Abstract
RNase III enzyme Drosha interacts with DGCR8 to form the Microprocessor, initiating canonical microRNA (miRNA) maturation in the nucleus. Here, we re-evaluated where Drosha functions in cells using Drosha and/or DGCR8 knock out (KO) cells and cleavage reporters. Interestingly, a truncated Drosha mutant located exclusively in the cytoplasm cleaved pri-miRNA effectively in a DGCR8-dependent manner. In addition, we demonstrated that in vitro generated pri-miRNAs when transfected into cells could be processed to mature miRNAs in the cytoplasm. These results indicate the existence of cytoplasmic Drosha (c-Drosha) activity. Although a subset of endogenous pri-miRNAs become enriched in the cytoplasm of Drosha KO cells, it remains unclear whether pri-miRNA processing is the main function of c-Drosha. We identified two novel in-frame Drosha isoforms generated by alternative splicing in both HEK293T and HeLa cells. One isoform loses the putative nuclear localization signal, generating c-Drosha. Further analysis indicated that the c-Drosha isoform is abundant in multiple cell lines, dramatically variable among different human tissues and upregulated in multiple tumors, suggesting that c-Drosha plays a unique role in gene regulation. Our results reveal a new layer of regulation on the miRNA pathway and provide novel insights into the ever-evolving functions of Drosha.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small non-coding RNAs about 22-nucleotide (nt) in length that collectively regulate more than half of the protein-coding genes in human cells (1,2). They play a key role in many, if not all, known biological pathways. As a master regulator, miRNA itself is also subject to regulation (3). Aberrant miRNA expression has been discovered in various diseases including cardiovascular disease, diabetes and cancer (4–7). This highlights the importance of clarifying the mechanisms by which biogenesis is controlled.
Typically, miRNA genes are transcribed by RNA polymerase II (Pol II) into primary miRNAs (pri-miRNAs), where one or more miRNAs hairpins are embedded (8). Some miRNAs share the same promoter with the host protein-coding gene while the others possess an independent transcriptional unit (9,10). Although the majority of miRNAs are encoded in the intron, about 10% of miRNA hairpins are found in the exon of host transcripts (9,11–13). The ribonuclease (RNase) III enzyme Drosha interacts with its cofactor DGCR8 (DiGeorge syndrome critical region gene 8) to form the Microprocessor, initiating miRNA maturation by cleaving pri-miRNAs to ∼60–70 nt precursor miRNAs (pre-miRNAs) in the nucleus (14–17). The pre-miRNA is then exported via Exportin-5 into the cytoplasm, where it is further processed by Dicer and subsequently loaded onto RISC (RNA-induced silencing complex) to exert its function (18–21). Depending on the degree of sequence complementarity between the miRNA and its target, RISC can induce either site-specific cleavage and/or non-cleavage repression, the latter of which consists of translational inhibition and/or enhanced mRNA degradation (22–24).
As the enzyme licensing miRNA production, Drosha is critical for miRNA biogenesis (25). Human Drosha contains two RNase III domains (RIIIDa and RIIIDb) and a dsRNA-binding domain (dsRBD) in the C-terminus, proline-rich and arginine/serine-rich domains in the N-terminus and a central domain in the middle (26,27). The N-terminal domain of Drosha is not required for pri-miRNA processing activity in vitro, but is essential for nuclear localization (16,26). Phosphorylation of Serine300 and Serine302 by glycogen synthase kinase 3 β (GSK3β) in the R/S-rich domain has been identified as its putative nuclear localization signal (NLS) (28,29).
Drosha was identified as a nuclear protein more than a decade ago (25). However, recent studies suggest that the subcellular localization of Drosha is subject to regulation. Drosha is redistributed to cytoplasm during Sindbis virus infection to cleave viral RNA independently of DGCR8 (30–33). Drosha and DGCR8 are colocalized with the adherens junctions of non-transformed polarized epithelial cells (34). Dynamic changes of Drosha subcellular localization have been observed during different stages of cancer progression, suggesting that the variation in subcellular distribution impacts Drosha function (35–37).
Here, by taking advantage of Drosha and/or DGCR8 knock out (KO) cells, we revisited the question of where Drosha functions in cells. To our surprise, the scope of Drosha activity is beyond the nucleus even in cells under physiological conditions. We found that alternative splicing generates cytoplasmic Drosha (c-Drosha) activity, which is tissue-specific and highly regulated during tumorigenesis.
MATERIALS AND METHODS
Plasmids
ORFs of Drosha isoforms and truncation were polymerase chain reaction (PCR) amplified with specific primers from HeLa cDNA and cloned into the pcDNA6 vector (Life Technologies) with a FLAG tag in the N-terminus. A similar approach was used to generate GFP-tagged Drosha isoforms in the pEGFP-C1 (Clontech) vector. Luciferase-based Drosha cleavage reporters were created based on the psiCHECK-2 vector (Promega). Pri-miRNA cassettes containing the pre-miRNA and ∼150 nt flanking sequences from both ends were PCR amplified from HEK293T genomic DNA and inserted into the 3′ untranslated region (3′UTR) of the Renilla gene. Plasmids of V5-DGCR8 (#51383), CMV-LUC2CP/ARE (#62857) and CMV-LUC2CP/intron/ARE (#62858) were obtained from Addgene. Primers used in cloning are listed in Supplemental Table S1.
Cell culture and transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (high glucose) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone), 100 U/ml penicillin and 100 pg/ml streptomycin. Transfections were performed using Lipofectamine 3000 (Life Technologies) or PolyJet™ DNA Tranfection Reagent (SignaGen) according to the manufacturers’ instructions.
Gene-editing by CRISPR/Cas-9 system
To generate KO, we applied multiple small guide RNAs (sgRNAs) complementary to the target gene to induce random sequence insertions and/or deletions. Both strands (sense and antisense) of sgRNA sequence targeting Drosha or DGCR8 were chemically synthesized, annealed, purified and inserted at the BbsI site downstream of U6 promoter in pX330 plasmids (Addgene #42229). Those oligo sequences are listed in Supplementary Table S1. CRISPR KO was performed according to a published protocol (38). In brief, pX330-gRNA plasmids were transfected into HEK293T cells. Forty-eight hours post-transfection, cells were split to 96-well plates with an average density of 0.5 cell/well. After single cell cloning, the genomic DNA was analyzed by Sanger sequencing to select the clones that contained frameshift mutations in all alleles. Drosha KO cells used in this study have a 5 bp deletion in exon 8 of Drosha gene. Drosha DGCR8 double KO (DKO) cells have two additional deletions in the exon 6 of DGCR8 (10 and 12 bp respectively). In both cases, these deletions cause frameshift, which abolishes the full-length protein production.
Northern blot
Total RNA was isolated using Trizol (Life Technologies) and then electrophoresed on 15% (w/v) acrylamide/8M urea gels. After being transferred onto Hybond-N1 membranes (Amersham Pharmacia Biotech), target miRNAs were detected using 32P-labeled oligonucleotides. Images were analyzed by Typhoon Trio Imaging System (GE Healthcare).
Dual luciferase reporter assay
A total of 100 ng of pri-miRNA reporters, Drosha and/or DGCR8 plasmids were co-transfected in triplicates into Drosha KO or Drosha/DGCR8 DKO cells in 24-well plates. Cells were harvested at 30–36 h post-transfection. Firefly luciferase and Renilla luciferase were then measured with Promega's dual-luciferase kit (catalog no. E1980) and detected by GloMax®-Multi Luminescence Module (Promega) according to the protocol.
Western blot
HEK293T or HeLa cells were lysed with M-PER mammalian protein extraction reagent (catalog no.78501; Thermo Scientific) with protease inhibitor cocktail (catalog no. 11836153001; Roche). About 30–80 μg protein samples were analyzed in 4–20% Mini-PROTEAN® TGX Stain-Free™ Gels and electro-transferred onto a PVDF membrane (Bio-Rad). Primary antibodies used in this study are rabbit anti-Drosha (Cell Signaling; #3364S), rabbit anti-DGCR8 (Proteintech; 10996-1-AP), mouse anti-α-tubulin (Sigma; T5168) and mouse anti-hnRNPC1/C2 (Abcam; ab10294). The images were obtained from the Chemidoc Touch Imaging System (Bio-Rad).
Immunofluorescence
HEK293T or HeLa cells were grown on glass coverslips in 6-well plates. About 24–30 h post-transfection, the cells were fixed using 4% paraphormaldehyde and permeabilized in phosphate buffered saline (PBS) containing 0.1% Tritonx X-100. After washing with PBS and saturation with 2% bovine serum albumin, the cells were immunostained with either mouse anti-V5 antibody (Life Technologies) or mouse anti-FLAG antibody (Sigma) for 1 h at room temperature. Subsequently, the cells were washed with PBS and incubated with DyLight 488 conjugated goat anti-mouse IgG H&L (Life Technologies) for 1 h at room temperature. Image was analyzed by Zeiss LSM 710 confocal microscope.
In vitro transcription of Pri-miRNAs and cell based Drosha cleavage assay
Plasmids containing the T7 promoter, including psiCHECK2, psiCHECK2/pri-miR-1, CMV-LUC2CP/ARE and CMV-LUC2CP/intron/ARE were used as templates after being linearized by restriction enzyme XhoI or BamHI. In vitro transcription was performed using mMESSAGE mMACHINE® T7 ULTRA Transcription Kit (Life Technologies) according to the manufacturer's protocol. RNAs produced were ARCA-capped and polyA-tailed. A total of 100 ng of transcribed mRNAs were used in transfection experiments with cells growing in 12-well plates and followed by dual luciferase assay or northern blot.
RT-PCR
Total RNA was isolated using Trizol (Life Technologies) and treated with DNase I by using TURBO DNA-free™ Kit (Life Technologies). Reverse transcription of mRNA was carried out with Superscript III or IV (Life Technologies) and Oligo dT. The PCR fragment used for cloning was amplified with AccuPrime™ high fidelity Taq DNA polymerase (Life Technologies). In reverse transcription of pri-miRNA, random hexamer primers were used to generate the cDNAs. The primer sequences are listed in Supplementary Table S1.
Quantitative real-time PCR (qPCR)
For quantification of Drosha isoforms, plasmids containing cDNA sequences of each isoform were used as templates to test the specificity of primers and establish standard curves. Quantitative real-time PCR of Drosha isoforms was performed with ZEN Double-Quenched Probes (Integrated DNA Technologies) on the StepOnePlus™ Real-Time PCR System (Life Technologies). The relative level of each isoform in HeLa, HEK293T and MCF-7 cells was determined with the cycle threshold (CT) value normalized to the standard curves. Quantification of pri-miRNAs was performed using PowerUp™ SYBR® Green Master Mix (Life Technologies) with specific primers flanking the hairpin structure. Of note, pri-miRNAs but not the corresponding pre-miRNA will be amplified by these primer sets. Sequences of primers and probe are listed in Supplementary Table S1.
Subcellular fractionation
The proteins of cytoplasmic and nuclear fractions were isolated using the Subcellular Protein Fractionation Kit (Thermo Scientific) according to the manufacturer's instructions. α-tubulin was used as a control for the cytoplasmic fraction and hnRNPC1/C2 was used as a control for the nuclear fraction. For RNA extraction, cytoplasmic fractions were separated with a buffer solution containing 10 mM Tris (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.3% (vol/vol) NP-40 and 10% (vol/vol) glycerol. The precursor mRNAs of ACTB and GAPDH were used as a control for cytoplasmic fraction.
Bioinformatic analysis
For analyzing the percentage of Drosha isoforms in human tissues, the single-end samples from Illumina BodyMap 2.0 (GSE30611) were downloaded and analyzed. First, all the raw reads were mapped to human reference genome hg19 with STAR_2.3.1t (39). Only the uniquely mapped reads were used for the downstream analysis. These reads were counted against the UCSC hg19 known Gene and the exon-level counts for all genes were calculated. The read counts for Drosha in the region from exon 4 to exon 8 were extracted. These counts were then used to estimate the isoform percentages with a Poisson generalized linear model (40).
For the frequencies of Drosha exon 5 or 6 skipping events in normal and tumor tissues, MISO (version 0.5.3) (41) with a Bayesian inference algorithm was used to analyze the RNA-seq data obtained from The Cancer Genome Atlas (TCGA). MISO computed the percentage of transcripts that were spliced to include exons, known as PSI (Percentage Spliced In) (Ψ) ratios, which is the number of reads supporting exon-inclusion divided by number of reads supporting exon-inclusion and exon-skipping. We obtained RNA-seq data from CGHub using TCGA's BAM-Slicer service, which allowed the download of slices of RNA-seq data (.BAM files) that only contained Drosha mRNA. Only RNA-seq files that mapped to the most popular genome reference (hg19) for each set of cancer RNA-seq files were downloaded and analyzed.
Statistical analysis
Statistical analysis was performed by the two-tailed, unpaired Student's t-test (GraphPad Prism 6). Differences are considered significant if P-value is <0.05.
RESULTS
Microprocessor cleavage assay
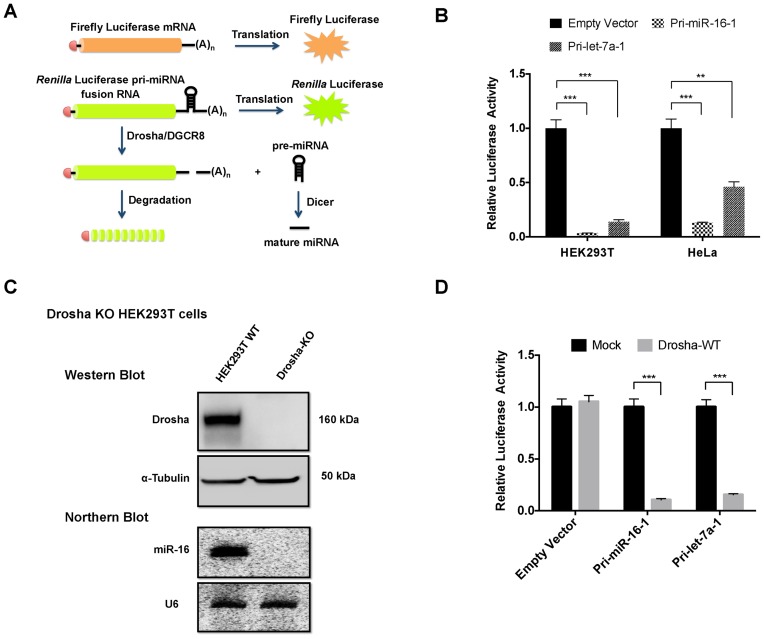
To directly measure Microprocessor cleavage activity in cells, we designed a dual-luciferase reporter in which pri-miRNA sequences were inserted into the 3′UTR of the Renilla luciferase gene (Figure 1A). The Microprocessor cleaves at the pri-miRNA structure, resulting in mRNA degradation and reduction of Renilla luciferase expression. Once normalized to the firefly luciferase level, the signal of Renilla luciferase serves as a faithful indicator of Microprocessor activity in cells (Figure 1A). As expected, reporters with pri-hsa-miR-16 or pri-hsa-let-7 inserted in the 3′UTR expressed at a much lower level (>10-fold) than the empty vector control in HEK293T cells. Similar results were observed in HeLa cells, indicating the robustness of the assay (Figure 1B).
Figure 1.
Microprocessor cleavage assay. (A) Schematic representation of Drosha cleavage reporters. (B) Empty reporter or reporter containing the sequence of either pri-hsa-miR-16-1 or pri-hsa-let-7a-1 was transfected into HEK293 or HeLa cells. Dual-luciferase assays were performed 36 h post-transfection. RL-luciferase activities were normalized with FF-luciferase and the percentage of relative enzyme activity compared to the negative control (empty reporter without any pri-miRNA sequence) was plotted. Error bars represent the standard deviation from three biological replicates. (C) Drosha KO cells were generated by CRISPR/Cas9 in HEK293T cells. Western blot and northern blot were used to assess the expression of Drosha proteins and miR-16 respectively. Probe-miR-16 and Probe-U6 (Supplementary Table S1) were used to detect miR-16 and U6 respectively. (D) Drosha cleavage reporters containing either pri-hsa-miR-16-1 or pri-hsa-let-7a-1 were co-transfected with empty vectors (mock treatment) or plasmids expressing Drosha protein into Drosha KO cells. Dual-luciferase assays were performed 36 h post-transfection. Results were plotted as illustrated in (A). **P < 0.01; ***P < 0.001.
Comparing reporters generates a bias due to variations in mRNA stability. There is a clear advantage to measure Microprocessor activity by comparing the expression from each reporter in the presence or absence of the Microprocessor. To this end, we generated Drosha KO in HEK293T cells by CRISPR. The loss of Drosha expression and abolishment of endogenous miRNA production were confirmed by western blotting and northern blotting, respectively (Figure 1C). As expected, co-expressing wild-type Drosha (Drosha-WT) from transfected cDNA in Drosha KO cells restored the repression on Drosha cleavage reporters. The same experiment had no effect on the reporter without pri-miRNA sequences, demonstrating the specificity of the assay (Figure 1D).
Together, we established a robust reporter system to monitor Drosha cleavage in cells, in which manipulations on both the enzyme (Drosha) and substrate (pri-miRNA) are possible.
Drosha is capable of cleaving pri-miRNA transcripts in the cytoplasm of human cells
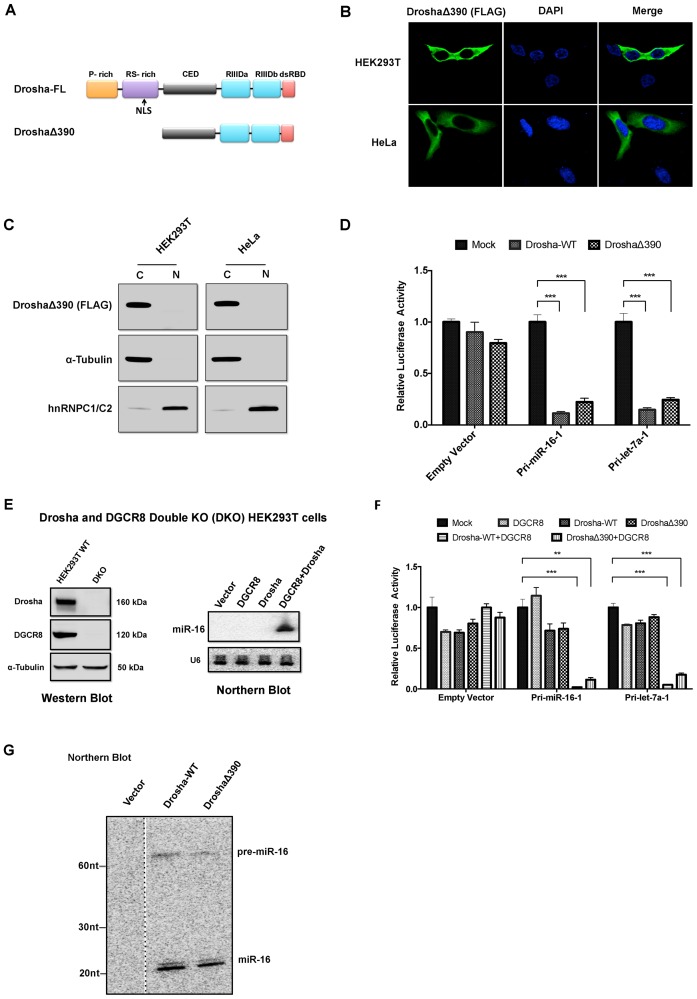
To understand how subcellular localization impacts Drosha activity, we created a truncated Drosha (DroshaΔ390) with a FLAG-tag and deletion of the first 390 amino acids in the N terminus where the NLS is located (Figure 2A). Of note, DroshaΔ390 retained the ability to form the Microprocessor and perform cleavage in vitro (16,26). Consistent with previous publications (28), immunostaining or subcellular fractionation followed by western blot with anti-FLAG antibody demonstrated that DroshaΔ390 was located exclusively in the cytoplasm of both HEK293T and HeLa cells (Figure 2B and C), presumably due to the loss of the NLS. We then tested its function by co-expressing Drosha cleavage reporters with either Drosha-WT or DroshaΔ390 in Drosha KO cells. Albeit to a lesser degree than Drosha-WT, DroshaΔ390 effectively repressed the reporter containing pri-hsa-miR-16 (∼20-fold). Similar results were obtained with the pri-hsa-let-7 reporter, while no reduction was observed with the empty reporter (Figure 2D). Together, these results demonstrated Drosha was able to specifically cleave pri-miRNA transcripts in the cytoplasm.
Figure 2.
Drosha is capable of cleaving pri-miRNA transcripts in the cytoplasm of human cells in a DGCR8 dependent manner. (A) Diagram of Drosha domains and DroshaΔ390 construct. Nuclear localization signal (NLS) is indicated by an arrow. (B) Subcellular localization of DroshaΔ390 in both HEK293T and HeLa cells detected by immunofluorescence with anti-FLAG antibodies. (C) Subcellular localization of DroshaΔ390 in both HEK293T and HeLa cells was detected by western blotting in cytoplasmic and nuclear fractions. A-Tubulin and hnRNPC1/2 were served as cytoplasmic and nuclear marker respectively. (D) The cleavage activity of DroshaΔ390 and Drosha-WT (full-length) were measured by dual luciferase assay in Drosha KO cells with a reporter containing either the pri-hsa-miR-16-1 or pri-hsa-let-7a-1 sequence. Results were plotted as illustrated in Figure 1 (B). (E) Drosha and DGCR8 Double KO (DKO) cells were generated by CRISPR/Cas9 in HEK293T cells. Western blot and northern blot were used to assess the expression of Drosha/DGCR8 proteins and miR-16, respectively. In northern blot, Probe-miR-16 and Probe-U6 (Supplementary Table S1) were used to detect miR-16 and U6 respectively. (F) The cleavage activity of DroshaΔ390 and Drosha-WT (full-length) with or without DGCR8 were measured by dual luciferase assay in DKO cells with reporter containing either pri-hsa-miR-16-1 or pri-hsa-let-7a-1 sequence. Results were plotted as illustrated before. (G) Pri-miR-16-1 reporter was co-transfected with either Drosha-WT or DroshaΔ390 into Drosha KO cells. Pre-miR-16-1 and mature miR-16-1 were detected by northern blot using Probe-miR-16. **P < 0.01; ***P < 0.001.
To investigate whether cytoplasmic Drosha activity requires DGCR8, we generated a Drosha and DGCR8 DKO in HEK293T cells by CRISPR. The loss of Drosha and DGCR8 expression was confirmed by western blotting (Figure 2E). The biogenesis of miRNA was abolished and could only be rescued by co-expression of both Drosha and DGCR8 from cDNAs (Figure 2E). Drosha cleavage reporters were co-transfected with various combinations of plasmids expressing Drosha-WT, DroshaΔ390 or DGCR8 in DKO cells. Similar to Drosha-WT, DroshaΔ390 failed to repress pri-miRNA cleavage reporter without DGCR8, suggesting c-Drosha still needed to form the Microprocessor to exert its cleavage function (Figure 2F). Supporting this idea, a minor but visible portion of DGCR8 was detected in the cytoplasm of both HEK293T and HeLa cells by both immunostaining and subcellular fractionation followed by western blot (Supplementary Figure S1).
To further examine the function of DroshaΔ390, we detected the cleavage products by northern blotting. When co-transfected with a reporter containing pri-hsa-miR-16 in Drosha KO cells, DroshaΔ390 generated a cleavage product (pre-miR-16) in the same size as that of Drosha-WT. In addition, the precursors were recognized by Dicer and further processed into mature miR-16, indicating the cleavage fidelity of DroshaΔ390 was comparable to that of Drosha-WT in cells (Figure 2G).
Taken together, these results demonstrated that Drosha, when artificially expressed in the cytoplasm, could process pri-miRNA transcripts in a manner similar to its nuclear counterpart.
Detection of endogenous Drosha activity in the cytoplasm of human cells
Encouraged by the finding that Drosha retained its pri-miRNA processing activity when ectopically expressed in the cytoplasm, we sought to determine whether such activity exists endogenously in human cells. One challenge of probing cytoplasmic Drosha activity is to distinguish it from the nuclear activity. After all, substrates expressed in cells are transcribed in the nucleus and therefore inevitably subjected to nuclear Drosha processing (42,43). To rule out nuclear interference, we decided to introduce in vitro transcribed pri-miRNA substrates directly into cytoplasm via transfection and test whether these can be processed without going into the nucleus.
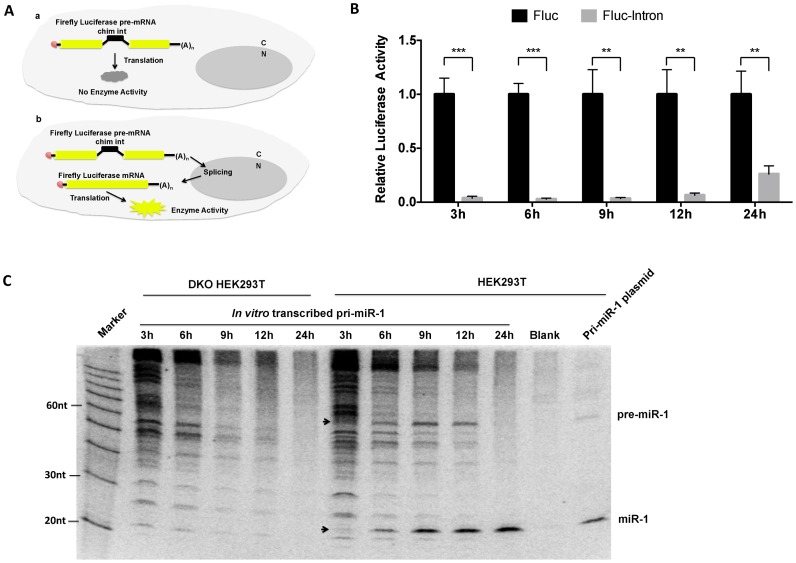
To track the location of RNA transcripts post-transfection, we used a firefly luciferase reporter in which a chimeric intron was inserted in the coding region (44). Expression of this reporter relies on localization to the nucleus, where the intron can be removed by the splicing machinery (Figure 3A). We made the mRNAs of firefly luciferase genes with or without the chimeric intron by in vitro transcription and transfected these mRNAs respectively into HEK293T cells. mRNA transcripts from Renilla luciferase gene were used as transfection control. Firefly luciferase activities were measured at various time points post transfection and normalized to Renilla. We were able to detect signals from a reporter without an intron (Figure 3B), indicating transfected mRNAs were translated in the cells as intended. To the contrary, signals from an intron-containing reporter were near background noise at first but became detectable after 6 h (Figure 3B and Supplementary Figure S2). Hence, we concluded that transfected mRNA transcripts were retained in the cytoplasm, at least during the first 6 h post-transfection.
Figure 3.
Detection of endogenous Drosha activity in the cytoplasm of human cells. (A) Schematic representation of experiment design: scenario a—no FF-luciferase signal will be detected if pre-mRNA of FF-luciferase reporter was trapped in the cytoplasm. Scenario b—FF-luciferase signal will be detected if its pre-mRNA has access to the nucleus. (B) mRNA of FF-luc reporter with (Fluc-intron) or without intron (Fluc) was co-transfected with mRNA of Renilla luciferase into HEK293T cells. Dual-luciferase assay was performed at various time points post transfection. After normalization to Renilla luciferase, levels of Fluc and Fluc-intron were compared to each other. The ratio of Fluc-intron level to Fluc level was plotted in the figure. Error bars represent the standard deviation from three biological replicates. (C) In vitro transcribed pri-miRNA or pri-miR-1 expression plasmid was transfected into DKO or wild-type HEK293T cells. Samples were collected at different time points post transfection as indicated. The pre-miRNA and mature miRNA were detected by Northern blot and indicated by arrows. Probe-miR-1 (Supplementary Table S1), which is the antisense sequence of miR-1, was used to detect miR-1 and pre-miR-1. **P < 0.01; ***P < 0.001.
Following this exact approach, we prepared mRNAs transcripts containing pri-hsa-miR-1 by in vitro transcription and transfected them into either DKO or HEK293T cells. Total RNA was harvested 3, 6, 9, 12 or 24 h post-transfection and subjected to northern blotting. As miR-1 is not expressed in HEK293T cells, we utilized the level of mature miR-1 to monitor the processing of pri-hsa-miR-1. While absent in the DKO cells, miR-1 was detected in HEK293T cells as early as 3 h post-transfection and gradually accumulated over time (Figure 3C). As transfected mRNAs were still in the cytoplasm 6 h post-transfection, these results demonstrated the existence of endogenous cytoplasmic Drosha activity, which was responsible for the processing of pri-hsa-miR-1 transcripts in the cytoplasm.
A subset of endogenous pri-miRNAs were detected in the cytoplasm of human cells
We next tested whether the pri-miRNA transcripts, which are endogenous substrates of Drosha, were present in the cytoplasm. Using pri-miRNA specific primers spanning Drosha cleavage sites, we performed RT-PCR on total RNA or RNA extracted from the cytoplasmic fraction of HEK293T cells. Out of nine pri-miRNAs tested, five could be detected in the cytoplasmic fraction, indicating that a certain portion of pri-miRNAs were located in the cytoplasm (Supplementary Figure S3A). mRNAs but not pre-mRNAs of either ACTB or GAPDH were detected in the cytoplasmic fraction of the same samples, demonstrating that this observation was not caused by nuclear fraction contamination (Supplementary Figure S3B).
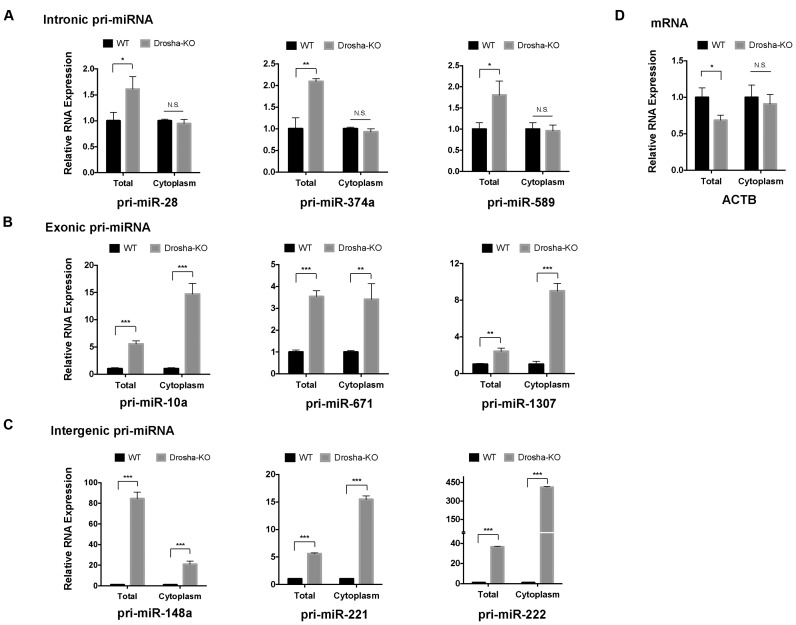
To investigate whether these cytoplasmic-localized pri-miRNAs were processed by Microprocessor, we performed the same experiment in Drosha KO cells. The levels of pri-miRNAs were quantified by RT-qPCR and normalized to that of GAPDH mRNA, which were compared to the same measurements in wild-type cells. As expected, upon Drosha removal the overall levels of pri-miRNAs increased across the board while no increase was observed with ACTB mRNA (Figure 4). The degree of increment varied, indicating that each individual pri-miRNA has a distinct stability. Interestingly, the extent to which pri-miRNA accumulated in the cytoplasm correlated with the genomic location of the pri-miRNAs: for pri-miRNAs coded in the intron of the host gene (pri-hsa-miR-589, pri-hsa-miR-28 and pri-hsa-miR-374a), there was little to no change of the cytoplasmic levels, indicating that most of these pri-miRNA transcripts were repressed in the nucleus (Figure 4A). On the contrary, for pri-miRNAs embedded in the exon of host gene (pri-hsa-miR-10a, pri-hsa-miR-1307 and pri-hsa-miR-671) as well as pri-miRNAs transcribed from the intergenic regions (pri-hsa-miR148, pri-hsa-miR-221 and pri-hsa-miR-222), dramatic increases of cytoplasmic levels were observed, suggesting these transcripts might be subject to c-Drosha cleavage (Figure 4B and C). However, we cannot exclude the possibility that these pri-miRNAs were processed in the nucleus. The elevated cytoplasmic level might be a result of exportation of uncleaved pri-miRNAs in the nucleus.
Figure 4.
A subset of endogenous pri-miRNAs were processed in the cytoplasm of human cells. The levels of pri-miRNAs in whole cell or cytoplasmic fraction were determined by real time RT-qPCR in Drosha KO cells. All values were normalized to the corresponding level in wild-type cells and plotted in the figure. (A) Pri-miRNAs encoded in the intron of host gene. (B) Pri-miRNAs encoded in the exon of host gene. (C) Pri-miRNAs encoded in the intergenic region. (D) ACTB mRNA level was used as a negative control for the effects of Drosha. Error bars represent the standard deviation from three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001. N.S.: non-significant.
Alternative splicing generates cytoplasmic Drosha activity
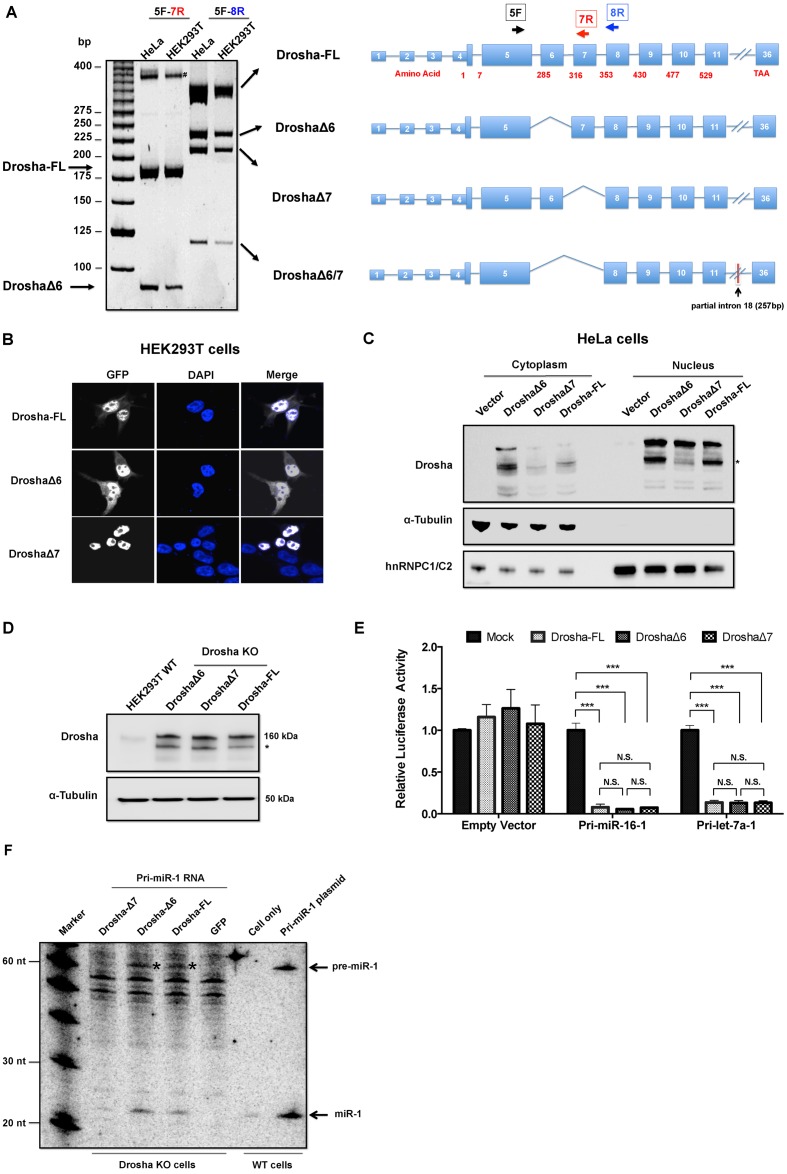
The putative NLS is encoded in the exon 6 of Drosha mRNA (28). Excluding the NLS via alternative splicing is a possible scenario to generate cytoplasmic Drosha. To test this directly, we designed two sets of primers flanking exon 6 and performed PCR on cDNAs from both HeLa and HEK293T cells. Interestingly, multiple amplicons were observed in both HeLa and HEK293T cells (Figure 5A). Sequencing of these PCR products revealed three novel mRNA isoforms in addition to the full-length (Drosha-FL): exon 6 skipping (DroshaΔ6), exon 7 skipping (DroshaΔ7) and skipping of both (DroshaΔ6/7). Of note, both exon 6 (96 nt) and 7 (111 nt) are relatively short and in frame. All three mRNA isoforms had the potential to generate functional Drosha with a small truncation in the N terminal domain. However, in efforts to clone the cDNA of each isoform, we found Δ6/7 skipping was always coupled with downstream out-of-frame insertions around exon 17, which made it unlikely to code a functional protein.
Figure 5.
Identification and characterization of Drosha isoforms in human cells. (A) RT-PCR was used to identify Drosha isoforms that were generated by alternative splicing in both HEK293T and HeLa cells. # indicates non-specific amplicon or hybrid of amplicons. (B) Subcellular localization of Drosha isoforms in HEK293T cells were detected by GFP infusion proteins. (C) Western blotting was used to detect subcellular localization of Drosha isoforms in HeLa cells after subcellular fractionation. Truncated Drosha protein was detected as previous reported (16,59) and marked with asterisk (*). (D) Western blotting was used to assess the level of Drosha isoforms after ectopically expression in the Drosha KO cells. (E) Drosha cleavage reporter containing either pri-hsa-miR-16-1 or pri-hsa-let-7a-1 was co-transfected with an empty vector (mock treatment) or plasmid expressing individual Drosha isoforms into Drosha KO cells. Dual-luciferase assays were performed 36 h post-transfection. Results were plotted as illustrated before. ***P < 0.001. N.S.: non-significant. (F) Drosha KO cells ectopically expressing individual isoforms or GFP were transfected with in vitro transcribed pri-miR-1 transcripts. Samples were collected 6 h post RNA transfection. Wild-type HEK293T cells and cells transfected with pri-miR-1 expression plasmid was used as negative and positive controls, respectively. Probe-miR-1 (Supplementary Table S1), which is the antisense sequence of miR-1, was used to detect miR-1 and pre-miR-1 (labeled with *) in the northern blot. See supplementary Figure S5 for results of a biological replicate of this experiment.
Next, we sought to investigate the subcellular distribution of Drosha isoforms by expressing GFP-tagged fusion proteins in HEK293T cells. As expected, GFP-DroshaΔ6, which lost the putative NLS, was observed in the cytoplasm. However, part of GFP-DroshaΔ6 was still in the nucleus, suggesting that other elements besides the identified NLS also contributed to the nuclear location of Drosha. In addition, although both GFP-DroshaΔ7 and GFP-Drosha-FL contain the putative NLS, only the former was detected exclusively in the nucleus while the latter had a minor portion in the cytoplasm, indicating exon 7 might contain nuclear exportation signals (NES) (Figure 5B). To eliminate the possibility that GFP fusion affected the subcellular localization of Drosha, we over-expressed each isoform without GFP tag in HeLa cells. Ectopically expressed Drosha was detected by western blotting in either cytoplasmic or nuclear fractions. Consistent with the observations in HEK293T cells, DroshaΔ6 and to some degree Drosha-FL, were found in the cytoplasmic fraction (Figure 5C and Supplementary Figure S4). Note, we cannot rule out the possibility that regions other than exon 6 and exon 7 also contribute to the subcellular localization of Drosha. Neither can we determine whether the putative NLS (in exon 6), putative NES (in exon 7) or both are responsible for the subcellular localization of Drosha isoforms.
To determine the cleavage efficiency of Drosha isoforms in cells, Drosha cleavage reporters were co-transfected with the cDNA plasmid of each isoform into Drosha KO cells. Western blot experiments established that these isoforms were expressed in nearly equivalent amounts (Figure 5D). Consistent with the observation that the N-terminus was dispensable for cleavage activity in vitro, DroshaΔ6 and DroshaΔ7 (two isoforms with small N-terminal truncation) had similar cleavage activity as Drosha-FL in processing both pri-hsa-miR-16 and pri-hsa-let-7 (Figure 5E).
Next, we measured the c-Drosha activity of each isoform by transfecting in vitro transcribed pri-miR-1 into Drosha KO cells where individual Drosha isoform was ectopically expressed. As expected, northern blotting results demonstrated that pri-miR-1 transcripts were processed into pre-miR-1 in the cytoplasm efficiently by cytoplasmic Drosha isoforms (DroshaΔ6 and Drosha-FL), but not by nuclear Drosha isoform (DroshaΔ7) (Figure 5F and Supplemental Figure S5).
Taken together, these results demonstrated that the endogenous c-Drosha activity could be, at least in part, explained by DroshaΔ6 generated via alternative splicing.
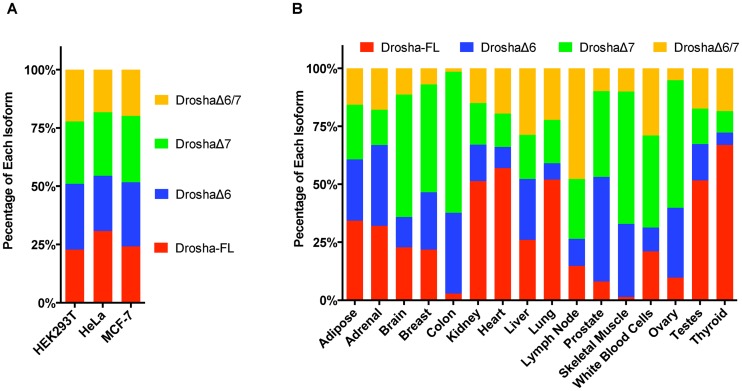
Quantification of Drosha isoforms in human cell lines and tissues
To quantify the abundance of these novel isoforms, we designed a set of exon–exon junction primers, which can be used to specifically amplify each isoform and establish standard curves (Supplementary Figures S6 and 7). Real-time PCR was performed on the cDNA of multiple human cell lines: HEK293T, HeLa and MCF-7. In all samples, about 70% of Drosha mRNAs were subject to alternative splicing around exon 6 and 7, missing at least one of these two exons. DroshaΔ6, which led to cytoplasmic location, was a major isoform in all three cell lines tested (Figure 6A).
Figure 6.
Quantification of Drosha isoforms in human cells and tissues. (A) The amounts of each isoform in HeLa, HEK293T and MCF-7 cells were measured by RT-qPCR with isoform specific primers and the corresponding standard curves (Supplementary Figure S7). The relative level of each isoform to that of Drosha-FL was plotted in the figure. (B) Percentage of each Drosha isoform in human tissues. The single end samples from Illumina BodyMap 2.0 were analyzed for the Drosha isoform compositions in different human tissues using a Poisson generalized linear model. See ‘Materials and Methods’ section for details.
Taking advantage of RNA-seq data from Illumina BodyMap 2.0, we analyzed the distribution of Drosha isoforms in human tissue. The landscapes of Drosha isoform profiles were vastly variable across human tissue. For instance, DroshaΔ6 is the dominant isoform in the prostate while the thyroid barely expresses any (Figure 6B). These results strongly suggest that c-Drosha activity is highly regulated via alternative splicing.
Cytoplasmic Drosha isoforms correlated with tumorigenesis
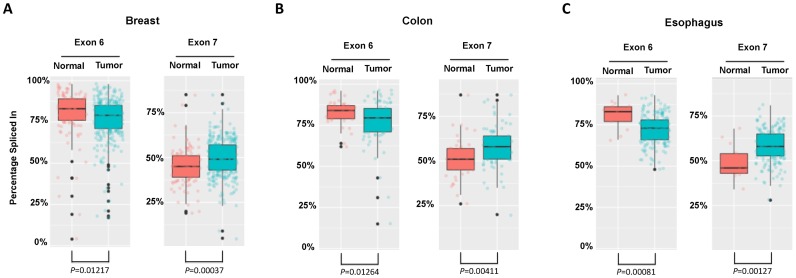
Previous studies have established that dysregulation of Drosha function plays a key role in cancer development (7). We hypothesize that the c-Drosha activity may also contribute to tumorigenesis. To test this, we compared the alternative splicing events of exon 6 and 7 between cancer and normal tissue based on RNA-seq data from TCGA. After analyzing over 300 breast cancer patient samples, we found that the percentage of Drosha exon 6 spliced-in was significantly lower than that of normal tissues. Interestingly, the percentage of Drosha exon 7 spliced-in was the opposite: higher in cancer tissues and lower in normal tissues (Figure 7). As skipping of exon 6 (DroshaΔ6) promotes cytoplasmic distribution and skipping of 7 (DroshaΔ7) enhances nuclear retention, these results strongly suggest that c-Drosha activity is enriched in breast cancer tissues. Remarkably, the same results were also obtained with colon cancer and esophagus cancer samples, indicating that c-Drosha activity is correlated with the tumorigenesis.
Figure 7.
The percentage spliced in (PSI) of either Drosha exon 6 or exon 7 in different normal and tumor tissues. The RNA-seq data of normal and tumor tissues was obtained from TCGA database and analyzed by MISO with a Bayesian inference algorithm. See ‘Materials and Methods’ section for details. (A) Breast tissues, including 113 normal tissues and 307 tumor tissues. (B) Colon tissues, including 41 normal tissues and 80 tumor tissues. (C) Esophagus tissues, including 13 normal tissues and 184 tumor tissues. P-values are labeled on the graphs.
DISCUSSION
Evidence from in vitro biochemical studies established that the first step of pri-miRNA processing via Microprocessor happens in the nucleus (25). Here, we provide multiple lines of evidence to support the existence of cytoplasmic Drosha activity: (i) a Drosha truncation (DroshaΔ390), which was exclusively located in the cytoplasm, had the ability to cleave pri-miRNA-like reporters in a DGCR8-dependent manner. (ii) In vitro transcribed pri-miRNA transcripts introduced into cells via transfection were processed into mature miRNAs without entering the nucleus. (iii) A subset of pri-miRNAs were detected in the cytoplasm and levels were enriched in the cytoplasm of Drosha KO cells.
Additionally, we identified a novel isoform of Drosha (DroshaΔ6), which lost its putative NLS through alternative splicing. This finding provides a mechanism to generate and regulate the c-Drosha activity. Alternative splicing is one of the key events in the regulation of gene expression. Sequencing of the transcriptome revealed that up to 90% of multi-exon human genes are subject to alternative splicing (45–47). Our findings demonstrate the connections between alternative splicing and the miRNA pathway, offering insights into the complex gene regulation networks in cells.
The abundance of the c-Drosha isoform (DroshaΔ6) is tissue-specific and highly regulated during tumorigenesis, suggesting that it has unique biological functions in gene regulation. One possible role for c-Drosha is to process certain pri-miRNAs in the cytoplasm. It could function as a safety mechanism by ensuring the production of miRNAs from pri-miRNAs that have escaped nuclear processing. Alternatively, the c-Drosha activity could be required for upregulating the levels of miRNAs whose pri-miRNAs are specifically transported into the cytoplasm. In support of this, we found that levels of exonic pri-miRNAs in the cytoplasm increased when c-Drosha activity was removed. Of note, more than one-third of intronic miRNAs have their own promoters, decoupling their transcriptions from the host gene (10,48–50). These intronic pri-miRNAs are not necessarily trapped in the nucleus and are potential substrates of c-Drosha as well. In such a scenario, the biogenesis of these miRNAs will no longer depend on the function of exportin-5 as long as the corresponding pri-miRNAs are exported out of the nucleus. In line with this, a recent report demonstrated that the levels of many miRNAs were reduced but not abolished in exportin-5 KO cells (51).
However, there is also evidence arguing that pri-miRNA processing is not the main function of c-Drosha. Given that microprocessor consists of one subunit of Drosha and two of DGCR8 (26), the modest amount of cytoplasmic DGCR8 (Supplementary Figure S1) is unlikely to be sufficient for all the c-Drosha to be in the form of the Microprocessor. In addition, as discussed before, the results that pri-miRNA increases upon Drosha KO are open to alternative interpretations. For exonic miRNAs, their pri-miRNAs are indistinguishable from the host mRNAs. Pri-miRNA increase may reflect an increase in mRNA resulting from the decrease in cleavage of the miRNA from exons in the Drosha KO cells. Finally, a recent study showed that the pri-miRNA processing efficiency is not correlated with the expression level of cytoplasmic Drosha isoform (52). We demonstrated that c-Drosha exists and is capable of processing pri-miRNA. However, it is unsettled whether or not a fraction of pri-miRNAs are processed in the cytoplasm. Further study is required to determine the degree to which the c-Drosha activity contributes to the cellular miRNA profile.
Genetic evidence indicates Drosha has additional roles beyond miRNA biogenesis (53). Recent reports suggest a function for Drosha in degradation of certain mRNAs that contain pri-miRNA-like stem-loop structures (54–58). It is intriguing to hypothesize that c-Drosha targets a novel set of RNA transcripts, mRNAs for example and regulates their stabilities. Our analysis of RNA-seq data from TCGA showed that c-Drosha expression is up regulated in multiple cancer tissues. c-Drosha might function as an oncogene by targeting mRNAs of tumor-suppressor genes for degradation. Further investigation on the function of c-Drosha may extend our understanding of the Drosha regulatory network and provide novel targets for cancer therapeutics.
Supplementary Material
Acknowledgments
We thank Dr Jun Zhu and Dr Yanqin Yang for their guidance in MISO data analysis. We would also like to thank Dr Markus Hafner, Dr Donald Court and members of the Gu lab for helpful discussion and critical reading of this manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by Intramural Research Program of National Cancer Institute, National Institutes of Health; ZG was supported by funding from TEES-AgriLife Center for Bioinformatics and Genomic Systems Engineering (CBGSE) at Texas A&M University. PY was supported by startup funding from the ECE department and Texas A&M Engineering Experiment Station/Dwight Look College of Engineering at Texas A&M University. Funding for open access charge: NIH/NCI Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell. Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 2.Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 4.Sun K., Lai E.C. Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arner P., Kulyte A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015;11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 6.Schober A., Nazari-Jahantigh M., Weber C. MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis. Nat. Rev. Cardiol. 2015;12:361–374. doi: 10.1038/nrcardio.2015.38. [DOI] [PubMed] [Google Scholar]
- 7.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsico A., Huska M.R., Lasserre J., Hu H., Vucicevic D., Musahl A., Orom U., Vingron M. PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 2013;14:R84. doi: 10.1186/gb-2013-14-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini H.K., Enright A.J., Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinske L.C., Franca G.S., Torres H.A., Ohara D.T., Lopes-Ramos C.M., Heyn J., Reis L.F., Ohno-Machado L., Kreth S., Galante P.A. miRIAD-integrating microRNA inter- and intragenic data. Database. 2014;2014:bau099. doi: 10.1093/database/bau099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 15.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 16.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landthaler M., Yalcin A., Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 20.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki S., Sasaki H.M., Sakaguchi Y., Suzuki T., Tadakuma H., Tomari Y. Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature. 2015;521:533–536. doi: 10.1038/nature14254. [DOI] [PubMed] [Google Scholar]
- 22.Gu S., Kay M.A. How do miRNAs mediate translational repression. Silence. 2010;1:11. doi: 10.1186/1758-907X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 24.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Kwon S.C., Nguyen T.A., Choi Y.G., Jo M.H., Hohng S., Kim V.N., Woo J.S. Structure of human DROSHA. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Tang X., Zhang Y., Tucker L., Ramratnam B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38:6610–6619. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X., Li M., Tucker L., Ramratnam B. Glycogen synthase kinase 3 beta (GSK3beta) phosphorylates the RNAase III enzyme Drosha at S300 and S302. PLoS One. 2011;6:e20391. doi: 10.1371/journal.pone.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro J.S., Langlois R.A., Pham A.M., Tenoever B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro J.S., Schmid S., Aguado L.C., Sabin L.R., Yasunaga A., Shim J.V., Sachs D., Cherry S., tenOever B.R. Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7108–7113. doi: 10.1073/pnas.1319635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouha H., Thurner C., Mandl C.W. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38:8328–8337. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro J.S., Varble A., Pham A.M., Tenoever B.R. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kourtidis A., Ngok S.P., Pulimeno P., Feathers R.W., Carpio L.R., Baker T.R., Carr J.M., Yan I.K., Borges S., Perez E.A., et al. Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat. Cell Biol. 2015;17:1145–1157. doi: 10.1038/ncb3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H., Hou Y., Xu L., Zeng Z., Wen S., Du Y.E., Sun K., Yin J., Lang L., Tang X., et al. Cytoplasmic Drosha is aberrant in precancerous lesions of gastric carcinoma and its loss predicts worse outcome for gastric cancer patients. Dig. Dis. Sci. 2015;61:1090–1080. doi: 10.1007/s10620-015-3986-0. [DOI] [PubMed] [Google Scholar]
- 36.Jafarnejad S.M., Sjoestroem C., Martinka M., Li G. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod. Pathol. 2013;26:902–910. doi: 10.1038/modpathol.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugito N., Ishiguro H., Kuwabara Y., Kimura M., Mitsui A., Kurehara H., Ando T., Mori R., Takashima N., Ogawa R., et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin. Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 38.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobson A.J., Barnett A.G. An Introduction to Generalized Linear Models. 3rd edn. Boca Raton: CRC press; 2008. [Google Scholar]
- 41.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlicki J.M., Steitz J.A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Younis I., Berg M., Kaida D., Dittmar K., Wang C., Dreyfuss G. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol. Cell. Biol. 2010;30:1718–1728. doi: 10.1128/MCB.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 48.Monteys A.M., Spengler R.M., Wan J., Tecedor L., Lennox K.A., Xing Y., Davidson B.L. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang T.C., Pertea M., Lee S., Salzberg S.L., Mendell J.T. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 2015;25:1401–1409. doi: 10.1101/gr.193607.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim Y.K., Kim B., Kim V.N. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Link S., Grund S.E., Diederichs S. Alternative splicing affects the subcellular localization of Drosha. Nucleic Acids Res. 2016;44:5330–5343. doi: 10.1093/nar/gkw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burger K., Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat. Rev. Mol. Cell Biol. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- 54.Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karginov F.V., Cheloufi S., Chong M.M., Stark A., Smith A.D., Hannon G.J. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knuckles P., Vogt M.A., Lugert S., Milo M., Chong M.M., Hautbergue G.M., Wilson S.A., Littman D.R., Taylor V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012;15:962–969. doi: 10.1038/nn.3139. [DOI] [PubMed] [Google Scholar]
- 57.Seong Y., Lim D.H., Kim A., Seo J.H., Lee Y.S., Song H., Kwon Y.S. Global identification of target recognition and cleavage by the Microprocessor in human ES cells. Nucleic Acids Res. 2014;42:12806–12821. doi: 10.1093/nar/gku957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johanson T.M., Keown A.A., Cmero M., Yeo J.H., Kumar A., Lew A.M., Zhan Y., Chong M.M. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat. Immunol. 2015;16:1134–1141. doi: 10.1038/ni.3293. [DOI] [PubMed] [Google Scholar]
- 59.Yang Q., Li W., She H., Dou J., Duong D.M., Du Y., Yang S.H., Seyfried N.T., Fu H., Gao G., et al. Stress induces p38 MAPK-mediated phosphorylation and inhibition of Drosha-dependent cell survival. Mol. Cell. 2015;57:721–734. doi: 10.1016/j.molcel.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.