Abstract
The topological state of DNA is important for replication, recombination and transcription, and is regulated in vivo by DNA topoisomerases. Gyrase introduces negative supercoils into DNA at the expense of ATP hydrolysis. It is the accepted view that gyrase achieves supercoiling by a strand passage mechanism, in which double-stranded DNA is cleaved, and a second double-stranded segment is passed through the gap, converting a positive DNA node into a negative node. We show here that gyrase with only one catalytic tyrosine that cleaves a single strand of its DNA substrate can catalyze DNA supercoiling without strand passage. We propose an alternative mechanism for DNA supercoiling via nicking and closing of DNA that involves trapping, segregation and relaxation of two positive supercoils. In contrast to DNA supercoiling, ATP-dependent relaxation and decatenation of DNA by gyrase lacking the C-terminal domains require both tyrosines and strand passage. Our results point towards mechanistic plasticity of gyrase and might pave the way for finding novel and specific mechanism-based gyrase inhibitors.
INTRODUCTION
Topoisomerases maintain and regulate the topological state of DNA in the cell by introducing or removing supercoils, and by resolving catenanes and knots (recently reviewed in (1,2)). The degree of supercoiling is described by the linking number (Lk), the integer number of turns of both DNA strands around each other (3). Changes in the linking number require DNA cleavage. Type I topoisomerases cleave one strand of their DNA substrate, and alter the linking number in increments of one (4). Type II topoisomerases are thought to cleave both strands of the DNA, and change the linking number in steps of two (5). Changes in the catenation, knotting and supercoiling state of DNA by type II topoisomerases are believed to be achieved by a strand passage mechanism (reviewed in (6)), in which one double-stranded DNA (dsDNA) segment, the G-segment, is cleaved, and a second, the T-segment, is passed through the gap. Strand passage leads to DNA supercoiling or relaxation when G- and T-segments are contiguous on the same molecule, and to decatenation or unknotting when they reside on different DNA molecules. DNA gyrase is the only member of the type II topoisomerase family that is able to introduce negative supercoils into DNA at the expense of ATP hydrolysis (7). The active form of gyrase is a heterotetramer, formed by two GyrB and two GyrA subunits (Figure 1; (8)). GyrB, a member of the GHKL phosphotransferase superfamily (for GyrB-Hsp90-histidine/serine protein kinases-MutL, reviewed in (9)), contains the active site for ATP binding and hydrolysis in its N-terminal domain. GyrB dimerizes upon ATP binding, and its N-terminal domains form the N-gate of gyrase that acts as an ATP-operated clamp ((10,11); Figure 1). The DNA-gate is formed by GyrB and GyrA, specifically by the topoisomerase-primase (TOPRIM) domains of GyrB and the winged helix domains (WHDs) of GyrA which harbor the catalytic tyrosines for strand cleavage ((12–14); Figure 1). The C-gate is formed by the GyrA subunits of the enzyme (12). The C-terminal domain (CTD) of the GyrA subunit forms six blades that are arranged in a propeller-like closed circular structure (15), with blades 1 and 6 connected by the conserved GyrA-box (16,17). According to the strand passage mechanism, gyrase achieves negative supercoiling by wrapping the DNA around the CTDs with a positive handedness (18), thereby fixing a positive node. Cleavage of both DNA strands in the G-segment, and passage of the adjacent T-segment through this break (19) then leads to sign-inversion of the node from positive to negative handedness, resulting in an overall decrease in linking number by two (5). The strand passage mechanism predicts coordinated opening and closing of the N-, DNA- and C-gate.
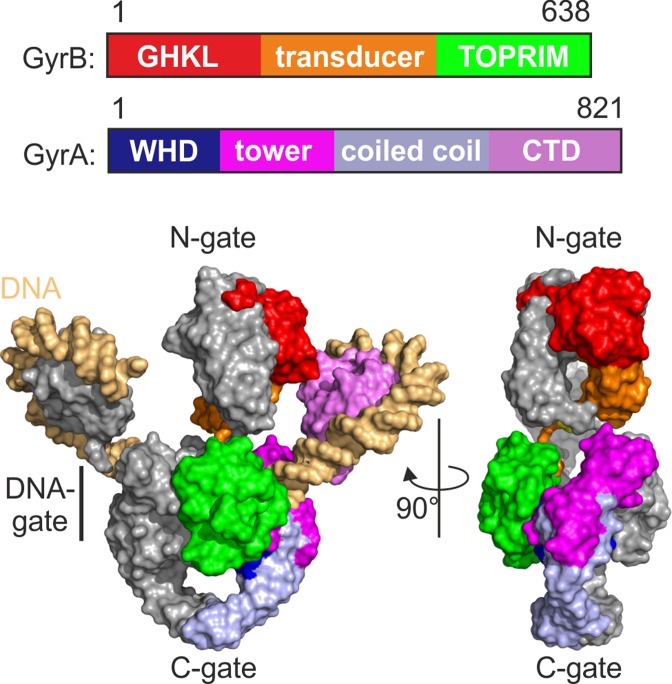
Figure 1.
Architecture of gyrase. Top: Domain architecture of GyrB and GyrA subunits. GHKL: GyrB-Hsp90-histidine/serine protein kinases-MutL, ATPase domain (GyrB); TOPRIM: topoisomerase-primase domain (GyrB); WHD: winged helix domain (GyrA); CTD: C-terminal domain (GyrA). Amino acid numbering refers to B. subtilis gyrase. Bottom: Model for the three-dimensional structure of gyrase, generated by superimposing the crystal structures of the GyrB ATPase domain (PDB-ID 1EI1; (60), the GyrA-NTD (PDB-ID 2XCR; (61)), and the GyrA-CTD (PDB-ID 3L6V; (62)) on the Cα coordinates of the cryo-EM model for T. thermophilus DNA gyrase in complex with DNA, ADPNP and ciprofloxacin (20). The domains in one GyrA and GyrB subunit are color-coded as in panel A, the second is depicted in gray.
Gyrase shares a common overall architecture with 2-fold symmetry with the type IIA family of topoisomerases, including eukaryotic topoisomerase II (topo II) and bacterial topoisomerase IV (topo IV) (20,21). Topo IV is also a heterotetrameric enzyme, formed by two ParE and two ParC subunits that are homologous to the gyrase GyrB and GyrA subunits, respectively (22,23). The CTDs of ParC comprise three to eight blades arranged in an open form (24), and lack the GyrA-box (17). Eukaryotic topo II forms a structure similar to gyrase and topo IV by dimerization of two subunits that each carry a region homologous to GyrB/ParE at the N-terminus, followed by a C-terminal region corresponding to the N-terminal domain of GyrA/ParC but lacking their CTD (25). Despite their common overall architecture, topo II, topo IV and gyrase catalyze different reactions in vivo, namely ATP-dependent relaxation (topo II) or introduction (gyrase) of negative supercoils, or ATP-dependent decatenation (topo IV). Deletion of their CTDs converts gyrase and topo IV into a topo II (26,27), suggesting a common topoisomerase IIA core mechanism that is differentially modulated by the CTDs.
Double-strand cleavage by the two catalytic tyrosines and strand passage are believed to be central features of the type IIA topoisomerase mechanism. The two DNA cleavage events are highly coordinated in topo II (28). Gyrase also catalyzes double-strand cleavage, but because of efficient religation cleavage complexes are not populated in equilibrium (29). Gyrase poisons lead to the accumulation of cleavage complexes, resulting in double-strand breaks that are dangerous for genome integrity (30).
We show here that gyrase containing only one catalytic tyrosine can still introduce negative supercoils into DNA, in contradiction to what the strand passage mechanism would predict. Although this variant can only cleave one strand of the DNA, the linking number is reduced in steps of two. Gyrase with one and two tyrosines undergoes the same sequence of DNA- and nucleotide-induced conformational changes up to the step where strand passage would follow. We present an alternative mechanism for negative DNA supercoiling by gyrase with only one catalytic tyrosine via trapping, segregating and relaxing two positive DNA supercoils, involving nicking and closing of the G-segment without strand passage. In contrast to ATP-dependent negative DNA supercoiling, gyrase-mediated DNA decatenation and ATP-dependent DNA relaxation, catalyzed by gyrase lacking the CTDs (26), require two tyrosines, double-strand cleavage and strand passage, suggesting that different reactions catalyzed by type IIA topoisomerases show different mechanistic plasticity. Our results proved a framework for the design of novel, mechanism-based gyrase inhibitors.
MATERIALS AND METHODS
Cloning, mutagenesis, protein production and purification
Bacillus subtilis gyrase: Mutations S7C (GyrB, N-gate, (31), E44Q (GyrB, ATP hydrolysis-deficient; (32), D75N (GyrB, ATP binding-deficient), Y123F (GyrA, cleavage-deficient) (33), T140C and D145C (GyrA, DNA-gate) (33) were introduced into gyrB, gyrA or gyrBA (Quickchange, Stratagene). GyrA and GyrB were produced in E. coli BL21(DE3)RP in autoinducing (34) (GyrA) or LB medium (GyrB, induction with 0.2 mM IPTG) at 37°C for 24 h (GyrA) or 4 h (GyrB). GyrA and GyrB were purified at 4°C as described (32,33). Heterodimeric (GyrA)2, GyrA·GyrBA and (GyrBA)2 were purified from Escherichia coli BL21(DE3) (autoinducing medium, 27°C, 24 h) by tandem-affinity chromatography (Supplementary Figure S1). Cells were harvested by centrifugation, resuspended in buffer A (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 2 mM β-mercaptoethanol) supplemented with 1 M NaCl, and disrupted in a Microfluidizer. The crude extract was applied to a StrepTactin sepharose column equilibrated in buffer A supplemented with 1 M NaCl to select for complexes that contain one or two Strep-tagged subunit. Proteins were eluted with buffer A with 1 M NaCl and 2.5 mM d-desthiobiotin (Sigma-Aldrich), dialyzed against buffer A with 200 mM NaCl, and applied to a Q-sepharose column equilibrated with buffer A with 200 mM NaCl. Fractions from elution with a linear gradient (200 mM to 1 M NaCl) in buffer A that contained DNA-free protein were pooled. Dimers containing a His-tagged subunit were selected for by chromatography on Ni2+-NTA sepharose, equilibrated with buffer A supplemented with 1 M NaCl and 20 mM imidazole, and eluted with buffer A with 1 M NaCl and 500 mM imidazole, followed by TEV protease cleavage and an additional purification step on Ni2+-NTA sepharose to remove the non-cleaved proteins and the His-tagged tobacco etch virus (TEV) protease. The final purification step was size exclusion chromatography on an S200 column in buffer A with 500 mM NaCl. For GyrAY123F_ΔCTD·GyrBAΔCTD, the selection steps on StrepTactin and Ni2+-NTA sepharose were inverted.
E. coli gyrase: GyrBA was constructed by linking GyrB to GyrA via a GAP linker. Mutation Y122F (GyrA, cleavage-deficient) (35,36) was introduced into GyrA according to the Quickchange protocol (Stratagene). The GyrBA·GyrAY122F heterodimer was produced in E. coli BL21(DE3) Star in TB medium at 30°C for 3 h, GyrB in E. coli BL21(DE3) in LB medium at 37°C for 3 h (induction with 0.2 mM IPTG). Cell disruption and protein purification were performed as described for B. subtilis gyrase by (a) Ni2+-NTA sepharose to select for complexes containing one His-tagged subunit, (b) thrombin cleavage, (c) Ni2+-NTA sepharose to remove non-cleaved protein, (d) StrepTactin sepharose to select for complexes containing Strep-tagged subunits, (e) TEV protease cleavage, (f) StrepTactin sepharose, (g) Q-sepharose to remove DNA, (h) size-exclusion chromatography on an S200 column (buffer A with 200 mM NaCl). Constructs used in this study are compiled in Supplementary Figure S1.
Topoisomerase reactions
Negatively supercoiled and relaxed pUC18 was prepared as described (31,33). The single pUC18 topoisomer (ΔLk = −1) was prepared after (5) by separation of pUC18 topoisomers on a 1.3% agarose gel in TEP buffer (36 mM Tris–HCl, pH 8.0, 30 mM NaH2PO4, 1 mM EDTA; 3.7 V/cm, 5 h), excision of the band for the single topoisomer with ΔLk = −1, gel extraction (Promega) and ethanol precipitation. Topoisomerase reactions were performed at 37°C with 6 nM (single topoisomer) or 20 nM (relaxed or supercoiled) pUC18 in 50 mM Tris–HCl, pH 7.5, 100 mM KCl and 10 mM MgCl2 (and 10% glycerol for E. coli gyrase). ATP-dependent reactions were performed in the presence of 1.5 mM ATP for 3 min if not indicated otherwise. Assembly of the gyrase/DNA complex occurs within <1 min, and supercoiling is completed after 3 min (Supplementary Figure S2). ATP-independent reactions were performed for 1 h. Concentrations were 200 nM GyrA and 800 nM GyrB, or 100 nM GyrA·GyrBA and 400 nM GyrB, if not stated otherwise. (GyrBAΔCTD)2 was used at a concentration of 500 nM, and GyrB·GyrAY123F_ΔCTD·GyrBAΔCTD was generated from 500 nM GyrAY123F_ΔCTD·GyrBAΔCTD and 1 μM GyrB. ATP-dependent decatenation was performed in 50 mM Tris–HCl, pH 7.5, 100 mM KCl and 10 mM MgCl2 as described (37), using 12.5 ng/μl of kDNA (Inspiralis Limited) as a substrate, and 100 nM GyrA or 50 nM GyrAY123F·GyrBA and 400 nM of GyrB. All topoisomerase reactions were stopped by addition of 0.5% SDS, 25 mM EDTA and 0.2 mg/ml proteinase K, incubated at 37°C for 10 min, and products were analyzed by electrophoresis on a 1.3% agarose gel in TEP buffer. To separate negative and positive topoisomers, a second electrophoretic separation in vertical direction to the first was performed in the presence of 10 μg/ml chloroquine. Gels were stained with ethidium bromide.
DNA cleavage reactions
DNA cleavage reactions were performed with 20 nM relaxed or negatively supercoiled pUC18, 200 nM GyrA and 800 nM GyrB or 100 nM GyrA·GyrBA and 400 nM GyrB in 50 mM Tris–HCl, pH 7.5, 100 mM KCl and 10 mM MgCl2 (and 10% glycerol for E. coli gyrase) at 37°C in the presence of 0 to 250 μM ciprofloxacine (CFX; 500 μM for E. coli gyrase) for 5 min (30 min for E. coli gyrase) if not specified otherwise, and products were separated by electrophoresis on a 1.3% agarose gel. Gels were stained with ethidium bromide, and band intensities were quantified by densitometry.
Fluorescent labeling, smFRET experiments and data analysis
Gyrase carrying two cysteines was labeled with maleimide derivatives of donor (AlexaFluor® 488) and acceptor (AlexaFluor® 546 for S7C, tetramethylrhodamine for T140C and D145C) fluorophores as described previously (31,33). smFRET experiments were performed at 37°C on a home-built confocal microscope on freely diffusing gyrase, and analyzed as described previously (31,33). A detailed description of the experimental set-up and data analysis can be found in (38,39).
RESULTS
B. subtilis gyrase with one catalytic tyrosine catalyzes negative DNA supercoiling
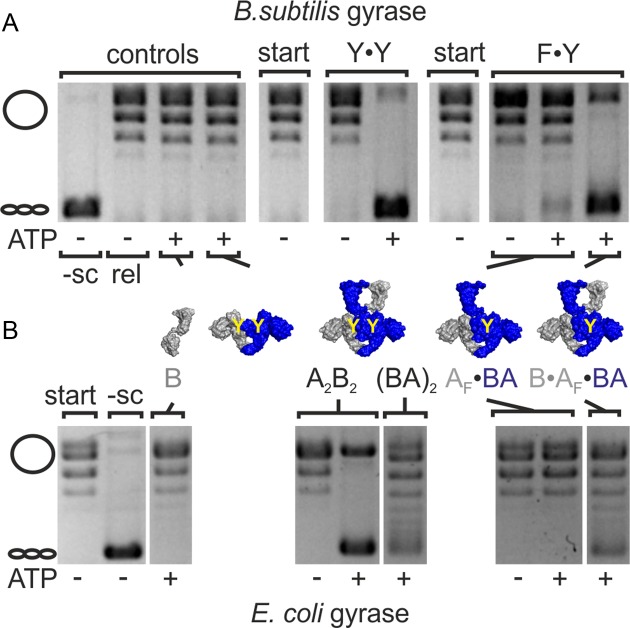
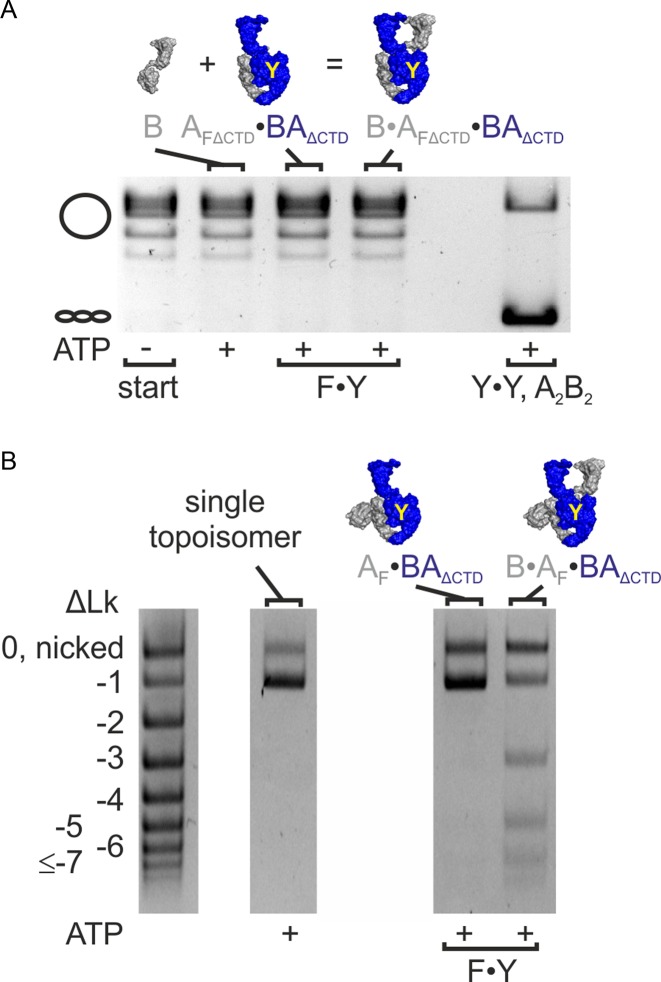
The strand passage model for DNA supercoiling predicts that gyrase lacking one of the two catalytic tyrosines will not be able to supercoil plasmid DNA due to its failure to generate a double-strand break in the G-segment. To test whether double-strand cleavage is a strict pre-requisite for DNA supercoiling by gyrase, we created a B. subtilis gyrase variant carrying only one catalytic tyrosine. We have previously established that gyrase formed by dimerization of a GyrB–GyrA fusion protein, (GyrBA)2, shows ATP-dependent supercoiling activity that is virtually identical to authentic, heterotetrameric gyrase (GyrA2GyrB2; (31)). To generate gyrase with only one catalytic tyrosine, we co-produced affinity-tagged versions of GyrBA and GyrAY123F in which the catalytic tyrosine is replaced by a phenylalanine, rendering it cleavage-deficient, and purified the GyrAY123F·GyrBA (AF·BA) heterodimer by tandem-affinity purification (see Methods; Supplementary Figure S1). AF·BA on its own should not display supercoiling activity because it lacks the second GyrB subunit and a functional N-gate. Therefore, any activity observed in the absence of additional GyrB must be due to contaminations. The tandem-affinity-purified AF·BA heterodimer showed very little background supercoiling activity in the presence of ATP without addition of external GyrB (Figure 2A, Supplementary Figure S3) that is caused by traces of (BA)2 that are not removed despite the rigorous purification procedure (see below). We next added GyrB to AF·BA to reconstitute complete heterotrimeric gyrase, B·AF·BA, that harbors only one catalytic tyrosine. While GyrB on its own did not display any supercoiling activity (Figure 2A, Supplementary Figure S3), B·AF·BA showed robust negative supercoiling activity and fully supercoiled DNA (Figure 2A, Supplementary Figures S3 and S4), although more slowly and less efficiently compared to wild-type gyrase (A2B2; Supplementary Figure S4). To determine the difference in supercoiling rates by gyrase with two and one catalytic tyrosine(s), we performed DNA supercoiling reactions with heterotetrameric gyrase (A2B2 and B·A·AF·B), and quantified the supercoiled DNA at different time points by densitometry of ethidium bromide-stained gels (Supplementary Figure S4). DNA gyrase with two tyrosines supercoiled DNA processively with a rate constant of product formation of k = 0.027 ± 0.003 s−1. Gyrase with only one catalytic tyrosine supercoiled DNA less processively. The rate constant of supercoiling was reduced 9-fold to k = 0.003 ± 0.0002 s−1.
Figure 2.
Gyrase containing one catalytic tyrosine negatively supercoils DNA. (A) ATP-dependent DNA supercoiling by B. subtilis gyrase containing one (F·Y) or two catalytic tyrosine(s) (Y·Y). Gyrase with only one tyrosine (B·AF·BA) shows similar ATP-dependent DNA supercoiling activity to wildtype gyrase (A2B2). The cartoons below the gel indicate the subunits present, the yellow Y marks the catalytic tyrosine(s). Control reactions containing the individual subunits (B, A2) show no supercoiling activity; the AF·BA heterodimer shows only little supercoiling activity (see main text). Concentrations were 50 nM AF·BA and 400 nM B, or 100 nM A and 400 nM B (gyrase:DNA ratio 2.5:1). (B) ATP-dependent DNA supercoiling by E. coli gyrase containing one (F·Y) catalytic tyrosine. Dimeric gyrase (BA)2 shows less ATP-dependent supercoiling activity than wild-type gyrase (A2B2; reaction time 30 min). Gyrase with only one catalytic tyrosine (B·AF·BA) shows DNA supercoiling activity, whereas no supercoiling is observed with AF·BA. Concentrations were 200 nM AF·BA and 800 nM B (gyrase:DNA ratio 5:1). -sc: negatively supercoiled DNA; rel: relaxed DNA; start: DNA substrate; A2B2: heterotetrameric gyrase; AF: cleavage-deficient GyrA subunit (B. subtilis: GyrAY123F, E. coli: GyrAY122F); B: GyrB; BA: GyrBA.
The observed supercoiling activity of B. subtilis gyrase with one catalytic tyrosine can unambiguously be assigned to the BA·AF·B species
DNA supercoiling by gyrase with one tyrosine that cannot catalyze double-strand cleavage cannot occur according to the strand passage mechanism. To be able to unambiguously assign the observed supercoiling activity to B·AF·BA, we ruled out (a) subunit exchange and in situ formation of active (BA)2 dimers, (b) contaminations of our preparations with E. coli gyrase subunits, and (c) artefacts of our construct design. To test for subunit exchange and formation of (BA)2 during the supercoiling reaction, we prepared a AF·BA variant containing cysteines for fluorescent labeling at the DNA-gate of both GyrA subunits (D145C), and labeled the heterodimer either with donor fluorophores or with acceptor fluorophores (Supplementary Figure S5). The donor-labeled and acceptor-labeled dimers were mixed in equimolar ratio, GyrB was added to reconstitute gyrase, and supercoiling reactions were performed. The FRET efficiency distribution was analyzed by single molecule FRET before and after supercoiling. Subunit exchange would generate donor/acceptor-labeled AF·BA, AF and BA dimers with high FRET efficiency (Supplementary Figure S5) (33). However, no high FRET species was observed, ruling out subunit exchange and in situ formation of (BA)2 on the time scale of the supercoiling experiments. The same was observed in analogous experiments with heterotetrameric gyrase (A2B2; Supplementary Figure S5). The observed negative supercoiling activity of B·AF·BA must thus be caused by the heterotrimeric gyrase with only one catalytic tyrosine. This conclusion is further supported by control experiments with BE44QA·AF, in which the ATPase activity of GyrB is abolished due to the E44Q mutation (32). Subunit exchange of BE44QA·AF·B would generate (BE44QA)2 and B2(AF)2 species, both of which are supercoiling-deficient (Supplementary Figure S5). Any supercoiling activity of BE44QA·AF·B therefore has to be ascribed to the BE44QA·AF·B species, and cannot be explained by subunit exchange. The purified BE44QA·AF heterodimer showed no detectable supercoiling activity (Supplementary Figure S5), ruling out contaminations of heterodimer purifications by E. coli gyrase. When BE44QA·AF was supplemented with GyrB to constitute complete gyrase (BE44QA·AF·B), negative supercoiling activity was observed (Supplementary Figure S5). This finding demonstrates that one ATP binding and hydrolysis event is sufficient to support DNA supercoiling, in agreement with earlier observations for E. coli gyrase (40). More importantly in this context, it further excludes subunit exchange as the reason for the observed supercoiling activity of gyrase with one catalytic tyrosine. A dimeric BE44QA·BAF, which can only generate supercoiling-deficient (BE44QA)2 and (BAF)2 by subunit exchange (and may contain residual amounts of these species as a contamination), also showed negative supercoiling activity (Supplementary Figure S5), further confirming that gyrase with one functional catalytic site for ATP hydrolysis and one catalytic tyrosine is responsible for the observed negative supercoiling of DNA. Separation of topoisomers by two-dimensional gel electrophoresis confirmed that the product was indeed negatively supercoiled (Supplementary Figure S5). As we have excluded the generation of active gyrase by subunit exchange during our supercoiling experiments, the residual activity of the BA·AF heterodimer (Figure 2) before addition of GyrB can be assigned to traces of (BA)2 that are not removed despite the rigorous purification procedure. Finally, we also tested for supercoiling activity upon addition of external GyrB to (BE44QA)2 (Supplementary Figure S5). Very little supercoiling activity was observed when adding GyrB to this enzyme, which excludes the possibility that the GyrB part of (BE44QA)2 can somehow be displaced by externally added GyrB in our experiments with a fusion protein subunit.
To exclude contaminations of B·AF·BA by E. coli gyrase subunits, we tested for supercoiling activity upon addition of GyrB from E. coli or B. subtilis to AF·BA. Mixing B. subtilis GyrA and E. coli GyrB gives rise to very little supercoiling activity. In contrast, E. coli GyrA forms active gyrase heterotetramers with B. subtilis GyrB (Supplementary Figure S6). Any contamination of AF·BA with E. coli GyrA would thus give rise to supercoiling activity with B. subtilis and E. coli GyrB. BB. subtilis·AF·BA negatively supercoiled DNA, but BE. coli·AF·BA showed no supercoiling activity (Supplementary Figure S6). Thus, the robust supercoiling activity of B·AF·BA is not caused by contaminations with E. coli GyrA subunits.
Lastly, supercoiling activity of gyrase containing only one tyrosine was also observed with heterotetrameric gyrase (constituted from tandem-affinity-purified AF·A heterodimers and GyrB; see also Supplementary Figure S4) and for an inverse heterotrimeric gyrase where the tyrosine was provided by GyrA (constituted from tandem-affinity-purified A·BAF and GyrB; Supplementary Figure S7). Supercoiling by gyrase with one catalytic tyrosine is thus observed independently of the design of the constructs.
Altogether, we can thus unambiguously assign the observed supercoiling activity of B·AF·BA to gyrase with one catalytic tyrosine.
E. coli gyrase also catalyzes negative DNA supercoiling when only one catalytic tyrosine is present
Gyrases from Gram-positive organisms, such as B. subtilis, lack an insertion in the TOPRIM domain in GyrB that is present in gyrase from Gram-negative bacteria (14,41,42) and contributes to DNA binding and supercoiling (14). To confirm that DNA supercoiling without strand passage is not a unique feature of the Gram-positive gyrase sub-family, we tested DNA supercoiling by E. coli gyrase containing only one catalytic tyrosine (Methods, Supplementary Figure S1). E. coli B·AF·BA also showed ATP-dependent DNA supercoiling activity (Figure 2B, Supplementary Figure S3), although reduced in comparison to wild-type E. coli gyrase that supercoils DNA rapidly and with high processivity (Figure 2B, Supplementary Figure S2). Thus, the potential to catalyze DNA supercoiling with only one catalytic tyrosine might be a more general feature of DNA gyrases.
Gyrase with one catalytic tyrosine cleaves one DNA strand and decreases the linking number by two
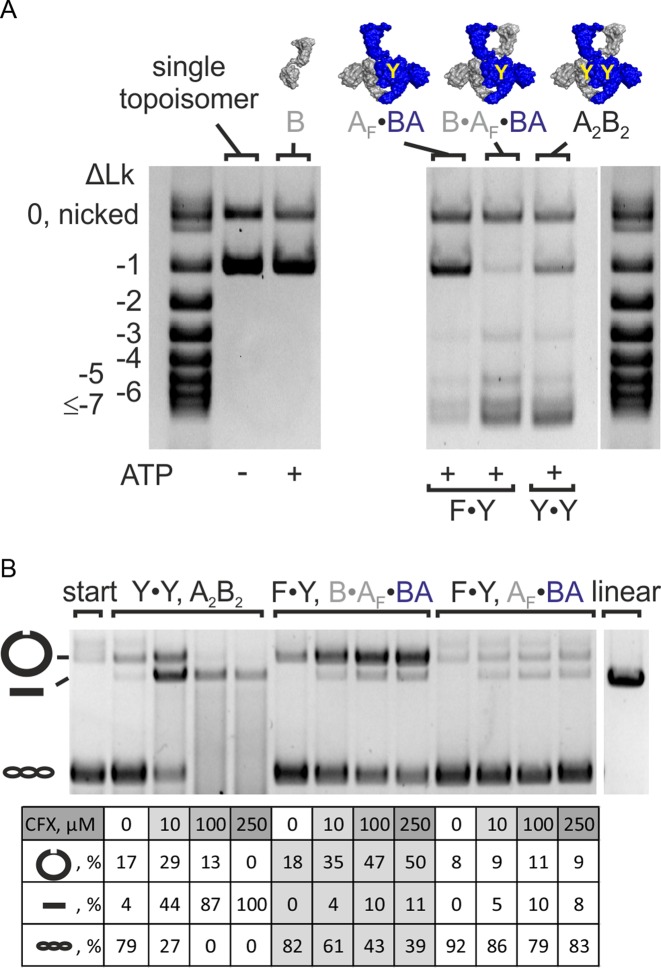
E. coli gyrase decreases the linking number of DNA in increments of two (5). A change in linking number by two is in agreement with negative supercoiling by sign-inversion, i.e. binding of DNA in a positive node (Lk = +1) and inversion to a negative node (Lk = −1; ΔLk = −2) by strand passage. Type I topoisomerases that contain a single catalytic tyrosine cleave only one strand of their DNA substrate, and change the linking number in steps of one (4). Our observation of negative supercoiling by B. subtilis (and E. coli) gyrase containing only one catalytic tyrosine raised the question whether the enzyme introduces one or two supercoils per catalytic cycle. To unambiguously determine the change in linking number, we prepared a single topoisomer as a substrate (Methods; Figure 3A). Strikingly, B. subtilis gyrase reduced the linking number of this plasmid in steps of two, independent of the number of tyrosines present (Figure 3A).
Figure 3.
Change in linking number and DNA cleavage. (A) Supercoiling of a single topoisomer (ΔLk = −1, right) by gyrase containing one (F·Y, B·AF·BA) or two (Y·Y, A2B2) catalytic tyrosine(s). Both enzymes change the linking number in steps of two. DNA supercoiling was performed for 4 min at 37°C. Concentrations were 20 nM GyrB, 20 nM GyrA and 20 nM GyrB (corresponding to 10 nM A2B2) or 25 nM AF·BA and 100 nM GyrB (corresponding to 25 nM BA·AF·B), respectively. Gyrase with two (A2B2) and one catalytic tyrosine(s) (B·AF·BA) introduces negative supercoils in steps of two. The AF·BA heterodimer serves as a negative control. AF·BA does not supercoil DNA, confirming that the observed supercoiling activity of B·AF·BA is not caused by contaminations in the preparation. (B) DNA cleavage in the presence of CFX. B. subtilis gyrase (A2B2) causes nicking and linearization, corresponding to single- and double-strandP cleavage. Gyrase containing only one catalytic tyrosine (B·AF·BA) shows an accumulation of nicked DNA, in agreement with single-strand cleavage. CFX concentrations are 0–250 μM (arrow). The gyrase:DNA ratio is 5:1. The table below the gel summarizes the fractions of nicked, linear and supercoiled DNA according to densitometric analysis. linear: pUC18 linearized by BamHI; start: DNA substrate; A2B2: heterotetrameric gyrase; A2: GyrA dimer; AF: cleavage-deficient GyrA subunit (GyrAY123F); B: GyrB; BA: GyrBA fusion protein; BAΔCTD: GyrBA fusion protein lacking the GyrA CTD; ΔLk: linking number difference.
We next tested whether DNA supercoiling by gyrase carrying only one catalytic tyrosine is achieved by single-strand cleavage. Quinolones inhibit gyrase activity by trapping covalently linked topoisomerase/DNA complexes (43–45). In the presence of ciprofloxacin (CFX), gyrase lacking catalytic tyrosines shows neither nicking nor linearization, confirming cleavage deficiency (Supplementary Figure S8) (36). B. subtilis gyrase nicks plasmid DNA at low CFX concentrations, but linearizes DNA at high CFX concentration (Figure 3B), indicating that the individual cleavage events are not tightly coupled. The B. subtilis AF·BA heterodimer already shows low levels of DNA nicking and linearization in the absence of GyrB, possibly due to single- and double-strand cleavage by (BA)2 traces in the preparation (see above). The fraction of linear DNA due to double-strand cleavage saturates at ∼10%. When GyrB is added to form B·AF·BA, the amount of nicked DNA is strongly increased (Figure 3B, Supplementary Figure S8). At the same time, the amount of double-strand cleavage is not affected by the addition of GyrB, and remains below 11%, at the level of double-strand cleavage by (BA)2 traces. Our results therefore confirm that gyrase with only one catalytic tyrosine performs single-strand cleavage. Thus, negative supercoiling of DNA with linking number changes in steps of two can be achieved with one catalytic tyrosine and cleavage of only one strand in the G-segment. Negative supercoiling in the absence of double-strand cleavage cannot follow the strand-passage model (5,19), which suggests the existence of an alternative pathway for DNA supercoiling by gyrase with only one catalytic tyrosine.
Gyrase with one and two catalytic tyrosines undergoes the same cascade of DNA- and nucleotide-induced conformational changes
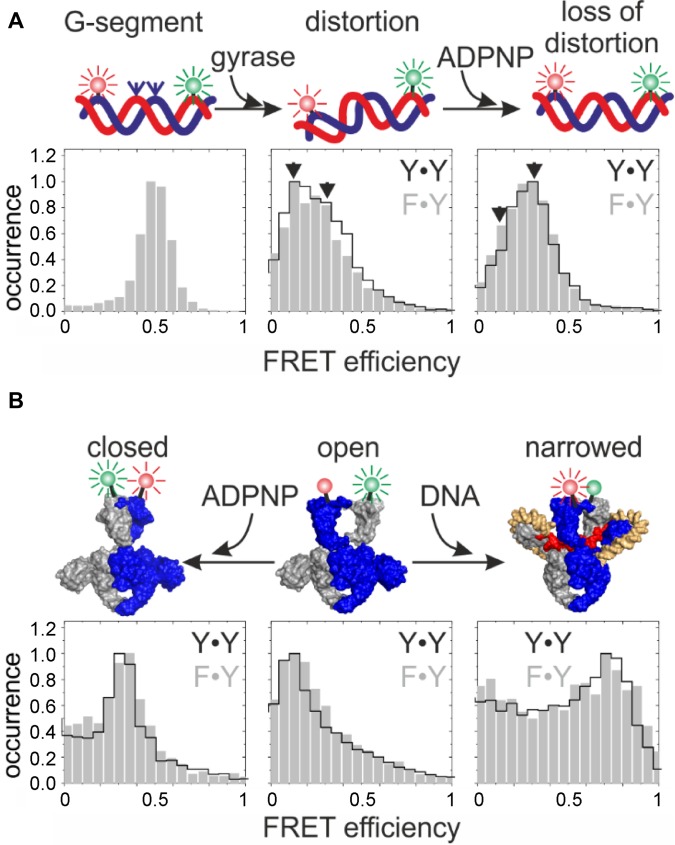
To further investigate similarities and possible differences in the mechanism of negative supercoiling by gyrase containing one or two catalytic tyrosine(s), we probed the sequence of conformational changes at the beginning of the supercoiling cycle (reviewed in (46)) for both enzymes. This sequence of events consists of DNA binding and distortion (33), a DNA-induced upward movement of the CTDs (47), DNA-induced narrowing of the N-gate (31), and nucleotide-induced N-gate closure (31,48). The conformation of the G-segment bound to gyrase was investigated in smFRET experiments using a 60 bp DNA (33) as a model G-segment (Figure 4A). This DNA contains a preferred gyrase binding site in the center of the sequence (49), flanked by donor and acceptor fluorophores (33). We have previously established that the 60 bp DNA exists in two different conformations when bound to gyrase (33). One state is moderately distorted from regular B-form geometry, the second is severely distorted ((33); Figure 4A). The severely distorted conformation is not induced by cleavage-deficient gyrase (33), suggesting that this state represents the G-segment in a conformation ready for cleavage or already cleaved. smFRET histograms of the 60 bp DNA bound to gyrase containing only one catalytic tyrosine is very similar to the histogram of 60 bp DNA bound to gyrase containing two catalytic tyrosines ((33); Figure 4A). The appearance of the severely distorted conformation of DNA (FRET efficiency 0.13; (33)) bound to gyrase with one tyrosine that can only cleave one of the DNA strands suggests that this state reflects DNA with one strand cleaved or prepared for cleavage. We have previously shown that addition of the non-hydrolyzable ATP analog ADPNP to the gyrase·DNA complex reduces the population of the severely distorted DNA bound to gyrase (33). The same effect was observed with gyrase containing one tyrosine (Figure 4A), indicating that the conformation of the G-segment before and after ATP binding and N-gate closure is similar for DNA bound to gyrase with one or two catalytic tyrosine(s).
Figure 4.
Conformational changes at the beginning of the catalytic cycle are independent of the number of tyrosines present. (A) Conformational changes of the DNA, probed by smFRET with a 60 bp DNA that contains a preferred cleavage site for B. subtilis gyrase (49), flanked by donor and acceptor fluorophores (green and red spheres). smFRET histograms of DNA in the presence of gyrase containing two catalytic tyrosines (A2B2) are outlined in black, histograms in the presence of gyrase containing one tyrosine (B·AF·BA) are shown in gray. Left: DNA only; center: DNA distortion upon binding to gyrase, right: loss of distortion upon ADPNP binding. Gyrase induces identical DNA conformations, irrespective of the number of tyrosines present. (B) DNA- and nucleotide-induced conformational changes of gyrase. N-gate conformation was probed in smFRET experiments using BAF·BA (one tyrosine) and (BA)2 (two tyrosines), carrying a S7C mutation for fluorescent labeling (31). FRET histograms show identical conformations for the proteins in the absence of DNA or nucleotide (center), and DNA-induced N-gate narrowing (right) and nucleotide-induced N-gate closure (left) independent of the number of tyrosines present.
When the G-segment binds to the DNA-gate of gyrase, flanking regions will extend to the CTDs, and cause their upward movement (47). This displacement of the CTDs is observed with cleavage-deficient gyrase that lacks both catalytic tyrosines, and is therefore independent of DNA cleavage. We can thus infer that gyrase with only one tyrosine also shows DNA-induced CTD displacement.
Wrapping of DNA flanking the G-segment around the CTDs induces narrowing of the N-gate (31). smFRET experiments monitoring N-gate conformation were performed with BA dimers to exclude dissociation of the active enzyme in smFRET experiments (31). (BA)2 carrying an S7C mutation for donor- and acceptor labeling at the N-gate shows low FRET efficiencies (Figure 4B) corresponding to an open N-gate (31). In the presence of plasmid DNA, a high-FRET state appears, previously assigned as gyrase with a narrowed N-gate ((31); Figure 4B). BAF·BA showed the same transition from low FRET (open N-gate) to high FRET (narrowed N-gate) upon DNA binding (Figure 4B). Thus, DNA-induced N-gate narrowing occurs in gyrase containing one or two catalytic tyrosine(s).
Finally, nucleotide-induced closure of the N-gate captures the T-segment in the upper cavity of gyrase (10,11). N-gate closure was also observed as an increase in FRET efficiency of BAF·BA (one tyrosine) and (BA)2 (two tyrosines; Figure 4B), consistent with our previous observation that nucleotide-induced closing of the N-gate is independent of DNA cleavage (31). Overall, the cascade of DNA- and nucleotide-induced conformational changes preceding a possible strand passage event is thus identical for gyrase containing one or two catalytic tyrosine(s).
Role of the CTDs for DNA supercoiling in the absence of double-strand cleavage
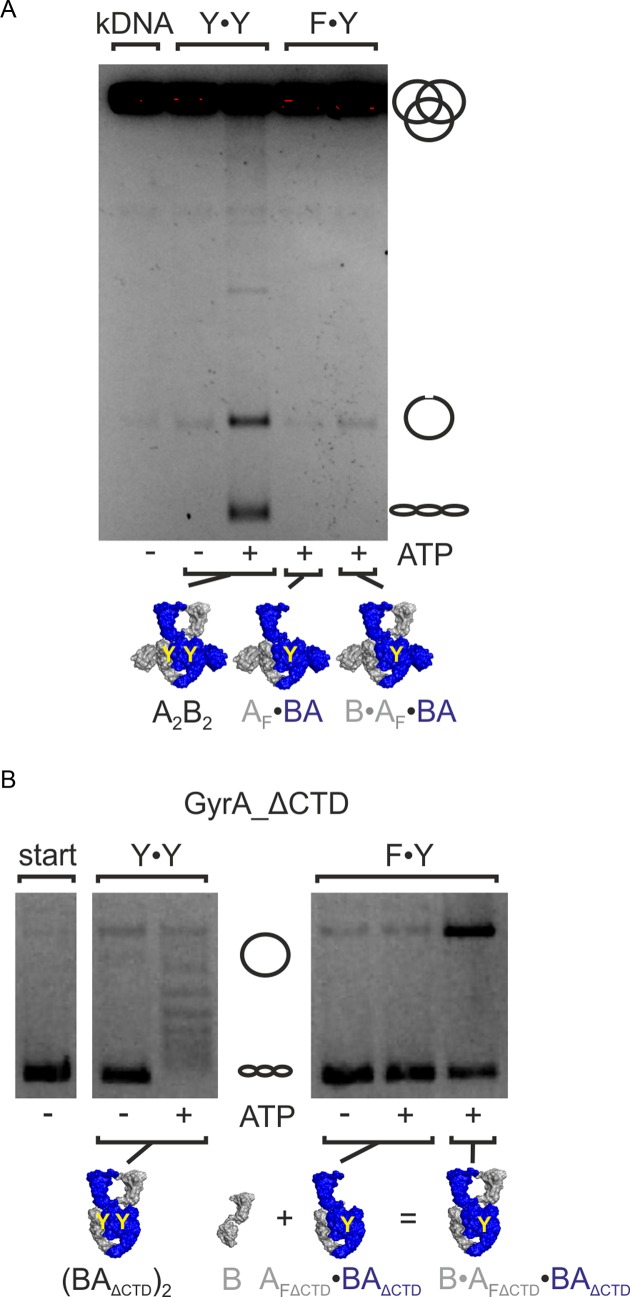
Removal of the CTDs abrogates the negative supercoiling activity of gyrase (26,33). To generate a gyrase lacking the CTDs and containing only one catalytic tyrosine, we purified an AF_ΔCTD·BAΔCTD heterodimer, and supplied GyrB in trans. B·AF_ΔCTD·BAΔCTD also does not show any ATP-dependent negative supercoiling activity (Figure 5A). In contrast, a B·AF·BAΔCTD variant of gyrase with one catalytic tyrosine and only one of the two CTDs was able to negatively supercoil DNA (Figure 5B, Supplementary Figure S9). Supercoiling occurred with a change in linking number in steps of two (Figure 5B). The AF·BAΔCTD heterodimer lacking one GyrB subunit did not show any supercoiling activity, excluding contaminations by E. coli gyrase. Other possible contaminations from the preparation, namely (AF)2 and (BAΔCTD)2, are supercoiling-deficient. Therefore, the negative supercoiling activity must be caused by B·AF·BAΔCTD that has only one catalytic tyrosine and one CTD. A linking number change in steps of two can thus be achieved without strand passage and involving only one CTD.
Figure 5.
Gyrase with one catalytic tyrosine and one CTD negatively supercoils DNA and changes the linking number in steps of two. (A) ATP-dependent negative supercoiling of DNA by gyrase lacking the CTDs that contains only one catalytic tyrosine (F·Y; B·AFΔCTD·BAΔCTD). GyrB, AFΔCTD·BAΔCTD or B·AFΔCTD·BAΔCTD do not show ATP-dependent negative supercoiling activity. Wildtype gyrase (A2B2, Y·Y) is used as a positive control. DNA supercoiling reactions were performed for 15 min at 37°C. Concentrations were 400 nM GyrB and 200 nM AFΔCTD·BAΔCTD (corresponding to 200 nM BAΔCTD·AFΔCTD·B) or 200 nM GyrA (corresponding to 100 nM A2B2). (B) Supercoiling of a single topoisomer (ΔLk = -1) by gyrase (F·Y, B·AF·BAΔCTD) containing one catalytic tyrosine and one CTD. The enzyme changes the linking number in steps of two. DNA supercoiling was performed for 15 min at 37°C. Concentrations were 400 nM GyrB and 200 nM AF·BAΔCTD (corresponding to 200 nM BAΔCTD·AF·B). The AF·BAΔCTD heterodimer serves as a negative control. AF·BAΔCTD does not supercoil DNA, confirming that the observed supercoiling activity of B·AF·BAΔCTD is not caused by contaminations in the preparation.
ATP-dependent DNA relaxation and decatenation require two catalytic tyrosines
We finally tested if other type II topoisomerase reactions, namely ATP-dependent DNA relaxation and decatenation, can also be catalyzed without double-strand cleavage and strand passage. Gyrase with two tyrosines catalyzed decatenation of kDNA (Figure 6A). In contrast, gyrase containing only one tyrosine failed to decatenate this DNA, suggesting that double-stranded DNA cleavage and strand passage are required for the decatenation reaction. To test the dependence of ATP-dependent DNA relaxation on strand passage, we used gyrase lacking the CTDs, which behaves as a topoisomerase II (26,31,47) (Figure 6B, Supplementary Figure S10). B·AF_ΔCTD·BAΔCTD did not show any ATP-dependent DNA relaxation (Figure 6B). Consequently, ATP-dependent relaxation of DNA also requires cleavage of both DNA strands and strand passage. Gyrase thus appears to be able to use different mechanisms for different topoisomerase reactions: Decatenation and ATP-dependent DNA relaxation by gyrase lacking the CTDs requires stand passage, but strand passage can be bypassed in ATP-dependent negative supercoiling of DNA when only one tyrosine is present.
Figure 6.
ATP-dependent DNA relaxation and decatenation. (A) ATP-dependent decatenation of kDNA by gyrase. Gyrase with two catalytic tyrosines (Y·Y, A2B2) catalyzes decatenation in the presence of ATP, but not in its absence. Gyrase containing only one catalytic tyrosine (B·AF·BA) shows no or very little decatenation activity. The AF·BA heterodimer control shows no significant background activity. Thus, decatenation requires two catalytic tyrosines and strand passage. (B) ATP-dependent relaxation by gyrase lacking the CTDs. Gyrase lacking the CTDs and containing one catalytic tyrosine (F·Y; B·AFΔCTD·BAΔCTD) shows no ATP-dependent DNA relaxation activity. Gyrase lacking the CTDs but containing two catalytic tyrosines (Y·Y, (BAΔCTD)2) serves as a positive control. Concentrations were 500 nM AF·BA and 1 μM B, or 500 nM (BAΔCTD)2. kDNA: catenated DNA; start: DNA substrate; A2B2: heterotetrameric gyrase; AF: cleavage-deficient GyrA subunit (GyrAY123F), AFΔCTD: cleavage-deficient GyrA carrying the Y123F mutation and lacking the C-terminal domain; B: GyrB; BA: GyrBA-fusion protein; BAΔCTD: GyrBA lacking the CTDs.
DISCUSSION
Negative supercoiling of DNA by a nicking-closing mechanism
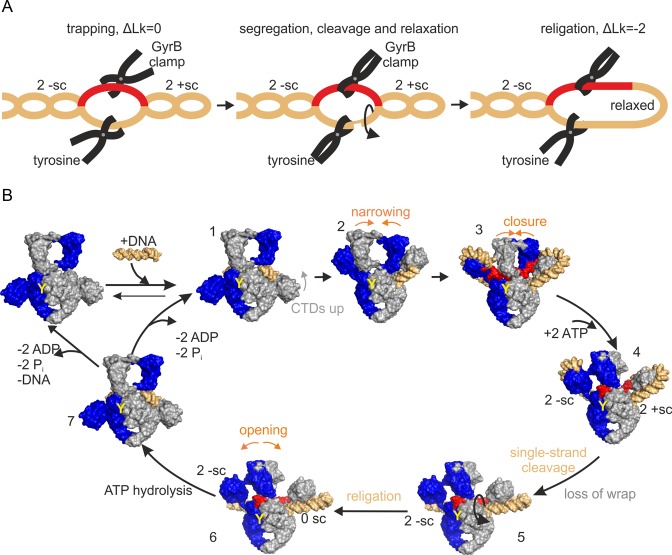
We have shown here that one catalytic tyrosine is sufficient for negative supercoiling of DNA by gyrase. This finding is in contradiction with the well-accepted strand passage and sign-inversion mechanism (5,19), in which a positive supercoil is captured and converted into a negative supercoil by passage of one duplex through the other (+1 → −1). Our results demonstrate that negative DNA supercoiling can be achieved by nicking and closing of a single DNA strand only, yet supercoils are introduced in steps of two. Conceptual models based on nicking-closing have been considered early on (8,50,51), but have been dismissed because they failed to explain the linking number change in steps of two. Negative supercoiling in increments of two by cleaving only a single strand can be achieved through a sequence of trapping, segregation, and selective relaxation of two positive supercoils by gyrase (+2 → 0; Figure 7A). Capture of two positive supercoils in a covalently closed DNA is accompanied by the formation of two compensatory negative supercoils in the rest of the DNA substrate. To achieve net DNA supercoiling, the resulting topological domains of the DNA have to be segregated, such that the negative and positive supercoils are fixed at both ends. Finally, the domain with the positive supercoils has to be liberated at one end to allow for relaxation by rotation, while the negative supercoils are maintained. This reaction sequence would lead to an overall decrease in linking number by two (Figure 7A).
Figure 7.
Negative DNA supercoiling by a nicking-closing mechanism. (A) Concept of negative DNA supercoiling by trapping, segregation and selective relaxation of two positive supercoils. Left: Trapping of two positive supercoils (2 +sc) leads to formation of two compensatory negative supercoils (2 -sc) in the rest of the DNA. Center: N-gate closure and nicking of the DNA by the catalytic tyrosine lead to fixation and segregation of the two topological domains. Release of the wrap liberates the DNA containing the positive supercoils, and they will relax by rotation (arrow). Right: Closing of the nick (religation) fixes the change in linking number (ΔLk) by −2. (B) Model for negative DNA supercoiling by DNA gyrase. (1): DNA binding, (2) CTD movement, (3) N-gate narrowing, positioning of T-segment, (4) ATP binding, N-gate closure, T-segment trapping and G-segment nicking establish topological segregation of two positive supercoils on one CTD, (5) ATP binding induces release of the wrap, freeing one end of the topological domain with the positive supercoils, and allowing for relaxation (arrow), (6), religation fixes the change in linking number by −2, and (7) ATP hydrolysis, N-gate opening, followed by product release, reset gyrase for subsequent catalytic cycles. Red: T-segment; orange: G-segment and rest of DNA; GyrA and GyrB subunits are depicted in gray and blue; Y: catalytic tyrosine that carries out nicking.
Combined with our current knowledge on conformational changes at the beginning of DNA supercoiling by gyrase, the concept of DNA supercoiling by nicking-closing can be integrated into the following model for DNA supercoiling by gyrase that carries only one catalytic tyrosine (Figure 7B): In the first step, the G-segment of the DNA binds to the DNA-gate. Contacts of flanking DNA regions with the CTDs induce their upward movement (step 1) (47), and the bound G-segment is distorted (step 2) (33). Wrapping of the neighboring DNA around the CTDs then induces N-gate narrowing (step 3) (31). Up to this step, the nicking-closing mechanism is formally identical to the strand passage mechanism. ATP binding, closing of the N-gate and cleavage of one DNA strand complete the topological segregation of positive supercoils on the enzyme, and compensating negative supercoils in the rest of the plasmid. The region containing the negative supercoils is now fixed at both ends, by GyrB dimerization and fixation of the T-segment, and by the covalent link of the 5′-end of the cleaved strand with the catalytic tyrosine, which also prevents rotation around the complementary region of the non-cleaved strand. On the opposite side of the nick, the domain with positive supercoils is prevented from rotating due to the fixation of the T-segment and by interactions with the CTD. ATP binding causes release of the wrap (52) (step 5). The topological domain of the DNA, bound to the GyrA subunit that has nicked the DNA, contains the negative supercoils, and remains fixed at both ends (covalent link to catalytic tyrosine/T-segment). The DNA containing the positive supercoils is still fixed on one end (T-segment), but is now free to rotate around the non-cleaved strand at the DNA-gate, and relaxes in an energetically downhill reaction, driven by its torsional energy. Religation will then close the nick in the G-segment (step 6). Once both ATP molecules have been hydrolyzed, re-opening of the N-gate (31) liberates the T-segment and relieves topological segregation. After product dissociation, gyrase is ready for further catalytic cycles (step 7→1).
DNA supercoiling by nicking-closing requires an intricate cooperation of the gyrase subunits. DNA supercoiling by nicking-closing can be performed by gyrase that contains only one tyrosine and one functional ATPase site. The nicking-closing model requires the capture of two positive supercoils by one CTD. In agreement with this prediction, we have shown that gyrase with only one catalytic tyrosine that has only one CTD still negatively supercoils DNA by changing its linking number in steps of two. Thus, one CTD is sufficient for DNA wrapping and for aligning the DNA for negative supercoiling.
Negative supercoiling of DNA by nicking-closing: an alternative pathway or a more general mechanism?
It is unclear if the nicking-closing mechanism is an alternative pathway that is only used once double-stranded cleavage is not possible, or if wild-type gyrase with two tyrosines can also use this mechanism. The supercoiling rate of gyrase with one tyrosine is 9-fold lower compared to wild-type gyrase, which might point to different mechanisms being used by these enzymes. On the other hand, it is conceivable that both enzymes use the nicking-closing mechanism, but gyrase with only one tyrosine is a less efficient catalyst. The probability of a DNA cleavage event to occur is reduced 2-fold if only one tyrosine is present, which would rationalize a 2-fold difference in supercoiling for mere statistical reasons. The exchange of catalytic tyrosines for phenylalanines not only abolishes DNA cleavage, but also increases DNA affinity, and leads to a failure of gyrase to distort the bound DNA (33), pointing to an altered mode of DNA binding. Tighter DNA binding may actually be counterproductive for rearrangements during supercoiling, and may affect cleavage and/or religation rates, and thereby further contribute to a reduced supercoiling velocity by gyrase with one tyrosine.
Several lines of evidence suggest that DNA supercoiling by nicking-closing may be more-widespread. This hypothesis is supported by (a) the identical sequence of DNA- and ATP-induced conformational changes of gyrase at the beginning of the supercoiling cycle, and (b) by the identical conformation of (severely distorted) DNA bound to gyrase, independent of the number of tyrosines present, and (c) by the capacity of both B. subtilis and E. coli gyrase to negatively supercoil DNA with one catalytic tyrosine only. The strongest evidence for the strand passage mechanism is provided by the observed loss of DNA supercoiling activity upon cross-linking of the DNA- and C-gates in E. coli gyrase, either with very short cross-linkers or directly by disulfide bonds (53,54). However, such a tight cross-linking will severely restrict gate dynamics, and thus presumably also prevent rotation of DNA in a gyrase/DNA complex that undergoes supercoiling by nicking-closing. Hence, the observed inhibition of DNA supercoiling by cross-linking does not strictly exclude supercoiling by nicking-closing. Future work will show if the nicking-closing mechanism is a back-up mechanism or might constitute a more widely used alternative pathway for negative DNA supercoiling by gyrase.
Nicking-closing versus strand passage: Different mechanistic plasticity of topo II, topo IV and gyrase
ATP-dependent negative DNA supercoiling, the hallmark reaction of gyrase, can be achieved in the absence of double-strand breaks by gyrase with only one catalytic tyrosine. In contrast, ATP-dependent relaxation of negative DNA supercoils by gyrase lacking the CTDs is only possible when two catalytic tyrosines are present, in accordance with the strand passage mechanism. This result suggests that DNA gyrase can use a different mechanism for DNA supercoiling, but topo II strictly depends on the strand passage mechanism for ATP-dependent relaxation. Mechanistic differences between topo II and gyrase are supported by (a) their different interactions with DNA (wrapping versus no wrapping (18)), (b) different effects of ATP binding on G-segment conformation (loss of DNA distortion with ATP binding for gyrase (33), increased DNA bending for topo II (55)), (c) different effects of ATP binding on DNA cleavage (nucleotide-independent DNA cleavage versus acceleration of DNA cleavage by ATP binding (56)), and (d) possible differences in DNA cleavage itself (coordination of cleavage events, favoring double-strand break formation by topo II (57,58), for gyrase only demonstrated in the presence of gyrase poisons (29)).
Decatenation reactions can only be achieved physically by passage of one duplex through the other (19,59), which is only possible when both strands of the DNA substrate are cleaved. In accordance, we have shown that decatenation by gyrase requires two catalytic tyrosines. Gyrase is a very inefficient decatenase (59), and catenane resolution in vivo is performed by topo IV (22). Our findings imply that decatenation by topo IV also strictly depends on the strand passage mechanism.
Removal of the CTDs abolishes the ATP-dependent negative DNA supercoiling activity of gyrase, and of gyrase with one tyrosine that performs nicking-closing. At the same time, it renders gyrase more efficient in catenane resolution (26,47) and confers ATP-dependent DNA relaxation activity, both of which strictly requires strand passage. The CTDs could thus be the distinctive elements that switch the common type II topoisomerase core from strand passage (topo II, topo IV) to the possibility of using a nicking-closing mechanism (gyrase). DNA supercoiling mediated by nicking-closing would circumvent the formation of double-strand DNA breaks and avoids life-threatening consequences for genome stability. The discovery of this alternative mechanism may have important ramifications for the identification and design of specific, mechanism-based gyrase inhibitors.
Supplementary Material
Acknowledgments
We thank Pavel Lulchev for providing positively supercoiled DNA, Andreas Schmidt and Jessica Guddorf for excellent technical assistance, Simon Hartmann and Markus Rudolph for critical reading of the manuscript, Tony Maxwell and Andy Bates for discussions, and Valerie Lamour for providing the Cα coordinates of T. thermophilus gyrase.
Author contributions: A.G., D.W. and D.K. designed research; A.G. and D.W. performed research; A.G., D.W. and D.K. analyzed data; and A.G. and D.K. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [KL1153/5-1]. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen S.H., Chan N.L., Hsieh T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 2.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick F.H. Linking numbers and nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 1976;73:2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J.C. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 5.Brown P.O., Cozzarelli N.R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 6.Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 7.Gellert M., Mizuuchi K., O'Dea M.H., Nash H.A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuuchi K., O'Dea M.H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl. Acad. Sci. U.S.A. 1978;75:5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta R., Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 10.Tingey A.P., Maxwell A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996;24:4868–4873. doi: 10.1093/nar/24.24.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams N.L., Howells A.J., Maxwell A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001;306:969–984. doi: 10.1006/jmbi.2001.4468. [DOI] [PubMed] [Google Scholar]
- 12.Morais Cabral J.H., Jackson A.P., Smith C.V., Shikotra N., Maxwell A., Liddington R.C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 13.Fu G., Wu J., Liu W., Zhu D., Hu Y., Deng J., Zhang X.E., Bi L., Wang D.C. Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoeffler A.J., May A.P., Berger J.M. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 2010;38:7830–7844. doi: 10.1093/nar/gkq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett K.D., Shultzaberger R.K., Berger J.M. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramlinger V.M., Hiasa H. The ‘GyrA-box’ is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 2006;281:3738–3742. doi: 10.1074/jbc.M511160200. [DOI] [PubMed] [Google Scholar]
- 17.Ward D., Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu L.F., Wang J.C. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 19.Mizuuchi K., Fisher L.M., O'Dea M.H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papillon J., Menetret J.F., Batisse C., Helye R., Schultz P., Potier N., Lamour V. Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type IIA DNA topoisomerase. Nucleic Acids Res. 2013;41:7815–7827. doi: 10.1093/nar/gkt560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt B.H., Osheroff N., Berger J.M. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol. 2012;19:1147–1154. doi: 10.1038/nsmb.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H., Marians K.J. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng H., Marians K.J. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 24.Hsieh T.J., Farh L., Huang W.M., Chan N.L. Structure of the topoisomerase IV C-terminal domain: a broken beta-propeller implies a role as geometry facilitator in catalysis. J. Biol. Chem. 2004;279:55587–55593. doi: 10.1074/jbc.M408934200. [DOI] [PubMed] [Google Scholar]
- 25.Barnes M.H., LaMarr W.A., Foster K.A. DNA gyrase and DNA topoisomerase of Bacillus subtilis: expression and characterization of recombinant enzymes encoded by the gyrA, gyrB and parC, parE genes. Protein Expr. Purif. 2003;29:259–264. doi: 10.1016/s1046-5928(03)00068-8. [DOI] [PubMed] [Google Scholar]
- 26.Kampranis S.C., Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbett K.D., Schoeffler A.J., Thomsen N.D., Berger J.M. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Deweese J.E., Burgin A.B., Osheroff N. Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIalpha. Biochemistry. 2008;47:4129–4140. doi: 10.1021/bi702194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison A., Cozzarelli N.R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979;17:175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- 30.Sugino A., Higgins N.P., Cozzarelli N.R. DNA gyrase subunit stoichiometry and the covalent attachment of subunit A to DNA during DNA cleavage. Nucleic Acids Res. 1980;8:3865–3874. doi: 10.1093/nar/8.17.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gubaev A., Klostermeier D. DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14085–14090. doi: 10.1073/pnas.1102100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottler T., Klostermeier D. Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J. Mol. Biol. 2007;367:1392–1404. doi: 10.1016/j.jmb.2007.01.055. [DOI] [PubMed] [Google Scholar]
- 33.Gubaev A., Hilbert M., Klostermeier D. The DNA Gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13278–13283. doi: 10.1073/pnas.0902493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Horowitz D.S., Wang J.C. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 1987;262:5339–5344. [PubMed] [Google Scholar]
- 36.Critchlow S.E., Maxwell A. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry. 1996;35:7387–7393. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 37.Lanz M.A., Klostermeier D. The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 2012;40:10893–10903. doi: 10.1093/nar/gks852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreou A.Z., Klostermeier D. Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 2012;511:75–109. doi: 10.1016/B978-0-12-396546-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 39.Gubaev A., Klostermeier D. In: RNA Structure and Folding. Klostermeier D, Hammann C, editors. Berlin: de Gruyter; 2013. pp. 181–213. [Google Scholar]
- 40.Kampranis S.C., Maxwell A. Hydrolysis of ATP at only one GyrB subunit is sufficient to promote supercoiling by DNA gyrase. J. Biol. Chem. 1998;273:26305–26309. doi: 10.1074/jbc.273.41.26305. [DOI] [PubMed] [Google Scholar]
- 41.Chatterji M., Unniraman S., Maxwell A., Nagaraja V. The additional 165 amino acids in the B protein of Escherichia coli DNA gyrase have an important role in DNA binding. J. Biol. Chem. 2000;275:22888–22894. doi: 10.1074/jbc.M001047200. [DOI] [PubMed] [Google Scholar]
- 42.Costenaro L., Grossmann J.G., Ebel C., Maxwell A. Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure. 2007;15:329–339. doi: 10.1016/j.str.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Gellert M., Mizuuchi K., O'Dea M.H., Itoh T., Tomizawa J.I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U.S.A. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hallett P., Maxwell A. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob. Agents Chemother. 1991;35:335–340. doi: 10.1128/aac.35.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilstermann A.M., Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 46.Gubaev A., Klostermeier D. The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair (Amst) 2014;16:23–34. doi: 10.1016/j.dnarep.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Lanz M.A., Klostermeier D. Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic Acids Res. 2011;39:9681–9694. doi: 10.1093/nar/gkr680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gubaev A., Klostermeier D. Potassium ions are required for nucleotide-induced closure of gyrase N-gate. J. Biol. Chem. 2012;287:10916–10921. doi: 10.1074/jbc.M111.308247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bashkirov V.I., Zvingila D.J. Sequence specificity of Bacillus subtilis DNA gyrase in vivo. Genetica. 1991;85:3–12. doi: 10.1007/BF00056101. [DOI] [PubMed] [Google Scholar]
- 50.Sugino A., Peebles C.L., Kreuzer K.N., Cozzarelli N.R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U.S.A. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L.F., Wang J.C. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2098–2102. doi: 10.1073/pnas.75.5.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kampranis S.C., Maxwell A. Conformational changes in DNA gyrase revealed by limited proteolysis. J. Biol. Chem. 1998;273:22606–22614. doi: 10.1074/jbc.273.35.22606. [DOI] [PubMed] [Google Scholar]
- 53.Williams N.L., Maxwell A. Locking the DNA gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry. 1999;38:14157–14164. doi: 10.1021/bi991478m. [DOI] [PubMed] [Google Scholar]
- 54.Williams N.L., Maxwell A. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999;38:13502–13511. doi: 10.1021/bi9912488. [DOI] [PubMed] [Google Scholar]
- 55.Lee S., Jung S.R., Heo K., Byl J.A., Deweese J.E., Osheroff N., Hohng S. DNA cleavage and opening reactions of human topoisomerase IIalpha are regulated via Mg2+-mediated dynamic bending of gate-DNA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2925–2930. doi: 10.1073/pnas.1115704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller-Planitz F., Herschlag D. Interdomain communication in DNA topoisomerase II: DNA binding and enzyme activation. J. Biol. Chem. 2006;281:23395–23404. doi: 10.1074/jbc.M604119200. [DOI] [PubMed] [Google Scholar]
- 57.Mueller-Planitz F., Herschlag D. DNA topoisomerase II selects DNA cleavage sites based on reactivity rather than binding affinity. Nucleic Acids Res. 2007;35:3764–3773. doi: 10.1093/nar/gkm335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deweese J.E., Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreuzer K.N., Cozzarelli N.R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980;20:245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- 60.Brino L., Urzhumtsev A., Mousli M., Bronner C., Mitschler A., Oudet P., Moras D. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 2000;275:9468–9475. doi: 10.1074/jbc.275.13.9468. [DOI] [PubMed] [Google Scholar]
- 61.Bax B.D., Chan P.F., Eggleston D.S., Fosberry A., Gentry D.R., Gorrec F., Giordano I., Hann M.M., Hennessy A., Hibbs M., et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 62.Hsieh T.J., Yen T.J., Lin T.S., Chang H.T., Huang S.Y., Hsu C.H., Farh L., Chan N.L. Twisting of the DNA-binding surface by a beta-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 2010;38:4173–4181. doi: 10.1093/nar/gkq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.