Abstract
Model enteric bacteria such as Escherichia coli and Salmonella enterica express hundreds of small non-coding RNAs (sRNAs), targets for most of which are yet unknown. Some sRNAs are remarkably well conserved, indicating that they serve cellular functions that go beyond the necessities of a single species. One of these ‘core sRNAs’ of largely unknown function is the abundant ∼100-nucleotide SdsR sRNA which is transcribed by the general stress σ-factor, σS and accumulates in stationary phase. In Salmonella, SdsR was known to inhibit the synthesis of the species-specific porin, OmpD. However, sdsR genes are present in almost all enterobacterial genomes, suggesting that additional, conserved targets of this sRNA must exist. Here, we have combined SdsR pulse-expression with whole genome transcriptomics to discover 20 previously unknown candidate targets of SdsR which include mRNAs coding for physiologically important regulators such as the carbon utilization regulator, CRP, the nucleoid-associated chaperone, StpA and the antibiotic resistance transporter, TolC. Processing of SdsR by RNase E results in two cellular SdsR variants with distinct target spectra. While the overall physiological role of this orphan core sRNA remains to be fully understood, the new SdsR targets present valuable leads to determine sRNA functions in resting bacteria.
INTRODUCTION
Bacteria encode a plethora of small regulatory RNAs (sRNAs), most of which act to control gene expression through base-pairing with target mRNAs (1–3). These base-pairing interactions commonly decrease the translation and/or stability of the target transcripts, although an increasing number of mechanisms of mRNA activation are known, too (4,5). In the well-studied Gram-negative bacteria Escherichia coli and Salmonella enterica serovar Typhimurium (henceforth Salmonella), an important co-factor of sRNA-based mRNA regulation is Hfq. This abundant RNA-binding protein not only protects sRNAs from endonucleolytic decay but also acts as a chaperone to facilitate base-pairing of sRNAs with multiple target mRNAs (6–8). Unsurprisingly, Hfq deficiency results in extensive phenotypic changes and complex regulatory defects including compromised stress responses and virulence gene expression (9–13).
Co-purification of cellular RNA with Hfq from E. coli and Salmonella revealed binding of 100–200 potential sRNA species (14–19). Since Hfq-dependent sRNAs typically operate by short, imperfect ‘seed-pairing’ interactions to regulate multiple targets, the sum of their post-transcriptional activities is expected to affect a large proportion of the mRNAs of these bacteria (20,21). Indeed, some sRNAs such as GcvB and RyhB alone may each regulate ∼1% of all mRNAs expressed in Salmonella or E. coli, respectively (22,23). Nevertheless, the target suites of most sRNAs have not been determined, and it is unclear to which extent these regulators contribute to gene control and microbial physiology.
While Salmonella expresses a repertoire of unique sRNAs some of which regulate virulence (24,25), there is a set of so-called ‘core sRNA’ genes which are present in the genomes of nearly all sequenced enterobacteria (26). Since these core sRNAs have been conserved in so many physiologically and ecologically different bacteria, one can assume that they serve central cellular functions that go beyond the necessities of a single species (27). Detailed analyses of several core sRNAs, for example the membrane stress-associated regulators CpxQ, MicA, MicL and RybB (15,28–31), the iron starvation-controlled RyhB sRNA (32), the carbon utilization regulators, Spot 42 and SgrS (33–37), and the amino acid metabolism-related GcvB, DapZ and SroC sRNAs (15,22,38,39), uncovered regulatory functions that may be conserved in many different bacteria. Disregarding those generated by mRNA cleavage (30,38), most core sRNAs carry conserved transcriptional control elements in their promoter regions, indicating linkage to common regulatory pathways (27).
The abundant ∼100 nt-long SdsR sRNA constitutes one of the most highly conserved enterobacterial sRNAs (40). We have recently shown that transcription of SdsR depends on σS (40) which is a general stress σ-factor whose association with RNA polymerase in stationary phase affects ∼10% of all E. coli genes (41,42). Accordingly, SdsR production is induced when cells enter stationary phase growth, and in response to various other σS-related stress conditions (40).
We further showed that in Salmonella SdsR base-pairs with and inhibits the translation of the mRNA coding for OmpD (40). OmpD is a highly abundant outer membrane porin (OMP) and its depletion by SdsR over-expression is readily detectable in standard SDS gels. Although no other proteins showed obvious regulation in these experiments, there are multiple lines of evidence to suggest the existence of additional SdsR targets. First, the sdsR gene is highly conserved amongst the enterobacteria, whereas expression of ompD is limited to few species such as Salmonella (43). Second, the sdsR gene contains an alternative putative seed-pairing domain, which is distinct from the region in SdsR that base-pairs with ompD. Third, SdsR appeared to be processed by the endoribonuclease, RNase E. In the resultant, shorter SdsR+31 species, the second seed sequence is located at the very 5′ end, similar to the seed sequences of several other characterized sRNAs (26,44–46).
In this study, we sought to determine the target suite of SdsR in Salmonella. In line with our prediction that this sRNA is a multi-target regulator, we find that SdsR can regulate many conserved mRNAs, including those coding for physiologically important regulators such as the transcriptional regulator, CRP, the global DNA-binding factors StpA and HupB, the antibiotic transporter protein, TolC and the RtsA/B two-component system (TCS). We have identified a total of 20 previously unknown targets, and extensively validated their post-transcriptional control by SdsR with reporter fusions. Using a series of sRNA mutants, we infer base-pairing interactions with either of two seed pairing domains in SdsR. Furthermore, we show that SdsR-mediated repression of the crp mRNA can alter the expression of genes involved in carbon source utilization. Our study presents the first global identification of SdsR target genes in the enterobacteria and suggests that SdsR, similarly to other highly conserved sRNAs, fulfills a global regulatory function by cross-connecting conserved stress response pathways and stationary phase physiology.
MATERIALS AND METHODS
DNA oligonucleotides
Sequences of all oligonucleotides employed in this study are listed in Supplementary Table S1.
Construction of plasmids
All plasmids used in this study are listed in Supplementary Table S2. Translational GFP fusions of SdsR target candidates were constructed as described before (47,48) using PCR products amplified from gDNA. Inserts were restricted with NheI/BfrBI (pKF119: NheI/XbaI), and ligated into an equally treated pXG10 plasmid backbone. Details on cloned inserts are summarized in Supplementary Table S2. Plasmid variant pKF226-1 (ydgT-M1::gfp) was constructed by PCR amplification of pKF127-1 (primer pair JVO-14343/JVO-14344), followed by direct transformation of the linear PCR product into competent E. coli. For plasmids expressing different SdsR mutant variants from the PL promoter, pKF68-3 served as a template for PCR amplification with primer pairs JVO-7161/JVO-7162 (pPL-SdsR C15G; pKF100-1), or JVO-9033/JVO-9034 (pPL-SdsR C38G; pKH6), and self-ligation was carried out as in (22,23). Competent E. coli TOP10 were used for all cloning purposes.
Bacterial strains and growth conditions
A complete list of bacterial strains employed in this study is provided in Supplementary Table S3. The S. enterica serovar Typhimurium strain SL1344 (JVS-0007) is referred to as wild-type strain and was used for mutant construction. Phage P22 was employed to transfer each single chromosomal modification to a fresh Salmonella wild-type background, as well as to obtain strains carrying multiple mutations. Strains expressing chromosomally encoded C-terminal 3XFLAG fusion proteins were constructed using a modified λRed approach as described in (49). Plasmid pSUB11 served as a template for PCR products to obtain rtsA::3XFLAG::kan (JVO-5673/JVO-5674), yhcB::3XFLAG::kan (JVO-5810/JVO-5811), yhaH::3XFLAG::kan (JVO-5808/JVO-5809), fbaB::3XFLAG::kan (JVO-9836/JVO-9837), and glgX::3XFLAG::kan (JVO-9839/JVO-9840). For standard cultures cells were grown aerobically in LB medium at 37°C unless stated otherwise. A final concentration of 0.2% l-arabinose was added to cultures to induce expression from pBAD-derived plasmids. Where appropriate, liquid and solid media were supplemented with antibiotics at the following concentrations: 100 μg/ml ampicillin; 50 μg/ml kanamycin; 20 μg/ml chloramphenicol.
Microarray experiment
Salmonella wild-type cells carrying either pKP8-35 (control) or pKP19-8 (pBAD-SdsR) were grown under standard conditions to an OD600 of 1.5. Expression of the sRNA was induced for 10 min, and RNA samples were collected in stop solution (95% ethanol, 5% phenol). Total RNA from three independent biological replicates was isolated using the SV Total RNA Isolation Kit (Promega); sample preparation, microarray hybridizations and analyses were carried out as described in (26). Raw values of the microarray analysis have been deposited in GEO (accession no. GSE77157). Genes were considered to be differentially expressed in the presence of SdsR if they displayed ≥2-fold-changes in all three replicates, and differences were statistically significant (Student's t-test; P-value ≤ 0.02).
In vitro RNA synthesis and electrophoretic mobility shift assays (EMSA)
For RNA in vitro synthesis, ∼200 ng of template DNA amplified from Salmonella genomic DNA using JVO-7023/JVO-7025 (SdsR) and JVO-7024/JVO-7025 (SdsR +31) were reverse transcribed employing the MEGAscript kit (Ambion) according to the manufacturer's guidelines. Correct size and integrity of the RNA were confirmed on a denaturing polyacrylamide gel. Formation of complexes between sRNAs and Hfq in vitro was analyzed by EMSA as described previously (43). Briefly, 5′ end-labelled RNA (4 pmol) was denatured (95°C, 2 min), chilled on ice for 5 min and supplemented with 1× structure buffer (Ambion) and 1 μg yeast RNA. Upon addition of purified Hfq (lab stock; concentration as indicated in the figure legend) or Hfq dilution buffer (control), samples were incubated at 37°C for 10 min. Prior to loading, reactions were mixed with native loading buffer, and separated by native PAGE. Gels were dried and signals were determined on a Typhoon FLA 7000 phosphorimager (GE Healthcare).
In vitro RNA structure probing
Structure probing was conducted on in vitro synthesized RNA as described before (46). In brief, the SdsR secondary structure was mapped by treating 0.4 pmol 5′ end‐labelled sRNA in the presence of 1× structure buffer (0.01 M Tris pH 7, 0.1 M KCl, 0.01 M MgCl2) and 1 μg yeast RNA with RNase T1 (0.1 U), or with lead(II) acetate (5 mM). Reactions were stopped at the indicated time-points, and samples were separated by denaturing PAGE on sequencing gels.
RNA isolation and Northern blot analysis
Total bacterial RNA was isolated from culture aliquots using TRIzol reagent (Invitrogen) or ‘Hot Phenol’ extraction as described before (46). Northern blotting was performed as in (11); briefly, for sRNA and mRNA detection, 5 or 10 μg of total RNA were separated on 6%/7M urea polyacrylamide gels and electroblotted. Membranes were hybridized with gene-specific 5′ end-labelled DNA-oligonucleotides or riboprobes at 42°C or 65°C, respectively, in Roti-Hybri-Quick hybridization solution (Roth) and washed in three subsequent steps with SSC wash buffers supplemented with 0.1% SDS (5×/1×/0.5× SSC or 2×/1×/0.5×, respectively). Signals were determined on a Typhoon FLA 7000 phosphorimager (GE Healthcare) and band intensities quantified with AIDA software (Raytest, Germany). SdsR sRNA and variants, CyaR and 5S RNA were detected by 5′ end-labelled oligonucleotides JVO-1032, JVO-1897 and JVO-0322, respectively. Riboprobes were synthesized by T7 in vitro transcription using ∼200 ng of template DNA amplified from gDNA with oligonucleotides JVO-0902/JVO-0997 (SdsR) or JVO-8855/JVO-8856 (crp mRNA) in the presence of 32P-α-UTP (50 μCi) with the MAXIscript kit (Ambion).
Protein sample analysis
To prepare total protein samples, bacterial cultures collected by centrifugation (2 min; 16 000 g; 4°C) were resuspended in 1× SLB (Fermentas) to a final concentration of 0.01 OD/μl. To analyze protein levels by Western blotting, 0.1 or 0.005 OD per lane were separated by SDS-PAGE and transferred onto PVDF membrane as described in (11). GFP fusion proteins, FLAG-tagged proteins, CRP or OmpX were detected using antibodies directed against GFP (1:5 000; mouse; Roche), FLAG M2 peptide (1:1 000; mouse; Sigma), an antiserum recognizing CRP (1:1 500; rabbit; Hiroji Aiba), an antiserum recognizing OmpX (1:10 000; rabbit; (50)), and either anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (1:10 000; GE Healthcare). Signals were visualized using Western Lightning reagent (PerkinElmer) and detected with an ImageQuant LAS 4000 CCD camera (GE Healthcare).
Sequence retrieval and alignments
Information for sequence alignments was collected using BlastN searches (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) of the following genome sequences (accession numbers are given in parentheses): Salmonella typhimurium LT2 (NC_003197); Salmonella typhi Ty2 (NC_004631); Citrobacter koseri ATCC BAA-895 (NC_009792); Escherichia coli K12 (NC_000913); Shigella flexneri 2a str 301 (NC_004337); Enterobacter Sp.638 (NC_009436); Cronobacter turicensis z30232 (NC_013282); Klebsiella pneumoniae 342 (NC_011283); Serratia proteamaculans 568 (NC_009832); Yersinia pestis KIM (NC_004088); Yersinia enterocolitica subsp. enterocolitica 8081 (NC_008800); Dickeya dadantii Ech 703 (NC_012880); Pantoea ananatis LMG 20103 (NC_013956); Sodalis glossinidius str. ‘morsitans’ (NC_007712); Erwinia pyrifoliae Ep1/96 (NC_003197); Photorhabdus luminescens subsp. laumondii TT01 (NC_005126); Xenorhabdus nematophila ATCC 19061 (NC_014228). Alignments were computed using MultAlin (51).
RESULTS
Identification of mRNAs regulated by SdsR in Salmonella
We have previously shown that SdsR base-pairs with and inhibits the expression of ompD mRNA (40). To systematically identify additional targets of SdsR in Salmonella, we grew wild-type bacteria carrying plasmid pBAD-SdsR to early stationary phase (OD600 of 1.5) and pulse-activated SdsR transcription by addition of the inducer, L-arabinose. Under these conditions SdsR is not expressed in Salmonella (40), indicating that competition with endogenous SdsR is not relevant to this experiment. Gene expression was terminated after ten minutes, when total RNA was extracted and subjected to whole genome microarray analysis. As listed in Table 1, we observed at least two-fold regulation of 34 genes from 28 operons. The majority of the potential SdsR targets were repressed by SdsR (32 targets): the strongest negative regulation was observed for ompD mRNA (-4.5-fold), which we previously identified as a direct target of SdsR (40). The other targets comprised additional membrane proteins (yhaH, yhcB, envE, tolC), DNA-binding proteins (rtsB, hupB, ydgT, stpA, crp), proteins involved in metabolic activities (yibF, glgB, artI, fbaB, dld, asd, dlhH, aphA, mpl, glgX), heat-shock proteins (mopA, mopB, ibpB, ibpA, dnaK, htpG) as well as the transcripts hslV, STM 4313, ydgH, STM 4312, yffB, hslU, phnA and stcD.
Table 1. Target genes of SdsR.
| Gene | ID | Microarray fold | Description | Reporter fusion (rel. to AUG) | Experimental | Target regulation by | Recovered in | |
|---|---|---|---|---|---|---|---|---|
| Regulation | confirmation 5) | SdsR | SdsR+31 | Hfq-CoIP | ||||
| nmpC1) | STM1572 | −4,50 | outer membrane porin D | - | WB | yes | no | (14,15,17) |
| mopA | STM4329 | −4,26 | co-chaperonin GroES | (−72 to +45 in mopB)::gfp | nc | – | – | – |
| yhaH | STM3234 | −3,70 | putative inner membrane protein | (−43 to +45 in yhaH)::gfp | FC | yes | yes | (14,15) |
| yhcB | STM3347 | −3,46 | cytochrome d ubiquinol oxidase subunit III | (−59 to +45 in yhcB)::gfp | FC | yes | yes | (14,15,17) |
| rtsB | STM4314 | −3,17 | putative regulatory protein | (−161 to +30 in rtsA)::gfp | FC | yes | yes | (15,17) |
| yibF | STM3684 | −3,13 | glutathione S-transferase | (−31 to +45 in yibF)::gfp | FC | yes | no | (15) |
| mopB | STM4330 | −3,10 | chaperonin GroEL | (−72 to +45 in mopB)::gfp | nc | – | – | (17) |
| ibpB2) | STM3808.S | −3,09 | heat shock chaperone IbpB | (−99 to +45 in ibpA)::gfp | nc | – | – | (17) |
| glgB3) | STM3538 | −2,99 | glycogen branching enzyme | (−154 to +45 in glgB)::gfp | nc | – | – | (14,17) |
| hupB | STM0451 | −2,87 | transcriptional regulator HU subunit beta | (−118 to +90 in hupB)::gfp | FC | yes | yes | (17,19) |
| envE | STM1242 | −2,78 | putative envelope protein | (−181 to +45 in envE)::gfp | FC | yes | yes | (15) |
| artI | STM0890 | −2,68 | arginine transport system | (−109 in artP to +45 in artI)::gfp | WB | yes | no | (14,15,17) |
| hslV | STM4092 | −2,67 | ATP-dependent protease peptidase subunit | (−61 to +45 in hslV)::gfp | nc | – | – | – |
| fbaB3) | STM2141 | −2,64 | fructose-bisphosphate aldolase | (−101 to +45 in fbaB)::gfp | nc | – | – | (14) |
| STM4313 | STM4313 | −2,63 | putative cytoplasmic protein | (−161 to +30 in rtsA)::gfp | FC | yes | yes | – |
| tolC | STM3186 | −2,61 | outer membrane channel protein | (−250 to +60 in tolC)::gfp | WB | yes | yes | (17) |
| ydgT | STM1461.S | −2,53 | oriC-binding nucleoid-associated protein | (−130 to +60 in ydgT)::gfp | FC | yes | no | (14,16) |
| ibpA2) | STM3809.S | −2,53 | heat shock protein IbpA | (−99 to +45 in ibpA)::gfp | nc | – | – | – |
| dnaK | STM0012 | −2,38 | molecular chaperone DnaK | (−145 to +45 in dnaK)::gfp | nc | – | – | (17) |
| dld | STM2167 | −2,29 | D-lactate dehydrogenase | (−42 to +45 in dld)::gfp | nc | – | – | – |
| stpA | STM2799 | −2,29 | DNA binding protein; nucleoid-associated | (−83 to +60 in stpA)::gfp | FC | yes | yes | (15) |
| asd | STM3539 | −2,28 | aspartate-semialdehyde dehydrogenase | (−225 to +60 in asd)::gfp | FC | yes | no | (14,15) |
| htpG | STM0487.S | −2,22 | heat shock protein 90 | (−45 to +60 in htpG)::gfp | FC | yes | no | – |
| ydgH | STM1478 | −2,18 | putative periplasmic protein | (−58 to +60 in ydgH)::gfp | WB | yes | no | – |
| crp | STM3466 | −2,16 | cAMP-regulatory protein | (−172 to +30 in crp)::gfp | FC | yes | no | (16,17) |
| dlhH | STM3967 | −2,15 | putative dienelactone hydrolase | (−31 to +120 in dlhH)::gfp | WB | yes | yes | (14,15) |
| aphA | STM4249 | −2,15 | acid phosphatase/phosphotransferase | (−59 to +60 in aphA)::gfp | nc | – | – | (15,17) |
| mpl | STM4416 | −2,06 | UDP-N-acetylmuramate/L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase | (−56 to +60 in mpl)::gfp | WB | yes | yes | (17) |
| STM4312 | STM4312 | −2,04 | hypothetical protein | (−161 to +30 in rtsA)::gfp | FC | yes | yes | (15) |
| yffB | STM2482 | −2,04 | hypothetical protein | (−97 to +75 in yffB)::gfp | WB | yes | no | – |
| glgX3) | STM3537 | −2,01 | glycogen debranching enzyme | (−154 to +45 in glgB)::gfp | nc | – | – | (17) |
| hslU | STM4091 | −2,00 | ATP-dependent protease ATP-binding subunit | (−61 to +45 in hslV)::gfp | nc | – | – | (14,15) |
| phnA | STM4289 | +2,47 | hypothetical protein | (−63 to +30 in phnA)::gfp | FC | yes | yes | (14) |
| stcD4) | STM2149 | +2,57 | putative outer membrane lipoprotein | - | nc | – | – | – |
1) Validated in (40); excluded from analysis.
2) GFP fusion is unstable; occurrence of spontaneous mutations; excluded from analysis.
3) Validation by analyzing protein expression from the Salmonella chromosome; compare Figure 5.
4) TSS not determined (54); excluded from analysis.
5) WB: Western blot; FC: flow cytometry; nc: not confirmed.
Validation of SdsR target genes using reporter fusions
Base-pairing sites of Hfq-associated sRNAs frequently reside in the 5′ region of mRNAs (52,53). To test post-transcriptional regulation of the new SdsR candidate targets (Table 1), we designed translational fusions to the green fluorescent protein (gfp) under the control of a constitutive promoter (48). For target genes organized in operons, we constructed reporter fusions to the first open reading frame. For all reporters, the 5′ UTR and up to 40 codons of the potential target gene were fused to the second codon of gfp (Table 1, Figure 1A and Supplementary Figure S1A). We excluded the previously validated target ompD (40), and did not construct a fusion for stcD since the transcriptional start site of the Salmonella stcABCD operon is unknown (54). Reporters for ibpAB and aphA were unstable or did not produce detectable GFP levels, respectively; these were omitted from further analysis.
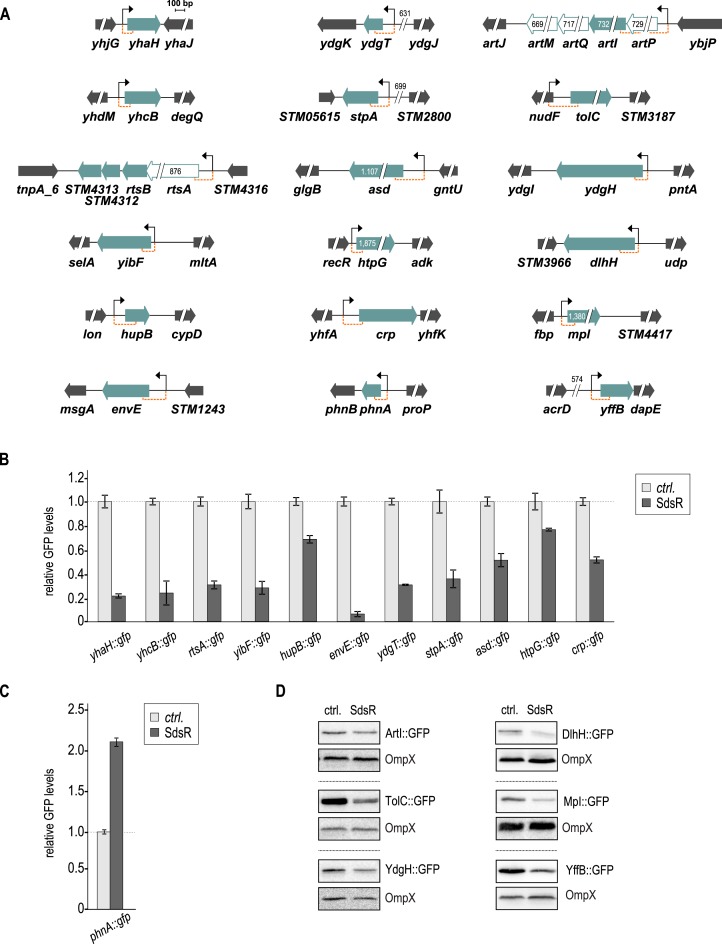
Figure 1.
Genomic context of SdsR target genes and validation with gfp reporter fusions. (A) Genomic location and flanking regions of confirmed SdsR targets. Target genes are shown as filled green arrows; genes within the same transcriptional unit as validated targets are represented as open green arrows. All genes are drawn to scale; where appropriate, numbers indicate fragment sizes. Transcriptional start sites (as determined by (54)) are marked by a black arrow, and sequence stretches represented in gfp reporter fusions are indicated by orange brackets. (B) Repression of target genes by SdsR. Salmonella ΔsdsR cells carrying the indicated gfp reporter fusion in combination with either a control, or a plasmid expressing Salmonella SdsR were grown to early stationary phase (OD600 of 2), fixed with PFA, and GFP fluorescence was determined by flow cytometry. For each GFP-fusion, fluorescence levels in the presence of the control plasmid were set to 1, and relative changes were determined for cells expressing SdsR. GFP levels were calculated from three biological replicates; error bars indicate the standard deviation. Detailed descriptions of all plasmids are provided in Supplementary Table S2. (C) Positive regulation of phnA::gfp in the presence of SdsR. Experimental procedure as in (B). (D) Validation of non-fluorescent target fusions by western blot analysis. Protein samples were collected from Salmonella ΔsdsR cells grown to early stationary phase (OD600 of 2) and carrying the indicated gfp reporter fusion in combination with either a control or a plasmid expressing Salmonella SdsR. OmpX was probed as loading control. A quantification of these results is provided in Supplementary Table S4.
All remaining gfp reporter fusions were co-transformed in a Salmonella ΔsdsR strain with either a plasmid constitutively over-expressing SdsR, or an empty vector control. A plasmid expressing heterologous luc::gfp (pXG-1) served as control. We used flow cytometry to determine SdsR-mediated regulation of the individual reporters, measuring GFP fluorescence of cells grown to early stationary phase (OD600 of 2). In some cases, fluorescence intensity of the fusion proteins was insufficient to be detected by flow cytometry (ArtI::GFP, TolC::GFP, YdgH::GFP, DlhH::GFP, Mpl::GFP and YffB::GFP). Instead, GFP levels were determined by Western blot analysis using an anti-GFP antibody.
When analyzing GFP expression of the 24 reporter fusions, we observed SdsR-dependent regulation for 18 constructs (Figure 1). Flow cytometry analysis confirmed that SdsR mediated repression of eleven targets (yhaH, yhcB, rtsA, yibF, hupB, envE, ydgT, stpA, asd, htpG, crp; Figure 1B), and the extent of regulation ranged from ∼1.4-fold (htpG) to ∼17-fold (envE). Our transcriptome analysis had predicted positive regulation of phnA mRNA (Table 1), and this result was confirmed with the target fusion phnA::gfp (Figure 1C). An additional six fusions were validated as negatively regulated using Western blot analysis (artI, tolC, ydgH, dlhH, mpl, yffB; Figure 1D and Supplementary Table S4). GFP expression from the control plasmid showed a basal increase in the presence of SdsR (∼1.4-fold; Supplementary Figure S2), and additional six target gene fusions were unaffected by the co-expression of the sRNA (mopB, glgB, hslV, fbaB, dnaK and dld; Supplementary Figure S1B).
Identification of potential SdsR base-pairing sequences
Bacterial sRNAs acting together with Hfq typically employ one or more seed-sequences to base-pair with trans-encoded mRNAs (55). The canonical seed sequence of an sRNA is an extended single-stranded region at the 5′ end or center of the regulator, which is energetically more favorable for base-pairing with target mRNAs, when compared to structured, obstructed regions of the sRNA. Post-transcriptional regulation of 18 novel target mRNAs by SdsR (Figure 1), as well as the existence of highly conserved sequence stretches in SdsR (Figure 2A) suggested that similar elements exist in this sRNA as well and prompted us to determine the secondary structure of SdsR through chemical and enzymatic probing. To this end, we synthesized Salmonella SdsR by in vitro transcription, and partially digested the 5′ end-labelled sRNA with lead(II) acetate (cleavage of single-stranded regions), or the endonuclease RNase T1 (specific cleavage of unpaired guanosine residues). Cleavage intermediates were separated on a polyacrylamide gel, and mapped using sequence specific ladders (Figure 2B). Based on the cleavage pattern, the SdsR structure contains a short hairpin structure at the 5′ end, and two additional stem-loops at the 3′ end of which the second one is followed by a U-run to facilitate ρ-independent transcription termination (Figure 2C). A 24 nt long (nt 18–42), unstructured region is located in between stem-loops 1 and 2, and an AU-rich sequence stretch upstream of the terminator might serve as a binding site for the chaperone Hfq.
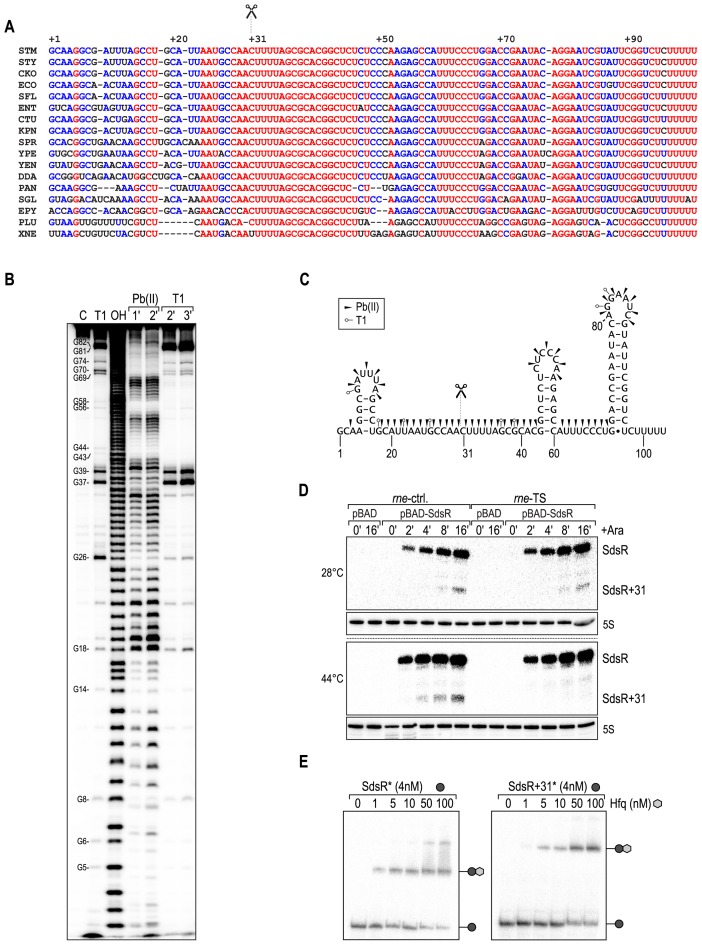
Figure 2.
SdsR sRNA is processed by RNase E, and associates with Hfq. (A) Multiple alignment of SdsR sRNA sequences of diverse enterobacteria. Fully, partially and poorly conserved nucleotides are indicated in red, blue and black, respectively. Scissors mark the RNase E processing site. Abbreviations correspond to the following species: STM, Salmonella Typhimurium; STY, Salmonella typhi; CKO, Citrobacter koseri; ECO, Escherichia coli; SFL, Shigella flexneri; ENT, Enterobacter sp. 638; CTU: Cronobacter turicensis; KPN, Klebsiella pneumoniae; SPR, Serratia proteamaculans; YPE, Yersinia pestis; YEN, Yersinia enterocolitica; DDA, Dickeya dadantii; PAN, Pantoea ananatis; SGL, Sodalis glossinidius; EPY, Erwinia pyrifoliae; PLU, Photorhabdus luminescens; XNE, Xenorhabdus nematophila. (B) Determination of the SdsR structure by in vitro probing. 5′ end-labelled SdsR was subjected to Pb(II) acetate (lanes 4 and 5) or RNase T1 cleavage (lanes 6 and 7), and reactions were stopped at indicated time-points. RNase T1 and alkaline (OH) ladders were used to map cleaved fragments. Positions of G-residues are indicated. (C) Secondary structure of SdsR. Cleavage sites as determined in (B) are indicated by arrowheads (Pb(II) acetate) or open circles (T1). Scissors mark the RNase E cleavage site. (D) SdsR processing is dependent on RNase E. Salmonella rne-TS and its isogenic control strain carrying either pBAD-SdsR or control plasmid pBAD were grown at the permissive temperature of 28°C to early stationary phase (OD600 of 1), when cultures were split, and growth was continued for 30 min at either 28°C, or 44°C to inactivate RNase E. Expression of SdsR was induced by the addition of l-arabinose, and RNA harvested at the indicated time-points was analyzed on Northern blots using a SdsR-specific probe. 5S rRNA served as loading control. (E) Full-length and processed SdsR associate with Hfq. Electrophoretic mobility shift assay (EMSA) of in vitro synthesized 5′-end-labelled SdsR or SdsR+31 RNAs (4 nM) in the presence of increasing concentrations of Hfq protein as indicated.
SdsR is present as two species both of which interact with Hfq
SdsR accumulates as two distinct species corresponding to the full-length molecule and a shorter version of ∼ 70 nt representing the 3′ end of the sRNA (40). Post-transcriptional maturation is not uncommon to RNAs, and two other highly conserved sRNAs, ArcZ and RprA, are also detected as two distinct sRNA species each (56). Processing of ArcZ and RprA depends on the activity of the major ribonuclease RNase E (26,57,58).
Cleavage of SdsR occurs within the conserved, single-stranded region at nt 31 (40,59), and in order to test the involvement of RNase E in sRNA processing we made use of a temperature-sensitive version (rne-TS) of the essential enzyme (60). When rne-TS cells are cultivated at permissive temperatures (28°C), RNase E retains its activity, but it is rapidly inactivated when the culture is shifted to a non-permissive temperature (44°C, (60,61)). Both the rne-TS as well as an isogenic wild-type strain were grown at 28°C to early stationary phase when the culture was split. Upon continued growth at either 28°C or 44°C for 30 min to inactivate RNase E in the temperature-sensitive strain, SdsR expression was induced from the pBAD promoter. At 28°C, SdsR accumulated to high levels within the monitored time of induction, and the processed fragment was detectable in both the control and the rne-TS strain (Figure 2D). In contrast, inactivation of RNase E at 44°C abrogated the accumulation of the ∼70 nt species (bottom panel, last four lanes), arguing that RNase E is required for SdsR maturation.
Two Hfq-binding sites on SdsR were identified in recent CLIP-seq studies (16,19): one immediately upstream of the processing site (covering nt 27) and a second binding site located between stem-loops 2 and 3 (covering nt 65). Therefore, both versions of SdsR could function as post-transcriptional regulators acting through Hfq. To determine the capability of the full-length as well as the processed form of the sRNA to interact with Hfq, we performed gel-shift experiments using purified Hfq protein and in vitro synthesized SdsR or SdsR+31, respectively. Since both sRNA variants formed complexes with Hfq (Figure 2E), we concluded that the full-length and processed forms of SdsR might interact with cellular mRNAs.
The processed form of SdsR is sufficient to regulate a subset of targets
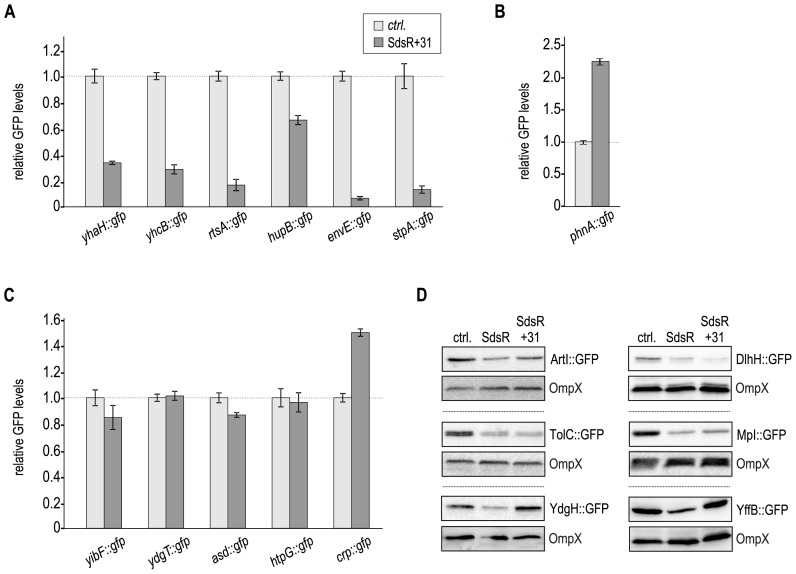
The association of both SdsR variants with Hfq indicated that processed SdsR could be sufficient to regulate target mRNAs. To test our hypothesis, we co-transformed all reporter fusions regulated by SdsR with a plasmid constitutively expressing the short variant of the sRNA. When comparing GFP expression levels in the presence of SdsR+31 to the control we found that a subset of target fusions was affected by sRNA expression (Figure 3). Reporter gene expression of yhaH::gfp, yhcB::gfp, rtsA::gfp, hupB::gfp, envE::gfp, stpA::gfp (Figure 3A) as well as the positively regulated phnA::gfp (Figure 3B) was similar to the levels observed for full-length SdsR (Figure 1C). In contrast, fusions yibF::gfp, ydgT::gfp, asd::gfp, htpG::gfp and crp::gfp were not repressed by SdsR+31 (Figure 3C). Western blot analysis of the non-fluorescent reporters confirmed that only tolC::gfp, dlhH::gfp and mpl::gfp, but not artI::gfp, ydgH::gfp and yffB::gfp were targeted by the SdsR+31 (Figure 3D and Supplementary Table S4).
Figure 3.
The processed form of SdsR sRNA regulates only a subset of target genes. (A) Repression of target genes in the presence of SdsR+31. Salmonella ΔsdsR cells carrying the indicated gfp reporter fusion in combination with either a control, or a plasmid expressing Salmonella SdsR+31 were grown to early stationary phase (OD600 of 2), fixed with PFA, and analyzed by flow cytometry as described in Figure 1B. (B) Positive regulation of phnA::gfp in the presence of SdsR+31. (C) Flow cytometry analysis of SdsR targets not repressed by SdsR+31. (D) Examination of non-fluorescent target fusions by Western blot analysis. Protein samples were collected from Salmonella ΔsdsR cells carrying the indicated gfp reporter fusion in combination with either a control plasmid or constructs expressing Salmonella SdsR or SdsR+31, respectively, grown to early stationary phase (OD600 of 2). A quantification of these results is provided in Supplementary Table S4.
The majority of sRNAs use a single, highly-conserved domain to interact with their target mRNAs. Howewer, several regulators including FnrS, Spot42, GcvB or VqmR have been shown to employ more than one sequence stretch to recognize their binding partners (22,39,62–64). The differential regulation of individual target fusions by SdsR and SdsR+31 (Figure 3A–D) suggested that the sRNA employs at least two distinct seed sequences: one located downstream of the processing site and another one upstream of, or overlapping the cleavage site.
SdsR employs two different domains to interact with individual targets
To confirm the involvement of different base-pairing sites within SdsR for target regulation, we introduced several mutations into the sRNA expression plasmid (Figure 4A). We took advantage of the previously established truncated SdsR+7 variant and the SdsR G26C mutant, which were used to probe base-pairing of SdsR with ompD (40). In addition, we introduced point mutations upstream (C15G) and downstream (C38G) of the SdsR processing site to test for additional base-pairing capacities of the two sRNA species. Expression of all these SdsR variants was verified on Northern blots (Figure 4B), and their effect on GFP production from the control plasmid pXG-1 was analyzed by flow cytometry. We observed a moderate increase in GFP levels in the presence of SdsR and SdsR+31 when compared to the sRNA control plasmid (∼1.4-fold and ∼1.6-fold, respectively; Supplementary Figure S2), while mutants SdsR+7, SdsR C15G and SdsR C38G did not significantly influence gfp expression. Through a mechanism currently unknown, SdsR G26C activated gfp expression of the control plasmid ∼2.4-fold.
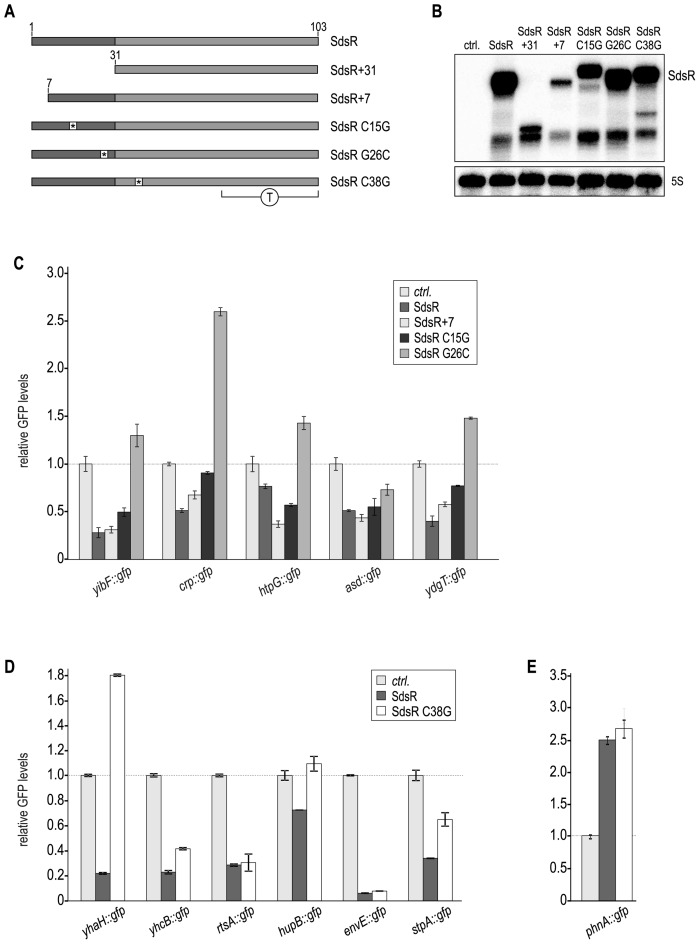
Figure 4.
SdsR employs different binding sites to control target gene expression. (A) Schematic representation of SdsR mutants. Asterisks mark locations of single point mutations; T denotes the terminator region. (B) Expression pattern of SdsR variants. RNA prepared from Salmonella ΔsdsR cells carrying either a control plasmid or expressing SdsR; SdsR+31; SdsR+7; SdsR C15G; SdsR G26C or SdsR C38G from the constitutive PL promoter was analyzed by Northern blotting. Detailed descriptions of all plasmids are provided in Supplementary Table S2. (C–E) GFP fluorescence of Salmonella ΔsdsR cells carrying the indicated gfp reporter fusion in combination with either a control plasmid, or a construct expressing a Salmonella SdsR variant was analyzed by flow cytometry. For each GFP-fusion, fluorescence levels in the presence of the control plasmid were set to 1, and relative changes were determined for cells expressing SdsR sRNA variants. (C) Effect of SdsR+7; SdsR C15G; SdsR G26C on target gene fusions. (D) Effect of SdsR C38G on target gene expression. (E) SdsR C38G activates phnA::gfp expression.
To allow high-throughput validation of target gene expression we restricted our analysis of the impact of SdsR mutants on fusions amenable to flow cytometry, i.e. we selected the 13 reporters that had shown sufficient fluorescence (Figure 1B and C). For the set of reporters regulated by SdsR, but not SdsR+31, we investigated the effects of three mutations at the 5′ end, namely a 5′ truncation (SdsR+7) and two point mutations (SdsR C15G and SdsR G26C) (Figure 4A). Mutation G26C affected the regulation of the majority of reporters: SdsR G26C was unable to repress yibF::gfp, crp::gfp, htpG::gfp and ydgT::gfp, and was less potent than the wild-type sRNA to down-regulate asd::gfp (Figure 4C). Mutation of position 15 of the sRNA (SdsR G15C) partially relieved regulation of yibF::gfp, crp::gfp and ydgT::gfp, but did not affect repression of htpG::gfp and asd::gfp. The very 5′ end of SdsR did not seem to contribute to any target interaction since SdsR+7 regulated all tested reporters.
We next determined the expression patterns of those fusions regulated both by the full-length as well as the processed version of SdsR when nucleotide C38 was mutated to G. This point mutation is positioned within the highly conserved, single-stranded stretch downstream of the processing site (Figure 2C). There was no or little regulation of yhaH::gfp, hupB::gfp and stpA::gfp in cells expressing SdsR C38G, suggesting that this nucleotide of SdsR was required for efficient base-pairing with these targets. In contrast, repression of yhcB::gfp, rtsA::gfp and envE::gfp was not affected by the single nucleotide exchange (Figure 4D). The positive effect on phnA::gfp expression that been observed both for SdsR and SdsR+31 was likewise maintained in strains over-expressing SdsR C38G (Figure 4E).
SdsR reduces protein expression of target genes
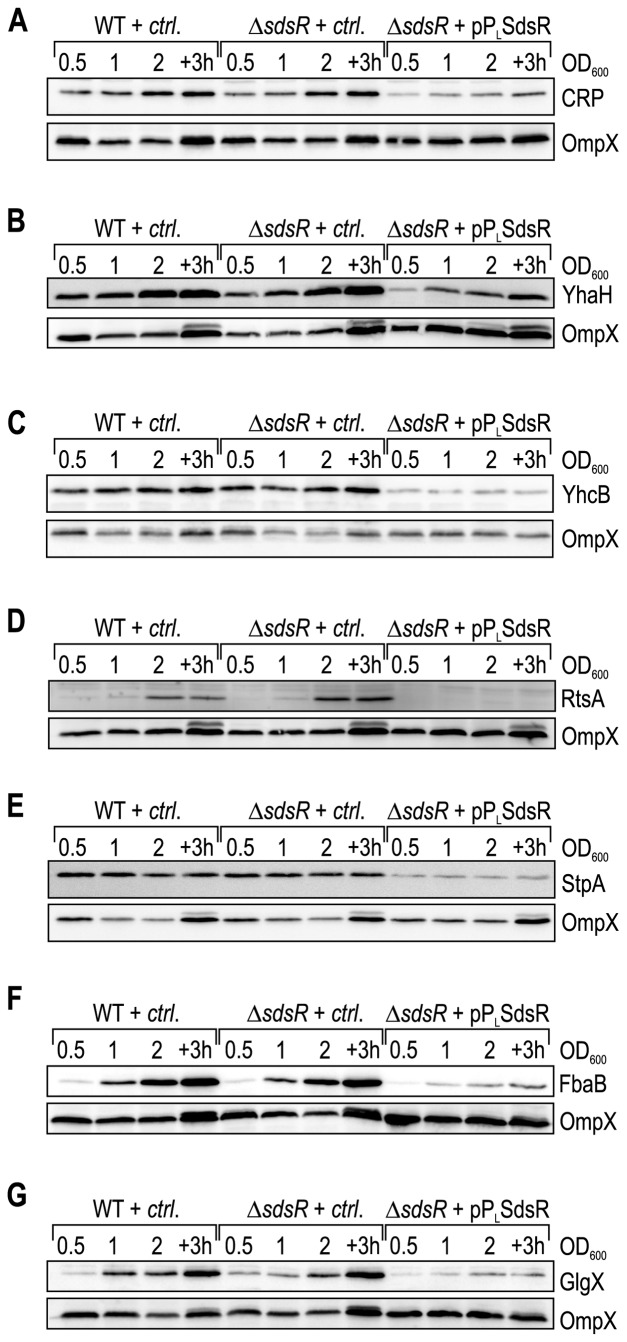
Using a series of SdsR mutants (Figure 4A) we were able to confirm that the regulation of target fusions was due to base-pairing interactions. We next wanted to test if the expression of SdsR also led to detectable changes in protein levels of the identified targets. To this end, we constructed chromosomal C-terminal fusions of the 3XFLAG affinity tag to a subset of targets confirmed in the reporter assay (yhaH, yhcB, rtsA, stpA), or probed protein production with a specific antibody (CRP). We monitored protein expression in wild-type and ΔsdsR Salmonella carrying either a control plasmid or the constitutive pPL-SdsR. Protein samples were collected from the three strains at different time-points of growth (OD600 of 0.5, 1, 2 and 3h after cells had reached an OD600 of 2). CRP levels increased in expression at the onset of stationary phase (OD600 of 2) (Figure 5A), and while the deletion of sdsR did not significantly affect CRP production, we observed strongly reduced CRP protein levels in the strain over-expressing SdsR at all time-points (Figure 5A, last 4 lanes).
Figure 5.
Protein expression patterns of SdsR target genes over growth. (A–E) Whole cell protein samples were prepared at different time-points over growth (OD600 of 0.5 (lanes 1, 5, 9); 1.0 (lanes 2, 6, 10); 2.0 (lanes 3, 7, 11); 3 h after cells had reached an OD600 of 2.0 (lanes 4, 8, 12)) from wild-type and ΔsdsR Salmonella carrying either a control vector or the constitutive SdsR-expression plasmid pPL-SdsR grown in LB. Expression of SdsR targets was monitored by Western blot analysis of (A) CRP; (B) YhaH-3XFLAG; (C) YhcB-3XFLAG; (D) RtsA-3XFLAG; (E) StpA-3XFLAG. (F) FbaB::3XFLAG; (G) GlgX::3XFLAG. CRP was detected using an anti-CRP antibody; for all other targets, chromosomal C-terminal 3XFLAG epitope tags were detected using a monoclonal anti-FLAG antibody. OmpX was probed as loading control.
A similar pattern was observed for targets YhaH (Figure 5B), YhcB (Figure 5C), RtsA (Figure 5D) and StpA (Figure 5E). Both YhaH and YhcB expression increased towards stationary growth, and deletion of SdsR did not affect steady-state protein levels (Figure 5B/C). The constitutive over-expression of SdsR, however, markedly decreased the levels of both YhaH and YhcB. When analyzing the protein expression pattern of RtsA we observed that RtsA was only detectable once cells reached early stationary phase (OD600 of 2), and was moderately increased in the SdsR mutant (Figure 5D). When SdsR was produced in the cell at even higher levels, i.e. in the over-expression strain, RtsA was no longer detectable (Figure 5D, last four lanes). Different from the other targets, StpA was expressed at all stages over growth, but again strongly repressed in the presence of high levels of SdsR (Figure 5E).
Comparison of protein patterns in the absence and presence of SdsR confirmed regulation of five target genes expressed from their native loci by the sRNA (Figure 5). We therefore asked whether the same approach was applicable to assess regulation of those potential targets from our initial microarray screen that had failed validation using gfp reporter plasmids (Supplementary Figure S1 and Table 1). Binding of SdsR to sequences downstream of canonical translation control sites (65) outside the cloned region could be one possible explanation for a discrepancy. We chose two potential targets, fbaB and glgX, to test this hypothesis, and inserted a 3XFLAG epitope tag to the C-terminus of both genes on the Salmonella chromosome. We again monitored expression of the fusion proteins over growth in wild-type or ΔsdsR Salmonella in the presence or absence of pPL-SdsR, respectively. Both FbaB and GlgX expression increased towards stationary phase, and protein levels were significantly reduced when SdsR was over-expressed (Figure 5F and G). The regulation of fbaB and glgX by SdsR at the protein level confirms the result of the microarray experiment, and hints at a base-pairing site outside of the sequence used in our post-transcriptional GFP-reporters.
SdsR expression interferes with downstream pathways of its direct targets
Monitoring the expression patterns of a subset of targets confirmed that SdsR was capable of reducing protein levels of its target genes (Figure 5). Consequently, we tested if pathways downstream of the sRNA's direct targets might likewise be affected by SdsR expression. Specifically, we focused on the conserved transcription factor, CRP, for which we had seen strong repression by SdsR.
CRP (cAMP receptor protein), also known as Cap (catabolite activator protein), is one of seven major transcription factors in Escherichia coli that together control >50% of the cell's transcription units (66). In complex with cyclic adenosine monophosphate (cAMP), CRP orchestrates carbon catabolite repression, and the usage of alternative carbon sources (67). Acting as a dual regulator, CRP-cAMP can either activate or repress its target genes. Within its large regulon, CRP also controls the two conserved sRNAs, CyaR and Spot42. Expression of CyaR requires CRP, and in turn, the sRNA represses the ompX, luxS, yqaE and nadE genes (50,68,69). In contrast, CRP-cAMP negatively regulates transcription of Spot 42, an abundant sRNA governing a set of at least 16 targets many of which are controlled by CRP themselves (62,70).
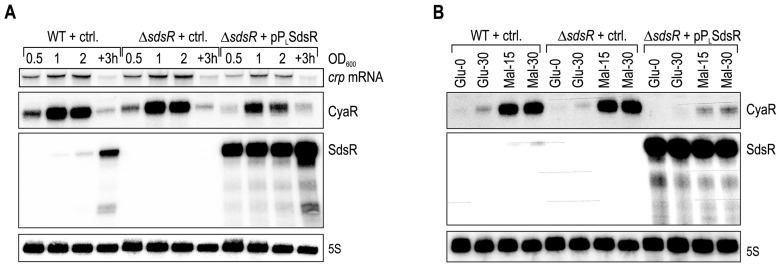
As a representative target of CRP, we monitored expression of the CyaR sRNA (50,68) over growth in the presence or absence of SdsR. To this end, RNA was prepared at four different time-points of bacterial growth in LB (ranging from exponential to stationary phase), and subjected to Northern blot analysis (Figure 6A). As expected, SdsR expression in WT cells increased towards stationary phase, but the sRNA was highly abundant at all phases of growth when transcribed from a constitutive promoter (Figure 6A, third panel). CyaR sRNA was expressed throughout growth, but was most abundant at late exponential and early stationary growth (OD600 of 1 and 2, respectively; second panel) when crp expression increased (Figure 6A, top panel). In the SdsR over-expression strain (last four lanes), CyaR was down-regulated in comparison to expression in wild-type cells during all growth phases. The inability of cells producing high amounts of SdsR to express CyaR sRNA paralleled the SdsR-mediated repression of crp mRNA and CRP protein (Figure 6A, top panel and Figure 5A, respectively).
Figure 6.
SdsR affects expression of genes within the CRP regulon. (A) Wild-type and ΔsdsR Salmonella carrying either a control vector, or the constitutive SdsR-expression plasmid pPL-SdsR were grown in LB, and RNA isolated at different time-points over growth (OD600 of 0.5 (lanes 1, 5, 9); 1.0 (lanes 2, 6, 10); 2.0 (lanes 3, 7, 11); 3 h after cells had reached an OD600 of 2.0 (lanes 4, 8, 12)) was analyzed on Northern blots. The two sRNAs Spot42 and CyaR were probed with sequence-specific oligos. SdsR and crp mRNA were detected using riboprobes. 5S rRNA served as loading control. (B) Wild-type and ΔsdsR Salmonella carrying either a control vector, or the constitutive SdsR-expression plasmid pPL-SdsR were grown in M9 minimal medium supplemented with 0.4% glucose. At OD600 of 0.5, cells were harvested by centrifugation, washed in M9, and resuspended in M9 supplemented either with 0.4% glucose or 0.2% maltose, respectively, as sole carbon source. RNA was prepared from samples collected prior to, and at 15 and 30 min post carbon source shift. Spot42, CyaR, SdsR and 5S rRNA were detected as described in (A).
We next wanted to assess whether the presence of SdsR could interfere with the cellular response to environmental changes. We again employed CyaR as a direct readout of CRP-cAMP activity, and monitored expression levels of the sRNA upon a sudden shift in nutrient availability. CRP acts as the master regulator of catabolite repression that optimizes nutrient consumption by the bacterial cell. Its activity is triggered when glucose levels drop, and the levels of the second messenger cAMP increase concomitantly. Using the same Salmonella strains as before, i.e. wild-type, and ΔsdsR carrying either a control plasmid or pPL-SdsR, we initially grew the cells in M9 minimal medium supplemented with 0.4% glucose to an OD600 of 0.5 (t = 0 min). We next washed the cultures once in M9 medium lacking nutrients, split the cultures and resuspended in either M9 containing glucose or 0.2% maltose as carbon sources. We collected RNA samples 15 and 30 min post nutrient shift to maltose, and analyzed sRNA expression levels on Northern blots. CyaR was rapidly induced upon carbon shift to maltose in both wild-type and ΔsdsR cells, however became barely detectable in Salmonella over-expressing SdsR (Figure 6B, first and second panel).
DISCUSSION
Small regulatory RNAs are now accepted to serve crucial functions for microbial lifestyle and physiology and RNA-seq studies have uncovered countless sRNA candidates in numerous bacterial genomes (2,71,72). While these ‘sRNA catalogues’ have permitted a global view on the non-coding repertoire of many microbes, the functional characterization of sRNAs, even in model bacteria, has lagged behind discovery. Bioinformatic predictions of sRNA targets have steadily improved over the past few years and now go beyond traditional thermodynamic models including features such as target-site accessibility (73–75) and base-pairing conservation (53). Unfortunately, in most cases, such in silico methods still suffer from low accuracy (76,77), reflecting the multifaceted process of target recognition by bacterial sRNAs (36,62).
In this study, we have determined the target profile of SdsR in Salmonella. SdsR is an enterobacterial core sRNA and an integral part of the large regulon controlled by the stationary phase sigma factor, σS (40). Additional bacterial regulators such as GadY, SraL and SdsN sRNAs are also controlled by σS (78–80), and recent experiments in E. coli identified σS-binding to the promoter sequences of the two Hfq-dependent sRNAs, SdsR and OmrA (81). OmrA is a regulator of membrane homeostasis and biofilm formation in E. coli and, in addition to σS, transcription of omrA is controlled by the EnvZ/OmpR TCS (82–84). Several mRNA targets of OmrA have been reported, all of which are repressed by this sRNA. Similarly, out of the 20 novel SdsR targets identified in this study, only a single transcript, phnA, is activated SdsR (Figure 1C). Indeed, previous studies suggested that sRNAs could function as the inhibitory arm of otherwise activating transcriptional regulators, which is well illustrated by those sRNAs in the σE regulon to envelope stress. The alternative σE factor employs three core sRNAs (MicA, MicL and RybB), to inhibit the synthesis of nearly all major OMPs when envelope integrity is compromised (29,44,85). SdsR could serve an analogous function in the σS regulon since it also inhibits the translation of membrane proteins (ompD, stcD, envE and tolC; Figure 7A). Different from the σE-dependent sRNAs, however, mRNAs encoding OMPs are not enriched amongst the SdsR targets indicating a regulatory role going beyond a single physiological demand.
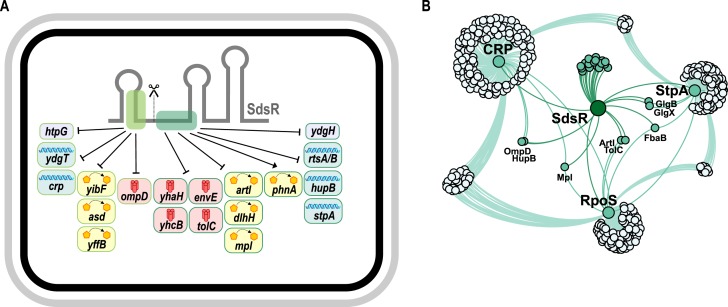
Figure 7.
The target spectrum of SdsR sRNA in Salmonella. (A) SdsR accumulates in the cell as two distinct species: a full-length and a processed version (SdsR+31; scissors indicate the sRNA processing site). SdsR employs two independent target sites (dark and light green boxes) to regulate 18 target mRNAs identified in this study from transcriptomic analysis; repression of ompD mRNA was confirmed previously (40). The target repertoire of SdsR includes genes encoding membrane proteins (red symbols), proteins with metabolic activity (yellow symbols), DNA-binding proteins (blue symbols) and factors not associated with any of these categories (gray). (B) Dense network of SdsR and the major transcriptional regulators, CRP, RpoS (σS) and StpA. Genes regulated by SdsR were obtained from Table 1, RpoS- and CRP-controlled genes (E. coli) were extracted from RegulonDB (106), and StpA-associated genes were collected from (100).
While several sRNAs perform dedicated functions within well-defined physiological pathways (86), there exists another group of sRNAs that seem to operate ‘in between’ regulons to harmonize cellular functions at the post-transcriptional level. The list of confirmed target genes (Table 1) indicates that SdsR may belong to this latter class of sRNAs. This list involves genes from diverse cellular pathways, including metabolic regulation (artI, asd and dlhH), membrane biogenesis (mpl), detoxification (yibF), generation of proton motive force (yhcB), and heat-shock response (htpG). In addition, the repression of global transcriptional regulators such as CRP, StpA, HupB and YdgT (Figures 1B and 7) hints at a pleiotropic role for SdsR in stationary phase. Indeed, Salmonella lacking sdsR display impaired stationary phase survival (87) and, possibly related, reduced biofilm formation (88). Interestingly, StpA also functions as an RNA-chaperone itself (89), adding another potential layer of regulation to the SdsR regulon.
Conversely, over-expression of SdsR renders E. coli cells more susceptible to levofloxacin and several other antibiotics (90,91). Repression of the tolC mRNA (Figure 3D), encoding part of a large multiple drug efflux pump (92), by SdsR is a key factor for this phenotype, however, regulation of additional target genes seems to matter as well (91). Of note, SdsR was recently shown in E. coli to base-pair with mutS mRNA, as part of a study presenting a potential role for SdsR in DNA repair in response to sub-inhibitory levels of beta-lactam antibiotics (93). However, our transcriptomic experiments did not indicate regulation of mutS by SdsR of Salmonella (Table 1), which we tentatively explain by a disruptive three-nucleotide difference in the predicted base-pairing region in mutS, as compared to E. coli. Whether or not SdsR targets identified in this study contribute to antibiotic resistance is yet to be established. One possible route would be regulation of crp by SdsR (Figure 1B), for CRP-deficient Salmonella display altered susceptibility to antibiotic challenge (94). The transcription regulators CRP and σS orchestrate two of the largest regulons in E. coli and related bacteria controlling sugar utilization and stationary phase growth, respectively (66). However, how these two factors interact is still not fully understood (95). Our data shows that SdsR connects the two key regulators at the post-transcriptional level (Figures 1B, 5A, 6 and 7B). Interestingly, cAMP-bound CRP also prevents transcription of rpoS (96), indicating a potential feed-forward loop where σS activates SdsR to reduce the inhibitory effect of CRP on rpoS transcription. Similarly, CRP controls OmpD expression, however, different from σS, CRP activates ompD transcription (97). In this case, SdsR will inhibit OmpD production by a dual mechanism: first, by inhibiting ompD translation through direct base-pairing with the mRNA (40), and second, by restricting transcriptional activation of ompD by CRP. Interestingly, two other SdsR-target mRNAs (hupB and mpl) have been reported to require CRP for transcriptional activation (98,99), indicating abundant cross-regulation between these two major regulons (Figure 7B). Repression of the conserved, nucleoid-associated factor StpA by SdsR (Figure 5E) adds yet another layer of complexity to the SdsR network. During exponential growth, StpA governs a large regulon of up to ∼5% of all Salmonella genes, many of which belong to the σS regulon (100). By restricting σS levels, StpA indirectly prevents premature expression of the σS regulon, and it is possible that SdsR helps override StpA-mediated repression of σS at the onset of stationary phase. In addition, StpA also promotes the expression of CRP-dependent genes (100,101). Hence, combined repression of crp and stpA mRNAs by SdsR fosters the expression of σS-dependent genes and endorses down-regulation of the CRP regulon.
Regulation by SdsR involves two seed-pairing domains for base-pairing with target mRNAs: one located in the distal sequence of the first stem-loop and another one located downstream of an RNase E-dependent cleavage site in the center of the molecule (Figures 2C and 7A). By investigating the effect of SdsR variants (Figure 4A) on the expression of gfp reporter fusions we tested the sRNA sequences required to regulate each individual target (Figures 1, 3 and 4). Based on our results, we predicted base-pairing interactions between SdsR and the cognate mRNA partners (Supplementary Figure S3). Alignments of the target genes revealed that in many cases, SdsR is anticipated to recognize conserved sequence elements within its mRNA counterparts, arguing that the regulon determined for Salmonella is at least partially overlapping the SdsR target sets in other species. To support this prediction, we chose two representative targets (ydgT and stpA) regulated either by the full-length or the processed sRNA, and experimentally validated their interaction with the SdsR. In our initial screen, we determined that two point mutations in SdsR, namely G26C and C38G, abrogated regulation of the ydgT::gfp and stpA::gfp fusions, respectively (Figure 4C and D). We introduced a single C to G mutation at position −24 relative to the translational start site in the ydgT::gfp reporter, obtaining fusion ydgT-M1::gfp. According to our prediction (Supplementary Figure S3E), this nucleotide base-pairs with guanosine 26 of SdsR, and its mutation should interfere with sRNA-mediated regulation. Indeed, ydgT-M1::gfp was fully resistant to SdsR (Supplementary Figure S4A). In contrast, regulation by SdsR G26C was restricted to the mutated target transcript; this successful compensatory base-pair exchange supports the predicted binding site of SdsR on the ydgT transcript.
Two possible interactions were predicted for base-pairing of SdsR and stpA mRNA (Supplementary Figure S3F). Therefore, we performed in vitro structure probing to identify which of the two sites was recognized by SdsR. Chemical probing using lead(II) acetate and RNase T1 in the presence and absence of either Hfq alone, or in combination with SdsR and SdsR G26C revealed protection of nucleotides −22 to −13 (relative to the translational start site of stpA) by the native SdsR variant. Given that no additional changes in the structure pattern were detectable in our assays, SdsR likely base-pairs with a single sequence element in the translation initiation region of stpA validating the first of our two predictions (Supplementary Figure S4B).
For a few reporters (yhcB::gfp, rtsA::gfp, envE::gfp, asd::gfp and phnA::gfp), none of the tested SdsR mutants was able to fully relieve regulation by the sRNA. However, close investigation of target site conservation and the potential interactions with SdsR indicated that the experimental data support the base-pairing predictions (Supplementary Figure S3). For example, the phnA::gfp mRNA is recognized by SdsR via an imperfect duplex excluding nt 38 of the sRNA (Supplementary Figure S3J). This finding could explain why SdsR C38G is functional as an activator of phnA::gfp expression. Except for asd::gfp, SdsR G26C fails to fully repress target gene expression in all target genes controlled by the full-length sRNA. In the predicted interaction forming between SdsR and asd mRNA (Supplementary Figure S3G), G26 is positioned at the distal end of one of the duplexes, contributing only marginally to RNA-duplex stability.
Processing by RNase E and accumulation of two sRNA species has also been observed for two other Hfq-dependent core sRNAs, i.e. ArcZ and RprA (26,56–58). Interestingly, for all three sRNAs, processing occurs upstream of the most conserved region which is employed as a seed for target recognition. In the case of RprA, we recently showed that the processed version of the molecule is an sRNA regulator in its own right (45). Likewise, processed SdsR is sufficient to regulate multiple target genes (Figure 3A). Whether or not processing is required to generate alternative sRNA regulators with target spectra different from their full-length isoforms, is currently unclear. Nonetheless, we note that full-length and processed sRNA regulators differ in their biochemical properties: whereas full-length sRNAs carry a tri-phosphate 5′ end, processed sRNAs bear a 5′ mono-phosphate which turns them into preferred substrates for RNase E-dependent degradation (102). However, recognition of 5′ mono-phosphorylated sRNAs by RNase E can also allosterically activate the enzyme, and thereby promote target degradation (103). The occurrence of mono- vs. tri-phosphorylated sRNA 5′ ends might also influence the mechanism by which sRNAs regulate their targets: as mono-phosphorylated sRNAs are more likely to recruit RNase E to the RNA duplex site, repression through target sequestration, as demonstrated for the Qrr sRNAs (57), might be incompatible with processed sRNA variants. Further experiments comparing natural, processed sRNAs with variants inactivated for processing will be required to pinpoint a functional role for sRNA processing in target selection and regulation.
Hfq-dependent sRNAs constitute the major class of post-transcriptional regulators in enteric bacteria (71). Whereas loss of Hfq results in drastic changes in protein expression in many species, mutants deficient for a single sRNA typically display only minor alterations in their proteomic profiles (3,104). Indeed, we found that deletion of sdsR had little or no effect on the protein levels of target genes (Figure 5 and Supplementary Figure S5). By contrast, SdsR over-expression clearly reduced protein production. The biological underpinnings of this conundrum are not understood, however, overlaying transcriptional control or redundantly acting sRNAs could be two possible explanations. Perhaps these regulations occur in a bacterial subpopulation, effects of which are too diluted to be observed in the current bulk cell analysis. In addition, sRNAs such as SdsR, which are expressed in late stationary phase cells, might be less likely to influence cellular protein levels as in cells that have stopped replicating the effect of protein dilution through cell division is reduced.
When is SdsR-mediated target regulation physiologically relevant? Documented phenotypes for sdsR deficient E. coli and Salmonella include long-term survival (87), biofilm formation (88) and antibiotic-induced mutagenesis (93). In all three cases, it is currently unclear if these phenotypes stem from the regulation of a single mRNA, or rather constitute the cumulative loss of multiple target regulation by SdsR. To address this, we compared the transcriptomes of wild-type and sdsR-deficient cells collected from late stationary phase (i.e. 10 h and 24 h post inoculation). Only a single gene (ybdL, encoding methionine aminotransferase) displayed significant deregulation (+3.2-fold) in the absence of sdsR (Supplementary Table S5). We did not identify ybdL as a target of SdsR in our target search (Table 1), and there is currently no evidence either that ybdL contributes to long-term survival, biofilm formation or response to antibiotics (105). Therefore, we propose that phenotypes associated with lack of sdsR might not be detectable in bulk cultures, but rather require analysis at the single cell level to minimize population-wide effects.
To summarize, the SdsR sRNA is a fascinating member of the group of Hfq-dependent sRNAs. Its broad conservation amongst the enterobacteria together with the global role of several of the here identified targets make SdsR a fascinating sRNA to study further. Indeed, SdsR has now become the subject of intensive research (40,81,87,88,91), and it is likely that a deeper understanding of this sRNA will provide important leads towards the mechanisms of target regulation by Hfq-dependent sRNAs and stationary phase physiology of Gram-negative bacteria.
Supplementary Material
Acknowledgments
We thank L. Barquist, E. Holmqvist and A. Westermann for helpful comments on the manuscript, and Barbara Plaschke for excellent technical assistance. Furthermore, we thank K. Förstner, J. C. Hinton and S. Lucchini for help with transcriptome analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [Vo875/2-2 to J.V., GRK2062 to K.P.]; Bavarian BioSysNet Program (to J.V.); Human Frontier Science Program [CDA00024/2016-C to K.P.]; Young Academy of the Bavarian Academy of Sciences. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wagner E.G., Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Waters L.S., Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz G., Vogel J., Wassarman K.M. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalaouna D., Simoneau-Roy M., Lafontaine D., Masse E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta. 2013;1829:742–747. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Papenfort K., Vanderpool C.K. Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 2015;39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lay N., Schu D.J., Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobrero P., Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 9.Ansong C., Yoon H., Porwollik S., Mottaz-Brewer H., Petritis B.O., Jaitly N., Adkins J.N., McClelland M., Heffron F., Smith R.D. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guisbert E., Rhodius V.A., Ahuja N., Witkin E., Gross C.A. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sittka A., Pfeiffer V., Tedin K., Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsui H.C., Leung H.C., Winkler M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 13.Oliva G., Sahr T., Buchrieser C. Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 2015;39:331–349. doi: 10.1093/femsre/fuv022. [DOI] [PubMed] [Google Scholar]
- 14.Bilusic I., Popitsch N., Rescheneder P., Schroeder R., Lybecker M. Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol. 2014;11:641–654. doi: 10.4161/rna.29299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sittka A., Lucchini S., Papenfort K., Sharma C.M., Rolle K., Binnewies T.T., Hinton J.C., Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 19.Tree J.J., Granneman S., McAteer S.P., Tollervey D., Gally D.L. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beisel C.L., Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papenfort K., Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Sharma C.M., Papenfort K., Pernitzsch S.R., Mollenkopf H.J., Hinton J.C., Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Rennie W., Liu C., Carmack C.S., Prevost K., Caron M.P., Masse E., Ding Y., Wade J.T. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 2015;43:10308–10320. doi: 10.1093/nar/gkv1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikumar S., Kroger C., Hebrard M., Colgan A., Owen S.V., Sivasankaran S.K., Cameron A.D., Hokamp K., Hinton J.C. RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015;11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westermann A.J., Forstner K.U., Amman F., Barquist L., Chao Y., Schulte L.N., Muller L., Reinhardt R., Stadler P.F., Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 26.Papenfort K., Said N., Welsink T., Lucchini S., Hinton J.C., Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 27.Updegrove T.B., Shabalina S.A., Storz G. How do base-pairing small RNAs evolve. FEMS Microbiol. Rev. 2015;39:379–391. doi: 10.1093/femsre/fuv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo M.S., Updegrove T.B., Gogol E.B., Shabalina S.A., Gross C.A., Storz G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gogol E.B., Rhodius V.A., Papenfort K., Vogel J., Gross C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao Y., Vogel J. A 3′UTR derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell. 2016;61:352–363. doi: 10.1016/j.molcel.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 31.Klein G., Raina S. Regulated Control of the Assembly and Diversity of LPS by Noncoding sRNAs. Biomed. Res. Int. 2015;2015:153561. doi: 10.1155/2015/153561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvail H., Masse E. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA. 2012;3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 33.Beisel C.L., Storz G. Discriminating tastes: physiological contributions of the Hfq-binding small RNA Spot 42 to catabolite repression. RNA Biol. 2011;8:766–770. doi: 10.4161/rna.8.5.16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desnoyers G., Masse E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 2012;26:726–739. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papenfort K., Sun Y., Miyakoshi M., Vanderpool C.K., Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papenfort K., Podkaminski D., Hinton J.C., Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E757–E764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bobrovskyy M., Vanderpool C.K. Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol. Microbiol. 2015;99:254–273. doi: 10.1111/mmi.13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyakoshi M., Chao Y., Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015;34:1478–1492. doi: 10.15252/embj.201490546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coornaert A., Chiaruttini C., Springer M., Guillier M. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013;9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fröhlich K.S., Papenfort K., Berger A.A., Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40:3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maciag A., Peano C., Pietrelli A., Egli T., De Bellis G., Landini P. In vitro transcription profiling of the σS subunit of bacterial RNA polymerase: re-definition of the σS regulon and identification of σS-specific promoter sequence elements. Nucleic Acids Res. 2011;39:5338–5355. doi: 10.1093/nar/gkr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber H., Polen T., Heuveling J., Wendisch V.F., Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. . 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer V., Sittka A., Tomer R., Tedin K., Brinkmann V., Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 44.Papenfort K., Bouvier M., Mika F., Sharma C.M., Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papenfort K., Espinosa E., Casadesus J., Vogel J. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4772–E4781. doi: 10.1073/pnas.1507825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fröhlich K.S., Papenfort K., Fekete A., Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C., Vogel J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012;84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 48.Urban J.H., Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papenfort K., Pfeiffer V., Lucchini S., Sonawane A., Hinton J.C., Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 51.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peer A., Margalit H. Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions. RNA. 2014;20:994–1003. doi: 10.1261/rna.043133.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright P.R., Richter A.S., Papenfort K., Mann M., Vogel J., Hess W.R., Backofen R., Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kroger C., Dillon S.C., Cameron A.D., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hebrard M., Handler K., et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunne T., Swarts D.C., Brouns S.J. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014;22:74–83. doi: 10.1016/j.tim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 57.Feng L., Rutherford S.T., Papenfort K., Bagert J.D., van Kessel J.C., Tirrell D.A., Wingreen N.S., Bassler B.L. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madhugiri R., Basineni S.R., Klug G. Turn-over of the small non-coding RNA RprA in E. coli is influenced by osmolarity. Mol. Genet. Genomics. 2010;284:307–318. doi: 10.1007/s00438-010-0568-x. [DOI] [PubMed] [Google Scholar]
- 59.Vogel J., Bartels V., Tang T.H., Churakov G., Slagter-Jager J.G., Huttenhofer A., Wagner E.G. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Apirion D., Lassar A.B. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- 61.Figueroa-Bossi N., Valentini M., Malleret L., Fiorini F., Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beisel C.L., Updegrove T.B., Janson B.J., Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durand S., Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papenfort K., Forstner K.U., Cong J.P., Sharma C.M., Bassler B.L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeiffer V., Papenfort K., Lucchini S., Hinton J.C., Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol .Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Antonio A., Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 67.Gorke B., Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 68.De Lay N., Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johansen J., Eriksen M., Kallipolitis B., Valentin-Hansen P. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and sigmaE-dependent CyaR-ompX regulatory case. J Mol Biol. 2008;383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 70.Moller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barquist L., Vogel J. Accelerating Discovery and Functional Analysis of Small RNAs with New Technologies. Annu. Rev. Genet. 2015;49:367–394. doi: 10.1146/annurev-genet-112414-054804. [DOI] [PubMed] [Google Scholar]
- 72.Fröhlich K.S., Papenfort K. Interplay of regulatory RNAs and mobile genetic elements in enteric pathogens. Mol. Microbiol. 2016 doi: 10.1111/mmi.13428. doi:10.1111/mmi.13428. [DOI] [PubMed] [Google Scholar]
- 73.Busch A., Richter A.S., Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eggenhofer F., Tafer H., Stadler P.F., Hofacker I.L. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011;39:W149–W154. doi: 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kery M.B., Feldman M., Livny J., Tjaden B. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014;42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peer A., Margalit H. Accessibility and evolutionary conservation mark bacterial small-rna target-binding regions. J. Bacteriol. 2011;193:1690–1701. doi: 10.1128/JB.01419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pain A., Ott A., Amine H., Rochat T., Bouloc P., Gautheret D. An assessment of bacterial small RNA target prediction programs. RNA Biol. 2015;12:509–513. doi: 10.1080/15476286.2015.1020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Opdyke J.A., Kang J.G., Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silva I.J., Ortega A.D., Viegas S.C., Garcia-Del Portillo F., Arraiano C.M. An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA. 2013;19:1253–1265. doi: 10.1261/rna.039537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hao Y., Updegrove T.B., Livingston N.N., Storz G. Protection against deleterious nitrogen compounds: role of sigmaS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw404. doi:10.1093/nar/gkw404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S., Landini P. Characterization of the Escherichia coli sigma(S) core regulon by Chromatin Immunoprecipitation-sequencing (ChIP-seq) analysis. Sci. Rep. 2015;5:10469. doi: 10.1038/srep10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guillier M., Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 83.Guillier M., Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holmqvist E., Reimegard J., Sterk M., Grantcharova N., Romling U., Wagner E.G. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J.C., Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richards G.R., Vanderpool C.K. Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta. 2011;1809:525–531. doi: 10.1016/j.bbagrm.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levi-Meyrueis C., Monteil V., Sismeiro O., Dillies M.A., Monot M., Jagla B., Coppee J.Y., Dupuy B., Norel F. Expanding the RpoS/sigmaS-network by RNA sequencing and identification of sigmaS-controlled small RNAs in Salmonella. PLoS One. 2014;9:e96918. doi: 10.1371/journal.pone.0096918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monteiro C., Papenfort K., Hentrich K., Ahmad I., Le Guyon S., Reimann R., Grantcharova N., Romling U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- 89.Doetsch M., Schroeder R., Furtig B. Transient RNA-protein interactions in RNA folding. FEBS J. 2011;278:1634–1642. doi: 10.1111/j.1742-4658.2011.08094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim T., Bak G., Lee J., Kim K.S. Systematic analysis of the role of bacterial Hfq-interacting sRNAs in the response to antibiotics. J. Antimicrob. Chemother. 2015;70:1659–1668. doi: 10.1093/jac/dkv042. [DOI] [PubMed] [Google Scholar]
- 91.Parker A., Gottesman S. Small RNA regulation of TolC, the outer membrane component of bacterial multidrug transporters. J. Bacteriol. 2016;198:1101–1113. doi: 10.1128/JB.00971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., Luisi B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gutierrez A., Laureti L., Crussard S., Abida H., Rodriguez-Rojas A., Blazquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J., et al. beta-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013;4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alper M.D., Ames B.N. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J. Bacteriol. 1978;133:149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Franchini A.G., Ihssen J., Egli T. Effect of Global Regulators RpoS and Cyclic-AMP/CRP on the Catabolome and Transcriptome of Escherichia coli K12 during Carbon- and Energy-Limited Growth. PLoS One. 2015;10:e0133793. doi: 10.1371/journal.pone.0133793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lange R., Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 97.Santiviago C.A., Toro C.S., Hidalgo A.A., Youderian P., Mora G.C. Global regulation of the Salmonella enterica serovar typhimurium major porin, OmpD. J. Bacteriol. 2003;185:5901–5905. doi: 10.1128/JB.185.19.5901-5905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Claret L., Rouviere-Yaniv J. Regulation of HU alpha and HU beta by CRP and FIS in Escherichia coli. J. Mol. Biol. 1996;263:126–139. doi: 10.1006/jmbi.1996.0564. [DOI] [PubMed] [Google Scholar]
- 99.Talukder A.A., Yanai S., Nitta T., Kato A., Yamada M. RpoS-dependent regulation of genes expressed at late stationary phase in Escherichia coli. FEBS Lett. 1996;386:177–180. doi: 10.1016/0014-5793(96)00426-7. [DOI] [PubMed] [Google Scholar]
- 100.Lucchini S., McDermott P., Thompson A., Hinton J.C. The H-NS-like protein StpA represses the RpoS (sigma 38) regulon during exponential growth of Salmonella Typhimurium. Mol. Microbiol. 2009;74:1169–1186. doi: 10.1111/j.1365-2958.2009.06929.x. [DOI] [PubMed] [Google Scholar]
- 101.Johansson J., Balsalobre C., Wang S.Y., Urbonaviciene J., Jin D.J., Sonden B., Uhlin B.E. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell. 2000;102:475–485. doi: 10.1016/s0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- 102.Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bandyra K.J., Said N., Pfeiffer V., Gorna M.W., Vogel J., Luisi B.F. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell. 2012;47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chao Y., Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 105.Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salgado H., Peralta-Gil M., Gama-Castro S., Santos-Zavaleta A., Muniz-Rascado L., Garcia-Sotelo J.S., Weiss V., Solano-Lira H., Martinez-Flores I., Medina-Rivera A., et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2013;41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.