Abstract
The ciliate protozoan Tetrahymena thermophila contains two types of structurally and functionally differentiated nuclei: the transcriptionally active somatic macronucleus (MAC) and the transcriptionally silent germ-line micronucleus (MIC). Here, we demonstrate that MAC features well-positioned nucleosomes downstream of transcription start sites and flanking splice sites. Transcription-associated trans-determinants promote nucleosome positioning in MAC. By contrast, nucleosomes in MIC are dramatically delocalized. Nucleosome occupancy in MAC and MIC are nonetheless highly correlated with each other, as well as with in vitro reconstitution and predictions based upon DNA sequence features, revealing unexpectedly strong contributions from cis-determinants. In particular, well-positioned nucleosomes are often matched with GC content oscillations. As many nucleosomes are coordinately accommodated by both cis- and trans-determinants, we propose that their distribution is shaped by the impact of these nucleosomes on the mutational and transcriptional landscape, and driven by evolutionary selection.
INTRODUCTION
In eukaryotic cells, nuclear DNA is packaged into chromatin, of which the nucleosome is the basic unit, formed by 147 bp of DNA wrapped around a protein core of histones (1). Nucleosome-associated DNA, relative to linker DNA, may have different access to protein factors, which can bind DNA, or catalyze reactions using DNA as the substrate or template (2). Accessibility of genetic information is affected by nucleosome distribution, which can be quantified by nucleosome occupancy and nucleosome positioning. Nucleosome occupancy represents the probability of a DNA sequence being associated with any nucleosome in a population, while nucleosome positioning, particularly translational positioning, focuses on the exact 147 bp of genomic DNA occupied by a subset of nucleosomes (3,4). Micrococcal nuclease (MNase) digestion is an established procedure for probing nucleosome distribution, based upon its preference to cut the linker DNA and release the mono-nucleosome (1). MNase-Seq, coupling the chromatin digestion with deep-sequencing technologies, has generated a wealth of information about genome-wide nucleosome distribution patterns in eukaryotic systems (3,5).
It has long been known that the nucleosome has intrinsic biases for certain DNA sequences (6). Poly (dA:dT) tracts are anti-nucleosomal due to their poor bendability (7–9), and are generally enriched in nucleosome depleted regions (NDR) (10,11). On the other hand, sequences with ∼10 bp phasing are often strongly associated with the nucleosome, as the periodic bending facilitates wrapping around the protein core (10–14). Furthermore, correlations between nucleosome occupancy and GC-content have been found in in vivo nucleosome distribution of yeast and worm, as well as in vitro nucleosome reconstitution (15).
Other than DNA sequence features, often referred to as cis-determinants, there are also various trans-determinants affecting nucleosome distribution. Transcription in particular has a pervasive influence on chromatin structure (16). Nucleosome association with energetically favored as well as disfavored DNA sequences can be driven by ATP-dependent chromatin remodelers (17). All these shape the nucleosome distribution in the gene body bounded by transcription start sites (TSS) and transcription end sites (TES), manifest in stereotypical arrays of well-positioned nucleosomes (referred to as +1, +2, etc., ordered 5′ to 3′) (18–21). The relative contributions of cis- and trans-determinants to nucleosome occupancy and nucleosome positioning are actively studied in various systems (2,22,23). Absence of stereotypical nucleosome arrays in yeast nucleosomes reconstituted without cell extracts and ATP favors trans-determinants as the predominant factor (10,11,24). However, this is contradicted by recent in vitro nucleosome reconstitution of the ciliate protozoan Tetrahymena thermophila, which exhibits stereotypical nucleosome arrays (25).
To resolve relative contributions of cis- and trans-determinants, we took advantage of the nuclear dimorphism in Tetrahymena (26), which features two types of structurally and functionally differentiated nuclei: the macronucleus (MAC) and the micronucleus (MIC). MAC are transcriptionally active, containing largely decondensed chromatin, as well as abundant RNA polymerases and transcription factors (27,28). By contrast, MIC are transcriptionally silent in asexually dividing cells, containing highly condensed chromatin and no transcription machinery (Figure 1A). Employing MNase-Seq to reveal global nucleosome distribution patterns, we found abundant well-positioned nucleosomes in MAC, downstream of TSS and flanking both the 5′ and 3′ splice sites in the gene body. By contrast, MIC nucleosomes were dramatically delocalized, attributable to a lack of transcription-associated trans-determinants. However, nucleosome occupancy in MIC was similar to MAC, and consistent with in vitro nucleosome reconstitution and theoretical predictions based upon DNA sequence features. In particular, well-positioned nucleosomes were often matched with oscillations in GC content, revealing unexpectedly strong contributions from cis-determinants. As many well-positioned nucleosomes are coordinately accommodated by both cis- and trans-determinants, we propose that their positioning is shaped by the impact of these nucleosomes on the mutational as well as transcriptional landscape, and driven by evolutionary selection.
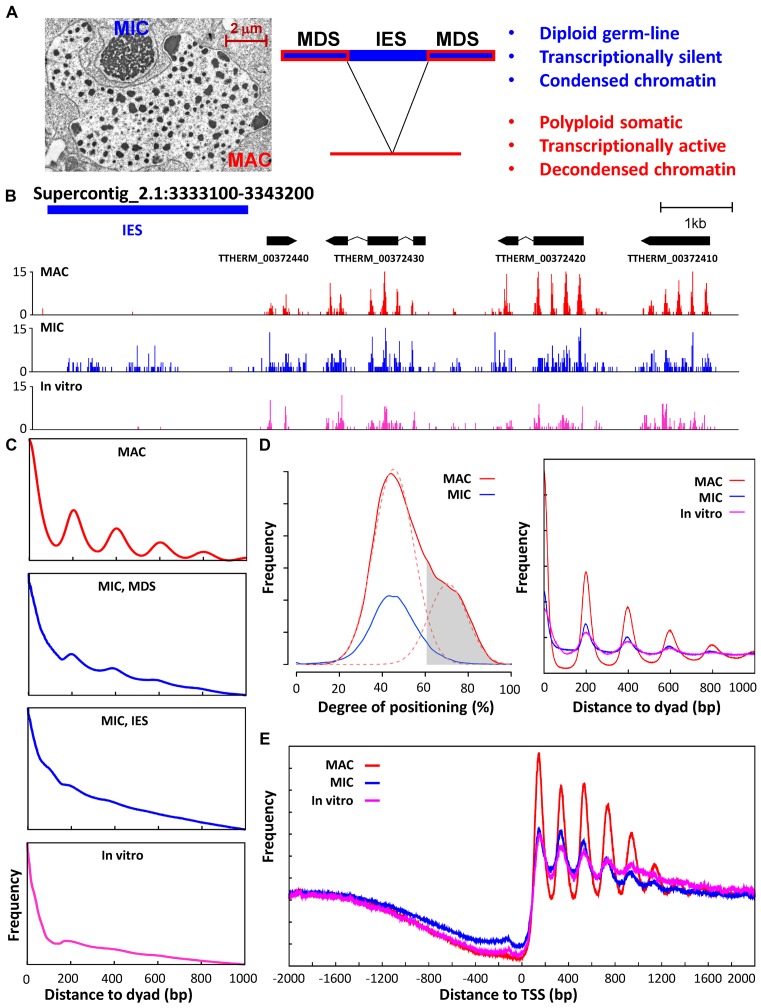
Figure 1.
Positioned nucleosomes are abundant in transcriptionally active MAC, but depleted in transcriptionally silent MIC. (A) Two types of structurally and functionally differentiated nuclei in Tetrahymena. Left, an electron micrograph showing the structurally differentiated macronucleus (MAC) and micronucleus (MIC), enclosed by their own nuclear envelope and contained in the same cytoplasmic compartment. Middle, DNA elimination accompanying MIC to MAC differentiation: IES, internal eliminated sequences, MIC-specific; MDS, MAC-destined sequences, shared between MAC and MIC. Right, functional differentiation of MAC and MIC. (B) Nucleosome distribution in a representative genome region. Paired-end MNase-Seq results from MAC (red), MIC (blue), and in vitro (magenta) (25) are mapped to the MIC genome. Distribution of fragment centers representing nucleosome dyads is plotted, together with models for genes and IES. (C) Phasogram of nucleosome distribution in MAC (red), MIC (blue), and in vitro (magenta) (25). x-axis: distance to fragment center (dyad); y-axis, frequency at a designated distance. MIC result is mapped to MDS and IES separately; MAC and in vitro results are mapped to MDS. (D) Nucleosome positioning and long-range order. Left, Nucleosome positioning in MAC and MIC. The distribution of degrees of nucleosome positioning in MAC (red solid line) can be decomposed into two peaks of normal distribution (red dashed line), with the left peak representing more delocalized nucleosomes and the right peak representing well-positioned nucleosomes. Well-positioned nucleosomes in MAC (gray area, 61% cutoff) are selected for further analysis. They have significantly reduced degrees of translational positioning in MIC (blue), similar to the more delocalized nucleosomes in MAC (red dashed line, left peak). See Methods for details and Supplemental File S1 for a compilation of properties of called nucleosomes. Right, composite analysis of nucleosome positioning in MAC (red), MIC (blue), and in vitro (magenta) (25), aligned to the dyads of well-positioned nucleosomes in MAC. (E) Composite analysis of nucleosome positioning in MAC (red), MIC (blue), and in vitro (magenta) (25), aligned to TSS. Distribution of fragment centers around TSS (±2kb) is aggregated over 15,841 well-modeled genes. See Methods for details and Supplemental File S2 for a compilation of properties of well-modeled genes.
MATERIALS AND METHODS
Cell culture, purification of nuclei and MNase digestion
T. thermophila wild-type strain CU428, obtained from the Tetrahymena Stock Center (https://tetrahymena.vet.cornell.edu/), was grown in SPP medium at 30ºC (29). After cell lysis, nuclei were isolated by differential centrifugation following established protocols (30,31). MAC fractions containing <5% MIC (count/count) and MIC fractions containing <0.1% MAC (count/count) were used for MNase digestion. MNase digestion was carried out at 25ºC for 15 min (50 mM Tris–HCl pH 8, 5 mM CaCl2, 1 mM β-mercaptoethanol, 0.1% NP-40, 0.1 mg/ml BSA, 1 U/μl MNase for heavy digestion of MAC and MIC, or 0.2 U/μl MNase for light digestion of MAC), stopped by adding 10 mM EGTA and 1 mM EDTA. Mono-nucleosome sized DNA was selected by agarose gel purification. Alternatively, mono-nucleosomes were purified by sucrose gradient ultracentrifugation.
Illumina sequencing and data processing
Illumina sequencing libraries were prepared using NEBNext® kit (New England Biolabs) to provide even coverage across GC%. Paired-end MNase-Seq results were mapped back to the Tetrahymena reference genome assemblies using Tophat2, with spliced alignments filtered out (32). The MAC reference is from the Tetrahymena genome database (TGD: http://ciliate.org) (33,34); the MIC reference is from the Tetrahymena Comparative Sequencing Project (http://www.broadinstitute.org/annotation/genome/Tetrahymena/MultiHome.html). Only one of any potential PCR duplicates was kept. The mapping results were visualized using GBrowse 2.0 (35).
Nucleosome calling and analyses of translational nucleosome positioning
Nucleosomes were called using the NucPosSimulator (36), which identifies non-overlapping nucleosomes using MNase-Seq fragment centers. We calculated the degree of translational positioning for each called nucleosome in MAC, defined as the number of fragment centers within ±20 bp of the called nucleosome dyad, relative to the number of all fragment centers within the 147 bp called nucleosome footprint (11). Only called nucleosomes supported by ≥50 fragments in the MAC sample were analyzed, to reduce noise in the calculated degree of translational positioning caused by random fluctuations. The MAC nucleosome distribution curve, plotted according to their degrees of translational positioning, was decomposed into two Gaussian distribution components, using the mixtools package in R (37).
Nucleosome distribution relative to TSS, TES, and splice sites
For analyses related to TSS, TES and splice sites, we focused on 15,841 gene models strongly supported by high depth RNA-Seq results (38), hereafter referred to as well-modeled genes (see Supplemental Methods for details). Selected TSS and TES are supported by at least five independent RNA-Seq reads. Selected splice sites are supported by at least one RNA-Seq read spanning the intron-exon junction. Composite analyses of MNase-Seq fragment centers, nucleosome occupancy, and called nucleosome dyads were performed after aligning to the selected landmarks.
Nucleosome occupancy from the MNase-Seq datasets or DNA sequence features
A uniform 147 bp nucleosomal footprint was assumed for all MNase-Seq fragments, extending 73 bp in both directions from their centers. Coverage for each genomic position was calculated based on these 147 bp footprints. Computational predictions were made using four models: (i) the Segal model built upon a dataset from nucleosomes reconstituted with yeast histones and genomic DNA in vitro (10); (ii) the 14-feature model that uses DNA sequence features selected for their dominant effects on nucleosome occupancy (15); (iii) a model based solely on GC% and (iv) a model based on rotational positioning. Software for the Segal model was downloaded from the Segal Lab website (http://genie.weizmann.ac.il/software/nucleo_prediction.html). These models and experimental occupancy data were compared in pairs using Spearman's rank correlations.
Nucleosome distribution heatmap
Nucleosome distribution heatmaps were plotted according to: (i) distances of called nucleosome dyads to TSS (x-axis) and degrees of translational positioning (y-axis); (ii) distances of called nucleosome dyads to TSS (x-axis) and splice sites (y-axis), using the Spatstat package in R (smoothing parameter sigma: 10) (39). Point densities—referred to as z values below—were represented in color scales. For the former, the following selections were made for further analysis: the +1 nucleosome, 114 ≤ x ≤ 168, 61.9% ≤ y ≤ 80.3%, z ≥ 0.296; the +2 nucleosome, 311 ≤ x ≤ 368, 67.6% ≤ y ≤ 85.3%, z ≥ 0.220; the +3 nucleosome, 506 ≤ x ≤ 569, 66.1% ≤ y ≤ 82.8%, z ≥ 0.174. The cutoff z values were at 1σ from the maximal z values, based upon a Gaussian distribution curve fitting of xz cross sections. For the latter, the following selections were made for further analysis: 1000 ≤ x ≤ 4000, 80 ≤ y ≤ 120.
ChIP-Seq
For analyzing Pol II distribution, endogenous RPB7 was tagged at the C-terminus with the hemagglutinin (HA) epitope. MAC from 2 × 108 growing Tetrahymena cells were purified and digested by MNase. The solubilized chromatin was incubated with anti-HA magnetic beads (Pierce). After repeated washing, recovered DNA was processed for Illumina sequencing. For analyzing H3K4 methylation distribution, Tetrahymena cells were fixed (1% paraformaldehyde) and lysed by sonication. The solubilized chromatin, after dilution, was incubated with H3K4me2-specific antibody (Abcam 07-030) and bound to protein A Dynabeads (Thermo Fisher). After repeated wash, recovered DNA was processed for Illumina sequencing. PCR duplicates were removed, before sequencing results were mapped to the Tetrahymena MAC reference genome.
RESULTS
Well-positioned nucleosomes are abundant in transcriptionally active MAC, but depleted in transcriptionally silent MIC
We purified the structurally and functionally differentiated MAC and MIC from growing Tetrahymena cells (Figure 1A). We performed paired-end Illumina sequencing of the mono-nucleosome DNA released by micrococcal nuclease digestion (MNase-Seq) (Supplementary Figure S1), and mapped the reads back to the Tetrahymena MAC (34) and MIC reference genomes (Tetrahymena Comparative Sequencing Project, Broad Institute of Harvard and MIT) (Supplementary Figure S2). MAC are derived from MIC by developmentally regulated genome reorganization, which removes thousands of internally eliminated sequences (IES) and religates the MAC-destined sequences (MDS) (26). MNase-Seq reads from the MAC sample were virtually all mapped to the MAC genome or MDS in the MIC genome (>99%), confirming its high purity, while 29% of MIC MNase-Seq reads were mapped to MIC IES (estimated at ∼30%), consistent with only minor contamination of the MIC sample by MAC.
In the MAC sample, well-positioned and regularly spaced nucleosomes were illustrated by the distribution of mapped MNase-Seq fragment centers—representing nucleosome dyads—in GBrowse views (Figure 1B, MAC). Phasogram analysis measuring the distribution of fragment centers relative to one another confirmed the presence of nucleosome arrays with 200 bp repeat length (NRL) and significant long-range order (Figure 1C, MAC). We focused on translational nucleosome positioning, defined as the 147 bp genomic DNA occupied by a significant nucleosome sub-population and represented by their exact dyad position. We calculated the degree of translational positioning for each called nucleosome in MAC (Figure 1D, left panel: solid red line), defined as the number of fragment centers within ±20 bp of the called nucleosome dyad, relative to the number of all fragment centers within the 147 bp called nucleosome footprint (11). The higher the number, the stronger is the nucleosome positioning (with 100% corresponding to perfectly positioned nucleosomes, seldom reached due to imprecision in MNase-Seq mapping as well as nucleosome positioning). This analysis revealed a sub-population of strongly positioned nucleosomes (Figure 1D, left panel: dashed red line, right peak), featuring prominent long-range order with 200 bp NRL (Figure 1D, right panel). These nucleosomes were highly enriched within the gene body (94%), which account for only 65% of the Tetrahymena MAC genome. Consistent with this observation, composite analysis of 15,841 well-modeled RNA polymerase II (Pol II) transcribed genes revealed stereotypical nucleosome arrays downstream of transcription start sites (TSS) (Figure 1E).
The nucleosome distribution pattern observed in MAC, with strong translational positioning and long-range order, degenerated in MIC. This was illustrated by a side-by-side comparison of the nucleosome dyad distribution in MAC and MIC (Figure 1B). Phasogram analysis showed much reduced amplitudes of periodic nucleosome distribution in MDS, and even more so, in IES (Figure 1C, MIC MDS). Compared with the strongly positioned nucleosomes in MAC (Figure 1D, left panel: red dashed line, right peak), degrees of translational positioning of their counterparts in MIC were dramatically reduced (Figure 1D, left panel: blue line), to levels very similar to the delocalized nucleosomes in MAC (Figure 1D, left panel: red dashed line, left peak). These nucleosomes showed reduced long-range order in MIC relative to MAC (Figure 1D, right panel). Composite analysis also revealed degeneration of nucleosome arrays in the gene body (Figure 1E).
We compared the MIC nucleosome distribution with the in vitro nucleosomes reconstituted with Tetrahymena histones and MAC genomic DNA (25), and found striking similarities (Figure 1B–E). Both showed delocalized distribution relative to MAC (Figure 1B), reflected in reduced nucleosome positioning (Figure 1D, left panel) as well as long-range order (Figure 1C and D, right panel). Stereotypical nucleosome arrays downstream of TSS also degenerated to similar degrees in both samples (Figure 1E). These results argue that strongly positioned and precisely phased nucleosome arrays in MAC, diminished in MIC and in vitro, are probably actively maintained by transcription-associated trans-determinants.
Nucleosome occupancy is affected by cis-determinants
Using the MNase-Seq datasets, we calculated nucleosome occupancy—the probability for a particular DNA base pair to be associated with any nucleosome—across Tetrahymena MAC and MIC genomes, using in vivo and in vitro data. Despite substantial differences between nucleosome positioning in MAC, MIC, and in vitro (Figure 1B), their nucleosome occupancy patterns were similar, as illustrated by a GBrowse view of the same genomic region (Figure 2A). In addition, a probabilistic model (referred to hereafter as the Segal model), based upon the yeast in vitro nucleosome distribution (10), closely matched nucleosome occupancy in MAC, MIC and in vitro (Figure 2A). Indeed, there were strong Spearman's rank correlation coefficients between all pairwise comparisons of MAC, MIC, and in vitro occupancy data and the Segal model (Figure 2B). We compared four different models: GC%, Segal (10), 14-feature (15), and rotational positioning, examining how they fit experimental data sets from yeast and Tetrahymena (Figure 2C). All four models fit the Tetrahymena data sets better than yeast (Figure 2C). While they fit the yeast in vitro data set much better than in vivo, they fit the Tetrahymena in vitro and in vivo data sets equally well (Figure 2C). In the Segal and 14-feature model, GC% was the predominant feature, while additional features slightly improved the prediction (Figure 2C and Supplementary Figure S3). A model based upon rotational nucleosome positioning, correlated with but substantially different from GC%, was equally predictive, supporting contributions from 10.4 bp periodicity in nucleosome-associated DNA (Figure 2C and Supplementary Figure S4). The result is consistent with strong contributions from cis-determinants, particularly GC%, to Tetrahymena nucleosome occupancy, in MAC, MIC and in vitro. This is in apparent contrast with the yeast in vivo nucleosome occupancy, which is more strongly affected by trans-determinants.
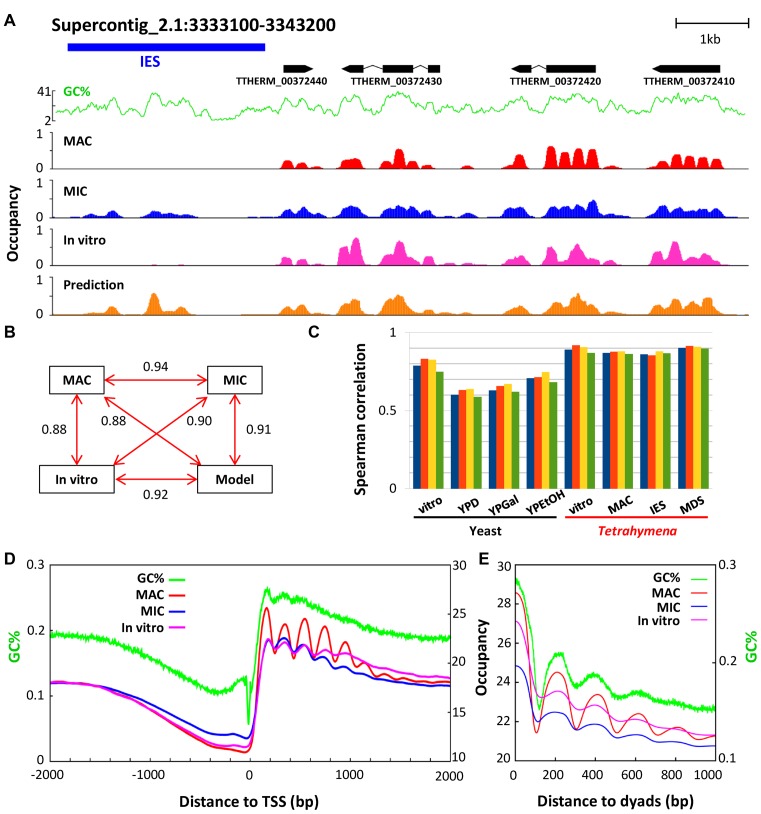
Figure 2.
Nucleosome occupancy is affected by cis-determinants. (A) Nucleosome occupancy in a representative genomic region (as in Figure 1B), based upon MNase-Seq of MAC (red), MIC (blue) and in vitro (magenta) (25), as well as computational modeling (beige) (10). GC% distribution (green) is superimposed. (B) Pair-wise Spearman's rank correlation coefficients between nucleosome occupancy levels of MAC, MIC (MDS), in vitro (25), and computational modeling (10). (C) Comparison of nucleosome occupancy predicted from different models and data from yeast and Tetrahymena, representing Spearman's rank correlations between the experimental occupancy data and the GC% model (blue), the Segal model (red) (10), the 14-feature model (yellow) (15), the rotational positioning model (green). Four yeast data sets and four Tetrahymena data sets are analyzed. See Methods for details. (D) Nucleosome occupancy in MAC (red), MIC (blue), and in vitro (magenta) (25), aligned to TSS. Nucleosome occupancy around TSS (±2 kb) is aggregated over 15,841 well-modeled genes. GC% distribution (green) is superimposed. (E) In phase oscillations of nucleosome occupancy and GC%. Nucleosome occupancy in MAC (red), MIC (blue) and in vitro (magenta) (25), is aligned to the dyads of the selected well-positioned nucleosomes (as in Figure 1D). GC% distribution (green) is superimposed.
We noted very similar decrease of nucleosome occupancy in nucleosome depleted regions (NDR) upstream of TSS and downstream of TES in MAC, MIC and in vitro (Figure 2D and Supplementary Figure S5A). Composite analysis revealed significantly decreased GC%, almost parallel to nucleosome occupancy (Figure 2D and Supplementary Figure S5A). There was also accumulation of poly (dA:dT) tracts around TSS and TES (Figure 4B and Supplementary Figure S5B, C). Tetrahymena NDR are therefore underlain by cis-determinants, particularly low GC% and prevalence of anti-nucleosomal sequences, consistent with observations in yeast (10,11).
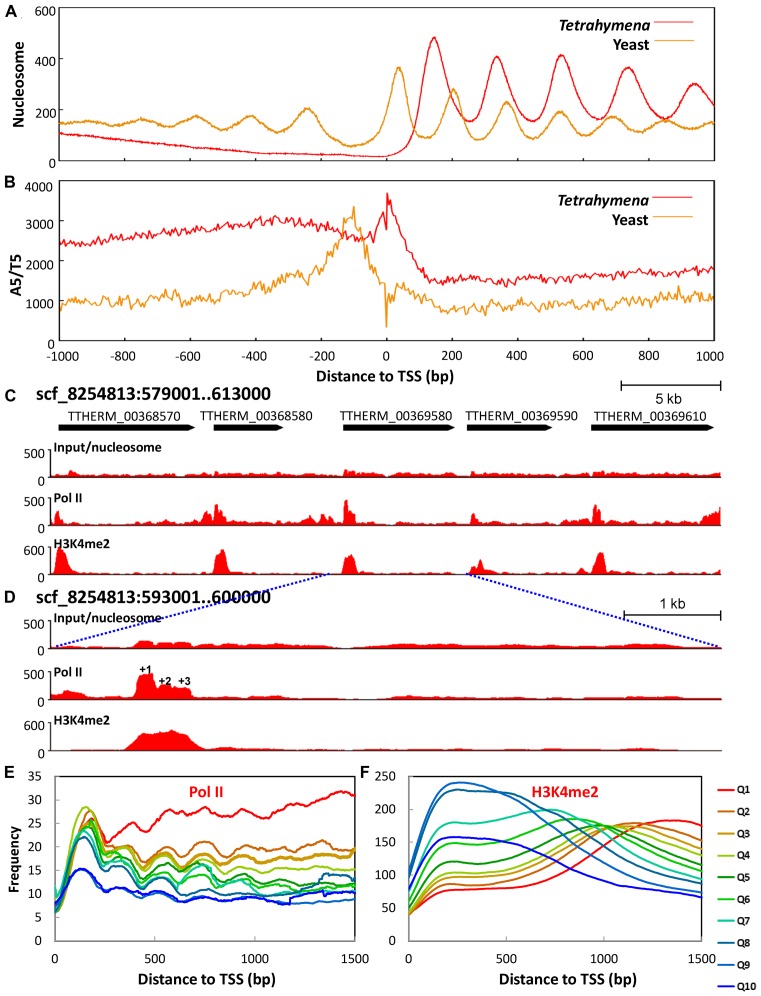
Figure 4.
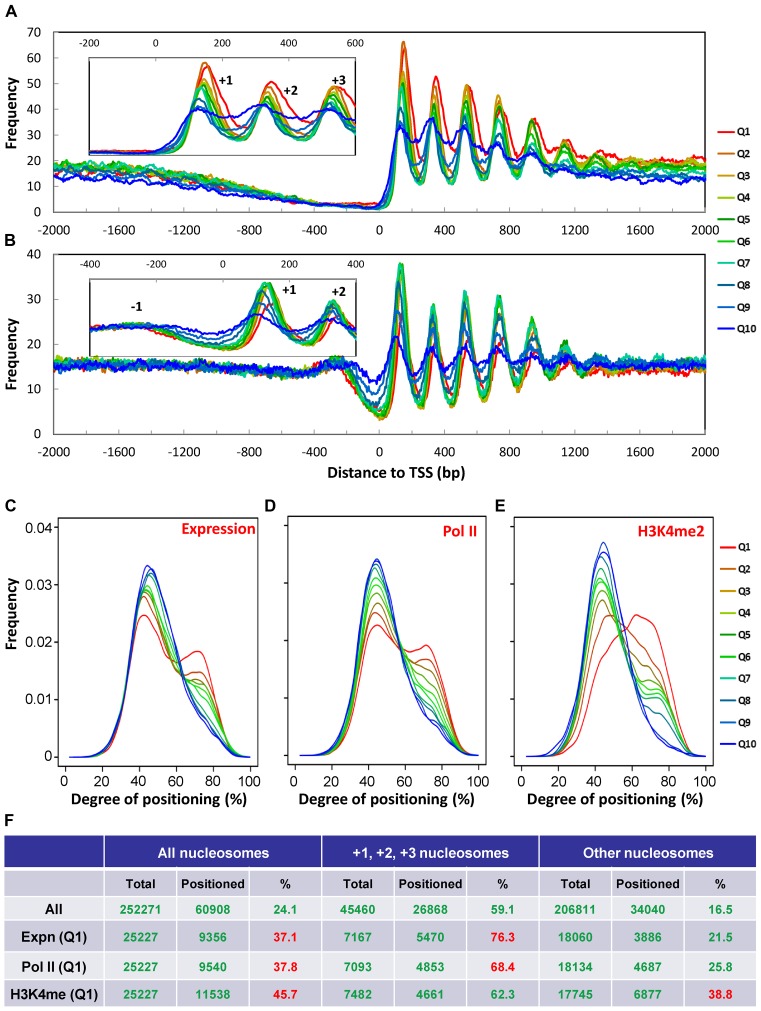
The +1 nucleosome is implicated in Pol II pausing. (A) The +1 nucleosome placement is further downstream of TSS in Tetrahymena than in yeast. Nucleosome distribution around TSS (± 1 kb) is aggregated over 15,841 well-annotated genes in Tetrahymena (Supplemental File S2) and 2231 genes in yeast (71). Note the downstream placement of the +1 nucleosome (yeast: 56 bp; Tetrahymena: 138 bp), increase in NRL (yeast: 167 bp; Tetrahymena: 200 bp), and a lack of nucleosome arrays upstream of TSS, when Tetrahymena is compared with yeast. (B) Poly (dA:dT) tract distribution relative to TSS in Tetrahymena (red) and yeast (beige). A5/T5 (AAAAA or TTTTT) distribution around TSS (± 1kb) is calculated using the 5 bp bin. C) Distribution of nucleosome, Pol II, and H3K4 methylation (from top to bottom) in a genomic region. Raw coverage of the input (nucleosome) and ChIP-Seq DNA fragments (Pol II and H3K4 methylation) was shown. Note the enrichment of Pol II and H3K4 methylation at the 5′ end of the gene body. (D) Zoom-in of a region containing a single gene. Note Pol II enrichment at the +1 nucleosome. (E) Composite analysis of Pol II distribution in the gene body. Well-modeled long genes (≥1.5 kb) are ranked from high to low by their expression levels and divided into 10 quantiles. Normalized Pol II distribution around TSS is aggregated over all genes in a quantile. (F) Composite analysis of H3K4 methylation in the gene body.
The relatively high GC% of nucleosomal DNA was consistent with, but not fully accounted for, by its enrichment in the gene body (GC%: 22%), especially exonic regions (28%), and its depletion in intergenic (18%) as well as intronic regions (16%). Composite analysis of nucleosome occupancy revealed in MAC the 200 bp nucleosome periodicity, which degenerated similarly in MIC and in vitro (Figure 2E). Composite analysis of GC% showed that the core of nucleosomes featured significantly higher GC%, with sharp boundaries defined by precipitous transitions in GC% (Figure 2E). Significantly, there were GC% oscillations in phase with the 200 bp NRL (Figure 2E). This suggests that nucleosome occupancy is affected by periodic changes in GC%.
Well-positioned nucleosomes in the gene body are coordinated by cis-determinants
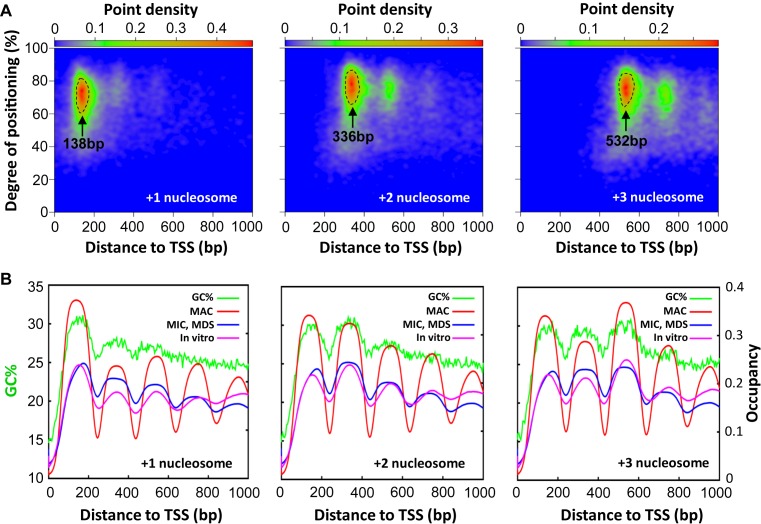
We further examined the nucleosome distribution in the gene body. Composite analysis of 15,841 well-modeled genes, aligned against TSS, showed a small peak of GC% coincident with the +1 nucleosome position (Figure 2D). We focused on well-positioned nucleosomes, which formed densely populated islands in nucleosome distribution maps plotted according to their distances from TSS (x-axis) and degrees of translational positioning (y-axis) (Figure 3A: circled by dashed lines). These well-positioned nucleosomes were therefore members of the stereotypical arrays. Strikingly, composite analysis of genes containing the well-positioned +1 nucleosome revealed a prominent GC% peak, tracking the +1 nucleosome occupancy; composite analysis of genes containing the well-positioned +2 nucleosome revealed two prominent GC% peaks, tracking the +1 and +2 nucleosome occupancy; composite analysis of genes containing the well-positioned +3 nucleosome revealed three prominent GC% peaks, tracking the +1, +2 and +3 nucleosome occupancy (Figure 3B). Furthermore, these well-positioned +1, +2 and +3 nucleosomes tended to be found together (Supplementary Figure S6A), contained in high GC% peaks of the gene body (Supplementary Figure S6B). For genes with these well-positioned nucleosomes, the peak-to-trough differences in GC%, at more than 5%, were much higher than average, at <1% (Figure 3B and Supplementary Figure S6B).
Figure 3.
Well-positioned nucleosomes in the gene body are coordinated by cis-determinants. (A) Distribution of called nucleosomes in the gene body, according to their distances from TSS (x-axes) and degrees of translational positioning (y-axes). Note the clustering of nucleosome distributions (point density in color scales). These strongly clustered +1, +2 and +3 nucleosomes, their peak positions relative to TSS as indicated, are selected for further analysis (enclosed by dashed lines). See Methods for details and Supplemental File S1 for a compilation of properties of called nucleosomes. (B) In phase oscillations of nucleosome occupancy and GC% downstream of TSS. Nucleosome occupancy in MAC (red), MIC (blue), and in vitro (magenta) (25) is aggregated over the genes containing the selected +1, +2 and +3 nucleosomes enclosed by dashed lines in the top panels. GC% distribution (green) is superimposed.
We also noted that stereotypical nucleosome arrays prominent in MAC were diminished but still detectable in MIC as well as in vitro (Figures 1E and 2F). They were even more obvious in genes with well-positioned +1, +2 and +3 nucleosomes (Figure 3B and Supplementary Figure S6C). In growing cells, MIC contain no detectable levels of RNA polymerases (27), transcription factors (28), or transcription associated histone variants like H2A.Z (40). This indicates that stereotypical nucleosome arrays in MIC are not preserved by these trans-determinants. Combined with the striking similarities between nucleosome distribution in MIC and in vitro, these results support strong contributions from cis-determinants. Importantly, well-positioned nucleosomes in MAC were often matched with GC% oscillations, as well as similar nucleosome occupancy profiles in MIC and in vitro, suggesting coordinate accommodation of these nucleosomes by cis- and trans-determinants.
The +1 nucleosome is implicated in Pol II pausing
The +1 nucleosome in Tetrahymena was separated from TSS by distances much larger than in yeast (Figure 4A). Well-positioned +1 nucleosomes clustered in a narrow band around 138 bp downstream of TSS (Figure 3A). Well-positioned +2 and +3 nucleosomes clustered at around 336 bp and 532 bp (Figure 3A), consistent with formation of stereotypical nucleosome arrays (nucleosome distances to TSS: 138 + 200n bp, n = 0, 1, 2 …). Nucleotide composition anomalies were observed in TSS (Supplementary Figure S7A) as well as TES (Supplementary Figure S7B). These anomalies may directly affect nucleosome distribution through cis-determinants. In contrast to yeast, poly (dA:dT) tracts were highly enriched around TSS in Tetrahymena (Figure 4B), disfavoring nearby placement of the +1 nucleosome. Alternatively but not mutually exclusively, these anomalies may represent DNA sequence motifs for recruiting other chromatin-associated proteins, thus affecting nucleosome distribution through trans-determinants. In support of this possibility, the TATA box was enriched around TSS (Supplementary Figure S7C), while the termination signal (AATAAA) was enriched near TES (Supplementary Figure S7D). Downstream placement of the +1 nucleosome in Tetrahymena is therefore at least partially underlain by DNA sequence features.
Downstream placement of the +1 nucleosome is more commonly observed in metazoa, associated with promoter proximal pausing of Pol II (41). To test this possibility, we examined the RNA polymerase II (Pol II) distribution in MAC by chromatin immuno-precipitation and Illumina sequencing (ChIP-Seq). It revealed in many genes high Pol II occupancy at the 5′ end, especially at the +1 nucleosome (Figure 4C–E). We performed composite analysis of Pol II distribution in 1.5 kb of the gene body, aligned to TSS (Figure 4E). Well-annotated genes longer than 1.5 kb were divided into 10 quantiles by their expression ranking in growing cells. Pol II enrichment at the +1 nucleosomes was most prominent in Quantiles 2–8; Pol II occupancy persisted at high levels in the most expressed genes (Quantile 1), while it remained at low levels in the least expressed genes (Quantiles 9 and 10) (Figure 4E). Pol II occupancy in the gene body generally increased with increasing expression levels, but less so at the 5′ than in the rest of the gene body (Figure 4E). This is consistent with Pol II elongation as a rate-limiting step of transcription, and the +1 nucleosome as a major site for Pol II pausing in Tetrahymena.
We also examined the distribution of H3K4 methylation, an epigenetic mark associated with transcription, abundant in MAC and absent from MIC (42). At many genes expressed in low to moderate levels, H3K4 methylation was enriched at the 5′ end (Figure 4C, D and F). Composite analysis revealed that H3K4 methylation distribution shifted from the 5′ towards the 3′ of the gene body, with increasing expression levels (Figure 4F). As H3K4-specific histone methyltransferases are associated with elongating Pol II (43), this result also supports that release of Pol II pausing promotes active transcription in Tetrahymena.
Transcription-associated trans-determinants promote nucleosome positioning
We compared nucleosome distribution patterns from MAC samples prepared by heavy and light MNase digestion (Figure 5A, B and Supplementary Figure S1). Light MNase digestion recovered more MNase-sensitive chromatin at intergenic regions (Figure 5B), revealing a broad peak 200–400 bp upstream of TSS corresponding to the −1 nucleosomes. With decreasing levels of gene expression, the NDR between the −1 and +1 nucleosome showed increasing levels of nucleosome occupancy in the light MNase digestion sample (Figure 5B), indicative of promoter occlusion by labile nucleosomes (16,44,45). Specific transcription factors, the general transcription machinery, and ATP-dependent chromatin remodelers can all compete with labile nucleosomes and bind promoters (46,47). Together with the cis-determinants like poly (dA:dT) tracts, these trans-determinants may act like nucleosome barriers to promote nucleosome positioning downstream of TSS.
Figure 5.
Transcription-associated trans-determinants promote nucleosome positioning. (A) Stereotypical nucleosome arrays with heavy MNase digestion. 15,841 well-modeled genes are ranked from high to low by their expression levels and divided into 10 quantiles. Nucleosome distribution around TSS is aggregated over all genes in a quantile. Inset: zoom in on the +1, +2 and +3 nucleosomes. (B) Stereotypical nucleosome arrays with light MNase digestion. (C) Nucleosome positioning with different levels of gene expression. Called nucleosomes in the gene body are ranked from high to low by the expression levels of their associated genes, and divided into 10 quantiles. Normalized distributions of degrees of nucleosome positioning are plotted for all 10 quantiles separately. (D) Nucleosome positioning with different levels of Pol II association. (E) Nucleosome positioning with different levels of H3K4 methylation. (F) Relationship between transcription-associated trans-determinants and well-positioned nucleosomes.
Nucleosome distribution patterns in the gene body were similar in heavy and light MNase digestion samples (Figure 5A and B). They were also similarly affected by transcription levels (Figure 5A and B). Stereotypical nucleosome arrays became less prominent with decreasing expression levels (Figure 5A and B), best illustrated by changing amplitudes of the +1, +2 and +3 nucleosomes (insets). There was dramatic and progressive degeneration of the nucleosome arrays in genes with little or no expression (Figure 5A and B). This was mainly attributed to reduced degrees of translational positioning (Figure 5C, Supplementary Figure S8A). In particular, the numbers of well-positioned +1, +2 and +3 nucleosomes decreased significantly in the least expressed genes (Supplementary Figure S8B and C), supporting a shift from well-positioned nucleosomes to delocalized nucleosomes with diminishing transcription activities. The result was reminiscent of the shift from well-positioned nucleosomes in MAC to delocalized nucleosomes in MIC, in the presence and absence of transcription, respectively (Supplementary Figure S8A). Taken together, our results support that transcription activities are required for maintaining strong positioning in stereotypical nucleosome arrays.
There was a dramatic shift towards stronger positioning for nucleosomes associated with highly expressed genes, high levels of Pol II, or high levels of H3K4 methylation (Figure 5C–E). For nucleosomes associated with highly expressed genes or high levels of Pol II, a large increase in the percentage of strongly positioned nucleosomes was observed in the +1, +2 and +3 nucleosomes, but much less so in the rest of the gene body (Figure 5F). On the other hand, for nucleosomes associated with high levels of H3K4 methylation, there was little increase in the strongly positioned +1, +2 and +3 nucleosomes, while a dramatic increase was observed in the rest of the gene body (Figure 5F). Transcription-associated trans-determinants therefore may differentially affect nucleosome positioning in the gene body. Consistent with dramatically different nucleosome positioning in Tetrahymena MAC and MIC, the transcription machinery, H3K4 methylation, histone variants H2A.Z and H3.3, SWI/SNF family of ATP-dependent chromatin remodelers are highly abundant in MAC, while completely absent or largely depleted in MIC (40,42,48,49).
Splice sites are flanked by positioned nucleosomes
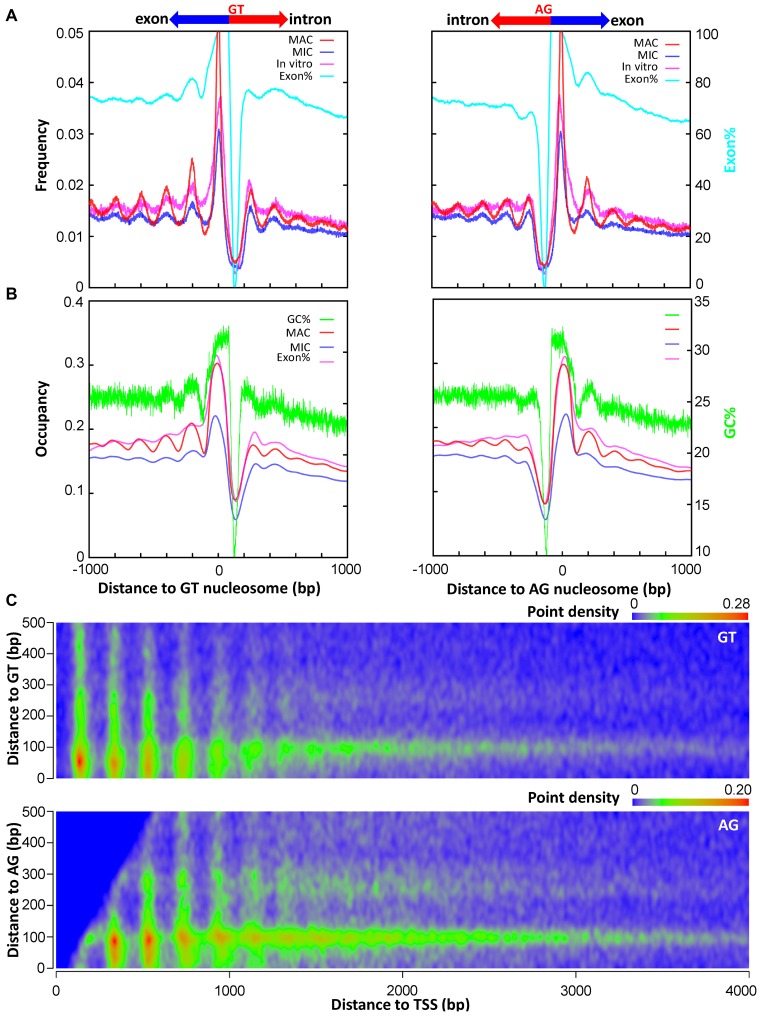
On average, there are 3.61 introns per gene in Tetrahymena, with introns significantly shorter than exons (intron: 141 bp; exon: 420 bp). Exons had much higher levels of nucleosome occupancy than intronic regions (Supplementary Figure S9A), consistent with GC% being much higher in exons (27.6%) than introns (16.3%) (Supplementary Figure S9B). This distribution is similar to that in metazoa and plants (50–52). Unexpectedly, composite analysis revealed positioned nucleosomes in exonic regions flanking the 5′ (GT) splice sites and, even more so, 3′ (AG) splice sites (Supplementary Figure S9C and D). Composite analysis of nucleosomes near splice sites revealed that they were strongly positioned in MAC, and their positioning was only moderately reduced in MIC (Figure 6A). Composite analysis also revealed that the occupancy levels of these nucleosomes coincided with a prominent GC% peak in exons, next to a stretch of DNA with very low GC% at the intron side (Figure 6B). Similar patterns were observed for in vitro nucleosomes flanking splice sites (Figure 6A and B), corroborating strong contributions from cis-determinants. Very low GC% intronic sequences may serve as a barrier, against which nucleosomes are placed in high GC% exonic sequences. Positioned nucleosomes flanking splice sites may provide epigenetic information to allow precise recognition of splice sites, or even modulate alternative splicing.
Figure 6.
Splice sites are flanked by positioned nucleosomes. (A) Composite analysis of nucleosome positioning in MAC (red), MIC (blue), and in vitro (magenta) (25), aligned to the dyads of the called nucleosomes flanking splice sites (left: GT, right: AG). The distribution of exon% (pale blue), calculated as the probability that a particular position containing exons, is superimposed. See Methods for details. (B) Composite analysis of nucleosome occupancy in MAC (red), MIC (blue), and in vitro (magenta) (25), aligned to the dyads of the called nucleosomes flanking splice sites (left: GT, right: AG). The distribution of GC% (green) is superimposed. Note the correlation between nucleosome arrays near splice sites and oscillations of GC%. (C) Distribution of called nucleosomes in the gene body, according to their distances from TSS (x-axes) and splice sites (y-axes; top: GT; bottom: AG). Note the clustering of nucleosome distribution along both the x- and y-axes (point density in color scales). See Supplemental File S1 for a compilation of properties of called nucleosomes.
We plotted heat maps of MAC nucleosome distribution relative to TSS and splice sites (Figure 6C and Supplementary Figure S9E). Nucleosomes positioned relative to TSS were manifest as a set of evenly spaced vertical streaks, which diminished gradually as the distances from TSS increased (Figure 6C). A conspicuous horizontal streak, representing nucleosomes positioned at ∼100 bp from splice sites, was also revealed (Figure 6C). Positioning of neighboring nucleosomes degenerated quickly away from splice sites, in apparent contrast with the long-range order downstream of TSS (Figure 6A and C). Nucleosome positioning around splice sites was also not much affected by transcription levels, compared with nucleosome positioning downstream of TSS (Supplementary Figure S9D). Nucleosome distribution downstream of TSS and around splice sites therefore most likely contributes independently to the chromatin environment of the gene body.
DISCUSSION
We have compared nucleosome distribution patterns in Tetrahymena MAC and MIC. MAC features a chromatin environment strongly affected by transcription-associated trans-determinants, manifest in stereotypical arrays of well-positioned nucleosomes in the gene body, while nucleosome distribution in MIC is predominantly shaped by cis-determinants in the absence of transcription, showing dramatic reduction in nucleosome positioning and long-range order. Despite differences in nucleosome positioning, there are remarkable similarities between nucleosome occupancy levels in MAC and MIC, as well as in vitro reconstitution and computational modeling based upon DNA sequence features. These results reveal unexpectedly strong contributions from cis-determinants, especially GC%. Importantly, cis- and trans-determinants often coordinately accommodate well-positioned nucleosomes downstream of TSS or flanking splice sites. This may be a result of evolution, driven by the impact of these nucleosomes on the mutational as well as transcriptional landscape.
Strong contributions to nucleosome distribution from cis-determinants
In vitro reconstitution of yeast nucleosomes, in the presence or absence of cell extracts and ATP, demonstrates the predominant role of trans-determinants in establishing the stereotypical nucleosome arrays downstream of TSS (10,11,21,24). By contrast, nucleosomes reconstituted in vitro with only Tetrahymena MAC genomic DNA and histones exhibit stereotypical nucleosome arrays, albeit at reduced amplitudes (25). To resolve relative contributions from cis- and trans-determinants, we have examined nucleosome distribution in Tetrahymena MAC and MIC, featuring actively transcribed and silenced chromatin, respectively. We find strong correlations between MAC and MIC nucleosome occupancy levels. In particular, well-positioned nucleosomes downstream of TSS and flanking splice sites in MAC are often reflected in nucleosome distribution of MIC, supporting strong contributions from cis-determinants. When aligned to these two landmarks in the gene body, the MIC and in vitro nucleosome distribution profiles are very similar to each other. This strongly suggests that MIC nucleosome distribution, in the absence of transcription-associated trans-determinants, is dominated by cis-determinants. Computational modeling based on DNA sequence features further reveals GC% as the dominant cis-determinant for nucleosome distribution. Well-positioned nucleosomes downstream of TSS and flanking splice sites in MAC are often matched with oscillations in GC%. In particular, regions upstream of TSS and inside introns have extremely low GC% in Tetrahymena, and correspond to NDR in both MAC and MIC. These anti-nucleosomal sequences serve as barriers, and when juxtaposed with high GC% sequences, can effectively anchor nucleosome positioning. The strong contribution of GC% to nucleosome distribution, manifest in Tetrahymena, has also been posited as container sites for nucleosomes in human cells (53). It may represent a general mechanism for chromatin organization.
We are aware of GC bias that may be introduced during library preparation for Illumina sequencing, which is potentially more severe for the AT-rich Tetrahymena genome. Our result is obtained after limiting GC bias by using NEBNext® kit (New England Biolabs) in DNA library preparation and removing PCR duplicates in sequencing data analysis. Indeed, nucleosome DNA upstream of TSS is covered at similar levels regardless of GC% in samples with light MNase digestion (comparing Figures 2D and 5B). Our data and conclusions are robust but will need to be confirmed in the future with improved sequencing protocols. It is important to note that our conclusions are not affected by MNase dosage, as the stereotypical nucleosome arrays in the gene body remain unchanged in samples with light and heavy MNase digestion (comparing Figure 5A and B). They also cannot be attributed to the sequence bias of MNase digestion, as demonstrated in general with an alternate nuclease (54), and specifically for Tetrahymena with the naked genomic DNA digestion control (25).
Nucleosome positioning promoted by transcription-associated trans-determinants
In transcriptionally active MAC, stereotypical arrays of well-positioned nucleosomes are conspicuous in the gene body, especially downstream of TSS, as found in yeast and other eukaryotes (23). In transcriptionally silent MIC, these nucleosome arrays degenerate, with dramatic reduction in nucleosome positioning and long-range order. The apparent discrepancy between the distinct nucleosome positioning and highly correlated nucleosome occupancy in MAC and MIC can be reconciled if nucleosome distribution in MIC is a more delocalized form of the MAC distribution. Nucleosome delocalization in MIC may be caused by variable nucleosome spacing, due to reduced ATP-dependent chromatin remodeling activities, which are often associated with transcription and play a significant role in shaping nucleosome arrays in the gene body (21,24). Alternatively, it may be attributed to shifting of nucleosome arrays in individual cells, due to insufficiency of nucleosome barriers, which, in addition to anti-nucleosomal sequences, are often defined by binding proteins including the general transcription machinery and specific transcription factors (55,56). Nucleosome distribution in MAC is most likely far from equilibrium and actively maintained by transcription-associated trans-determinants.
Chromatin landscape for transcription
Well-positioned nucleosomes in the gene body shape the chromatin environment for Pol II transcription, and are involved in its regulation (2). Transcription initiation from cryptic sites occurs when stereotypical nucleosome arrays are perturbed (57). The +1 nucleosome, when placed at large distances from TSS, is implicated in promoter-proximal pausing of Pol II (58). The +1 nucleosome in Tetrahymena is placed much further away from TSS than in yeast, in positions similar to metazoa. In yeast, gene promoters are often wrapped up in the +1 nucleosome (22). In metazoa, transcription initiation and promoter proximal pausing are regulated separately, with the latter affected by the +1 nucleosome (41,59). In Tetrahymena, Pol II is preferentially associated with the +1 nucleosome. With increasing expression levels, more Pol II is detected in the rest of the gene body. Concomitantly, peaks of H3K4 methylation, a mark associated with elongating Pol II (43), also move further downstream. These results implicate Pol II pausing, particularly at the +1 nucleosome, in transcription regulation in Tetrahymena. Promoter proximal pausing is mediated by the negative elongation factors (NELF) in some unicellular eukaryotes (60) and in metazoa in general (41). However, similar patterns have also been observed in Arabidopsis (61) as well as Tetrahymena, which have no apparent NELF homologues. This suggests the existence of an evolutionarily ancient, NELF-independent mechanism for promoter proximal pausing of Pol II.
The somatic MAC is derived from the germ-line MIC during sexual reproduction, accompanied by dramatic changes in the chromatin landscape as well as extensive genome reorganization (26). Transcription activation occurs in developing MAC, establishing de novo mRNA transcription in a euchromatin environment in MDS, while IES are destined for heterochromatin formation, and ultimately deletion, in a nuclear RNAi and Polycomb repression-dependent pathway (62–65). By delineating the nucleosome distribution patterns in MAC and MIC, our work sets the stage for characterizing the transition state(s) during MIC to MAC differentiation, which may reveal novel mechanisms reshaping the chromatin landscape upon transcription activation and repression.
Coordinate accommodation of nucleosomes by cis- and trans-determinants
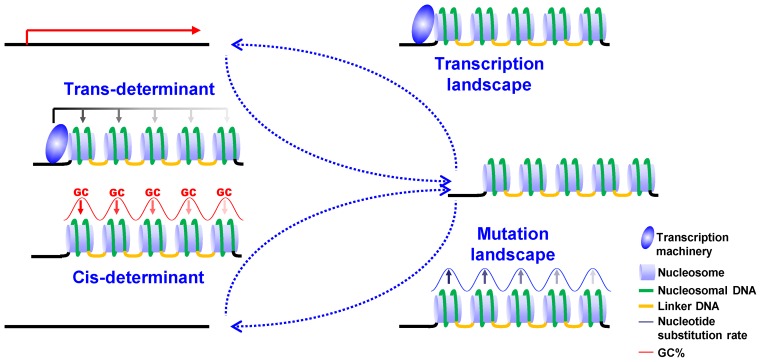
The similarities between nucleosome distribution in MAC and MIC reflect the coordination between cis- and trans-determinants, especially for well-positioned nucleosomes in the gene body. Here we propose an evolutionary pathway that can lead to this outcome (Figure 7). Effects of the chromatin environment on transcription may provide the selection pressure to optimize cis-determinants: those in conflict with trans-determinants and disturbing stereotypical nucleosome arrays are selected against, while those in coordination with trans-determinants and reinforcing well-positioned nucleosomes are selected for. Importantly, DNA sequences can be evolutionarily optimized for nucleosome placement through a positive feedback loop. Cis-determinants, particularly oscillations in GC%, promote nucleosome placement; reciprocally, nucleosome association changes the mutational landscape. Specifically, significantly elevated nucleotide substitution rates are found around nucleosome dyads (66–69). Nucleosome association may even alter mutation spectra, leading directly to higher GC% (66,70). The unexpectedly strong contributions from cis-determinants manifest in Tetrahymena may be attributed to two factors: first, nucleosome distribution, and by inference, the mutational landscape, is predominantly affected by cis-determinants in the germ-line MIC; second, the overall low GC% of the genome provides an anti-nucleosomal background, in which nucleosome placement can be reinforced by mutations increasing GC% of nucleosomal DNA. We further speculate that coordination of cis- and trans-determinants may be a general phenomenon in eukaryotic systems, driven by a positive feedback loop of evolutionary processes, in which nucleosome distribution shapes the mutational landscape, while cis-determinants in return reinforce nucleosome distribution.
Figure 7.
Coordinate accommodation of well-positioned nucleosomes in the gene body by cis- and trans-determinants. A positive feedback loop allows optimization of cis-determinants, and their coordination with trans-determinants to accommodate well-positioned nucleosomes in the gene body. See Discussion for details.
Supplementary Material
Acknowledgments
WT Tetrahymena strain CU428 was obtained from the Tetrahymena Stock Center (https://tetrahymena.vet.cornell.edu/). The compiled annotations for Tetrahymena MAC genome sequence were obtained from the Tetrahymena Genome Database (http://ciliate.org). Tetrahymena MIC genome sequence was obtained from the Tetrahymena Comparative Sequencing Project (Broad Institute of Harvard and MIT: http://www.broadinstitute.org/annotation/genome/Tetrahymena/MultiHome.html). Illumina sequencing was performed at the DNA Sequencing Core of the University of Michigan.
ACCESSION NUMBERS
All MNase-Seq and ChIP-Seq data have been deposited at the NCBI Gene Expression Omnibus (GEO: http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE77660 and GSE77583.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31301930] (to J.X.); Knowledge Innovation Program of the Chinese Academy of Sciences (J.X.); Youth Innovation Promotion Association, Chinese Academy of Sciences (J.X.); Natural Science Foundation of China [31522051] (to S.G.); Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, China (S.G.); NIH [R01 GM106024] (to S.D.T.); Canadian Institutes of Health Research [MOP13347] (to R.E.P.); Natural Sciences and Engineering Research Council of Canada [53909]; Natural Sciences and Engineering Research Council of Canada (to W.A.); Projects of International Cooperation and Exchanges, Ministry of Science and Technology of China [2013DFG32390] (to W.M.); NSF [MCB 1411565] (to Y.L.); NIH [R01 GM087343]; Department of Pathology at the University of Michigan (Y.L.). Funding for open access charge: NSF [MCB 1411565] (to Y.L.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Hughes A.L., Rando O.J. Mechanisms underlying nucleosome positioning in vivo. Annu. Rev. Biophysi. 2014;43:41–63. doi: 10.1146/annurev-biophys-051013-023114. [DOI] [PubMed] [Google Scholar]
- 3.Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R.L., Schuster S.C., et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Pugh B.F. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson R.T., Stafford D.W. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. U.S.A. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segal E., Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson H.C., Finch J.T., Luisi B.F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 9.Drew H.R., Travers A.A. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Moqtaderi Z., Rattner B.P., Euskirchen G., Snyder M., Kadonaga J.T., Liu X.S., Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satchwell S.C., Drew H.R., Travers A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 13.Gaffney D.J., McVicker G., Pai A.A., Fondufe-Mittendorf Y.N., Lewellen N., Michelini K., Widom J., Gilad Y., Pritchard J.K. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brogaard K., Xi L., Wang J.P., Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tillo D., Hughes T.R. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 19.Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 20.Lantermann A.B., Straub T., Stralfors A., Yuan G.C., Ekwall K., Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 21.Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieleg C., Krietenstein N., Walker M., Korber P. Nucleosome positioning in yeasts: methods, maps, and mechanisms. Chromosoma. 2015;124:131–151. doi: 10.1007/s00412-014-0501-x. [DOI] [PubMed] [Google Scholar]
- 23.Struhl K., Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Wippo C.J., Wal M., Ward E., Korber P., Pugh B.F. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beh L.Y., Muller M.M., Muir T.W., Kaplan N., Landweber L.F. DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome. Genome Res. 2015;25:1727–1738. doi: 10.1101/gr.188516.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karrer K.M. Nuclear dualism. Methods Cell Biol. 2012;109:29–52. doi: 10.1016/B978-0-12-385967-9.00003-7. [DOI] [PubMed] [Google Scholar]
- 27.Mochizuki K., Gorovsky M.A. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryotic Cell. 2004;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stargell L.A., Gorovsky M.A. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol. Cell. Biol. 1994;14:723–734. doi: 10.1128/mcb.14.1.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorovsky M.A., Yao M.C., Keevert J.B., Pleger G.L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 30.Sweet M.T., Allis C.D. Isolation and purification of tetrahymena nuclei. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4500. doi:10.1101/pdb.prot4500. [DOI] [PubMed] [Google Scholar]
- 31.Papazyan R., Voronina E., Chapman J.R., Luperchio T.R., Gilbert T.M., Meier E., Mackintosh S.G., Shabanowitz J., Tackett A.J., Reddy K.L., et al. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. eLife. 2014;3:e02996. doi: 10.7554/eLife.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stover N.A., Punia R.S., Bowen M.S., Dolins S.B., Clark T.G. Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database. 2012:bas007. doi: 10.1093/database/bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinform. 2013;14:162–171. doi: 10.1093/bib/bbt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schopflin R., Teif V.B., Muller O., Weinberg C., Rippe K., Wedemann G. Modeling nucleosome position distributions from experimental nucleosome positioning maps. Bioinformatics. 2013;29:2380–2386. doi: 10.1093/bioinformatics/btt404. [DOI] [PubMed] [Google Scholar]
- 37.Benaglia T., Chauveau D., Hunter D.R., Young D.S. mixtools: an R package for analyzing finite mixture models. J. Stat. Softw. 2009;32:1–29. [Google Scholar]
- 38.Xiong J., Lu X., Zhou Z., Chang Y., Yuan D., Tian M., Zhou Z., Wang L., Fu C., Orias E., et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using Deep RNA sequencing. PLoS One. 2012;7:e30630. doi: 10.1371/journal.pone.0030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baddeley A., Turner R. spatstat: An R package for analyzing spatial point patterns. J. Stat. Softw. 2005;12:1–42. [Google Scholar]
- 40.Stargell L.A., Bowen J., Dadd C.A., Dedon P.C., Davis M., Cook R.G., Allis C.D., Gorovsky M.A. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 41.Kwak H., Lis J.T. Control of transcriptional elongation. Annu. Rev. Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl B.D., Ohba R., Cook R.G., Allis C.D. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fromm M., Avramova Z. ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 2014;21:75–82. doi: 10.1016/j.pbi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi Y., Yao J., Chen R., Li W., He X. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 2011;21:718–724. doi: 10.1101/gr.117101.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent N.A., Adams S., Moorhouse A., Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henikoff J.G., Belsky J.A., Krassovsky K., MacAlpine D.M., Henikoff S. Epigenome characterization at single base-pair resolution. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui B., Liu Y., Gorovsky M.A. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell. Biol. 2006;26:7719–7730. doi: 10.1128/MCB.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fillingham J.S., Garg J., Tsao N., Vythilingum N., Nishikawa T., Pearlman R.E. Molecular genetic analysis of an SNF2/brahma-related gene in Tetrahymena thermophila suggests roles in growth and nuclear development. Eukaryotic Cell. 2006;5:1347–1359. doi: 10.1128/EC.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz S., Meshorer E., Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 51.Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcarcel J., Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 52.Chodavarapu R.K., Feng S., Bernatavichute Y.V., Chen P.Y., Stroud H., Yu Y., Hetzel J.A., Kuo F., Kim J., Cokus S.J., et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valouev A., Johnson S.M., Boyd S.D., Smith C.L., Fire A.Z., Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allan J., Fraser R.M., Owen-Hughes T., Keszenman-Pereyra D. Micrococcal nuclease does not substantially bias nucleosome mapping. J. Mol. Biol. 2012;417:152–164. doi: 10.1016/j.jmb.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhee H.S., Pugh B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee H.S., Pugh B.F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 58.Adelman K., Lis J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang G.S., Noegel A.A., Mavrich T.N., Muller R., Tomsho L., Ward E., Felder M., Jiang C., Eichinger L., Glockner G., et al. Unusual combinatorial involvement of poly-A/T tracts in organizing genes and chromatin in Dictyostelium. Genome Res. 2012;22:1098–1106. doi: 10.1101/gr.131649.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li G., Liu S., Wang J., He J., Huang H., Zhang Y., Xu L. ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. Cell Mol. Biol. 2014;78:706–714. doi: 10.1111/tpj.12499. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y., Mochizuki K., Gorovsky M.A. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 64.Taverna S.D., Coyne R.S., Allis C.D. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y., Taverna S.D., Muratore T.L., Shabanowitz J., Hunt D.F., Allis C.D. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prendergast J.G., Semple C.A. Widespread signatures of recent selection linked to nucleosome positioning in the human lineage. Genome Res. 2011;21:1777–1787. doi: 10.1101/gr.122275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sasaki S., Mello C.C., Shimada A., Nakatani Y., Hashimoto S., Ogawa M., Matsushima K., Gu S.G., Kasahara M., Ahsan B., et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith D.J., Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reijns M.A., Kemp H., Ding J., de Proce S.M., Jackson A.P., Taylor M.S. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Chen Z., Chen H., Su Z., Yang J., Lin F., Shi S., He X. Nucleosomes suppress spontaneous mutations base-specifically in eukaryotes. Science. 2012;335:1235–1238. doi: 10.1126/science.1217580. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z.H., Dietrich F.S. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.