Abstract
The exosome-independent exoribonuclease DIS3L2 is mutated in Perlman syndrome. Here, we used extensive global transcriptomic and targeted biochemical analyses to identify novel DIS3L2 substrates in human cells. We show that DIS3L2 regulates pol II transcripts, comprising selected canonical and histone-coding mRNAs, and a novel FTL_short RNA from the ferritin mRNA 5′ UTR. Importantly, DIS3L2 contributes to surveillance of maturing snRNAs during their cytoplasmic processing. Among pol III transcripts, DIS3L2 particularly targets vault and Y RNAs and an Alu-like element BC200 RNA, but not Alu repeats, which are removed by exosome-associated DIS3. Using 3′ RACE-Seq, we demonstrate that all novel DIS3L2 substrates are uridylated in vivo by TUT4/TUT7 poly(U) polymerases. Uridylation-dependent DIS3L2-mediated decay can be recapitulated in vitro, thus reinforcing the tight cooperation between DIS3L2 and TUTases. Together these results indicate that catalytically inactive DIS3L2, characteristic of Perlman syndrome, can lead to deregulation of its target RNAs to disturb transcriptome homeostasis.
INTRODUCTION
The lifetimes of different RNA species vary significantly. Although rRNAs, tRNAs and snRNA are very stable molecules that are degraded only under particular conditions such as stress (1), protein-coding transcripts (mRNAs) have much shorter life spans. In eukaryotes, degradation of RNA polymerase II transcripts, especially mRNAs, has been extensively studied (reviewed in (2)). mRNA degradation usually begins with shortening of the poly(A) tail or endonucleolytic cleavage, followed by exonucleolytic decay, which may proceed from either the 5′-end or the 3′-end. Degradation from the 5′-end requires cap removal that is mediated by the Xrn1 exoribonuclease, while the exosome complex governs degradation from the 3′-end. Recent progress in high-throughput sequencing technologies allowed the discovery of many non-coding RNAs (ncRNAs) and other variable RNAs that result from pervasive transcription and post-transcriptional misprocessing events. However, little is known about the processing and degradation mechanisms for these RNAs. More detailed investigation of this issue is needed, since ncRNAs have important influences on cell physiology (3,4).
The human genome encodes three processive 3′ to 5′ exoribonucleases: DIS3, DIS3L and DIS3L2, each equipped with a RNB catalytic domain (5). Through their N-terminal PIN domains, DIS3 and DIS3L associate with either the nuclear or the cytoplasmic exosome complexes, respectively. The nucleoli-enriched ribonuclease EXOSC10 (RRP6) is also a part of the exosome complex (5–7). Meanwhile, DIS3L2 lacks a PIN domain and is not known to be a subunit of any macromolecular complex (8). DIS3L2 localizes to the cytoplasm (8,9) where it contributes to an evolutionarily conserved, exosome-independent, cytoplasmic RNA degradation pathway. Biochemical analyses showed that DIS3L2 is a robust 3′ to 5′ exoribonuclease, capable of degrading single- and double-stranded RNA substrates in vitro (8). Recent structural analyses of S. pombe and mouse DIS3L2 indicated that the wide channel leading to the catalytic site allows this protein to degrade structured substrates (10,11). siRNA-mediated DIS3L2 depletion changes the expression of many mRNAs, and increases the half-lives of selected mRNAs and P-body amounts (8). Additionally, DIS3L2 interacts with XRN1 suggesting these proteins cooperate to degrade mRNAs from both termini, which was shown to occur, at least partially, on polysomes (8). S. pombe DIS3L2 also participates in mRNA metabolism and deletion of its gene is synthetically lethal with xrn1Δ, since presumably both proteins act redundantly by degrading transcripts from either the 5′- or 3′-end (12).
Several lines of evidence point to a cooperative function of DIS3L2 and the cytoplasmic terminal uridyltransferases TUT4 and TUT7. DIS3L2 mediates mRNA decay during apoptosis through degradation of uridylated intermediates (13). Similarly, in S. pombe and in mouse embryonic stem cells (ESC), DIS3L2 activity is preferentially enhanced by 3′-terminal uridine tails (11,12). DIS3L2 degrades uridylated let-7 family pre-miRNAs and thus plays a role during cell differentiation (14,15).
The possible association between abnormalities in DIS3L2-mediated RNA metabolism and diseases suggests that DIS3L2 also plays important roles in maintaining cell homeostasis. Indeed, DIS3L2 mutations are involved in Perlman syndrome (a rare overgrowth disorder) and Wilms’ tumor (kidney cancer that typically occurs in children) (9). DIS3L2 knockdown in HeLa cells results in aneuploidy, mitotic errors and deregulation of mitotic control proteins and their transcripts (9). Gene ontology (GO) analysis of high-throughput RNA-Seq data showed that loss of DIS3L2 expression resulted in an increased abundance of transcripts related to cell-cycle regulation, which partially explains the elevated cell proliferation that occurs with DIS3L2 dysfunction (8). However, in order to understand how widespread the role of DIS3L2-mediated RNA metabolism is, further research is needed to identify additional DIS3L2 substrates and to examine the role of terminal uridylation in their degradation.
Here, we generated a comprehensive human cellular model to study the role of DIS3L2 and performed global transcriptomic analyses to investigate the genome-wide consequences of DIS3L2 dysfunction. Our results revealed novel DIS3L2 substrates, including selected mRNAs, vault and Y RNAs, BC200 (an Alu-like element RNA), a novel FTL_short RNA from the ferritin mRNA 5′ UTR and extended snRNAs, all of which are degraded by DIS3L2 in a uridylation-dependent manner. Our data suggest that DIS3L2 dysfunction manifested through deregulation of its substrate RNAs likely abolishes cell homeostasis and may contribute to disease development.
MATERIALS AND METHODS
Human cell cultures and generation of stable cell lines
Human Flp-In T-REx 293 (HEK293) (Invitrogen) cells were cultured in monolayers in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in a 5% CO2 incubator.
The stable inducible HEK293 cell lines producing DIS3L2 variants were obtained using ‘pAL_01’ and ‘pAL_02’ plasmid constructs and the Flp-In™ T-REx™ system according to the manufacturer's guidelines. For WT and mut variants of DIS3L plasmids, ‘pAL_03’ and pAL_04’ were used, respectively. Plasmid generation is described in detail in the Supplementary Data. Lipofectamine2000 (Invitrogen) was used for transfection. After establishment of stable cell lines, cells were cultured as described above, but hygromycin B (Invitrogen, 100 μg/ml) was added to the medium. To induce exogenous gene expression, doxycycline (100 ng/ml) was added to the medium. For experiment with inducer titration, the following concentrations (ng/ml) of tetracycline were used: 0.2, 1, 2, 5, 10 and 100. Generation of the stable inducible HEK293 cell lines producing different versions of DIS3 and RRP6 proteins as well as their culture and induction were described previously (16).
RNA isolation
For RNA isolation, stable HEK293 cell lines producing DIS3L2 and DIS3L variants were treated with doxycycline (100 ng/ml) for 3 days before RNA was isolated using TRI Reagent (Sigma-Aldrich) according to the manufacturer's guidelines.
cDNA synthesis and quantitative PCR (qPCR)
Total RNA (10 μg) isolated from cells was treated with 4 U TURBO™ DNase (Ambion) in the presence of RiboLock RNase Inhibitor (Thermo Fisher Scientific), according to the manufacturer's protocol. Phenol:chloroform extraction was then performed and RNA was precipitated with isopropanol. DNase-treated RNA (2 μg) was used for cDNA synthesis. Reverse transcription was carried out using a mixture of 50 pmol of an oligo(dT) primer, 250 ng of random primers (Invitrogen) and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's guidelines, in a final volume of 20 μl. Samples were diluted 10-fold and mixed with 2.5 pmol of each qPCR primer (see Supplementary Table S2 for sequences), 0.5 μg bovine serum albumin and Platinum® Quantitative PCR SuperMix-UDG (Invitrogen), in a final volume of 10 μl. qPCR was carried out in a Roche LightCycler® 480 system. Negative controls (-RT) were included for each experiment and showed an insignificant background. GAPDH mRNA was used for normalization. Reaction specificity was confirmed by melting curve analysis. Analyses were conducted in triplicate.
Northern blotting
Total RNA (10 μg) was separated by electrophoresis in a 1% agarose/0.4 M formaldehyde gel in NBC buffer (50 mM boric acid; 1 mM sodium acetate; 5 mM NaOH) and transferred onto a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) by overnight capillary transfer in 20× SSC buffer (3 M NaCl; 0.3 M sodium citrate). The RNA was then fixed on membranes by UV-crosslinking. The blots were hybridized with probes in PerfectHyb Plus hybridization buffer (Sigma-Aldrich) at 42°C overnight. Oligonucleotide probes (10 pmol/reaction) were labelled with T4 PNK (NEB) and 1 μl [γ-32P]ATP (Hartmann Analytic; 3000 Ci/mmol) in a total volume of 10 μl according to the manufacturer's guidelines (see Supplementary Table S2 for sequences). After hybridization the membranes were washed twice with 2× SSC, 0.1% SDS (40 min each) at 42°C. The membrane was exposed in a PhosphorImager cassette (Fuji Film) and analyzed by phosphorimaging [FLA 7000 scanner (Fuji Film)]. Between successive hybridizations, the membranes were stripped using boiling 0.1% SDS followed by incubation at 65°C.
For separation of shorter fragments, total RNA (10 μg) was fractionated in a polyacrylamide-urea denaturing gel (6%, 0.5 or 1.0 mm thick) and transferred onto a Hybond-N+ nylon transfer membrane by electroblotting (0.5× TBE at 4°C, for 90 min or 180 min, respectively). RNA was then fixed on membranes by UV-crosslinking and blots were handled as described above.
RNA-Seq
For RNA-seq we used RNA samples treated with DNase (performed as described above) and rRNA-depleted using a Ribo-Zero Kit (Epicentre) according to the manufacturer's protocol. Strand-specific RNA libraries were prepared in triplicate using a TruSeq RNA Sample Preparation Kit (Illumina) as per the manufacturer's protocol with several previously described modifications (16). The libraries were sequenced using an Illumina HiSeq sequencing platform to an average number of 18.8 million reads per sample in the 75-nt paired-end mode.
Small-RNA-Seq
Following DNase treatment and ribodepletion of 10 μg total RNA that was performed as described above, RNA molecules underwent TAP (Epicentre) treatment (5 U enzyme, 40 U RiboLock RNase Inhibitor) at 37°C for 2 h. After phenol:chloroform extraction and ethanol precipitation, 30 pmol of pre-adenylated linker RA3 (Illumina) was ligated using 200 U of T4 RNA Ligase 2 truncated (NEB, M0242) in the presence of 12% PEG 8000 (NEB) and 20 U RiboLock RNase Inhibitor in T4 RNA Ligation buffer (NEB) for 1 h at 25°C in a final volume of 20 μl, preceded by denaturation at 70°C for 2 min in the absence of enzyme. In the next step, RTP primer (Illumina) was annealed by incubating 30 pmol of the oligo with RNA according to following scheme: 5 min at 75°C, 30 min at 37°C and 15 min at 25°C. Afterward, 30 pmol of RA5 linker (Illumina) was ligated with 10 U T4 RNA Ligase 1 (NEB, M0204) in the presence of 1 mM ATP in T4 RNA Ligation buffer (NEB) for 1 h at 25°C, in a final volume of 30 μl. One-third of the ligation mixture was used for reverse transcription, conducted with SuperScript III Reverse Transcriptase in the presence of 20 U RiboLock RNase Inhibitor, 0.5 mM dNTPs and 0.01 M dithiothreitol (DTT) for 1 h at 42°C, in a final volume 20 μl, following incubation at 42°C for 2 min in the absence of enzyme. Finally, libraries were amplified in a PCR reaction with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) using 20 pmol of RP1 and RPI_X - primers (where X represents index number) (Illumina) in a volume of 40 μl. The following PCR profile was used: initial denaturation at 98°C for 30 s, then 10 cycles of denaturation (98°C, 10 s), annealing (60°C, 30 s) and elongation (72°C, 30 s), followed by 5 min of final elongation at 72°C. Following PCR, libraries were purified with Magnetic Beads (Agencourt AMPure XP; Beckman Coulter), according to the manufacturer's guidelines, eluted in 12 μl RSB Buffer (Illumina) and separated in a 6% native polyacrylamide gel. Products ranging in size from 140 to 320 bp were excised and retrieved from the acrylamide by a crush-and-soak method and overnight incubation with elution buffer (0.5 M ammonium acetate, 1 mM EDTA and 0.1% SDS). Libraries were then precipitated and purified again with Magnetic Beads. The quality of prepared libraries was checked using the Bioanalyzer and High Sensitivity DNA kit (Agilent Technologies).
RNA-Seq and small RNA-Seq analysis
RNA-Seq reads were mapped to a reference human genome (hg38) using the STAR short read aligner with default settings (version STAR_2.4.0b) to yield on average 83.1% and 79.4% uniquely mapped reads for WT and mutant samples, respectively.
Reads from small RNA-Seq were aligned to the genome using the STAR short read aligner with the outFilterScoreMinOverLread and outFilterMatchNminOverLread options set to 0.20. The mapping yielded on average 1.54 million and 1.21 million uniquely aligned reads in mutant and WT samples, respectively. The quality control, read processing and filtering, visualization of the results and counting of reads for the GENECODE v22 comprehensive annotation was performed using custom scripts and elements of the RSeQC, BEDtools and SAMtools packages. Differential expression analyses were performed using the DESeq2 Bioconductor R package. GO analysis was performed using DAVID. To calculate the score for GENCODE gene type enrichment, the number of significantly upregulated genes of a given type was divided by the total number of expressed genes of the group (defined as at least 10 reads of coverage). The statistical significance of this enrichment was estimated with Fisher's exact test.
RACE-Seq
RNA for 3′-RACE-Seq was treated with DNase (performed as described above) and 2 μg of DNase-treated RNA was used to prepare libraries via the following steps: (i) RA3 linker ligation, (ii) RTP primer hybridization, (iii) reverse transcription (carried out as described in the Small RNA-Seq protocol), (iv) PCR amplification (different primers compared to Small RNA-Seq; forward: gene specific primer with RA5 sequence at the 5′ end (see Supplementary Table S2 for sequences), reverse: RTP), (v) Clean-Up purification (A&A Biotechnology, according to the manufacturer's protocol) and (vi) PCR amplification (different primers than the previous PCR, forward: RP1, reverse: RPI_X). For PCR, 10 cycles of amplification were used except for U4atac (RNU4atac) and U5 (RNU5A-1), when 30 cycles were performed. After PCR, libraries were purified following electrophoresis in a native, 6% polyacrylamide gel and then additionally with Magnetic Beads (in the case of VTRNA1-2 and RNY4 transcript) or with Magnetic Beads alone (snRNA transcripts).
RACE-Seq analysis
Sequences were clipped off the adapters using cutadapt, followed by R1 reads mapping to reference transcript sequences using bowtie2. R2 reads that had mates mapping to references were mapped to reference transcripts with the soft-clipping option enabled (bowtie2 option—very-sensitive-local). Transcript ends were determined based on the mapping, whereas tails were analyzed based on soft-clipped fragments of reads using in-house Perl and R scripts. Soft-clipped fragments were treated as having a polyU tail if they contained >80% uridines. For vtRNA1-2 and Y4 transcripts, terminal uridines were included in the soft-clipped part of the read. Due to high level of mature transcript reads in U1 and U11 samples, tails were analyzed only for fragments mapped downstream of mature transcript (positions 902 and 741 for U1 and U11, respectively).
Altering iron levels
To change cellular iron levels, cells were treated with either 50 μg/ml ferric ammonium citrate (FAC) or 100 μM deferoxamine (DFO, both from Sigma) for 24 h, after which RNA and proteins were isolated for northern blot and western blot experiments, respectively.
Analysis of FTL_short uridylation status
Reads from the shortRNA library preparation aligning to the FLT 3′ end were stripped of adapter sequences using cutadapt, examined for the presence of untemplated sequences using the SamExtractClip tool of the JVarkit package, and the nucleotide composition was assessed with a custom script.
TUT4 and TUT7 knockdown and overexpression
To achieve efficient knockdown of TUT4 and TUT7 expression in HEK293 cell lines producing different DIS3L2 variants, cells were seeded in six-well plates at 0.3 × 106 cells per well in a medium lacking antibiotics. On the next day (∼20 h post seeding), the cells in a single well were transfected with 5 μl RNAiMAX transfection reagent, combined with either 30 pmol control siRNAs (medium GC or high GC control stealth siRNA; Invitrogen ref. nos. 462000 and 462002) or a pool of siRNAs containing 15 pmol siRNAs against either TUT4 or TUT7 (both Stealth siRNA from Invitrogen ref. nos. HSS177328 and HSS149224) in 300 μl OPTI-MEM serum-free medium. Triplicates for each condition were prepared. Sixteen hours post-transfection, the medium was exchanged with fresh medium containing penicillin+streptomycin. Cells were harvested 72 h after transfection. Around one-third of the cells were scraped with medium, centrifuged and used for western blotting to verify knockdown of protein expression. The remaining cells were harvested with TRI Reagent for RNA retrieval.
For TUT4 and TUT7 overexpression, HEK293 cells were seeded in six-well plates at 0.5 × 106 cells per well. On the next day (∼20 h post seeding), cells from a single well were transfected with 7 μl Lipofectamine 2000 (Invitrogen) and 2.5 μg pcDNA5/FRT/TO plasmid DNAs comprising a cassette encoding N-terminally eGFP-tagged TUT4, TUT7 or their catalytically-inactive mutant versions, D1011A and D1060A, respectively. Triplicate transfections for each condition were performed. At 12 h post-transfection, the medium was exchanged for DMEM+FBS containing 100 ng/ml tetracycline and penicillin+streptomycin. Transfection efficiency and eGFP-tagged protein expression were tested by flow cytometry and western blotting (data not shown). Cells were harvested 72 h post transfection with TRI Reagent, and RNA was isolated as described above.
Production of recombinant DIS3L2
DIS3L2 CDS was cloned into the pET28 expression vector with a C-terminal 6xHis tag. The plasmids were transformed into BL21-RIL Escherichia coli cells and plated onto LB agar plates supplemented with kanamycin (50 μg/ml) and chloramphenicol (37.5 μg/ml). A single colony was used to start a 50 ml inoculum in LB + kanamycin and chloramphenicol and the cells were allowed to grow for 12 h at 37°C with constant rocking. The inoculum was then used to start 1 l of culture in an autoinducing medium (AIM-Formedium). The cells were allowed to proliferate for 48 h at 18°C with constant rocking before they were harvested by centrifugation. The cell pellet was suspended in 150 ml Buffer 1 (20 mM Tris–HCl pH 8, 500 mM NaCl, 10 mM imidazole supplemented with protease inhibitors and 50 μg/ml lysozyme). The cells were lysed with Emulsiflex for 15 min following the manufacturer's guidelines. The lysate was cleared by centrifugation at 37 000 rpm in a Thermo FL37 8 × 100 rotor at 4°C for 45 min. Recombinant DIS3L2 was bound to a 5 ml NiNTA FastFlow column (Qiagen) using Äkta Xpress, and the column was washed with 20 column-volumes of Buffer 1 and 2 column volumes of Buffer 2 (20 mM Tris–HCl pH 8, 1 M NaCl). DIS3L2 protein was eluted with Buffer 3 (20 mM Tris–HCl pH 8, 500 mM NaCl, 300 mM imidazole). Collected fractions were analysed by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and those containing DIS3L2 were diluted to reach 100 mM NaCl followed by final purification on a RESOURCEQ column (GE Healthcare). The purified protein was analysed by SDS-PAGE, and the concentration was determined using a Nanodrop apparatus at 280 nm and the respective molar extinction coefficient. Samples were supplemented with glycerol to 20%, frozen and stored at −80°C.
TUT4 production
TUT4 was expressed in FreeStyle 293-F cells (Invitrogen). The cells were cultured in 30 ml 5× FreeStyle 293 Expression Medium (Gibco) to a density of 1 × 106 cells/ml. The cells were transfected using 90 μg linear PEI at 1 μg/μl (Mw = 25 000; Polysciences Inc.) and 30 μg plasmid pZW_GZ11 for eGFP-TEV-TUT4 expression (Supplementary Materials and Methods). Following transfection, the cells were cultured for 48 h at 37°C, 5% CO2, with constant rocking at 120 rpm. The cells were then centrifuged for 5 min at 500 g and the resulting cell pellet was suspended in 5 ml buffer 1 (20 mM Tris–HCl pH 8, 500 mM NaCl 0.1% Triton X-100 supplemented with 100 U viscolase (nuclease; A&A Biotechnology) and protease inhibitors. The mixture was rotated head-over-tail at 10°C for 15 min and then sonicated at 10°C for 30 min using a Bioruptor sonicator (Diagenode) operating in the heavy duty setting with 15 s pulses followed by 45 s pauses. The lysate was cleared by centrifugation at 12 000 g at 4°C for 20 min. The resulting supernatant was bound to magnetic beads (CNBr-activated SepFast Mag4F; Biotoolomics) coupled to an anti-GFP nanobody (home-made) for 2 h at 10°C with head-over-tail rotation. The beads were then washed with 40 bed-volumes buffer 1. TUT4 was released from the beads by TEV protease digestion in 300 μl buffer 1 supplemented with 10 μg TEV protease (home-made) for 12 h. The released and bead-retained proteins were analysed by SDS-PAGE. Aliquots were supplemented with glycerol to 20%, frozen and stored at −80°C.
In vitro vault RNA and Y RNA degradation assays
DNA sequences corresponding to RNY4, VTRNA1-1 and VTRNA1-2 were retrieved from a nr (non-redundant) nucleotide database (respective accession numbers: NR_004393.1, NR_026703.1 and NR_026704.1) and chemically synthesized with additional sequences from the T7 RNA polymerase promoter (agatgtaatacgactcactatagg) at the 5′-ends. RNY4, VTRNA1-1, VTRNA1-2 sequences and variants of the latter comprising 2, 4 and 6 Us were amplified by PCR, and the RNAs were produced by standard in vitro transcription with a home-made T7 RNA polymerase. The transcripts were purified after electrophoresis in 6% denaturing PAGE. Transcripts (2 μg) were dephosphorylated by FastAP (Thermo Fisher Scientific) and purified by phenol:chloroform extraction and precipitation. Dephosphorylated RNAs (10 pmol) were 5′-labelled with [γ-32P]ATP using PNK (NEB) in 0.5× NEB2 buffer and purified by 6% denaturing PAGE, elution in 300 mM NaOAc pH 5.3, 2 mM EDTA, 0.5 vol/vol phenol:chloroform:isoamyl 25:24:1 at 10°C overnight and precipitation. Two master mixes, one containing 50 nM ‘cold’ RNA and trace amounts of 32P-labelled RNA in 0.5× NEB2 buffer with RNase inhibitor (RiboLock; Thermo Fisher Scientific), and the other containing 50 nM recombinant DIS3L2 in 0.5× NEB2 buffer alone were adjusted to 37°C for 2 min and then rapidly combined in a 1-to-1 volumetric ratio to start a time-course in a total volume of 20 μl. Time-point aliquots (3 μl) at 3, 6, 9, 12, 22 and 40 min were withdrawn and rapidly quenched in 5 μl ice-cold formamide-RNA loading dye containing 10 mM ethylenediaminetetraacetic acid (EDTA). 0 and 40 min time point aliquots without proteins were withdrawn either before mixing RNA (0 min) or after a 40 min incubation at 37°C without proteins. RNAs were separated by 18% (19:1) denaturing PAGE in 40 cm × 40 cm gels at 600 V for 2h. The gels were exposed in Phosphorimager cassettes for 40 min and scanned using a Fuji PhosphorImager (FLA7000). Full-length substrates and degradation products were quantified using Multigauge software and quantification results were plotted using GraphPad Prism software.
In experiments in which hTUT4 was added, 2 nM of wild type or catalytically inactive mutant (D1011A) proteins in 20 mM Tris–HCl pH 8, 0.5 M NaCl and 25% glycerol were included in the reactions (added to the protein master mix). The protein preparation or respective buffer (control w/o protein) constituted 1/10th of the final reaction volume. UTP (1 mM) was added to both master mixes before combining them and commencing the time-course. The presence of UTP slowed down the DIS3L2 degradation kinetics (data not shown) in a concentration dependent manner, which accounts for the difference in the calculated half-times of the vault RNA1-2 in degradation assays without and with TUTs and UTP.
In vitro vault RNA and Y RNA uridylation assays
RNY4, VTRNA1-1 and VTRNA1-2 (50 nM) prepared as described above were combined with 2 nM wild type or catalytically inactive (D1011A) hTUT4 as described for the degradation assays, but without DIS3L2. RNAs were separated by 10% sequencing PAGE (20 cm × 50 cm gels) at 2100 V for 3 h 15 min, exposed to a Phosphorimager screen, scanned and quantified as described above. Control reactions were carried out with 50 nM 5′-FAM-labelled 44-mer RNA (CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA). The RNAs were separated by 6% denaturing PAGE and scanned using a FLA-9000 fluorescence scanner (GE Healthcare).
Cell fractionation
Stable inducible human HEK293 cell lines were cultured in 100 mm dishes and treated with doxycycline (100 ng/ml) for 3 days. To obtain RNAs from different cellular compartments, cells were fractionated after induction using a previously described procedure (17) with slight modifications (see Supplementary Materials and Methods).
4sU labelling
Stable inducible human HEK Flp-In T-REx cell lines producing WT/mut versions of DIS3L2 were cultured in 100 mm dishes and treated with doxycycline (100 ng/ml) for 3 days. 4sU labelling was carried out as previously described (18) with minor modifications (see Supplementary Materials and Methods).
RESULTS
Global analysis of DIS3L2 substrates
For global analyses of DIS3L2 functions, we established a human cellular model based on stable cell lines derived from parental HEK293 Flp-In T-Rex cells expressing sh-miRNAs to silence endogenous DIS3L2, and simultaneously producing sh-miRNA-insensitive versions of DIS3L2 that were either wild type (WT) or the catalytically-inactive mutant D391N (mut) (8). We chose the D391N mutation because mutations leading to inhibition of DIS3L2 ribonuclease activity were identified in patients with the Perlman syndrome (9). Validation experiments showed efficient silencing of endogenous DIS3L2 and potent production of exogenous proteins (Supplementary Figure S1), which confirmed that the system was working properly.
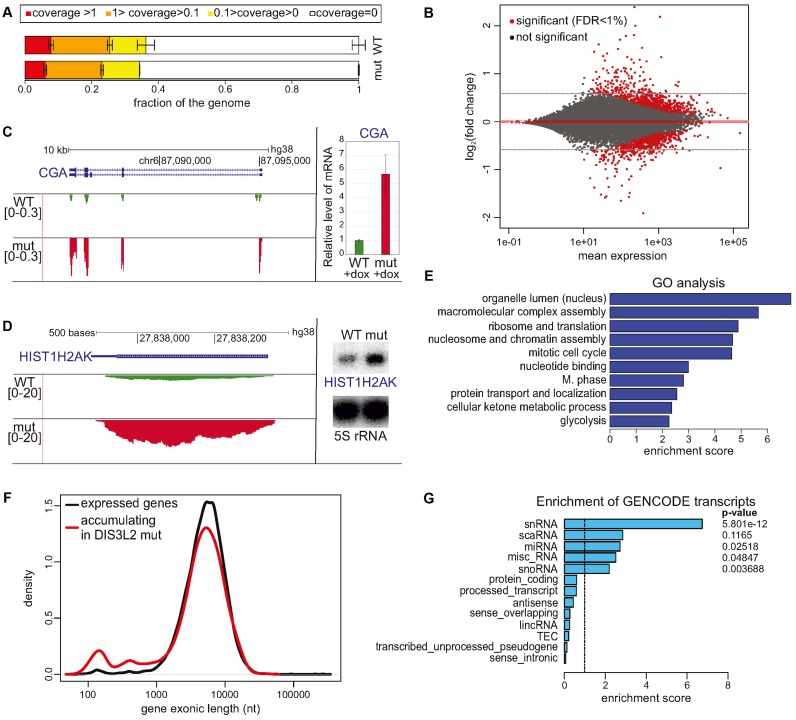
To investigate DIS3L2 functions genome-wide, total RNA samples were collected from model cell lines producing either WT or mut DIS3L2 three days after induction with doxycycline. The RNA samples were rRNA-depleted before preparation of strand-specific total RNA libraries according to the standard TruSeq (Illumina) protocol. The samples were sequenced and mapped to an average depth of 15.3 million uniquely aligning reads. Since DIS3L2 is a cytoplasmic nuclease, there were no effects on spurious, nascent transcription degradation, as visualized by the unaltered fraction of the genome portion that was poorly covered (Figure 1A) and the majority of reads (on average 79.2%) that mapped to exons of known genes.
Figure 1.
Global analysis of DIS3L2 substrates. (A) RNA-Seq showed no significant difference between the genome fractions covered by RNA-Seq reads to given depths in WT and mutant DIS3L2 expressing cells. Bars represent the standard deviation for three biological replicates. (B) MAplot of total RNA sequencing results representing differential expression of DIS3L2 WT compared to DIS3L2 mut. Statistically significant hits (FDR < 1%) are color-coded red. Transcripts accumulating in the DIS3L2 mut have a positive value on the y-axis. (C and D) mRNAs are DIS3L2 substrates. Screenshots from Genome Browser showing genomic regions encoding CGA mRNA (C) or HIST1H2AK mRNA (D), with reads from deep sequencing for WT and mut DIS3L2. Number denotes the normalized expression measured by RNA-Seq (left); validation of CGA mRNA (C) and HIST1H2AK mRNA (D) accumulation in mut DIS3L2 by qPCR (bars represent the standard deviation for three biological replicates) and northern-blot (5S rRNA was used as an internal control), respectively (right). (E) Clustered GO analysis of genes accumulating in the DIS3L2 mutant. (F) Exon size distribution in genes that were upregulated in mutant DIS3L2 relative to all expressed genes is skewed towards a minor gene population with exons shorter than 1 kb and a maximum of approximately 150 nt. (G) Enrichment analysis of GENCODE transcript types among transcripts accumulating in DIS3L2 mutant cells compared to all expressed genes reveals a significant overrepresentation of snRNA. Other classes of short and non-protein coding RNA molecules, including miRNA, miscRNA and snoRNA, also showed moderate enrichment. Right, Fisher exact test P-value.
RNA-Seq data analysis summarized in a MAplot showed differential gene expression between cells producing WT and mut DIS3L2, which had global deregulation of mRNA (Figure 1B). Differential expression analysis with a 1% FDR cut-off revealed that there were 809 upregulated genes and 764 downregulated genes in DIS3L2 mutant cells compared to WT cells. However, the extent of deregulation was moderate, with only 58 genes showing more than 2-fold changes in expression (42 up- and 16-downregulated in the DIS3L2 mutant). Selected mRNA targets were validated by qPCR (Figure 1C, Supplementary Figure S2A). In agreement with a previous report (8), mRNAs encoding selected replication-dependent histones were also identified as DIS3L2 substrates (Figure 1D, Supplementary Figure S2B). Importantly, the effect was not related to general changes in the cell cycle, which is known to have a huge impact on the histone mRNA expression, as verified by flow cytometry analysis (Supplementary Figure S2C), suggesting that histone-coding mRNAs are direct DIS3L2 targets. Furthermore Gene Ontology (GO) analysis of statistically significant hits showed no deregulation of a specific pathway, but rather moderate deregulation of general homeostasis processes. Clustered GO analysis of transcripts accumulating in the presence of mutated DIS3L2 using DAVID indicated that DIS3L2 substrates are involved in chromatin assembly and nuclear organization, ribosome structure and translation, mitotic cell cycle and the M-phase regulation (Figure 1E).
Although most deregulated RNAs followed the size distribution characteristic of all expressed transcripts, among the upregulated genes in DIS3L2 mut cells there was an overrepresentation of transcripts shorter than 1kb, with a clear peak at about 100 nt (Figure 1F). Similarly, the GENCODE gene types representing snRNA were significantly enriched (Figure 1G), even though standard TruSeq library preparation favours RNA molecules longer than 200 nt, and shorter transcripts are suboptimal for sequencing via this protocol. Thus, to obtain information about potential DIS3L2 RNA substrates with lengths between 20 and 220 nt, another RNA-Seq was carried out in parallel (see Materials and Methods).
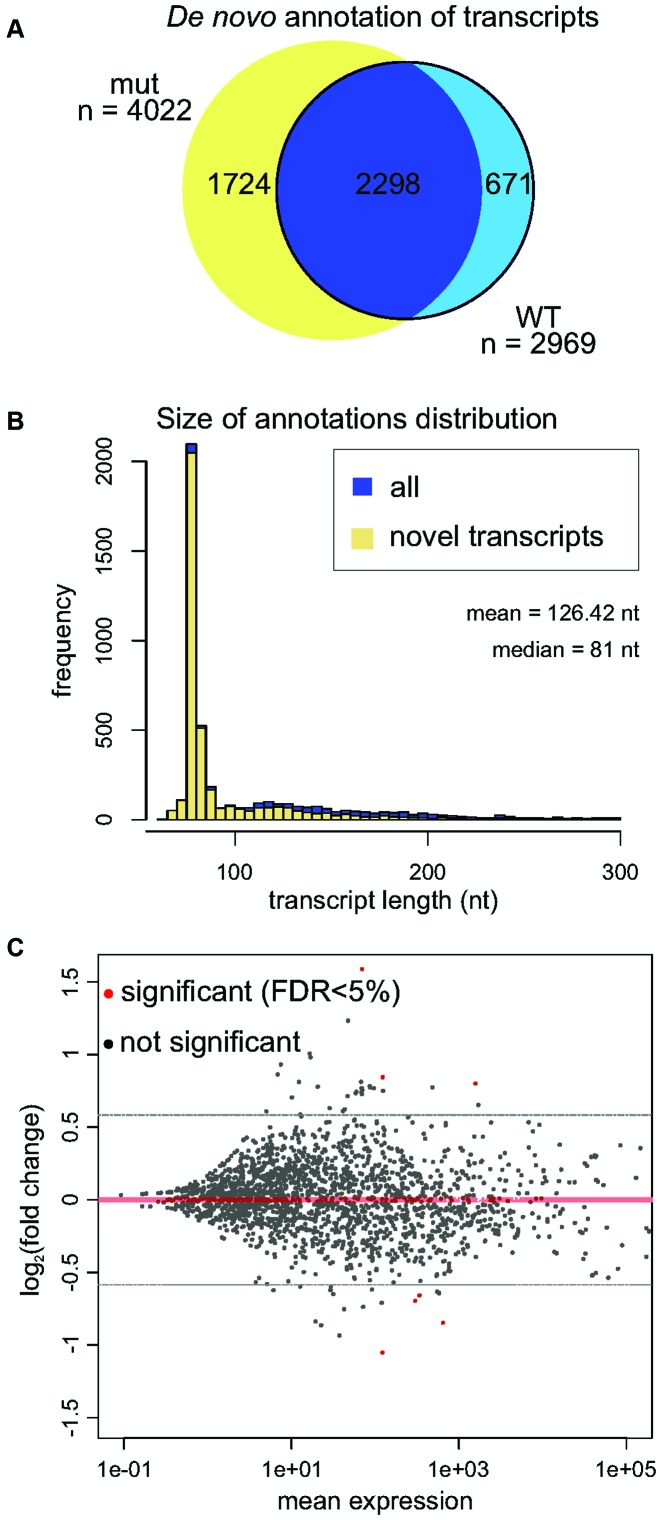
For small RNA-Seq analysis, 4802 transcripts were assembled de novo by Cufflinks (Figure 2A), which showed that 3,941 did not belong to any GENCODE annotation (Figure 2B). A custom transcript annotation was then prepared by integrating the de novo assembled transcriptome into the GENCODE annotated short RNAs. The expression changes in the DIS3L2 mutant were moderate, with only 12 genes and 9 genes showing at least 1.5-fold up- and down-regulation, respectively (Figure 2C). The top hits accumulating with DIS3L2 dysfunction that had a FDR threshold lower than 5% were vault RNAs, Y RNAs and snRNAs.
Figure 2.
Analysis of short (20-200 nt) RNA DIS3L2 substrates. (A) Venn diagram summarizing de novo annotation of transcripts in the DIS3L2 mutant and WT samples. (B) Histogram representing distribution of size annotations in Cufflinks de novo transcriptome assembly. Potentially novel transcripts are shown in yellow. (C) MAplot of small RNA sequencing results demonstrates differential expression in DIS3L2 WT compared to DIS3L2 mutant. Statistically significant hits (FDR < 5%) are color-coded red. Transcripts accumulating in the DIS3L2 mut have a positive value on the y-axis.
Specific classes of non-coding RNA: vault RNAs and Y RNAs are DIS3L2 substrates
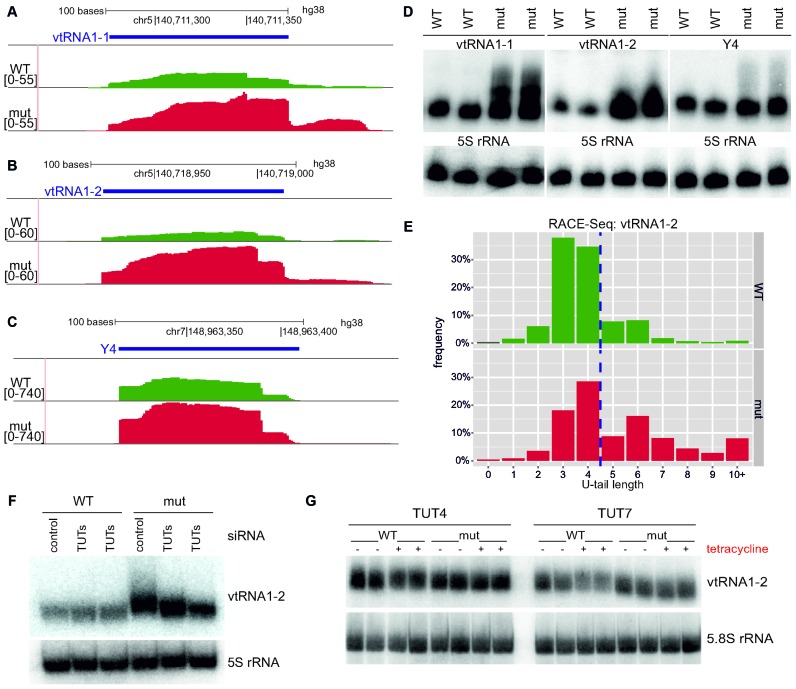
Analysis of the small RNA-Seq data showed that DIS3L2 dysfunction leads to a significant upregulation of certain non-coding RNAs transcribed by polymerase III. As shown in selected Genome Browser views, some vault RNAs (vtRNA1-1, vtRNA1-2, vtRNA1-3) and, to a lesser extent, Y RNAs (Y1, Y3 and Y4), were among the most prominently upregulated (Figure 3A–C). These results were validated by northern blot (Figure 3D, Supplementary Figure S4A). Curiously, we observed smearing of the RNAs towards longer species in DIS3L2 mut cells (Figure 3D), indicating that the elongated vault and Y RNAs are preferential DIS3L2 substrates in vivo. To ensure that the observed length increase is not due to changes in transcription start sites, primer extension analysis for vtRNA1-1 and Y4 transcripts was performed, which showed no change in the 5′-end position (Supplementary Figure S3). Given that uridylation often constitutes signal enhancing DIS3L2-mediated decay (13–15), we hypothesized that the observed smearing is a consequence of the addition of non-templated uridines to the 3′-ends of transcripts. This hypothesis was tested for both analyzed RNAs by RACE and massive parallel sequencing on Illumina NextSeq (RACE-Seq). Since vault1-2 and Y4 are polymerase III transcripts, more than 30% ended in three and four or five uridines, respectively. Importantly, the results showed an accumulation of longer vtRNA1-2 and Y4 RNA with 3′ non-templated nucleotide additions in cells expressing mutant DIS3L2 and the nucleotides added to the 3′-end were almost exclusively uridines (Figure 3E, Supplementary Table S1 and Supplementary Figure S4B). For vtRNA1-2, nearly 48% and 19% comprised non-templated 3′ uridines in mutant and wild type cells, respectively (Supplementary Table S1). The potential candidates that mediate ncRNA uridylation were cytoplasmic TUTases, especially TUT4 and TUT7, which are known to uridylate another DIS3L2 substrate, let-7 pre-miRNA (14,15,19–21). To investigate whether TUTases are responsible for uridylation of vtRNAs and Y RNAs in our experimental cell lines in vivo, TUT4 and TUT7 were silenced with respective siRNAs and vtRNA1-2 and Y4 transcripts were then inspected by northern blot. Importantly, smears representing elongated vault and Y RNAs disappeared upon TUTase knockdown (Figure 3F and Supplementary Figure S5B). To test whether TUT4 or TUT7 overexpression impacts vaultRNA1-2 and Y4 abundance in vivo, HEK293 cells were transfected with plasmids for tetracycline-inducible overexpression of wild type or catalytically inactive TUT4 or TUT7. Northern blot analysis demonstrated that overexpression of wild type TUT7 and, to a lesser extent, wild type TUT4 (but not mutant variants of both TUTases) led to a decrease in the amounts of vtRNA1-2 (Figure 3G, Supplementary Figure S5C). A similar, but less pronounced, effect was visible for Y4 transcripts (Supplementary Figure S5D). These data indicated that uridylation likely promotes DIS3L2-mediated degradation of vault and Y RNAs.
Figure 3.
Vault RNAs and Y RNAs are DIS3L2 substrates after TUT4/TUT7 uridylation (A–C). Screenshot from Genome Browser showing genomic region encoding vtRNA1-1 (A), vtRNA1-2 (B) and Y4 RNA (C), with reads from deep sequencing for WT and mut DIS3L2. Number denotes the normalized expression measured by RNA-Seq (left); (D) Validation of transcript accumulations shown in panels a-c in DIS3L2 mut as assessed by northern blot. 5S rRNA was used as an internal control. (E) RACE-Seq results showing that vtRNA1-2 transcripts bearing 3′ non-templated nucleotide additions accumulated in cells expressing catalytically inactive DIS3L2. Since vtRNA1-2 is a polymerase III transcript, it has four uridines encoded in the genome (left side of dashed line). The distribution of reads with the selected U-tail length is shown (bars represent frequency of reads with selected U-tail length normalized to total read counts). (F) Northern blot analysis of vtRNA1-2 in WT and mut DIS3L2 cell lines after siRNA-mediated TUTase repression. 5S rRNA was used as an internal control. (G) Northern blot analysis of vtRNA1-2 RNA in HEK293 parental cell line after transfection with plasmids for tetracycline-inducible overexpression of wild type or catalytically inactive TUT4 or TUT7. ‘+’ indicates tetracycline induction and ‘-’ represents no induction. 5.8S rRNA was used as an internal control.
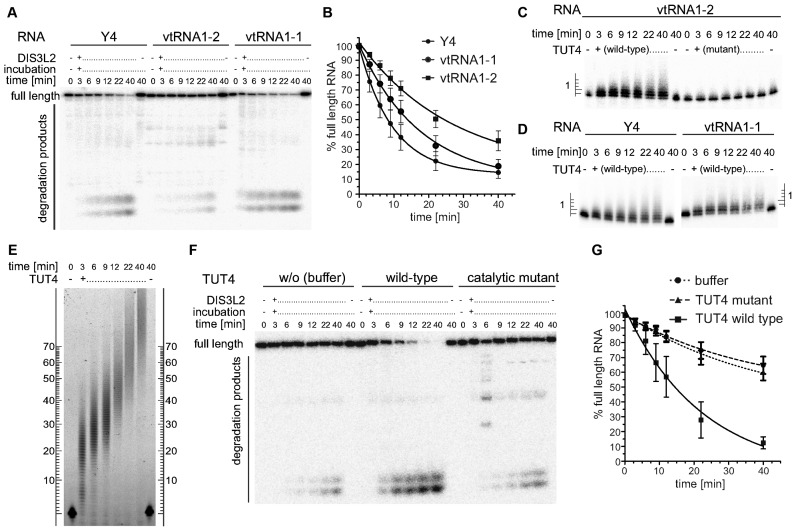
To gain deeper insight into the kinetics of DIS3L2-mediated vtRNA and Y RNA degradation and to better assess how uridylation regulates this process, in vitro biochemical time-course assays were performed. In vitro transcribed 5′ 32P-labelled vtRNA1-1, vtRNA1-2 or Y4 RNA were combined in a 1:1 molar ratio with purified recombinant DIS3L2. Each RNA served as a substrate for DIS3L2 degradation (Figure 4A), and DIS3L2 acted processively to produce short oligonucleotide degradation products and few longer degradation intermediates, which faded as time progressed. Among these RNA substrates, Y4 was most readily degraded by DIS3L2 (the half-life of full-length RNA was ∼7 min), while vtRNA1-2 seemed less prone to degradation (half-life 17 min) (Figure 4B). The degradation kinetics of vault and Y RNAs resembled those obtained previously with duplex RNAs comprising few unpaired 3′ nucleotides (8). This outcome is consistent with the strong secondary structure of vault and Y RNAs and a short stretch of 3–5 genomically-encoded unpaired Us at the 3′-ends. Furthermore, a significant fraction (around 20%) of the full-length vault and Y RNAs was resistant to DIS3L2-mediated degradation.
Figure 4.
DIS3L2 preferentially degrades TUTase-uridylated substrates in vitro. (A) A typical time-course experiment visualized by denaturing PAGE and phosphorimaging. In vitro transcribed, 5′ radioactively labelled Y4, vtRNA1-2 and vtRNA1-1 were incubated with purified recombinant DIS3L2 protein. Aliquots were withdrawn at the indicated time points and reactions were stopped rapidly. (B) Results of 6 independent time-course experiments were quantified by Multi Gauge software, plotted and fit with a single exponential decay equation using Graph Pad Prism software. (C) In vitro transcribed, 5′ radioactively labelled vtRNA1-2 was incubated in the presence of either wild type or catalytically inactive (D1011A) TUT4. Reactions were rapidly stopped, separated by 10% sequencing PAGE and visualized by phosphorimaging. The ladder on the left indicates the number of uridines added to the 3′ end of the RNA. (D) As in panel C with Y4 and vtRNA1-1 used as substrates in the presence of WT TUT4. (E) As in panels C and D but a 5′ fluorescently labelled (FAM) 44 mer RNA oligonucleotide was the substrate. The RNAs were separated by 6% PAGE and fluorescent signals were visualized with a Typhoon FLA-9000 imaging scanner (GE Healthcare). (F) Representative PAGE analysis of a combined extension-degradation time-course assay of vtRNA1-2 in the presence of DIS3L2 and either wild-type or catalytically inactive mutant TUT4 or buffer (control w/o TUT). The assay was visualized as in panel A. (G) Plot summarizing results of four independent extension-degradation time-course assays (as in panel F). The graph was generated as in panel B.
Since RACE-Seq analyses (Supplementary Table S1) revealed the preferential addition of 1–2 uridines to the 3′-ends of vtRNA1-2 and Y4 in vivo, we tested whether vault and Y RNAs can be uridylated in vitro by purified TUT proteins, and whether TUTs would add single U, oligo- or poly-U tails. The tests were performed with vtRNA1-1, vtRNA1-2 or Y4 and full-length wild type or catalytically inactive mutant (D1011A) TUT4 proteins that were affinity-purified from human cells. We observed preferential addition of 1–2 uridine residues to the 3′-end of either RNA type by wild type TUT, but not the mutant variant (Figure 4C and D). At the final time-point of our assay (40 min), those RNA substrates extended by 1–2 uridines constituted ∼70% of the total, while those extended by ≥3 uridines constituted ∼15%. In contrast, by the final time-point a single-stranded, generic, fluorescently-labelled RNA substrate (FAM44mer) acquired 80 to 100+ uridines under similar conditions (Figure 4E). This finding that a limited number of uridines can be added to the 3′ ends of vault and Y RNAs was in agreement with in vivo data. The reason why TUTases act more efficiently on a linear substrate than on a structured one remains to be determined. We further asked whether the low number of uridines added to the 3′-ends of vtRNA1-2 would influence the kinetics and efficiency DIS3L2-mediated degradation in vitro. To this end, we performed time-course assays that combined vtRNA1-2 with DIS3L2 and, optionally, with either wild type or catalytically inactive TUT4. Indeed, vtRNA1-2 was degraded by DIS3L2 faster and significantly more effectively in the presence of wild type TUT relative to either the mutant TUT or the absence of TUT (Figure 4F and G) as evidenced by the ∼16 min half-life of full-length vtRNA1-2 in the presence of wild type TUT and the ∼26 min half-life for the two controls. The enhancement of DIS3L2-mediated vtRNA degradation by uridine addition was also obvious with synthetic RNA substrates that had different lengths of non-templated U tails (see Supplementary Figure S6A,B and Supplementary Note).
Together these analyses demonstrate that vault and Y RNA are degraded by DIS3L2 in an uridylation-dependent manner.
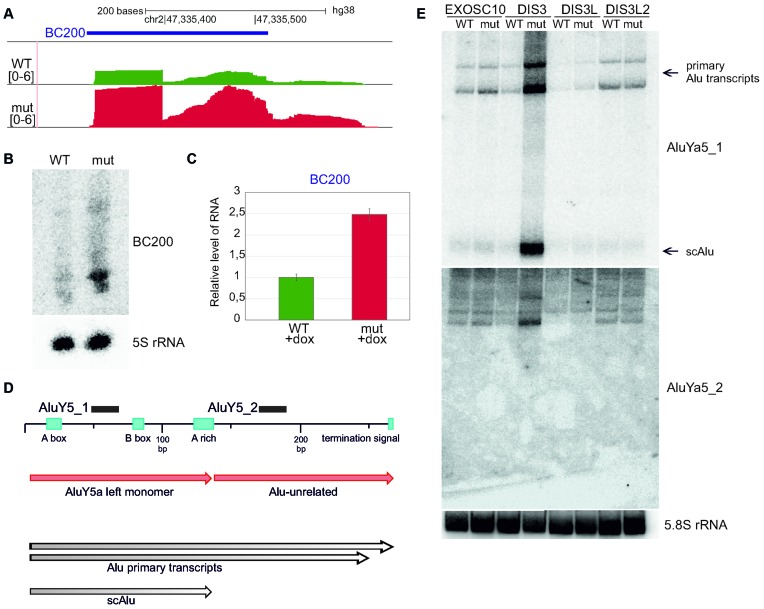
DIS3L2 is involved in the degradation of the Alu-homologue-derived RNA BC200, but not other Alu-like elements
Another ncRNA that is transcribed by polymerase III and accumulates in the presence of mutated DIS3L2 is BC200 (also known as BCYRN1), a 200 nt-long non-coding RNA (lncRNA) (Figure 5A). BC200 is enriched in the brain and is thought to repress translation of several mRNAs that have strong secondary structures at the 5′-end (22). Both northern blot and qPCR analyses confirmed that BC200 is indeed strongly upregulated in DIS3L2 mutant cells (Figure 5B and C).
Figure 5.
BC200, an Alu-like element RNA, is a DIS3L2 substrate, while DIS3 degrades transcripts from Alu-repeats. (A) Screenshot from Genome Browser showing the genomic region encoding BC200 RNA with reads from deep sequencing for WT and mut DIS3L2. Number denotes the normalized expression measured by RNA-Seq. (B) Validation of BC200 RNA accumulation in DIS3L2 mut by northern blot. 5S rRNA was used as an internal control. (C) Validation of BC200 accumulation in mut DIS3L2 by qPCR. Bars represent the standard deviation for three biological replicates. (D) Example of transcripts arising from Alu repeats (AluYa5) accumulating in DIS3 mut, but not in EXOSC10, DIS3L or DIS3L2 mutants. Schematic representation of the AluYa5 transcript. Northern blot probe binding sites are shown for AluY5_1 and AluY5_2. (E) Northern blot showing accumulation of AluYa5 transcripts in DIS3 mut cells, including scAlu (small cytoplasmic Alu) (produced from AluY5a left monomer). 5.8S rRNA was used as an internal control.
BC200 is transcribed from an Alu-like element composed of a monomeric Alu repeat and a non-repetitive domain (23). To find out if it is a specific DIS3L2 substrate, or whether it represents a wider spectrum of Alu-derived transcripts, northern blot experiment was conducted with a probe complementary to another transcriptionally active Alu element - AluYa5 (24) (Figure 5D). There was no accumulation of AluYa5 in the presence of mutated DIS3L2 (Figure 5E), in agreement with RNA-Seq data, in which we haven't seen increased levels of any specific Alu elements. In contrast, DIS3 dysfunction led to strong upregulation of AluYa5 (Figure 5E). Moreover, short RNA arising from Alu repeats accumulated, which appeared to be a left monomer with an Alu sequence, known as scAlu (small cytoplasmic Alu) (25) (Figure 5E; Supplementary Figure S7).
These results indicate that BC200 is a specific DIS3L2 substrate, while AluYa5 levels are controlled by DIS3.
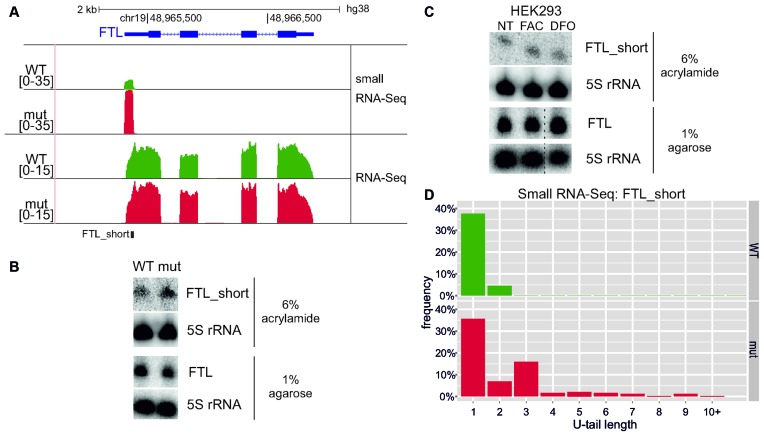
A stem loop at the 5′-end of ferritin mRNA accumulates as a short RNA species in DIS3L2 mutant cells (FTL_short)
Small RNA-Seq data analysis revealed that the catalytic mutation in DIS3L2 led to significant accumulation of a previously unknown, ∼80 bp long transcript originating from the 5′ UTR of ferritin (FTL) pre-mRNA (Figure 6A). Interestingly, this transcript corresponds to the short conserved stem-loop iron-responsive element (IRE) that regulates ferritin mRNA translation. When iron concentrations are low and ferritin-mediated iron storage is not needed, specific RNA binding proteins bind the ferritin IRE to inhibit translation (26,27). The existence and accumulation of such novel short RNA species in mutated DIS3L2 was confirmed by northern blot (‘FTL_short’ in Figure 6B), which showed that full-length ferritin transcript levels did not change upon DIS3L2 mutation (‘FTL’ in Figure 6B).
Figure 6.
Identification of FTL_short, a novel transcript from the 5′-UTR of ferritin mRNA. (A) Screenshot from Genome Browser showing genomic region encoding FTL mRNA with RNA-Seq and small RNA-Seq reads (see Materials and Methods) for WT and mut DIS3L2 cells. Number denotes the normalized expression measured by RNA-Seq. (B) Validation of FTL_short by northern blot after 6% PAGE. The amount of full-length ferritin mRNA remained constant, as shown by northern blot after electrophoresis in 1% agarose. 5S rRNA was used as an internal control for both cases. (C) FTL_short possibly does not function in iron metabolism. Parental HEK293 cells were treated for 24 h with substances that alter environmental iron levels: FAC (ferric ammonium citrate) and DFO (deferoxamine). The amounts of FTL_short and full-length ferritin transcript were then analysed by northern blot. 5S rRNA was used as an internal control in both cases. Dashed lines indicate degraded RNA samples that were excised (second biological replicate). (D) RNA-Seq results showing that FTL_short transcripts bearing 3′ non-templated nucleotide additions accumulated in cells expressing catalytically inactive DIS3L2. The distribution of reads with the selected U-tail length is shown (bars represent frequency of 3′ end reads with selected U-tail length normalized to the total number of reads maping to 3′ end).
To test whether the short FTL transcript (FTL_short) functions in iron homeostasis, we treated parental HEK293, WT and mut cells with ferric ammonium citrate (FAC) or deferoxamine (DFO) to respectively increase or decrease iron concentrations. After treatment, the amounts of FTL_short did not change with regard to the untreated cells (Figure 6C, Supplementary Figure S8A). Simultaneously, the abundance of the FTL protein grew upon FAC-treatment and dropped upon DFO-treatment, confirming proper activity of these compounds (Supplementary Figure S8B). Thus, under these conditions, we were not able to demonstrate any role of the FTL_short transcript in iron homeostasis regulation. Further investigation would reveal whether it has a biological function or is a transcription by-product.
We next examined the uridylation status of FTL_short, and found from RNA-Seq data that 39.6% of reads mapping to the FTL_short 3′-end with DIS3L2 WT had a non-templated U, compared to 63.3% of reads in DIS3L2 mut samples. Furthermore, in mutant DIS3L2 cells a fraction of FTL_short had stretches of >2 Us, which were absent in the WT control. This outcome strongly suggested that DIS3L2 is involved in degradation of uridylated FTL_short (Figure 6D).
In summary, we identified a novel DIS3L2 substrate that is a short transcript of unknown function derived from the 5′-UTR of ferritin mRNA.
RNA surveillance of maturing snRNA by DIS3L2
DIS3L2 dysfunction led to the accumulation of transcripts originating from small nuclear RNA (snRNA) loci, but longer than mature snRNAs (‘extended snRNAs’). This outcome suggests that DIS3L2 may be involved in snRNA processing during their biogenesis and/or surveillance (Figure 7A). In a typical biogenesis pathway of snRNAs, primary transcripts from snRNA loci are capped and the integrator complex is responsible for initial endonucleolytic 3′-end processing (28) that produces shorter precursors for export to the cytoplasm (29,30), where they associate with survival motor neuron complex (SMN) (31) to coordinate further processing of snRNAs and their re-import into the nucleus. These steps include modification of the 5′-end to form the 2,2,7-trimethylguanosine cap (TMG) (32) and maturation following 3′-end trimming that is mediated by an unknown nuclease. The mature snRNAs function in nuclear splicing.
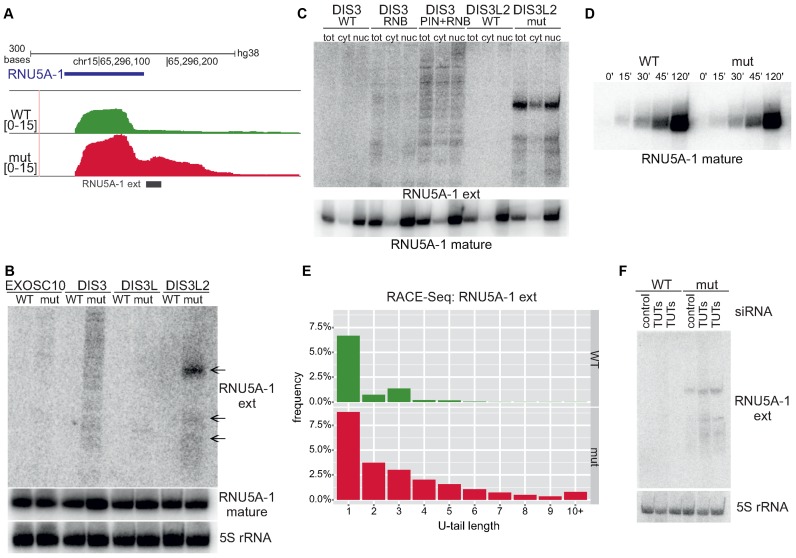
Figure 7.
DIS3L2-mediated surveillance of maturing U5 snRNA. (A) Screenshot from Genome Browser showing genomic region encoding U5 snRNA with deep sequencing reads for WT and mut DIS3L2. Number denotes the normalized expression measured by RNA-Seq. (B) Northern blot verification of extended snRNAs (‘ext’) in model cell lines producing wild-type or mutated EXOSC10, DIS3, DIS3L or DIS3L2. The level of mature snRNAs was also analyzed. 5S rRNA was used as an internal control. (C) Extended snRNAs accumulating in the presence of mutated DIS3 and DIS3L2 are present in the cytoplasm and the nucleus. The localization of extended snRNAs in different cellular compartments was assessed by cell fractionation and northern blot analysis (tot – total RNA, nuc – nuclear RNA, cyt – cytoplasmic RNA). Analysis of mature U5 snRNA localisation after cell fractionation was also conducted. Control of cell fractionation is identical as in Supplementary Figure S7. (D) Accumulation of extended U5 snRNAs does not influence production of mature snRNA species. 4-thiouridine (4sU) labelling was used to study the kinetics of mature snRNA production. RNA was collected after labelling and terminating reactions at the indicated time points. RNA labelled with 4sU was retrieved by biotinylation and separation on streptavidin beads. (E) Extended U5 snRNA transcripts bearing 3′ non-templated nucleotide additions accumulate in cells expressing catalytically inactive DIS3L2 as shown by RACE-Seq. The distribution of reads with selected U-tail lengths is shown (bars represent the frequency of reads with selected U-tail length normalized to total read counts). (F) Uridylation enhances degradation of extended snRNAs. TUT4 and TUT7 were silenced in model cell lines using siRNA (lanes TUTs) and the presence of extended snRNA transcripts was monitored by northern blot. 5S rRNA was used as an internal control.
To gain insight into how DIS3L2 could contribute to snRNA metabolism, we conducted northern blot hybridisations with probes complementary to selected snRNAs, which were located beyond 3′ end of mature transcripts. We confirmed that extended snRNAs accumulate in DIS3L2 mutant cells. These transcripts were visible as distinct bands that migrated more slowly than mature snRNAs, which was demonstrated for U5 (RNU5A-1) (Figure 7B) and several other snRNA species (Supplementary Figure S9A–D). These extended forms could constitute snRNA precursors (pre-snRNAs), however their relative size is bigger than so far identified canonical pre-snRNAs (33).
To assess possible roles of other 3′–5′ exonucleases in snRNA metabolism, RNA samples isolated from model cell lines analogous to those described herein for DIS3L2, but corresponding to EXOSC10, DIS3 or DIS3L exosome catalytic subunits, were analysed in parallel. While dysfunctional DIS3L and EXOSC10 had no effect on snRNAs, in cells producing a mutant variant of DIS3 we noted an accumulation of distinct, longer snRNA transcripts of variable lengths that are presumably ‘read-through’ snRNAs that missed integrator-mediated processing and extend to the region downstream of the termination site. These longer forms accumulated particularly when both DIS3 PIN and RNB domains were mutated, indicating that both endo- and exoribonucleolytic activities of DIS3 contribute to snRNA degradation (Figure 7B, Supplementary Figure S9A–D).
Importantly, and in contrast to accumulation of elongated snRNA species, northern blot hybridisations for mature snRNA molecules revealed that in most cases there was no change in their levels upon DIS3L2 dysfunction, while a slight increase was observed for DIS3 (Figure 7B, Supplementary Figure S9A-D and Supplementary Figure S10A). These results indicate that neither DIS3L2, nor DIS3 are essential for snRNA maturation. DIS3 could however participate in the decay of mature snRNA. To show that the effects of DIS3L2 mutation are specific for snRNA, we performed northern blot analysis for another small RNA: snoRNA, the maturation of which was previously shown to depend on nuclear EXOSC10 and DIS3 (16). The lack of changes in pre- and mature snoRNA levels in cells expressing cytoplasmic DIS3L and DIS3L2 indicates that these proteins likely do not participate in snoRNA maturation (shown on the example of SNORD13, see Supplementary Figure S10B). These results are in agreement with the nuclear localisation of snoRNAs through their entire life cycle. To further analyse the subcellular location of accumulating extended snRNAs and read-through forms, we carried out cell fractionations and isolated RNAs from the nuclear and cytoplasmic fractions that were then subjected to northern blotting with probes specific to U5 (RNU5A-1) or U4atac (RNU4atac) (Figure 7C, Supplementary Figure S11A). The blot showed that extended snRNAs accumulated upon DIS3L2 mutation and the read-through forms of snRNAs accumulated upon DIS3 mutations, are both present in the nucleus and the cytoplasm. The localization of the read-through forms indicates their ability to leave the nucleus when mutated DIS3 is present.
To resolve whether DIS3L2 is involved in snRNA processing or quality control, we treated cells with 4-thiouridine (4sU) to investigate the production of newly synthesized snRNAs in a time-dependent manner and assessed how mutated DIS3L2 and accompanying accumulation of extended snRNAs influenced the kinetics of mature snRNA production. There was no visible difference between WT and mut versions of DIS3L2 in the production of both mature U5 snRNA and U1 snRNA 15 min to 120 min after the 4sU pulse (Figure 7D; Supplementary Figure S11B). This result suggests that accumulation of extended snRNA species in the presence of mutated DIS3L2 (Supplementary Figure S11C), does not influence production of mature snRNA. This implies that extended snRNAs accumulating upon DIS3L2 mutation are non-functional (meaning that DIS3L2 is involved in surveillance rather than processing). In the second scenario, extended snRNAs may still represent functional snRNA precursors in a novel snRNA biogenesis pathway, and they are processed by an as yet unknown nuclease(s) in place of DIS3L2 (when mutated).
Given that in the case of previously described ncRNAs degraded by DIS3L2, uridylation constituted signal enhancing DIS3L2-mediated decay, we used RACE-Seq to examine whether the elongated U5 snRNA is uridylated in cells producing WT or mut versions of DIS3L2 or DIS3. DIS3L2 mut cells showed a marked increase in uridylated U5 and several other extended snRNA species relative to WT cells (Figure 7E, Supplementary Figure S11D–F). Importantly, RACE-Seq data analysis revealed that, relative to previously described transcripts from snRNA biogenesis pathways, in the presence of DIS3L2 mutations, the accumulating transcripts were longer and extended up to 70 nt, while canonical snRNA precursors have relatively short (5–10 nt) overhangs (33). It agrees well with sizes observed in northern blot analyses and further confirms a role for DIS3L2 in snRNA surveillance rather than processing. Additionally, in agreement with northern blot data, we observed that ‘read-through’ transcripts accumulating in the presence of DIS3 mutations are longer than snRNA species seen when DIS3L2 is mutated (Supplementary Figure S11G).
We next investigated the possible role of TUT4 and TUT7 in extended snRNAs degradation using DIS3L2 model cell lines and simultaneous siRNA silencing of selected TUTases followed by northern blot quantification of U5 extended snRNA. We observed a relatively modest, but clearly additive effect of the DIS3L2 catalytic mutation and TUTase depletion (Figure 7F), which supports the hypothesis that TUT4 and TUT7-mediated uridylation marks extended snRNAs for degradation by DIS3L2, and that the 3′-terminal uridylation promotes their degradation.
Taken together, DIS3L2 and TUTs cooperate in surveillance and degradation of maturing snRNA transcripts while DIS3 is involved in the decay of snRNA ‘read-through’ transcripts and mature snRNA molecules.
DISCUSSION
Previous studies showed that the Perlman syndrome nuclease DIS3L2 participates in the decay of selected mRNA and pre-miRNA. In this study, we identified novel physiological substrates of DIS3L2 and showed that DIS3L2 dysfunction disturbs transcriptome homeostasis.
snRNA surveillance by DIS3L2
The complicated process of snRNA biogenesis begins with the production of transcripts with a monomethyl-guanosine (m7GpppG) cap and transcription-linked cleavage by the integrator complex (28) to produce nuclear precursor forms. These precursors are then transported to the cytoplasm, where they assemble with SMN complexes (31) that coordinate hypermethylation of the m7GpppG cap (32), exonucleolytic trimming, and formation of snRNP particles. Finally, snRNPs are reimported to the nucleus, where they undergo additional remodeling. However, the nuclease that is involved in cytoplasmic processing of snRNAs remains elusive. We showed that DIS3L2 dysfunction promotes the accumulation of RNA species that are longer than mature snRNAs, but without a concomitant change in mature snRNA levels. These transcripts are typically longer than known cytoplasmic snRNA precursors - e.g. for U1 and U4 snRNAs, the cytoplasmic pre-snRNAs are only several nucleotides longer than the corresponding mature species (34,35), whereas the transcripts accumulating upon DIS3L2 mutation are up to 50 nt or 70 nt longer for U1 and U5, respectively. Thus, DIS3L2 likely governs a surveillance pathway for misprocessed precursors, which is also supported by the lack of change in snRNA production kinetics that was visualized by pulse-labelling with 4sU. Nonetheless, because of its potential redundancy with other nucleases that has been documented for other processing events, we cannot completely exclude the possibility that DIS3L2 is also involved in snRNA processing (36).
Besides the involvement of DIS3L2 in snRNA biology, our data indicate that the DIS3L2 homologue DIS3 participates in the decay of mature snRNA and also degrades ‘read-through’ snRNAs that extend downstream of the termination sites. Assuming that DIS3L2 has a quality control but not processing function, and taking into account the finding that DIS3L and EXOSC10 mutations do not influence either form of snRNA, then the nuclease involved in cytoplasmic processing of pre-snRNAs remains to be discovered. However, we did demonstrate that both DIS3 and DIS3L2 play roles in the surveillance of maturing snRNA molecules, with the former overseeing their nuclear biogenesis, and the latter removing unwanted extended forms from the cytoplasm. This function is particularly important, given that these elongated transcripts could enter the normal snRNA biogenesis pathway (based on their dual localization), which could sequester the processing machinery from canonical precursors.
DIS3L2 surveillance of short cytoplasmic RNA species
Here, we identified several short cytoplasmic RNA species as novel DIS3L2 substrates. The unifying feature of these RNAs is their strong secondary structure that is similar to the previously described DIS3L2 substrate, let-7 miRNA precursors (37). Importantly, given known functions of most of these transcripts, their dysregulation may have significant physiological consequences.
Vault RNAs are non-coding RNAs that constitute part of large vault ribonucleoprotein particles that are associated with multidrug resistance (38,39). Moreover, vtRNA1-1 overexpression was recently shown to protect cells from apoptosis (40). Given that DIS3L2 knockdown exerted a similar effect, inhibition of programmed cell death in cells that are defective in DIS3L2 activity (13) could be mediated at least in part through this particular vtRNA.
Y RNAs play a role in DNA replication, RNA processing and quality control, and are other members of the non-coding RNA repertoire that are targeted by DIS3L2 (41). The Y3 isoform (Y3**) was recently implicated in proper 3′-end processing of histone pre-mRNAs since Y3** depletion leads to misprocessed histone mRNAs without a change in their total abundance (42). In the presence of mutated DIS3L2, Y3 levels increased, but we did not observe misprocessed histone mRNAs (Supplementary Figure S2D), only their accumulation.
Both vault RNAs and Y RNAs are thought to be a source of smaller RNAs, namely svRNAs (43) and YsRNAs (44), respectively. The findings that YsRNAs were upregulated in breast cancer patients, which would make them potential prognostic biomarkers (45), and that upregulation of a specific svRNAs is widespread in Parkinson's disease, thus suggesting potentially novel therapeutic approaches (46), significantly enhanced the interest in these small RNAs. In terms of DIS3L2, catalytic mutants could lead to svRNA and YsRNA overproduction that in turn abolishes cell homeostasis.
Our RNA-Seq data showed that another consequence of DIS3L2 dysfunction is upregulation of BC200, which is an ncRNA that is produced by polymerase III and is normally expressed in primate brain (23). BC200 is produced from a monomeric Alu repeat and was shown to inhibit translation initiation and possibly contribute to synaptic plasticity (22), although whether DIS3L2 is involved in this mechanism remains to be determined. In particular, it would be interesting to explore whether DIS3L2 plays tissue-specific roles in the brain. This possibility is consistent with unpublished data by Nowak (personal communication), which showed that pre-miR-9, a miRNA precursor that is important for neuronal differentiation, is degraded by DIS3L2. Additionally, we show that DIS3, but not DIS3L2, contributes to removal of transcripts derived from Alu repeats. Since these repeats are present in the human genome in more than 1 million copies that together represent up to 10% of DNA (47), controlling the levels of the transcripts that are derived from them may be highly relevant for maintaining genome stability.
DIS3L2 and histone mRNAs
A prominent class of DIS3L2 mRNA substrates are replication-dependent histone mRNAs. Since these mRNAs are quickly degraded after replication and lack poly(A) tails, a unique mechanism exists to remove these transcripts. A stem loop structure (SL) present in the 3′ UTR of histone mRNAs interacts with the SLBP protein and Eri1 exoribonuclease (48). To initiate degradation, SLBP recruits TUT4 (ZCCHC11), which adds an oligo(U) tail to serve as a signal for Lsm1 (49) that in turn triggers histone mRNA degradation - either from the 5′- (by XRN1 after decapping) or the 3′-end through activation of ERI1. This ERI1 activation involves consecutive rounds of degradation and TUT4-mediated uridylation to remove the SL structure, leading to subsequent degradation by the RNA exosome complex (48,49). Since our data indicate that DIS3L2 mutations lead to accumulation of replication-dependent histone mRNAs, DIS3L2 may represent an alternate pathway for the degradation of these mRNAs. DIS3L2 is known to be present in polysomes (8), which is in agreement with the fact that histone mRNA degradation also occurs during ongoing translation (50). Conversely, participation of DIS3L2 in this process was questioned by Slevin et al. (50), who showed that there is no change in histone mRNA degradation upon DIS3L2 silencing. However, these authors used a siRNA approach to downregulate DIS3L2 expression, which is not always sufficient, as was shown previously by our laboratory (5). Furthermore, even if DIS3L2 expression was silenced efficiently, histone mRNAs might still be removed by other pathways, suggesting the existence of functional redundancy. Furthermore, DIS3L2 may only degrade specific replication-dependent histone mRNAs.
The role of uridylation in DIS3L2-mediated decay
Earlier studies (13–15) showed that uridylation constitutes a signal that enhances DIS3L2 activity towards let-7 pre-miRNA. Moreover, the role of uridylation in the decay of replication-dependent histone mRNAs, which are known to accumulate upon DIS3L2 mutation, has been well-documented (49). Here we found that the newly identified DIS3L2 substates - vault RNAs, Y RNAs, FTL_short and extended snRNAs are all uridylated.
Our biochemical experiments with in vitro transcribed vtRNA1-2 let us unequivocally conclude that uridylation enhances DIS3L2 activity towards this substrate. On the other hand, in vivo studies suggest that it is not indispensable for degradation, because we do not observe accumulation of VTRNA1-2 and RNY4 transcripts after downregulation of the enzymes responsible for uridine tail synthesis (TUTases), which, however, can be due to residual TUTase activity after inefficient silencing (this is especially important considering the fact that there is a functional redundancy between TUTases, depending on the nature of the substrate (21)). Furthermore, after overexpression of wild-type DIS3L2 we still observe a fraction of vault RNAs and Y RNAs without additional uridines, which points to preferential degradation of uridylated substrates in vivo. A structure of mouse DIS3L2 (11) shows a significant network of interactions between the DIS3L2 entrance channel and an artificial oligo(U) substrate that explains the role of the oligo(U) tail in enhancing DIS3L2-mediated RNA decay. Additionally, structural data suggest that the DIS3L2 channel is sufficiently wide to allow for processing of structured substates (11). This hypothesis is in concordance with our in vitro data, which demonstrated that DIS3L2 degrades unmodified Y4 with preference over vtRNA1-2 that, in constrast, showed enhanced uridylation relative to Y4 in a RACE-Seq experiment. This result suggests that uridylation may be particularly important for substrates that are otherwise poorly degraded by DIS3L2 unless they are additionally modified at the 3′ terminus.
Given recent information concerning widespread uridylation (51) and the broad spectrum of DIS3L2 substrates, we not only demonstrate that oligouridylation plays a prominent role in the decay of these newly discovered DIS3L2 substrates, but we also expand DIS3L2 exoribonucleolytic activity functions from simply contributing to mRNA decay to controlling the levels of cytoplasmic non-coding RNAs.
Supplementary Material
Acknowledgments
We acknowledge Joanna Kufel for critical reading of this manuscript, Roman Szczesny for critical reading of the manuscript and help with the flow cytometry analysis, and Katarzyna Kowalska for help with cloning.
ACCESSION NUMBERS
RNA-seq data supporting the conclusions of this article are available in the NCBI GEO repository [accession number GSE82336, http://www.ncbi.nlm.nih.gov/geo/].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Centre for Research and Development [NCBR LIDER/35/46/L-3/11/NCBR/2012 to R.T.]; ERC Starting grant [309419-PAP&PUPs to A.D.]; Foundation for Polish Science fellowships (Master and Ideas for Poland to A.D.); National Science Centre [Fuga; UMO-2012/04/S/NZ1/00036 to Z.W.]; Experiments were carried out with the use of CePT infrastructure that was financed by the European Union via the European Regional Development Fund [Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00]. Funding for open access charge: ERC Starting Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mroczek S., Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 2008;36:2874–2888. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siwaszek A., Ukleja M., Dziembowski A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014;11:1122–1136. doi: 10.4161/rna.34406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu X.-D. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomecki R., Kristiansen M.S., Lykke-Andersen S., Chlebowski A., Larsen K.M., Szczesny R.J., Drazkowska K., Pastula A., Andersen J.S., Stepien P.P., et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler J.S., Mitchell P. Rrp6, Rrp47 and Cofactors of the Nuclear Exosome. Adv. Exp. Med. Biol. 2010;2010:91–104. [PubMed] [Google Scholar]
- 7.Januszyk K., Liu Q., Lima C.D. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lubas M., Damgaard C.K., Tomecki R., Cysewski D., Jensen T.H., Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astuti D., Morris M.R., Cooper W.N., Staals R.H.J., Wake N.C., Fews G.A., Gill H., Gentle D., Shuib S., Ricketts C.J., et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat. Genet. 2012;44:277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 10.Lv H., Zhu Y., Qiu Y., Niu L., Teng M., Li X. Structural analysis of Dis3l2, an exosome-independent exonuclease from Schizosaccharomyces pombe. Acta Crystallogr. Sect. D. 2015;71:1284–1294. doi: 10.1107/S1399004715005805. [DOI] [PubMed] [Google Scholar]
- 11.Faehnle C.R., Walleshauser J., Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malecki M., Viegas S.C., Carneiro T., Golik P., Dressaire C., Ferreira M.G., Arraiano C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas M.P., Liu X., Whangbo J., McCrossan G., Sanborn K.B., Basar E., Walch M., Lieberman J. Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Rep. 2015;11:1079–1089. doi: 10.1016/j.celrep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ustianenko D., Hrossova D., Potesil D., Chalupnikova K., Hrazdilova K., Pachernik J., Cetkovska K., Uldrijan S., Zdrahal Z., Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.-M., Triboulet R., Thornton J.E., Gregory R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczepinska T., Kalisiak K., Tomecki R., Labno A., Borowski L.S., Kulinski T., Adamska D., Kosinska J., Dziembowski A. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015 doi: 10.1101/gr.189597.115. doi:10.1101/gr.189597.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagnon K.T., Li L., Janowski B.A., Corey D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borowski L.S., Szczesny R.J. Measurement of mitochondrial RNA stability by metabolic labelling of transcripts with 4-thiouridine. In: Rorbach J, Bobrowicz AJ, editors. Polyadenylation: Methods and Protocols. Humana Press; 2014. [DOI] [PubMed] [Google Scholar]
- 19.Heo I., Joo C., Kim Y.-K., Ha M., Yoon M.-J., Cho J., Yeom K.-H., Han J., Kim V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornton J.E., Chang H.-M., Piskounova E., Gregory R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA N. Y. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D., Pestova T.V., Hellen C.U.T., Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell. Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martignetti J.A., Brosius J. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015;43:817–835. doi: 10.1093/nar/gku1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia R.J., Driscoll C.T., Bilyeu T., Hsu K., Darlington G.J. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol. Cell. Biol. 1993;13:4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziz N., Munro H.N. Iron regulates ferritin mRNA translation through a segment of its 5′ untranslated region. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8478–8482. doi: 10.1073/pnas.84.23.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentze M.W., Caughman S.W., Rouault T.A., Barriocanal J.G., Dancis A., Harford J.B., Klausner R.D. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 28.Baillat D., Hakimi M.-A., Näär A.M., Shilatifard A., Cooch N., Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W. PHAX, a mediator of U snRNA nuclear export whose activity Is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 30.Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche N.E.H., Gostan T., Vandermoere F., Robert M.-C., Cusack S., et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013;20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- 31.Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 32.Mouaikel J., Narayanan U., Verheggen C., Matera A.G., Bertrand E., Tazi J., Bordonné R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003;4:616–622. doi: 10.1038/sj.embor.embor863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 34.Madore S.J., Wieben E.D., Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J. Cell Biol. 1984;98:188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madore S.J., Wieben E.D., Kunkel G.R., Pederson T. Precursors of U4 small nuclear RNA. J. Cell Biol. 1984;99:1140–1144. doi: 10.1083/jcb.99.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Nam Y., Chen C., Gregory R.I., Chou J.J., Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha N.L., Rome L.H. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol. 1986;103:699–709. doi: 10.1083/jcb.103.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossink M.H., van Zon A., Scheper R.J., Sonneveld P., Wiemer E.A. Vaults: a ribonucleoprotein particle involved in drug resistance. Oncogene. 2003;22:7458–7467. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]
- 40.Amort M., Nachbauer B., Tuzlak S., Kieser A., Schepers A., Villunger A., Polacek N. Expression of the vault RNA protects cells from undergoing apoptosis. Nat. Commun. 2015;6:7030. doi: 10.1038/ncomms8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kowalski M.P., Krude T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015;66:20–29. doi: 10.1016/j.biocel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köhn M., Ihling C., Sinz A., Krohn K., Hüttelmaier S. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 2015;29:1998–2003. doi: 10.1101/gad.266486.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson H., Kvist A., Vallon-Christersson J., Medstrand P., Borg Å., Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 44.Nicolas F.E., Hall A.E., Csorba T., Turnbull C., Dalmay T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586:1226–1230. doi: 10.1016/j.febslet.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 45.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Mote P., Martin D.I.K. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics. 2013;45:990–998. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 46.Miñones-Moyano E., Friedländer M.R., Pallares J., Kagerbauer B., Porta S., Escaramís G., Ferrer I., Estivill X., Martí E. Upregulation of a small vault RNA (svtRNA2-1a) is an early event in Parkinson disease and induces neuronal dysfunction. RNA Biol. 2013;10:1093–1106. doi: 10.4161/rna.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 48.Hoefig K.P., Rath N., Heinz G.A., Wolf C., Dameris J., Schepers A., Kremmer E., Ansel K.M., Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 49.Mullen T.E., Marzluff W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slevin M.K., Meaux S., Welch J.D., Bigler R., Miliani de Marval P.L., Su W., Rhoads R.E., Prins J.F., Marzluff W.F. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol. Cell. 2014;53:1020–1030. doi: 10.1016/j.molcel.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang H., Lim J., Ha M., Kim V.N. TAIL-seq: genome-wide determination of poly(A) tail Length and 3′ end modifications. Mol. Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.