Abstract
Myoblasts are precursor skeletal muscle cells that differentiate into fused, multinucleated myotubes. Current single-cell microfluidic methods are not optimized for capturing very large, multinucleated cells such as myotubes. To circumvent the problem, we performed single-nucleus transcriptome analysis. Using immortalized human myoblasts, we performed RNA-seq analysis of single cells (scRNA-seq) and single nuclei (snRNA-seq) and found them comparable, with a distinct enrichment for long non-coding RNAs (lncRNAs) in snRNA-seq. We then compared snRNA-seq of myoblasts before and after differentiation. We observed the presence of mononucleated cells (MNCs) that remained unfused and analyzed separately from multi-nucleated myotubes. We found that while the transcriptome profiles of myoblast and myotube nuclei are relatively homogeneous, MNC nuclei exhibited significant heterogeneity, with the majority of them adopting a distinct mesenchymal state. Primary transcripts for microRNAs (miRNAs) that participate in skeletal muscle differentiation were among the most differentially expressed lncRNAs, which we validated using NanoString. Our study demonstrates that snRNA-seq provides reliable transcriptome quantification for cells that are otherwise not amenable to current single-cell platforms. Our results further indicate that snRNA-seq has unique advantage in capturing nucleus-enriched lncRNAs and miRNA precursors that are useful in mapping and monitoring differential miRNA expression during cellular differentiation.
INTRODUCTION
Approximately 40% of the human body consists of skeletal muscle (1). The minimum functional unit of skeletal muscle is the multinucleated myotube, which originates from fusing myoblasts. Muscle cell differentiation (myogenesis) entails activation of muscle-specific transcription network governed by four partially-redundant muscle-specific regulatory factors (MRFs) (Myf5, MyoD1, Myogenin and MRF4/Myf6) working together with E proteins and MEF2 family members (2). Molecular and genetic experiments in vivo in mice, chicken and Drosophila over the last decades have uncovered the genetic and epigenetic networks critical for skeletal muscle differentiation (3–5). In vitro, the precise process of myogenesis has been extensively studied using the murine immortalized C2C12 myoblast cell line as a model system because of its high proliferation and differentiation capacities (for examples of genome-wide studies, see (6–9) among many others). However, similar progress in human myogenesis has been limited by the lack of a convenient human myogenesis model system and the high heterogeneity of primary human muscle cells.
Scalable single-cell RNA sequencing (scRNA-seq) using platforms such as Fluidigm (10,11) and drop-seq (12,13) has revolutionized the study of cell-to-cell transcriptome heterogeneity. These methods have enabled large-scale characterization of cell-to-cell heterogeneity and distinct subpopulations of cells in tissues as well as in the defined intermediary states during cell differentiation (10–14). In particular, methods such as Monocle (15) have been developed to deconvolve time courses of development in differentiating cells where collected time points can consist of mixtures of cells in different states or stages of differentiation. The power of Monocle was demonstrated elegantly using human skeletal myogenesis on the Fluidigm platform, in which primary human myoblasts were differentiated into myocytes and myotubes (15). However, a critical limitation of the Fluidigm microfluidic chips is the size restrictions on cells that can be captured for single-cell analysis. Mature multinucleated myotubes, which are typically longer than 100μm, are too large to be captured on the largest-size chip designed for cells up to 30μm. This issue limited the previous analysis to unfused cells or smaller, less mature myotubes. A potential solution to this problem is to isolate cell nuclei and perform RNA-seq on single nuclei as has previously been done manually in a dozen neurons (16).
We report the successful use of single-nucleus RNA sequencing (snRNA-seq) to analyze gene expression profiles of large myotubes compared to myoblasts and differentiated MNCs on the Fluidigm platform using the immortalized human Hu5/KD3 myoblast cell line (KD3). This line was derived from a single clone of abdominal muscle cells that were immortalized using hTERT with p16INK4a-resistant R24C mutant CDK4 (mtCDK4) and cyclin D1 (17). KD3 exhibits a normal karyotype and the phenotypic characteristics of its primary parent cells (including doubling time) as well as the ability to undergo myogenic terminal differentiation in vitro and in vivo with no evidence of transformation (i.e. anchorage-independent growth in vitro or tumor formation in vivo) (17). KD3 has all of the attributes for serving as an ideal system to study human myogenesis using functional genomics. Comparison of scRNA-seq and snRNA-seq in myoblasts indicate that we can recover comprehensive single-nucleus transcriptomes that are comparable to single-cell transcriptome with higher enrichment of long-noncoding RNAs (lncRNAs) in snRNA-seq. We then compared single-nuclei from myoblasts as well as myotubes and MNCs that did not fuse after 72 h in differentiation medium. We found relative uniformity of transcriptional profiles in myoblast and myotube nuclei. In contrast, greater heterogeneity was observed in the MNC nuclei, with the majority of them showing a non-myoblast, mesenchymal profile, providing the evidence for alternative differentiation. We also analyzed the expression of lncRNAs and found that some of them represent primary miRNAs (pri-miRNAs) that are differentially expressed between the three samples. Corresponding miRNA expression was validated using the NanoString platform. Thus, snRNA-seq has distinct advantages in characterizing nuclear lncRNAs and is highly instrumental in deconvolving fate heterogeneity during myoblast differentiation.
MATERIALS AND METHODS
Human myoblast culture
Hu5/KD3 (KD3) cells were grown on dishes coated with collagen in high glucose DMEM (Gibco) supplemented with 20% FBS (Omega Scientific, Inc.), 1% Pen-Strep (Gibco), and 2% Ultrasor G (Crescent Chemical Co.) (17). Upon reaching 80% confluence, we induced differentiation using high glucose DMEM medium supplemented with 2% FBS and ITS supplement (insulin 0.1%, 0.000067% sodium selenite, 0.055% transferrin; Invitrogen). Fresh differentiation medium was changed every 24hrs. Myotubes and MNCs were harvested at 72 h after induction of differentiation.
Separation of myotubes and MNCs, and single nucleus extraction
In order to separate MNCs from multinucleated myotubes, 6 cm tissue culture plates were washed with PBS twice followed by the addition of 1.5 ml trypsin/EDTA (Invitrogen). We monitored the plate with a microscope at room temperature and tapped the plate repeatedly. After 1–2 min, the MNCs detached first, and we took the supernatant and washed the plate gently with PBS twice. The PBS was combined with the supernatant to recover MNCs. Myotubes were still attached on the plate and we used a cell scraper to scrape them out. After the cells were lifted by trypsin/EDTA or cell scraper, they were pelleted at 1000 x g, 4°C for 5 min. The cell membrane was ruptured with cold cell lysis buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.005% IGEPAL CA-630) for 4–5 min and the lysis efficiency was checked by Trypan blue (Lonza) staining. The nuclei were then pelleted at 500 × g, 4°C for 5 min and resuspended in cold PBS buffer. We used less IGEPAL CA-630 than usual for cell lysis in order to preserve intact MNC nuclei. We found that the KD3 myotube nuclei recovery efficiency after lysis is 50–60%, compared to 70–80% from myoblast or MNC, but believe that the amount of IGEPAL will need to be optimized for specific cell samples. Suspended nuclei were passed through Falcon 40 μm cell Strainer (Fisher Scientific) to remove cell debris.
Single nucleus capture, RNA-seq library construction and sequencing
Single cells and nuclei were isolated using the Fluidigm C1 System. We used the largest integrated fluidic circuits (IFCs) of 17–25 μm for KD3 cells. Single nuclei C1 runs were completed using the smallest IFC (5–10 μm). Cells were loaded at an approximate concentration of 400 cells/μl in a total of 50 μl. Each individual capture site was visualized to ensure single cell or single nucleus capture. After visualization, the IFC was loaded with Clontech SMARTer kit lysis, RT, and PCR amplification reagents. Lysis, RT and PCR amplification was completed overnight and harvested the following day. After harvesting, all cDNA was normalized between 0.1 and 0.3 ng/μl and libraries were constructed using Illumina's Nextera XT library prep kit per Fluidigm's protocol. Constructed libraries were multiplexed and purified using AMPure beads. The final multiplexed library was analyzed on an Agilent 2100 Bioanalyzer for fragment distribution and quantified using Kapa Biosystem's universal library quantification kit. The library was normalized to 2 nM and sequenced using the Illumina NextSeq500 system to an average depth of 1.0–6.0 million reads per sample using paired 75bp reads.
Read alignment
KD3 single-cell RNA-seq reads were aligned using STAR v.2.4.2a (18) with parameters ‘–outFilterMismatchNmax 10 –outFilterMismatchNoverReadLmax 0.07 –outFilterMultimapNmax 10’ to the reference genome GRCh38/hg38. We used STAR to then convert to transcriptome-based mapping and gene expression was measured using RSEM v.1.2.12 (19) with expression values normalized into transcript per million (TPM).
Gene filtering for protein-coding genes and lncRNA analysis
Libraries with mapped read number <300 000 or mapping percentages <50% were filtered out as were cells with <1000 genes expressed with TPM ≥ 1.0. The TPM and count of each sample were merged into matrices, respectively. The mitochondrial genes were filtered out to limit the differential effects of varying mitochondrial contamination in the nuclei pellet after cell lysis. Genes with multiple Ensembl IDs were further processed to keep the ID from the gene with the highest TPM. The average TPM of each gene in the single-nuclei matrix was calculated and only those genes with average TPM ≥ 1.0 were kept. The number of nuclei expressing a given gene served as the final filter. Genes with TPM ≥ 1.0 in at least five nuclei (for lncRNA analysis, 13 182 genes) or 10 nuclei (for other analyses, 11 004 genes) were included in the final matrices. A pseudocount of 0.1 was added to each value for logarithmic calculation. The merged count matrix was used in differential gene analysis and the merged TPM matrix was used to make graphs. Fastq files and processed data matrices were deposited in GEO with the accession ID GSE79920.
Differential gene expression analysis
Differential expression analysis was performed using edgeR v.3.2.2 (20). For the differential expression analysis of all genes (protein-coding genes and lncRNAs), differentially expressed genes were selected using fold change (FC) ≥4 and false discovery rate (FDR)<0.001. For our lncRNAs-centric analysis, differentially expressed ones were selected using FC ≥4 and FDR <0.05, which is less restrictive.
qPCR verification of scRNA-seq and snRNA-seq results
The harvested cDNA from Fluidigm C1 system after pre-amplification but before library construction was used to test individual cells by qPCR. 5μl cDNA was diluted by four times and 1μl of diluted cDNA was used for each qPCR reaction. We used SsoAdvanced SYBR Green Supermix (Bio-Rad) to set up the assay on the Bio-Rad CFX96 real-time system. Each reaction was set up with 10 μl supermix, 1 μl template, 1μl forward and reverse primer (10 μM) each and 7 μl ddH2O to bring up the volume to 20 μl. The program was 95°C for 1 min, followed by 95°C for 15 s and 58°C for 20 s for 40 cycles, then finalized by melting curve generation. The qPCR data was processed with Bio-Rad CFX Manager program. Ten cells were selected randomly from each cell type. We used two reference housekeeping genes GAPDH and UBC to normalize the variation of input material amount (21,22). The geometric average of GAPDH and UBC levels was applied as internal control for this normalization (22,23). Primer pairs were designed for the 3′-end of the genes in order to match the SMART-seq results. Most of the primers do not span an exon junction and we therefore designed a primer pair that covers the last exon of NEUROD1 as a negative control to determine whether any remaining trace amount of genomic DNA could contaminate the cDNA. Primer sequences are in Supplementary Table S1.
LncRNA association with neighboring protein-coding gene
For protein-coding genes, we merged the coordinates of transcripts from GENCODE GRCh38 version 23 annotations (24) for the same genes. We defined lncRNA and protein-coding gene as neighbors when the maximal distance between their gene bodies is <10 kb. We ignored the strandedness of the lncRNAs and protein-coding genes because the full set of neighboring protein-coding genes are needed to annotate the function of differentially expressed lncRNAs that are convergently or divergently transcribed (25,26). We found no significant difference with our unstranded results for gene expression and gene ontology when categorizing lncRNAs and protein-coding gene neighbors based on strand. We kept neighboring protein-coding genes that shared the same regulation direction (up or down) with differentially expressed lncRNAs in the final list for downstream Gene Ontology (GO) analysis. GO analysis was performed with Metascape (27) using the hypergeometric test corrected P-value lower than 0.05. In order to test the significance of the correlation in expression between lncRNAs and neighboring genes, we performed 1000 random permutations of lncRNA and gene pairs, and calculated the Pearson correlation coefficient for each permutation and P-values based on the distribution.
NanoString miRNA assay and SMART-seq coverage analysis of pri-miRNA
Total RNA samples were extracted with QIAGEN RNeasy Plus Micro kit from pools of KD3 myoblast, myotube and MNC cells. 100ng of total RNA was used for each NanoString assay and two biological replicates from each differential stage were included. The human miRNA NanoString probe set version 2.1 was used for all the assays, which were performed using the standard NanoString protocol (28). The raw counts were normalized by the total counts of all the tested miRNA and housekeeping genes in order to compensate for variations introduced by experimental procedure and counts were averaged between replicates. MiRNAs with counts lower than 50 in myoblast, myotube and MNC were filtered out and the remaining 490 miRNAs were used to calculate corresponding SMART-seq coverage. We used coordinates of miRNAs and lncRNAs from GENCODE GRCh38 version 23 (24).
Messenger RNA SMART-seq coverage was measured within 3kb of miRNAs by using bedtools/2.19.1 (29). We defined a lncRNA as neighboring a miRNA when (i) the lncRNA and miRNA are transcribed from the same strand and (ii) the lncRNA overlaps with the miRNA or the lncRNA is within 3 kb of the miRNA. We considered the lncRNA to be the pri-miRNA when overlapping the miRNA and being transcribed from the same strand.
RESULTS
Single nucleus RNA-seq faithfully recapitulates the transcriptome of single whole cells
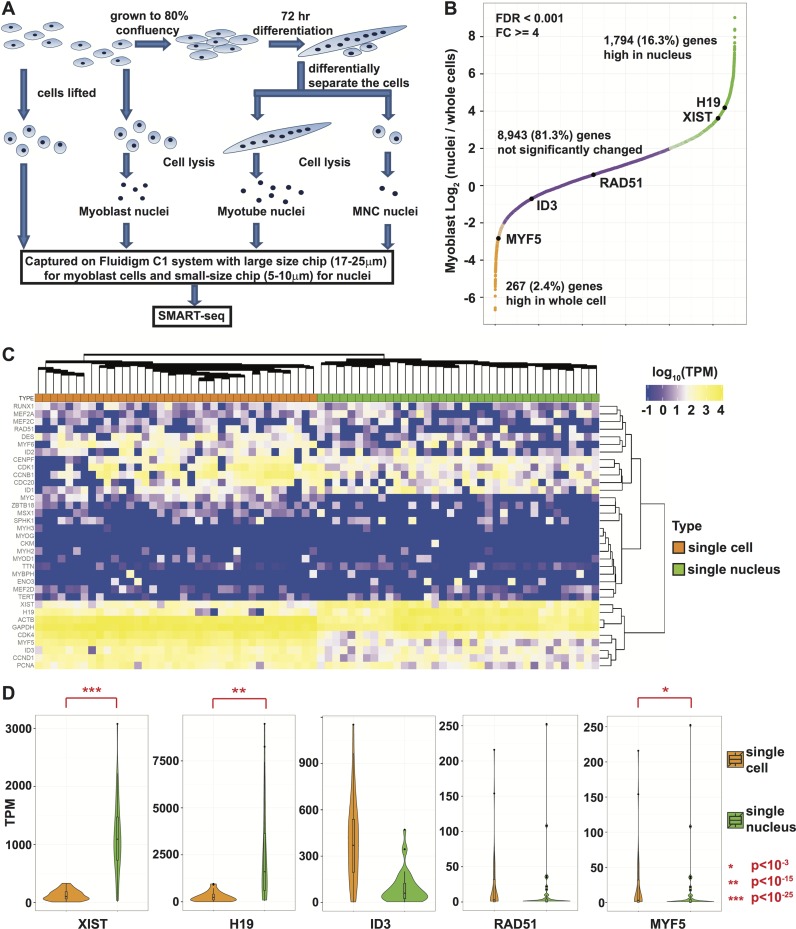
The large-size Fluidigm C1 chip can capture cells with diameter up to 30 μm. However, the length of differentiated myotubes are usually far above this upper limit as large myotubes can reach several hundred micrometers in length (Supplementary Figure S1). In order to study the transcriptome of full-size single myotubes with Fluidigm, we investigated the possibility of capturing single nuclei on the smallest microfluidic chip. In order to confirm that this approach is viable, we first compared single-cell and single-nucleus transcriptomes of undifferentiated KD3 myoblasts (Figure 1A; Supplementary Figure S2). The myoblasts were grown at low confluence (30%) before harvest to prevent premature differentiation. Nuclei were released with cell lysis buffer and ∼80–90% of nuclei were recovered based on calculating the ratio of nuclei to myoblasts. The diameter of myoblast nuclei is 5–10 μm, which fits well on the small-size Fluidigm chip. We constructed snRNA-seq libraries from the cDNA produced by the Fluidigm C1 using the SMART-seq protocol. As control, KD3 myoblast single cells with diameter of 15–20 μm were captured with the large-size chip and built into libraries. Both single cell and single nucleus myoblast libraries were sequenced at depths of 1–3.0 million reads and the percentage of reads mapping to the genome averaged between 80 and 90% (Supplementary Figure S3A).
Figure 1.
KD3 myoblast snRNA-seq faithfully captures the transcriptome identified by scRNA-seq. (A) Workflow of snRNA-seq assays for KD3 myoblast, myotube, and MNCs after differentiation, together with the scRNA-seq on myoblast. (B) Gene expression Fold change (FC) curve of single-nucleus versus single-cell RNA-seq data. We used maximum FDR 0.001 and minimum FC of 4 to identify differential genes. Genes with higher expression in single-nucleus data are highlighted in green and those higher in single-cell data are highlighted in orange. Purple genes pass neither FDR nor FC threshold and are considered as non-differential. (C) Expression of selected myogenic, cell cycle and lncRNA genes (orange for single-cell and green for single-nucleus). (D) Violin plots of representative genes measured in TPM. P-values are calculated with edgeR.
The median numbers of genes with TPM ≥ 1.0 averaged 7091 in single cells and 4550 in single nuclei (Supplementary Figure S3B). There were 13 371 genes with TPM ≥ 1.0 in at least five single cells and 11 414 genes with TPM ≥ 1.0 in at least 10 single cells, compared to 10 703 and 8112 respectively in single nuclei. Two-way hierarchical clustering of all the data identified two outliers in the 75 myoblast single cell samples (Supplementary Figure S4A). These outliers were the only two myoblast cells with <4000 genes with TPM ≥ 1.0, suggesting that reverse transcription could have been less successful for these cells. We excluded these cells from downstream analyses. We further eliminated from analysis myoblast single nuclei with <4000 expressed genes, leaving 37 of 58 myoblast single nuclei for analysis. Since we had more cells than nuclei, we picked 37 cells with the highest MYF5 expression level to compare with the single nucleus samples (Supplementary Figure S4B).
The expression level of 8943 (81.3%) genes showed no significant difference between single cell and single nucleus data using a FDR ≤ 0.001 and minimum fold change of 4 (Figure 1B). 1794 (16.3%) genes were enriched in single nuclei, 395 (22.0% of 1794 genes) of which were lncRNAs. In contrast, only 267 (2.4%) genes were enriched in the single whole cell samples, out of which only 10 genes (3.7% of 267 genes) are lncRNAs. Some well-studied lncRNAs involved in genome organization such as XIST (30) are at least 10-fold enriched on average in single nuclei, suggesting that a comparison of single cells and single nuclei can identify the subset of lncRNAs with nuclear functions (Figure 1B). We further found that H19 is enriched in our snRNA-seq. The enrichment of XIST and H19 in snRNA-seq over scRNA-seq was further confirmed by qPCR (Supplementary Figure S5A). H19 is highly expressed during fetal development but is repressed in adult tissues except skeletal muscles (31). Although H19 RNA has been reported to play a cytoplasmic role (32,33), H19 lncRNA also recruits methyl-CpG-binding domain protein 1 (MBD1) to mediate epigenetic silencing at imprinted gene loci and also interacts with polycomb repressive complex 2 (PRC2), indicating its role in the nucleus (30–35). Thus, H19 RNA appears to play multiple roles in both the cytoplasm and the nucleus. Myogenesis and cell cycle-related genes showed similar expression patterns in myoblast single cells and single nuclei (Figure 1C). Factors that control myoblast proliferation such as CDC20 and ID1 were highly expressed at similar levels in both single nuclei and single cells (Figure 1C) (36,37). Genes involved in DNA replication and repair such as PCNA and RAD51 also showed similar levels of expression, indicating the normal progression of cell cycle in KD3 myoblasts (Figure 1C and D) (38). The MRF MYF6 showed no difference whereas MYOD1 was expressed at a relatively low level in a few samples whether in cells or nuclei. Myogenin (MYOG), which is the MRF whose expression is associated with myoblast differentiation (5), is not expressed in any of the myoblast samples. This indicates that our myoblasts were undifferentiated. MYF5 was the most highly expressed MRF in KD3 myoblasts. It was enriched more in the single whole cell samples (Figure 1C and D) partly because we selected single cell samples with the highest MYF5 levels in order to have comparable numbers of single cell and single nucleus samples. Taken together, the majority of the transcriptome characterized by scRNA-seq can also be accurately recapitulated by snRNA-seq.
Isolation of nuclei from multinucleated myotubes and mononucleated cells (MNCs) that fail to undergo cell fusion
We optimized our differentiation protocol to achieve greater than 80% of nuclei found in multi-nucleated myotube after 3 days of differentiation (Supplementary Figures S1 and S2). We observed significant upregulation of MYOG and myosin heavy chain by immunofluorescent staining correlating with myotube formation (Supplementary Figure S1). The rest of the cells (<20%) remained unfused (i.e. MNCs) and resided between myotubes (Supplementary Figures S1 and S2). In order to study the differences between myotubes and unfused MNCs, we used a short trypsin/EDTA treatment to separate them prior to nucleus isolation (Figure 1A; Supplementary Figure S2). While the length of KD3 myotubes is typically longer than 100 μm, MNCs are even smaller than undifferentiated myoblasts. However, their nucleus sizes were all within 5–10 μm and we therefore used the same Fluidigm chip that we used for myoblast nuclei (Supplementary Figure S2). Libraries were sequenced to depths of 1–6 million reads (Supplementary Figure S3A).
Comparison of single-nucleus transcriptomes of myoblasts and myotubes
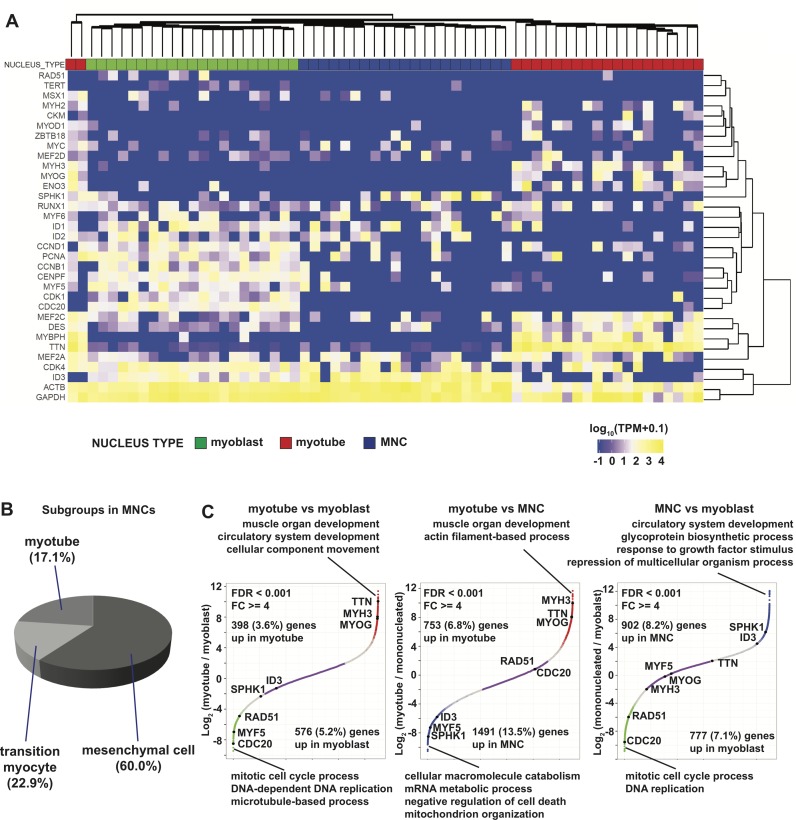
Hierarchical clustering of 11 004 genes expressed above 1 TPM in at least 10 single nuclei collected from myoblasts (54 out of 58 nuclei passing filter), myotubes (44 out of 50 nuclei), and MNCs (35 out of 70 nuclei) showed the relative homogeneity of myotubes and myoblasts in contrast to the marked heterogeneity of the MNCs (Figure 2A, B; Supplementary Figure S6A). Nuclei from a cluster of 41 myoblasts (‘cluster 5′ in Supplementary Figure S6B) expressed high levels of CDK1, CDC20, ID1, ID3 and PCNA, which are hallmarks of proliferating cells, but exhibited no or low expression of MYOG and Titin (TTN) that are associated with myogenic differentiation (Figure 2C) (39). The gene expression pattern of myogenic and cell cycle-related genes was also relatively uniform in myotubes (cluster 1 in Supplementary Figure S6B) and showed decreased expression of the cell proliferation genes described above as well as MYF5, along with significant upregulation of myogenic differentiation marker genes, such as MYOG, TTN, myosin heavy chain 3 (MYH3), and MYBPH, as expected of mature myotubes (Figure 2A, C). The expression profiles of the 13 myoblast nuclei clustered with myotubes (2 in cluster 1 and 11 in cluster 2) were similar to that of the main myoblast cluster (cluster 5) including no MYOG expression, except that their CDC20 levels were low. Further analysis reveals that these myoblast single nuclei have fewer than 4000 genes expressed above 1 TPM, which may contribute to their clustering with myotube nuclei that have fewer genes expressed on average (Supplementary Figure S3B). We also checked the expression levels of the three genes hTERT, CDK4 and CCND1 used to immortalize KD3. We found that hTERT is only expressed at a modest level in KD3. CDK4 was expressed at twice the level in KD3 as in HSMM, while CCND1 levels were comparable, which is consistent with the lack of carcinogenicity or interference with myotube differentiation (17). Nevertheless, all three genes are slightly down-regulated in differentiated myotubes as well as MNCs compared to myoblasts even though this does not reach the level of statistical significance (Figure 2A; Supplementary Figures S6B and S7; Tables S2–S4).
Figure 2.
Differentially expressed genes between nuclei of KD3 myoblast, myotube and MNC. (A) Expression heatmap of selected myogenic and cell cycle genes for 63 snRNA-seq cells. The color bar on top indicates the sample origin (green for myoblast, red for myotube and blue for MNC). Sample clustering tree on top is based on 11 004 expressed genes. (B) Percentage of different subgroups in MNCs based on clustering result in Supplementary Figure S6B. (C) Gene expression FC curves between different cell types with enriched GO terms of differential genes. The FDR and FC thresholds used to identify differential genes are the same as those used in Figure 1B. The up-regulated genes in each cell type are labeled with the same color codes used in Figure 2A. Non-differential genes are in purple.
snRNA-seq of MNCs reveal cell fate heterogeneity
Differentiated MNCs showed the highest expression heterogeneity (Figure 2A, B; Supplementary Figure S6). The largest MNC cluster (cluster 6) consisted of 21 nuclei with no expression of CDC20, CDK1, or MYOG together with low expression of MYF5. All these characteristics implied that they were distinct from proliferating myoblasts and showed no evidence of myotube differentiation. These cells showed distinct upregulation of ID1 and ID3, which are basic helix-loop-helix (bHLH) repressors normally silenced during myotube differentiation (36,37). Ten of these nuclei were SPHK1-positive, which is a hallmark of mesenchymal cells (MSCs) (15) (Figure 2A, C; Supplementary Figures S6B and S8A). SPHK1 was expressed higher in MNCs than in myoblasts (Figure 2A, C; Supplementary Figure S6B). These results are similar to the mesenchymal cluster found in HSMM with Monocle, where these cells were thought to be the contaminants of interstitial MSCs in their primary muscle cell culture (15). However, since the KD3 line is monoclonal and we did not find any cell population in our myoblast data that exhibited a similar phenotype, our results raise the possibility that these MNCs were derived from myoblasts by ‘alternative differentiation’.
A smaller cluster of six MNC nuclei (cluster 4 in Supplementary Figure S6B) were more closely related to myoblasts with intermediate characteristics of both myoblast and myotube. Although they no longer expressed CDC20 and had lower expression levels of CDK1 as well as ID1, their levels of PCNA, MYF5 and ID3 were still comparable to those of proliferating myoblasts. At the same time, despite the low expression of mature myotube markers such as TTN and MYOG, and the absence of MYH3 and MYH2, these clustered nuclei have upregulated expression of enolase 3 (ENO3) and myocyte enhancer factor 2C (MEF2C), which mark the onset of myogenic differentiation (15). These MNC nuclei are most likely myocytes in the transition state between myoblast and myotube (Figure 2B). Another eight MNC nuclei are dispersed in the myotube cluster 1 (Supplementary Figure S6B). Further analysis revealed that these eight nuclei have also the lowest number of expressed genes and a couple of them do not even have detectable GAPDH expression. These may be either dying cells or nuclei with poor RNA capture rates. This is in contrast to the MNC nuclei that we identified as myocytes in cluster 4, which have among the highest detected gene numbers.
Comparative analysis of mesenchymal MNCs with myoblasts and myotubes
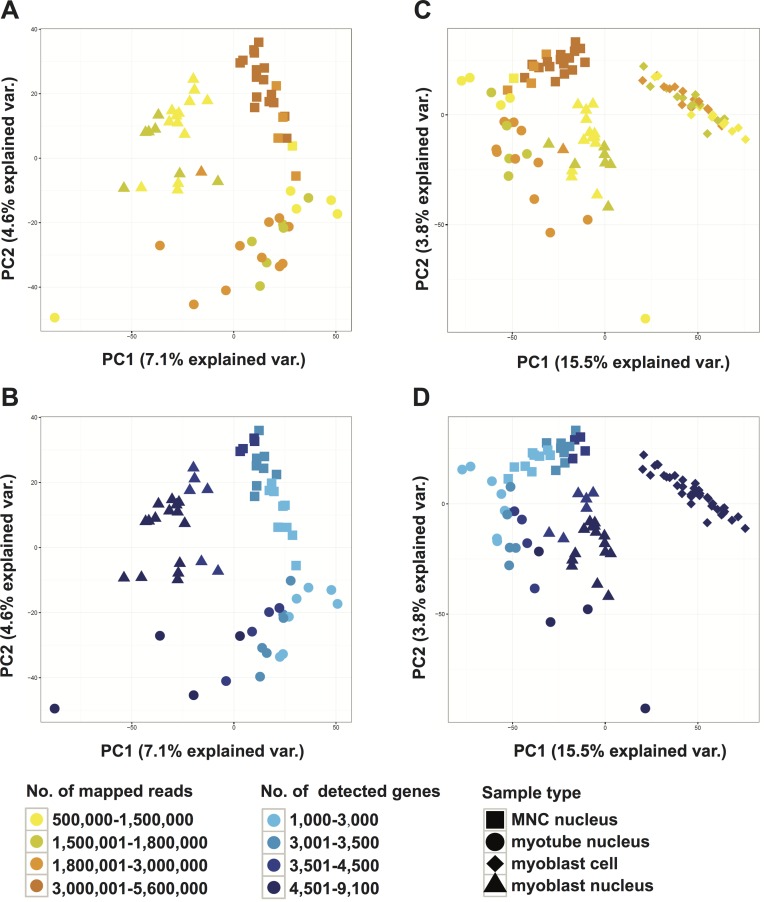
We selected a subsample of nuclei that best represented the myoblast, myotube, and mesenchymal MNCs with a high number of expressed genes. We selected the myoblast nuclei with the highest CDC20 expression, the 21 MNC nuclei that are mesenchymal-like, and the 21 myotube nuclei with the highest TTN expression for comparison with similar power (Figure 2A). Principal Component Analysis (PCA) with all expressed genes showed that the three nuclei populations clustered distinctly (Figure 3A and B). We find that PC1 corresponds to the number of genes detected while PC2 separates multinucleated myotubes from mononucleated myoblasts and MNCs (Figure 3B). This result is consistent with our previous finding that upon differentiation into myotube and MNC, the number of expressed genes decreased in comparison to that in undifferentiated myoblast (Supplementary Figure S3). Even with myoblast scRNA-seq data included, the clustering pattern of the three snRNA-seq samples remains similar (Figure 3C and D). Interestingly, the PCA of the snRNA-seq data is the mirror image of those in Figure 3A and B minus the scRNA-seq data as the myoblast scRNA-seq cluster is separated from snRNA-seq clusters. However, the myoblast snRNA-seq cluster is still the closest to the myoblast scRNA-seq (Figure 3). We identified differentially expressed genes with FDR <0.001 and minimum fold change of 4 and called 9–20% of the genes as differentially expressed in one or more conditions (Figure 2C; Supplementary Tables S2–S4). GO analysis of differentially expressed genes between myotube and myoblast nuclei reflected different stages of myogenesis (Supplementary Tables S2 and S5). Genes upregulated in myotube are enriched in skeletal muscle development and functions, while the genes highly expressed in myoblast are mainly involved in cell cycle progression and mitosis, which is characteristic of proliferating myoblasts as described above. The differences between myotubes and MNCs stand out (Supplementary Tables S3 and S6). The 1491 genes upregulated in MNC nuclei compared to myotube nuclei mainly function in mRNA processing, which indicates that these cells followed a distinct developmental pathway (Figure 2C). Platelet-derived growth factor a (PDGFA), which is the ligand for mesenchymal cell marker PDGFRA, and other genes related to PDGF binding contribute to the GO terms enriched in MNC nuclei compared to myoblast nuclei such as ‘circulatory system development’ and ‘response to growth factor stimulus’ (Figure 2C; Supplementary Table S7). Particularly interesting GO terms that are enriched in comparisons of MNCs versus myoblasts or myotubes point to processes such as growth factor response, glycoprotein biogenesis, mitochondrial organization and negative regulation of apoptosis. Histone deacetylase 4 (HDAC4) interacts with MEF2C and suppresses myoblast differentiation. The dissociation of HDAC4 from MEF2C is important for MEF2C activation of myoblast differentiation (40). However, both HDAC4 and MEF2C are transcriptionally down-regulated in MNCs compared to myoblasts or myotubes (Supplementary Figure S8A). All these suggest that MNCs have adopted a fundamentally different identity from myoblasts and myotubes as mesenchymal cells. TP53 and ID3 have been shown to regulate the proliferation and differentiation of MSC and epithelial-mesenchymal transition (41–43). The upregulation of TP53 and ID3 in MNCs also matches with their putative mesenchymal characteristic (Figure 3A; Supplementary FigureS8A). The repression of MYOG and activation of ID3 in MNCs were further confirmed by qPCR (Supplementary Figure S5B). Furthermore, several genes upregulated in MNCs are involved in the mTOR pathway as well as TAF7 that are important for cellular proliferation (44,45). However, we also observed the upregulation of several signaling related genes such as beta–catenin (CTNNB1), SMAD1 and SMAD2, as well as RELA from the NF-κB pathway (Supplementary Figure S8A) (46,47). We considered whether the upregulation of RELA in MNCs could be due to stress and MNCs mounted a proinflammatory response due to handling (48). However, the expression of proinflammatory genes is either negative or not specifically increased in MNCs (Supplementary Figure S8B). These results suggest that these MNCs are not fully differentiated and maintain high proliferation capacity while waiting for additional signaling input to adopt a terminal fate.
Figure 3.
Single nuclei from three different cell types show distinct clustering profiles in Principal Component Analysis. (A, B) PCA plots of the 63 single nuclei from Figure 2A, labeled by cell type and number of mapped reads (A) or number of expressed genes detected (B). (C, D) PCA plot of the 63 nuclei together with the 37 myoblast single-cell samples, colored by number of mapped reads (C) or number of expressed genes detected (D).
In order to validate our myotube and MNC separation procedures and the efficacy as well as relevance of KD3 differentiation at the 72 h time point, we compared our single-nucleus data with data from the previously published monocle analysis of scRNA-seq from primary human skeletal muscle myoblasts (HSMM) (15). We compared the 0hr undifferentiated HSMM data with KD3 myoblast data, 72hr differentiated HSMM MYOG-positive cells with KD3 myotubes, and 72h differentiated HSMM SPHK1-positive cells with KD3 MNCs (Supplementary Figure S9). Interestingly, both myoblast samples exhibited high MYF5 and spotty MYOD1 expression before differentiation, which is different from C2C12 mouse myoblasts, which have much higher levels of MyoD1 than Myf5. Thus, this is not an immortalized myoblast-specific phenomenon. In both SPHK1-positive differentiated HSMMs and KD3 MNCs, MYOG and TTN expression is low, supporting the similarity between these two groups of cells. The results indicate that these cells are distinct from the myocytes that will further differentiate into myofibers.
Differentially expressed lncRNAs include primary transcripts of microRNAs that play important roles in myogenesis
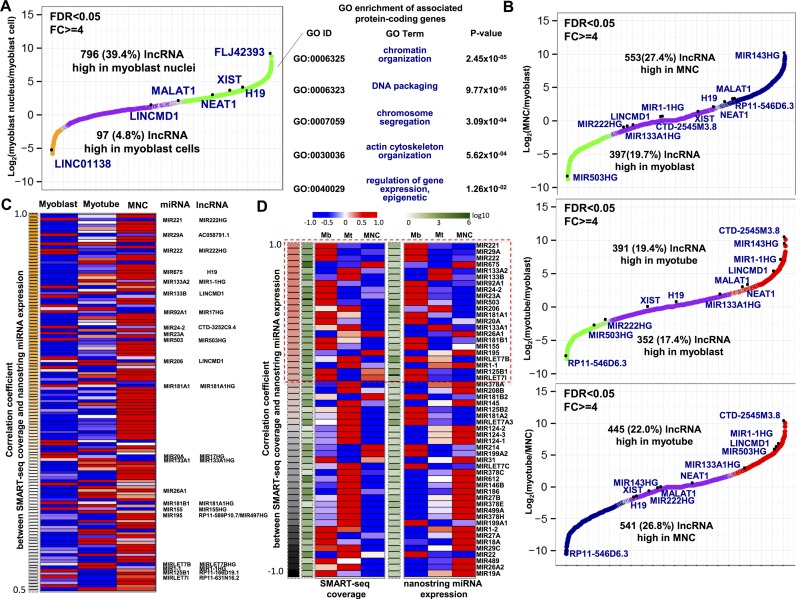
Both lncRNAs and miRNAs are known to be key regulators in muscle development and have been associated with various muscle disorders (49–51). An increasing number of lncRNAs have been found in human (more than 15 000 in GENCODE GRCh38.p3 v23) and many of them are able to regulate gene expression in cis or in trans in muscle (52,53). Since we found evidence for distinct enrichment of nuclear lncRNAs in snRNA-seq compared to scRNA-seq (Figure 1B and D), we decided to further analyze lncRNA expression levels in nuclei and cells. We found 796/2020 (39.4%) lncRNAs using FDR <0.05 and FC ≥ 4 that were highly expressed in nuclei while only 97/2020 (4.8%) that were enriched in cells (Figure 4A). We detected neighboring protein-coding genes for our differentially expressed lncRNAs in nuclei as well as cells and identified 243 lncRNAs-protein-coding genes pairs (Supplementary Table S12). These lncRNA and neighboring protein-coding gene pairs showed marginally significant greater correlated expression compared to randomized gene pairs using single-cell expression (P-value = 0.008) but not when using single-nucleus expression (P-value = 0.063). We associated neighboring protein-coding genes with nucleus-enriched lncRNAs and found that genes with chromatin organization and chromosome segregation annotations were enriched (Figure 4A, Supplementary Tables S8 and S12), raising the possibility that these nucleus-enriched lncRNAs may regulate gene expression by coordinating chromatin structure. Furthermore, lncRNAs that are highly enriched in myoblast nuclei are associated with genes specific for cytoskeleton development, such as TTN, FMNL1, SSH1 and DAAM2 (Figure 4A, Supplementary Tables S8 and S12).
Figure 4.
lncRNAs have differential enrichment in myoblast, mononucleated and myotube nuclei. (A) Differential expression analysis of lncRNAs between myoblast nuclei and cells (green, high expression in nuclei; yellow, high expression in cells; purple, no significant change). FC ≥ 4 and FDR < 0.05. (B) Differential expression analysis between myoblast, MNC and myotube nuclei (green, high expression in myoblast; blue, high expression in MNC; red, high expression in myotube; purple, no significant change). Fold change ≥ 4 and FDR < 0.05. (C) SMART-seq coverage and NanoString expression of miRNAs with Pearson correlation coefficient greater than 0.5. Myogenic miRNA and corresponding pri-miRNAs are labeled. (D) SMART-seq coverage and NanoString expression of 53 myogenic miRNAs. Both coverage and NanoString expression are normalized between -1 and 1 (red, high level; blue, low level). Myogenic miRNAs with correlation coefficient greater than 0.5 are boxed with red dash line. Absolute reads counts for coverage and absolute expression for NanoString are averaged for each of three nuclei types and normalized by log10.
We next analyzed differentially expressed lncRNAs in myoblast, myotube and MNC nuclei. We found that hundreds of lncRNAs have significantly higher expression in the nuclei of one or more of the three cell types (Figure 4B). GO analysis of neighboring protein-coding genes showed that myoblast-enriched lncRNAs are mainly associated with DNA replication and cell division while myotube-enriched lncRNAs exhibit high enrichment in muscle organ development. In contrast, MNC-enriched lncRNAs are highly associated with metabolic process and regulation of gene expression (Supplementary Tables S9–S11, S13–S15). Muscle-related lncRNAs, such as LINCMD1 (54), are only highly expressed in myotube nuclei while NEAT1 (55) and MALAT1 (56) had higher expression levels in both MNC and myotube nuclei (Figure 4B; Supplementary Figure S10; Tables S19 and S20).
lncRNAs and pri-miRNA transcripts are enriched in human nuclear transcription factories (57). We observed that several lncRNAs serving as primary microRNA (pri-miRNA) transcripts were among the top differentially expressed lncRNAs between myoblasts, myotube, and MNC nuclei (Figure 4B; Supplementary Table S16–S18). Many miRNAs have been identified and validated to promote or inhibit muscle differentiation (52) and lncRNAs that serve as pri-miRNA transcripts should be associated with the regulation of those miRNAs. We therefore tested whether lncRNA expression correlated with mature microRNA expression using the NanoString platform for validation. We measured NanoString expression level for miRNAs in myoblast, myotube and MNC nuclei and found that 25.10% (123/490) of miRNAs show positive correlation (r > 0.5) between NanoString expression and SMART-seq coverage over 3kb within miRNAs (Figure 4C, Supplementary Figure S11A; Tables S21 and S22). Among these miRNAs, 36.6% (45/123) have neighboring lncRNAs transcribed from the same strand. Interestingly, we found that about 44.4% (20/45) of these miRNAs are known to be involved in muscle differentiation (out of 53 myogenic miRNAs in total) and their neighboring lncRNAs serve as pri-miRNAs (Figure 4C and D, Supplementary Figure S11B). For example, muscle-specific miRNAs, MIR1-1/1-2 and MIR133A1/A2 play pivotal roles in skeletal muscle differentiation, which are organized in bicistronic clusters on two different chromosomes (58,59). MIR1-1/133A2 are embedded in MIR1-1HG and MIR1-2/133A1 are located in MIR133A1HG, MIR133A1, MIR133A2 and MIR1-1 are well known to promote muscle differentiation and expression during myogenesis (52,60,61). They show exclusively high level in both expression and coverage in myotube nuclei, which is consistent with the upregulation of their pri-miRNAs MIR1-1HG and MIR133A1HG in myotube nuclei compared to myoblast nuclei (Figure 4B and C). Similarly, MIR206/133B is known to be upregulated during skeletal muscle development. This well studied locus also encodes Lincmd1 in mouse, which expresses jointly with Mir206/133b during myogenesis (54,62–64). However, the human LINCMD1 locus only partially overlaps with MIR206/133B locus and they are transcribed in the reverse strand, which is different from the locus in the mouse genome. This raises the possibility that LINCMD1 may be misannotated in the human genome. Nevertheless, we found significantly high positive correlation between expression and coverage in MIR206/133B and the apparently high enrichment in myotube nuclei is consistent with the high expression of LINCMD1 in myotube nuclei. Furthermore, we found many miRNAs that show exclusively high expression in myoblasts compared to myotubes and MNCs, such as MIR503 (65). Several miRNAs that repress muscle differentiation such as MIR221/222 (66), MIR125B1 (67) and MIR155 (68) also show exclusively high expression in myoblast nuclei. We confirmed the differential expression of the MIR222HG and MIR503HG pri-miRNA by qPCR (Supplementary Figure S5C). Correspondingly, their expression levels are positively correlated with SMART-seq coverage in their pri-miRNAs locus. Overall, we observed that approximately 25% of miRNAs show strong positive correlation (r > 0.5) between coverage and expression. About half of the myogenic miRNAs have a strong correlation and their patterns of expression and coverage on pri-miRNAs are consistent with their known roles during myogenesis.
DISCUSSION
We report here that snRNA-seq using the Fluidigm C1 platform is a practical method to faithfully capture the transcriptome of single cells from isolated nuclei. In comparison to the dramatic variation of cell size in different tissues and at different developmental stages, changes in nucleus sizes are usually within a tighter range. Thus snRNA-seq is a practical alternative for the transcriptome analysis of cells that would be otherwise difficult to capture on standard-size Fluidigm microfluidic chips while at the same time enriching for a set of lncRNAs that are more prevalent in the nucleus. Our work differs from previously published sequencing methods on single nuclei such as single-nucleus genome sequencing (nuc-seq) and single nucleus exome sequencing (SNES) that focus on characterizing mutations in the genomic DNA of single nuclei isolated by flow sorting (69,70). In particular, these methods did not examine the transcriptome from these nuclei and the hour-long procedures between cell lysis and the end of nuclei isolation could result in altered gene expression. Manual micromanipulation of single nuclei is difficult to scale up, as demonstrated by an earlier publication solely based on only three individual single-nucleus RNA-seq libraries (16). Our snRNA-seq method with the Fluidigm microfluidic system is both time and labor saving and higher throughput. The time from cell lysis to the end of single-nucleus capture and the start of cDNA synthesis is about 30 minutes for up to 96 nuclei at a time.
Using snRNA-seq, we characterized myoblast differentiation, which allowed us to sample multinucleated myotubes and recover purer expression profiles that match previous reports of genes differentially regulated during myogenic differentiation. Indeed, MYF5 is more clearly downregulated and the TTN level was much higher as well as distributed in a more uniform pattern in KD3 myotube nuclei compared to the monocle 72 h MYOG-positive HSMM. This could be the result of our single-nucleus sequencing strategy capturing nuclei from more mature myotubes. Furthermore, we were able to separately characterize gene expression signatures for a small percentage of myoblasts that failed to form multi-nucleated myotubes (i.e. MNCs) and found that they are a heterogeneous mixture of mesenchymal cells and differentiation intermediate myocytes. Since KD3 is a clonal cell line, MNCs with mesenchymal features cannot be a mere contamination as previously suspected (15) but are more likely a subset of cells that have adopted an alternate or reversal cell fate in the mesenchymal pathway. Similarly, SPHK1-positive differentiated HSMMs that are thought to be contaminating interstitial mesenchymal cells (15) might have also been derived from undifferentiated HSMMs. We observed that the ratio of myotubes to MNCs can vary depending on the growth conditions (data not shown). Thus, our results directly captured and provide important insight into the plasticity of myoblast cell fate determination. It would be interesting to apply this approach to primary muscle cells of both normal and disease conditions, which may allow us to capture the extent of cell to cell variation and subtle pathogenic changes of myotubes/myofibers with greatly enhanced accuracy.
The snRNA-seq method also gives us the opportunity to assess with greater sensitivity transcripts that are primarily found in the nucleus. Our analysis of lncRNAs reveals that nucleus-enriched lncRNAs may be actively associated with regulation of gene transcription and coordination of chromatin structure in myoblast. Three-way nuclei comparisons show that the expression changes of lncRNAs encoding pri-miRNA transcripts can be used to infer miRNAs expression levels at specific stages during muscle differentiation. Our NanoString validation of miRNA expression in the three different cell sub-types shows that we can indeed correlate approximately half of myogenic miRNAs to the expression of their primary transcripts. Interestingly, we found that MIR675 encoded in the exons of H19 and critical for muscle differentiation and regeneration (31) show high level in expression and coverage in MNC nuclei by NanoString, strongly correlating with H19 lncRNA by snRNA-seq. The significance of this upregulation is currently unclear. Since H19 was shown to play an important role in adult stem cell maintenance (71,72) and also activates Wnt signaling by upregulation of β-catenin (73), it is possible that this H19 upregulation may be associated with the observed MSC-like phenotype of MNCs. Intriguingly, there is a subset of miRNAs that are perfectly anti-correlated to the expression of their primary lncRNA. This could be due to errors in lncRNA quantitation due to the unstranded nature of the SMART-seq protocol or due to incomplete annotations in those loci. However, this could also be the result of the nature of the NanoString assay, which cannot distinguish between identical family members, whereas we can potentially distinguish between different lncRNA/primary transcripts that encode for the same mature miRNA. More work will be necessary to identify the set of miRNAs whose pri-miRNA expression can faithfully recapitulate their expression levels.
The large-scale analysis of single nuclei also opens up the opportunity to analyze biological systems where the biology is nucleus-centric, such as during early Drosophila development when the embryo is a multinucleated syncytium where different nuclei transcribe different genes or when cells are simply too large to capture intact as in the case of some mature motor neurons or large cells such as in the early Xenopus embryo (16,74). Even in the case of vertebrate muscle, the subset of nuclei in myotubes that are directly under the neuromuscular junction (NMJ) express different set of transcripts encoding NMJ-specific proteins (75). We expect that large-scale snRNA-seq will take its place alongside scRNA-seq to answer appropriate biological questions.
Supplementary Material
Acknowledgments
We thank Alessandra Breschi and Rabi Murad with help in making heatmaps and PCA plots. We thank the UCI GHTF for access to the Fluidigm C1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [DP2 GM111100 to A. M. and AR067636 to K. Y.]. Funding for open access charge: NIH New Innovator funds (to A.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Schroder E.A., Esser K.A. Circadian rhythms, skeletal muscle molecular clocks and exercise. Exerc. Sport Sci. Rev. 2013;41:224–229. doi: 10.1097/JES.0b013e3182a58a70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckingham M., Rigby P.W.J. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Moncaut N., Rigby P.W.J., Carvajal J.J. Dial M(RF) for myogenesis. FEBS J. 2013;280:3980–3990. doi: 10.1111/febs.12379. [DOI] [PubMed] [Google Scholar]
- 4.Bryson-Richardson R.J., Currie P.D. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- 5.Bentzinger C.F., Wang Y.X., Rudnicki M.A. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., Macquarrie K.L., Davison J., Morgan M.T., Walter L., et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon A.T., Chou A.Y., Arenillas D.J., Wasserman W.W. Validation of skeletal muscle cis-regulatory module predictions reveals nucleotide composition bias in functional enhancers. PLoS Comput. Biol. 2011;7:e1002256. doi: 10.1371/journal.pcbi.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum R., Vethantham V., Bowman C., Rudnicki M., Dynlacht B.D. Genome-wide identification of enhancers in skeletal muscle: Tte role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollen A. a, Nowakowski T.J., Shuga J., Wang X., Leyrat A. a, Lui J.H., Li N., Szpankowski L., Fowler B., Chen P., et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014;32:1–37. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalek A.K., Satija R., Shuga J., Trombetta J.J., Gennert D., Lu D., Chen P., Gertner R.S., Gaublomme J.T., Yosef N., et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;509:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grindberg R. V., Yee-Greenbaum J.L., McConnell M.J., Novotny M., O'Shaughnessy A.L., Lambert G.M., Arauzo-Bravo M.J., Lee J., Fishman M., Robbins G.E., et al. RNA-sequencing From Single Nuclei. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiomi K., Kiyono T., Okamura K., Uezumi M., Goto Y., Yasumoto S., Shimizu S., Hashimoto N. CDK4 and cyclin D1 allow human myogenic cells to recapture growth property without compromising differentiation potential. Gene Ther. 2011;18:857–866. doi: 10.1038/gt.2011.44. [DOI] [PubMed] [Google Scholar]
- 18.Dobin A., Davis C.a, Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:4. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 22.Vandesompele J., De Preter K., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT -PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.1–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jian B., Liu B., Bi Y., Hou W., Wu C., Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008;9:59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G. Divergent transcription of long noncoding RNA / mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2012;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K., Huang K., Luo Y., Li S. Identification and functional analysis of long non-coding RNAs in mouse cleavage stage embryonic development based on single cell transcriptome data. BMC Genomics. 2014;15:845. doi: 10.1186/1471-2164-15-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C.F., et al. Resource meta- and orthogonal integration of influenza “OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dey B.K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Heesch S., Van Iterson M., Jacobi J., Boymans S., Essers P.B., De Bruijn E., Hao W., Macinnes A.W., Cuppen E., Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monnier P., Martinet C., Pontis J., Stancheva I., Ait-si-ali S., Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diao Y., Guo X., Li Y., Sun K., Lu L., Jiang L., Fu X., Zhu H., Sun H., Wang H., et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Wei Q., Paterson B.M. Regulation of MyoD function in the dividing myoblast. FEBS Lett. 2001;490:171–178. doi: 10.1016/s0014-5793(01)02120-2. [DOI] [PubMed] [Google Scholar]
- 38.Kohn K.W. Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol. Biol. Cell. 1999;10:2703–2734. doi: 10.1091/mbc.10.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herzog W., Powers K., Johnston K., Duvall M. A new paradigm for muscle contraction. Front. Physiol. 2015;6:1–11. doi: 10.3389/fphys.2015.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J., McKinsey T.a, Zhang C.L., Olson E.N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 41.Armesilla-Diaz A., Elvira G., Silva A. P53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp. Cell Res. 2009;315:3598–3610. doi: 10.1016/j.yexcr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Chang C.-J., Chao C.-H., Xia W., Yang J.-Y., Xiong Y., Li C.-W., Yu W.-H., Rehman S.K., Hsu J.L., Lee H.-H., et al. p53 regulates epithelial–mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowanetz M., Valcourt U., Bergström R., Heldin C.-H., Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol. Cell. Biol. 2004;24:4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gegonne A., Tai X., Zhang J., Wu G., Zhu J., Yoshimoto A., Hanson J., Cultraro C., Chen Q.-R., Guinter T., et al. The general transcription factor TAF7 is essential for embryonic development but not essential for the survival or differentiation of mature T cells. Mol. Cell. Biol. 2012;32:1984–1997. doi: 10.1128/MCB.06305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laplante M., Sabatini D.M. MTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terada K., Misao S., Katase N., Nishimatsu S.-I., Nohno T. Interaction of Wnt signaling with BMP/Smad signaling during the transition from cell proliferation to myogenic differentiation in mouse myoblast-derived cells. Int. J. Cell Biol. 2013:616294. doi: 10.1155/2013/616294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakkar N., Wang J., Ladner K.J., Wang H., Dahlman J.M., Carathers M., Acharyya S., Rudnicki M.a., Hollenbach A.D., Guttridge D.C. IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L.-F., Greene W.C. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 49.Sharma M. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. 2014;5:1–9. doi: 10.3389/fphys.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neguembor M., Jothi M., Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet. Muscle. 2014;4:8. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nie M., Deng Z., Liu J., Wang D. Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed Res. Int. 2015:676575. doi: 10.1155/2015/676575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2015;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 54.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sunwoo H., Dinger M.E., Wilusz J.E. MEN ϵ/β nuclear retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watts R., Johnsen V.L., Shearer J., Hittel D.S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am. J. Physiol. Cell Physiol. 2013;304:C995–C1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 57.Caudron-Herger M., Cook P.R., Rippe K., Papantonis A. Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res. 2015;43:e95. doi: 10.1093/nar/gkv390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams A.H., Liu N., Van Rooij E., Olson E.N. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Townley-tilson W.H.D., Callis T.E., Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int. J. Biochem. 2011;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q., Guo J., Lin X., Yang X., Ma Y., Fan G.-C., Chang J. An intragenic SRF-dependent regulatory motif directs cardiac-specific microRNA-1-1/133a-2 expression. PLoS One. 2013;8:e75470. doi: 10.1371/journal.pone.0075470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Mandel E.M., Thomson J.M., Wu Q., Thomas E. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sweetman D., Goljanek K., Rathjen T., Oustanina S., Braun T., Dalmay T., Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 64.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 65.Sarkar S., Dey B.K., Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cardinalli B., Castellani L., Fasanaro P., Basso A., Alemà S., Martelli F., Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ge Y., Sun Y., Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seok H.Y., Tatsuguchi M., Callis T.E., He A., Pu W.T., Wang D.Z. miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J. Biol. Chem. 2011;286:35339–35346. doi: 10.1074/jbc.M111.273276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Waters J., Leung M.L., Unruh A., Roh W., Shi X., Chen K., Scheet P., Vattathil S., Liang H., et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:1–15. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung M.L., Wang Y., Waters J., Navin N.E. SNES: single nucleus exome sequencing. Genome Biol. 2015;16:55. doi: 10.1186/s13059-015-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venkatraman A., He X.C., Thorvaldsen J.L., Sugimura R., Perry J.M., Tao F., Zhao M., Christenson M.K., Sanchez R., Yu J.Y., et al. adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinet C., Monnier P., Louault Y., Benard M., Gabory A., Dandolo L. Stem cells and regeneration H19 controls reactivation of the imprinted gene network during muscle regeneration. Development. 2016;143:962–971. doi: 10.1242/dev.131771. [DOI] [PubMed] [Google Scholar]
- 73.Liang W., Fu W., Wang Y., Sun Y., Xu L. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jevtić P., Levy D.L. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr. Biol. 2015;25:45–52. doi: 10.1016/j.cub.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaeffer L., De Kerchove D'Exaerde A., Changeux J.P. Targeting transcription to the neuromuscular synapse. Neuron. 2001;31:15–22. doi: 10.1016/s0896-6273(01)00353-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.