (Chen and Meyerowitz, 1999). The levels of ectopic AP1 expression were consistently lower than those found in the outer two whorls of hua1-1 hua2-1 flowers (Fig. 4C and data not shown). In hua1-1 hua2-1 hen1-1 flowers, ectopic AP1 expression in the inner two whorls appeared more precocious and extensive. In some stage 3 flowers, the center of the floral meristem expressed low levels of AP1 RNA (Fig. 4E). Most stages 6–7 flowers contained AP1 RNA in stamens and carpels (Fig. 4F). By stages 9–10, AP1 RNA was detected in the fourth whorl gynoecia at levels comparable to those in the outer two whorls (Fig. 4G). In mature flowers, all cells in the ovary walls expressed AP1 RNA, whereas only some cells in hua1-1 hua2-1 ovaries expressed AP1 (Fig. 4H,D).
Fig. 4.
AP1 and AG RNA accumulation patterns in hua1-1 hua2-1 and hua1-1 hua2-1 hen1-1 flowers. (A–H) AP1 RNA localization in hua1-1 hua2-1 (A–D) and hua1-1 hua2-1 hen1-1 (E–H) flowers of various stages. (A,E) Stage 3 flowers; (B,F) stage 6–7 flowers; (C,G) stages 9–10 flowers. (D,H) Mature flowers. Arrows indicate ectopic AP1 expression. In hua1-1 hua2-1 hen1-1 flowers, the ectopic AP1 expression is more precocious (arrows in E and F) and extensive (G and H). (I–L) AG RNA localization in (I,K) hua1-1 hua2-1 and (J,L) hua1-1 hua2-1 hen1-1 flowers. (I,J) AG expression in young flowers (stages indicated by numbers). (K,L) Stage 9 flowers. The arrow in L indicates a fourth whorl organ with little AG RNA. Se, sepals; St, stamens; C, carpels. Numbers in G and L indicate floral whorls. Scale bars, 50 µm.
Class C: AG
AG starts to be expressed in the incipient stamen and carpel primordia at stage 3 and continues to be expressed in the two organ types throughout flower development (Bowman et al., 1991a; Drews et al., 1991). This expression pattern is preserved in hua1-1 hua2-1 flowers (Chen and Meyerowitz, 1999) (Fig. 4I,K). In hua1-1 hua2-1 hen1-1 flowers, the initiation and the domain of AG expression were unaltered in stages 1–6 flowers (Fig. 4J). However, AG signals were barely detectable and often absent from some third and fourth whorl organs in stage 7 and older flowers (Fig. 4L and data not shown).
Genetic interactions of hen1-1 with ap1 and ap2 mutations
To investigate if the homeotic transformation in the third whorl of hua1-1 hua2-1 hen1-1 flowers is due to ectopic activities of A function genes, we introduced severe loss-of-function mutations in the A function genes into the hua1-1 hua2-1 hen1-1 background.
hua1-1 hua2-1 hen1-1 ap1-1
In ap1-1 flowers, second whorl petals are missing and first whorl sepals are transformed to leaves with additional flowers in their axils (Bowman et al., 1993; Irish and Sussex, 1990). The ap1-1 mutation is largely additive with the hua1-1 and hua2-1 mutations: the hua1-1 hua2-1 ap1-1 flowers resemble ap1-1 flowers in the outer two whorls and hua1-1 hua2-1 flowers in the fourth whorl, although the bulge in the gynoecium is somewhat reduced (Chen and Meyerowitz, 1999). When introduced into the hua1-1 hua2-1 hen1-1 triple mutant, ap1-1 partially suppresses the stamen-to-petal transformation in the third whorl (compare Fig. 5A and B with C and D, respectively). The third whorl organs in the quadruple mutant resemble stamens in pigmentation and in overall shape, except that the anthers are flattened and enlarged compared to wild-type anthers (Fig. 5D).
Fig. 5.
Third whorl identity and A function genes. (A) a hua1-1 hua2-1 hen1-1 flower, (C) a hua1-1 hua2-1 hen1-1 ap1-1 flower and (E) a hua1-1 hua2-1 hen1-1 ap2-2 flower. (B,D,F) Dissected third whorl organs from the flowers in A, C and E, respectively. Scale bars: 1 mm.
hua1-1 hua2-1 hen1-1 ap2-2
In ap2-2 flowers, the first whorl organs are leaves or are absent in the lateral positions and in the medial positions are carpelloid with stigmatic papillae all along the edges (Bowman et al., 1991b). The carpelloidy of the first whorl organs is largely due to ectopic AG activities (Bowman et al., 1991b; Drews et al., 1991). Additional phenotypes of ap2-2 include suppression of petal development, reduction in stamen number and often unfused carpels (Bowman et al., 1991b). ap2-2 is largely additive with hua1-1 and hua2-1 in the inner three whorls, such that hua1-1 hua2-1 ap2-2 flowers exhibit reduced organ numbers in the second and third whorls like ap2-2 and have bulged gynoecia like hua1-1 hua2-1 (Chen and Meyerowitz, 1999). The first whorl organs in hua1-1 hua2-1 ap2-2 flowers have greatly reduced carpelloidy: stigmatic papillae are much reduced in number, suggesting that the HUA genes play a role in carpel identity specification (Chen and Meyerowitz, 1999).
Flowers of the hua1-1 hua2-1 hen1-1 ap2-2 genotype resemble ap2-2 flowers in overall morphology (Fig. 5E). Petals are missing, stamen number is reduced (Fig. 5E), and carpels are often unfused (data not shown). The first whorl organs resembled those in hua1-1 hua2-1 ap2-2, in that the number of stigmatic papillae is greatly reduced compared to that in ap2-2 (Fig. 5E and data not shown). The most noticeable phenotype of the quadruple mutant is that the third whorl organs resemble stamens (Fig. 5F) instead of petals, which are found in hua1-1 hua2-1 hen1-1 flowers (Fig. 5A,B). However, the third whorl stamens in the quadruple mutant are not completely wild type: the anthers often have reduced number of thecae and are green on the abaxial side (Fig. 5F and data not shown).
Cloning of HEN1
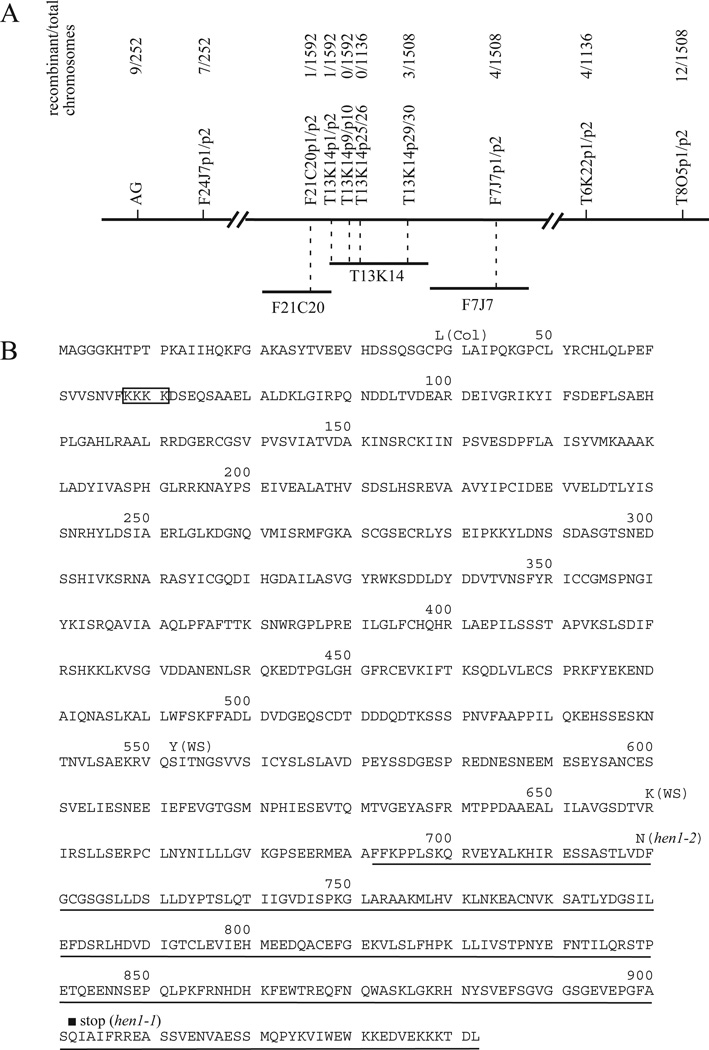
A map-based strategy was used to clone HEN1. HEN1 was first mapped to a few centimorgans south of AG on chromosome IV (Fig. 6A). Detailed mapping with CAPS markers developed from the Cereon SNPs localized HEN1 to a single BAC, T13K14. Three genes from this BAC were sequenced from hen1-1. A C-to-T mutation was identified in one of the genes, T13K14.80. This mutation would cause a premature stop codon in the predicted protein sequence (Fig. 6B). This gene was then sequenced from hen1-2, and a G-to-A mutation was found, which would cause a D-to-N amino acid substitution in the open reading frame (Fig. 6B).
Fig. 6.
Cloning of HEN1. (A) Mapping of HEN1 to the BAC T13K14 on chromosome IV. The HEN1 genomic region is represented by the long horizontal line. The markers used for mapping are shown above the line with the numbers of recombination break points between HEN1 and those markers shown. (B) The HEN1 protein sequence from Ler. HEN1 codes for a novel protein with a C-terminal domain (underlined) showing similarity to yeast, C. elegans and Drosophila proteins. The hen1-1 and hen1-2 mutations cause truncation and amino acid substitution, respectively, in the C-terminal domain. A potential nuclear localization signal (NLS) is boxed. The amino acids from Col and WS that differ from those in Ler are indicated above the sequence.
A genomic fragment, HEN1p10/p11, which contained the entire T13K14.80 gene but excluded neighboring genes, was introduced into hen1-1. In eight of the nine T1 transgenic lines, the hen1-1 defects in leaf size, leaf shape, length of the stem, floral organ size, and fertility were fully rescued (Fig. 1D; data not shown). The increased number of cauline leaves in hen1-1 was largely rescued, but the increase in rosette leaf number was not (Table 1). Analyses of T2 populations showed that the T-DNA was responsible for the rescue (data not shown). The same genomic clone was also used to transform hua1-1 hen1-1 hua2-1/+ plants (the hua1-1 and hua2-1 genotypes were determined with molecular markers). Among 7 T1 transformants, only one was homozygous for hua2-1. This plant exhibited floral phenotypes similar to those in hua1-1 hua2-1 (Fig. 2I), suggesting that the clone also rescued the floral homeotic phenotype of the hua1-1 hua2-1 hen1-1 triple mutant. Therefore, T13K14.80 is HEN1.
A cDNA corresponding to T13K14.80 from the WS ecotype was published in a study on the function of its neighboring gene, POLLENLESS3 (Sanders et al., 1999). We obtained and sequenced the full-length HEN1 cDNA from Ler. The predicted HEN1 protein sequences from Ler and WS are nearly identical, with only conservative changes at two amino acids (Fig. 6B). The annotated T13K14.80 sequence from Col differs significantly from the Ler and WS sequences in a small region (between amino acids 87 and 117). Sequencing of this region in a Col HEN1 cDNA confirmed that the Col sequence is identical to those from Ler and WS. Therefore, the difference was the result of mistakes in annotation.
The predicted HEN1 protein does not contain any motifs of known function except for a putative nuclear localization signal (Fig. 6B). One putative protein in the Arabidopsis genome shows approximately 50% amino acid identity to HEN1. In fact, this putative gene resides right next to HEN1: the initiation codon of HEN1 and the putative stop codon of this gene are approximately 2.1 kb apart. A transcript from this gene was detected in Col inflorescences by RT-PCR (J. L. and X. C., unpublished result). BLAST searches with the HEN1 protein sequence revealed a region in the HEN1 protein (underlined in Fig. 6B) that shows similarities to predicted proteins from Caenorhabditis elegans, Drosophila melanogaster, Schizosaccharomyces pombe and Streptomyces coelicolor. Both hen1 mutations affect this region of the HEN1 protein.
Expression of HEN1
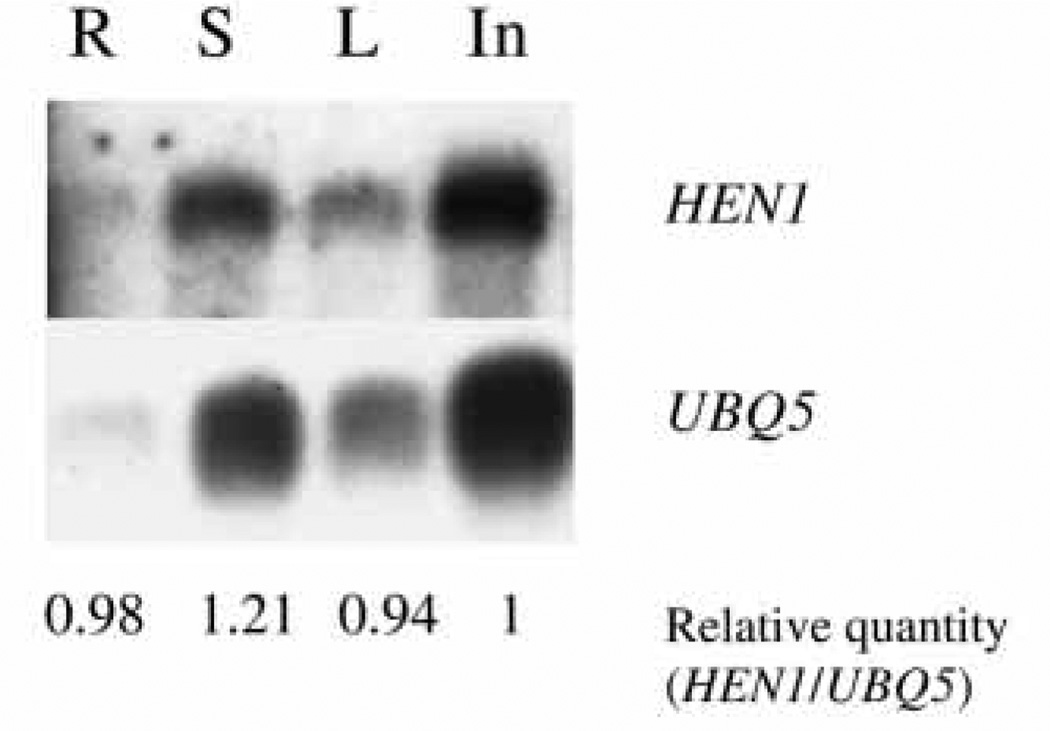
RNA filter hybridization with HEN1 as the probe detected a single RNA species of the expected size in roots, leaves, stems and inflorescences (Fig. 7). The abundance of HEN1 RNA is comparable among the different organs (Fig. 7).
Fig. 7.
HEN1 RNA accumulation. HEN1 RNA can be detected in roots (R), stems (S), leaves (L) and inflorescences (In). UBQ5 was used as a loading control. The relative abundance of HEN1 RNA in these tissues was calculated based on quantification with a phosphorimager.
In situ hybridization with radioactively labeled HEN1 probes showed that HEN1 RNA is present in all cells of the inflorescence meristem, the inflorescence stem, and flowers of all stages (data not shown).
DISCUSSION
HEN1 plays multiple roles in plant development
In addition to causing homeotic transformation in the hua1-1 hua2-1 background, the two recessive mutations in HEN1 result in similar and pleiotropic phenotypes during most stages of Arabidopsis development in the HUA1 HUA2 background. The earliest detectable difference between wild-type and hen1 plants occurs at the end of the 2-leaf stage, when the first pair of hen1 true leaves do not reach the size of their wild-type counterparts. From this point on, HEN1 appears to be required for normal growth and development. HEN1 performs three major functions as revealed by the hen1-1 and hen1-2 single mutant phenotypes.
First, HEN1 is required for most organs, such as leaves, stems, and all four floral organs to achieve their normal size. In leaves and petals, the smaller size in hen1-1 and hen1-2 appears to be at least partially due to smaller cells. Therefore, HEN1 promotes cell expansion. The size of cotyledons appears unaffected by hen1 mutations.
Second, HEN1 is required for male and female fertility. This is evident because pollen production in hen1 mutants is greatly reduced and they exhibit reduced silique elongation and seed set after pollination with wild-type pollen. It remains to be determined what processes in gametophyte development and reproduction are affected by hen1 mutations.
Third, HEN1 promotes the transition to flower production on the inflorescence stem. Similar to leafy mutants (Weigel et al., 1992), hen1 mutants make more secondary inflorescences subtended by cauline leaves before producing single flowers on the inflorescence stem. Therefore, like LEAFY, HEN1 instructs lateral meristems produced from the inflorescence meristem of their floral fate. HEN1 plays a minor role in floral meristem identity compared to LEAFY, since the hen1-1 phenotype is much weaker than that of leafy null mutants. However, hen1-1 may not be a null allele and therefore the hen1-1 phenotypes may not fully reflect the function of HEN1.
While most aspects of the vegetative and floral phenotypes caused by hen1-1 and hen1-2 are consistent, with hen1-1 being consistently slightly stronger than hen1-2, the two mutants differ in the number of rosette leaves. hen1-2 plants have fewer rosette leaves while hen1-1 plants have more rosette leaves than wild type. The hen1-1 rosette leaf number phenotype, however, was not rescued by the HEN1 genomic clone, suggesting that this phenotype may be due to another mutation still present despite the three backcrosses. If the hen1-2 rosette leaf number defect is indeed the real hen1 loss-of-function phenotype, HEN1 plays a role in delaying the transition from vegetative phase 1 (rosette leaf producing stage) to vegetative phase 2 (cauline leaf producing stage). This conclusion can be verified by further backcrosses of hen1-1 to wild type and by transforming hen1-2 with the HEN1 genomic clone to determine if the rosette leaf number phenotype can be rescued.
HEN1 promotes C function and floral determinacy in flower development
HEN1’s role in promoting stamen and carpel identities was revealed only in the hua1-1 hua2-1 background, in which C function is already affected to some extent. The stamen-to-petal transformation in the third whorl of hua1-1 hua2-1 hen flowers is largely due to ectopic AP2 and AP1 activities, suggesting that HEN1 antagonizes A function genes. HEN1 plays a positive role in organ identity specification in addition to its role in repressing A function because the hua1-1 hua2-1 hen1-1 ap2-2 and hua1-1 hua2-1 hen1-1 ap1-1 third whorl organs are not completely stamens. Therefore, HEN1 promotes C function in the flower, which specifies reproductive organ identities and antagonizes A function.
HEN1 plays a minor role in conferring floral determinacy: late-arising hua1-1 hua2-1 hen1 flowers can be indeterminate. A number of genes, such as AG, HUA1, HUA2 and CRC, have been implicated in controlling floral determinacy. AG is essential for floral determinacy, as indicated by the indeterminate nature of ag-2 flowers (ag-2 is a potential null allele) (Bowman et al., 1991b). That HUA1 and HUA2 also play a role in conferring floral determinacy was revealed by the indeterminate nature of ag-1/+ hua1-1 hua2-1 but not ag-1/+ flowers (Chen and Meyerowitz, 1999). Similarly, ag-1/+ crc-1 flowers also exhibit defects in floral determinacy (Alvarez and Smyth, 1999). The fact that ag-1 hua1-1 hua2-1 flowers are nearly identical to ag-1 flowers suggests that ag-1 is epistatic to the hua mutations (Chen and Meyerowitz, 1999). In contrast, mutations in CLAVATA1 and SUPERMAN greatly enhance the floral determinacy defect of ag mutants (Clark et al., 1993; Schultz et al., 1991). These data suggest that HUA1 and HUA2 act in the AG pathway to influence floral determinacy. It is unclear whether CRC act in the AG pathway or in a parallel pathway regarding floral determinacy. Owing to the close linkage between HEN1 and AG, we have not obtained an ag-1 hen1-1 double mutant to assess if hen1-1 enhances the floral determinacy phenotype of ag-1. An ag-4 hen1-1 double mutant, however, has been constructed. hen1-1 does not appear to enhance the floral determinacy defect of ag-4 (X. C. and J. L., unpublished results). Since ag-4 and ag-1 flowers are similar with regard to floral determinacy, it is likely that HEN1 acts in the AG pathway.
Although HEN1 is involved in the three activities known to require AG (specification of reproductive organ identity, repression of A function and control of floral determinacy), hen1 single mutants do not exhibit any phenotypes that reflect these roles. Severe loss-of-function mutations in AG, however, cause complete loss of reproductive organ identity and floral determinacy. If hen1-1 is a null allele, we can conclude that HEN1 plays a minor role in reproductive organ identity and floral determinacy specification or that other genes with similar functions can compensate for the loss of HEN1 function. In fact, in the HEN1 genomic region, there exists another potential gene that is 50% identical to HEN1 at the protein level. This potential gene may be partially redundant with HEN1. However, it is likely that hen1-1 is not a null allele and therefore its phenotypes do not fully reflect the functions of HEN1. A complete loss-of-function mutation in HEN1 is necessary to assess the full spectrum and extent of HEN1’s functions.
HEN1 and AG
The observation that AG RNA is reduced in stage 7 and older flowers of the hua1-1 hua2-1 hen1-1 but not the hua1-1 hua2-1 genotype suggests that HEN1 acts to maintain AG expression during later stages of flower development. This may be the molecular basis for HEN1’s functions in organ identity. In fact, little is known about how AG expression is maintained after its initial activation by LEAFY and WUS (Busch et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001). WUS RNA disappears by stage 6 (Schoof et al., 2000), while LFY expression is greatly reduced in the center of the flower after stage 3 (Weigel et al., 1992). HEN1 is apparently involved in maintaining AG expression after stage 6. Since reduction in AG expression is not detected in hen1 single mutants (data not shown), HEN1 may not be the only gene acting to maintain AG expression.
HEN1 may also function as a floral meristem identity gene because hen1 mutants exhibit delayed transition to making single flowers on the inflorescence stem. We hypothesize that HEN1 plays a minor role in the initial activation of AG early in flower development as well as acting in the maintenance of AG expression after the initial activation. The requirement for HEN1 to activate AG expression in early flowers may somehow increase during plant development, such that floral determinacy is lost in late-arising hua1-1 hua2-1 hen1-1 flowers. This early role of HEN1 was suggested but not unequivocally demonstrated by in situ hybridization experiments in early flowers (Fig. 4I,J). In summary, we propose that the effects of hen1 mutations on reproductive organ identity, repression of A function, and floral determinacy are secondary to their effects on AG expression. However, it is also possible that the loss of AG expression in stages 7 and older flowers in hua1-1 hua2-1 hen1 flowers is secondary to changes in organ identities.
The fact that most hua1-1 hua2-1 hen1 flowers exhibit only organ identity but not determinacy defects is apparently contradictory to the previous hypothesis that floral determinacy is more sensitive to the reduction in AG expression than organ identity (Mizukami and Ma, 1995; Sieburth et al., 1995). However, this may be explained by the relative timing of reduction of AG expression in hua1-1 hua2-1 hen1-1 and the specification of organ identity and floral determinacy. Owing to the negative regulation by AG, WUS RNA disappears from the center of the floral meristem by stage 6 in wild-type plants (Lenhard et al., 2001; Lohmann et al., 2001). Therefore, it is possible that floral determinacy is specified by stage 6, at which point reduction of AG expression in hua1-1 hua2-1 hen1-1 has not commenced or is just about to begin. Conversely, identity specification of the reproductive organs may occur later. Little is known about when AG is needed for stamen and carpel identities, but experiments with the temperature sensitive allele ap3-1 demonstrated that AP3 function is needed till beyond stage 6 to specify stamen identity (Bowman et al., 1989). Similar temperature shift experiments with the Antirrhinum def-101 mutant showed that DEF, an AP3 homologue, is required throughout development to maintain petal and stamen identities (Zachgo et al., 1995).
The HEN1 protein
The HEN1 protein sequence does not indicate any molecular function. The fact that the C-terminal region in HEN1 shows similarity to predicted proteins from other organisms and that the two hen1 mutations both lie in this region suggests that this domain is important for the function of HEN1. The apparently ubiquitous pattern of HEN1 expression and the pleiotropic phenotypes associated with hen1 mutations suggest that HEN1 likely performs a basic molecular function throughout the plant. The specific loss-of-C-function defects in the flower conferred by hen1 mutations in the hua1-1 hua2-1 background indicate that either HEN1 performs a specific function in the flower or that the floral homeotic C function pathway is more sensitive to alterations in the molecular process involving HEN1.
Acknowledgments
We thank Yevgeniy Turovskiy for his assistance in the map-based cloning of HEN1. The nucleotide polymorphisms between Ler and Col discovered by Cereon greatly facilitated the establishment of markers for the map-based cloning of HEN1. We are grateful to Dr Hugo Dooner, Dr Tamara Western, Dr Wenming Wang and Junjie Li for their comments on the manuscript. This work was supported by grants from March of Dimes (5-FY99-760) and National Institutes of Health (1 R01 GM61146-01) to X. C
REFERENCES
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, Sciences de la vie/Life sciences. 1993;316:1194–1199. [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell. 1991a;3:749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991b;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Chen X, Meyerowitz EM. HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Molecular Cell. 1999;3:349–360. doi: 10.1016/s1097-2765(00)80462-1. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 1994;26:581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF. Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell. 1994;76:131–143. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Cortines ME, Davies B. Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci. 2000;5:471–476. doi: 10.1016/s1360-1385(00)01761-1. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;128:1089–1098. doi: 10.1242/dev.128.7.1089. [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the apetala1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Montagu MV, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA-binding protein. Plant Cell. 2001;13:2269–2281. doi: 10.1105/tpc.010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1–AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandrel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandrel MA, Yanofsky MF. The Arabidopsis AGL9 MADS-box gene is expressed in young flower primordia. Sex. Plant Reprod. 1998;11:22–28. [Google Scholar]
- Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D. A genetic and molecular model for flower development in Arabidopsis thaliana. Development. 1991;(Suppl. 1):157–167. [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol. Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Szeto W, Lotys-Prass C, Jofuku KD. Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell. 1997;9:37–47. doi: 10.1105/tpc.9.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-Lopez R, Alvarez-Buylla ER, Yanofsky MF. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001;11:182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reprod. 1999;11:297–322. [Google Scholar]
- Savidge B, Rounsley SD, Yanofsky MF. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell. 1995;1995:721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Pickett FB, Haughn GW. The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell. 1991;3:1221–1237. doi: 10.1105/tpc.3.11.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H, Okada K, Shimura Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993;4:385–398. doi: 10.1046/j.1365-313x.1993.04020385.x. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowtiz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Saedler H. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zachgo S, Silva ED, Motte P, Tröbner W, Saedler H, Schwarz-Sommer Z. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development. 1995;121:2861–2875. doi: 10.1242/dev.121.9.2861. [DOI] [PubMed] [Google Scholar]