Abstract
Genome-wide mechanisms that coordinate expression of subsets of functionally related genes are largely unknown. Recent studies show that receptor tyrosine kinases and components of signal transduction cascades including the extracellular signal-regulated protein kinase (ERK), once thought to act predominantly in the vicinity of plasma membrane and in the cytoplasm, can be recruited to chromatin encompassing transcribed genes. Genome-wide distribution of these transducers and their relationship to transcribing RNA polymerase II (Pol2) could provide new insights about co-regulation of functionally related gene subsets. Chromatin immunoprecipitations (ChIP) followed by deep sequencing, ChIP-Seq, revealed that genome-wide binding of epidermal growth factor receptor, EGFR and ERK pathway components at EGF-responsive genes was highly correlated with characteristic mitogen-induced Pol2-profile. Endosomes play a role in intracellular trafficking of proteins including their nuclear import. Immunofluorescence revealed that EGF-activated EGFR, MEK1/2 and ERK1/2 co-localize on endosomes. Perturbation of endosome internalization process, through the depletion of AP2M1 protein, resulted in decreased number of the EGFR containing endosomes and inhibition of Pol2, EGFR/ERK recruitment to EGR1 gene. Thus, mitogen-induced co-recruitment of EGFR/ERK components to subsets of genes, a kinase module possibly pre-assembled on endosome to synchronize their nuclear import, could coordinate genome-wide transcriptional events to ensure effective cell proliferation.
INTRODUCTION
Mitogen-activated protein kinases (MAPKs) are a family of highly conserved serine/threonine (Ser/Thr) kinases that play an essential role in multiple cellular processes including activation of gene expression in response to a variety of extracellular stimuli. Importantly, altered MAPK signaling has been recognized as a key contributor to inflammation, cancer and other disorders (1). There are multiple MAPK cascades and among those the extracellular signal-regulated protein (ERK1/ERK2) kinase pathways are perhaps the most extensively studied (2). In a canonical view, following stimulation by mitogens, cell surface receptor tyrosine kinase (RTKs), such as epidermal growth factor receptors (EGFR), activate small GTPases of the Ras and Rho family in the vicinity of plasma membrane through specific guanine-nucleotide exchange factors. Ras and Rho, in turn, control the activity of down-stream kinase cascades (e.g. BRAF→MEK→ERK) that reach their targets in multiple subcellular compartments including the nucleus. For example, ERK is translocated to the nucleus where it phosphorylates nuclear proteins including transcription factors (TFs), Pol2, chromatin remodeling complexes, histones and histones modifying enzymes (3). Phosphorylation of these nuclear proteins regulates their functions by altering affinity to nucleic acids, stability, cellular localization, interactions with other proteins together leading to changes in gene expression (4). Remarkably, there is increasing evidence that several RTKs themselves can translocate to the nucleus and take part in transcriptional regulation directly at gene loci (5). In some cases, RTKs and their known interacting partners co-localize along target genes (6). Until now, genome-wide co-localization of nuclear RTKs and their downstream kinases has not been investigated.
Here, for the first time we used ChIP-Seq to simultaneously interrogate constitutive and mitogen-induced genome-wide chromatin binding of EGFR and its downstream ERK pathway components.
MATERIALS AND METHODS
Cell culture
Hela-S3 cells were purchased from American Type Culture Collection and grown in Dulbecco's modified Eagle's medium F-12 Ham media (Sigma; D8437) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin-streptomycin at 50 IU/ml in a humidified 5% CO2 atmosphere at 37°C. Prior to EGF stimulation, cells were starved for 48 h in serum-free medium and then were stimulated for the indicated times with EGF (100 ng/ml) (Sigma; E9644).
Total RNA extraction and quantitative(q)-PCR
Total RNA was extracted from cells using TRIzol® Plus RNA Purification Kit (Invitrogen) followed by on-column PureLink™ DNase (Invitrogen) treatment. RT reaction was performed using Superscript III (Invitrogen) with 1 μg of total RNA according to manufacturer's protocol. qPCRs of cDNA and ChIP samples were carried out using Sensimix SYBR kit (Bioline) on Applied Biosystems 7900HT Fast Real-Time PCR System as described (7). RT-qPCR data were normalized to RPLP0 mRNA expression using ΔΔCt method. Primer pairs used are provided in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP) assay
About 1 × 107 cells were crosslinked with 1% formaldehyde for 10 min followed by formaldehyde quenching with 125 mM glycine for 5 min at room temperature. FBS was added to 5% final concentration prior to harvest to avoid cells aggregation and sticking to pipette tips. After harvest cells were spun down at 1600 × g then washed with 5 ml phosphate buffered saline (PBS), re-spun and cell pellets were stored at −80°C. Frozen cell pellets were resuspended in a 2 ml of hypotonic buffer A [10 mM HEPES, pH 7.9, 2 mM MgCl2, 2 mM KCl and NP-40 0.5% vol/vol] supplemented with protease and phosphatase inhibitors (Thermo; 78441) and suspension was kept for 5 min on ice then spun down at 10 000 × g/4°C for 3 min. Nuclei pellets were resuspended in lysis buffer [12.5 mM Tris–HCl, pH 8.0, 2.5 mM ethylenediaminetetraacetic acid (EDTA) and 0.1% Sodium dodecylsulphate (SDS) vol/vol] containing protease and phosphatase inhibitors (Thermo, 78441). Chromatin was sheared in a Bioruptor Plus (Diagenode) using a 30 s on-off cycle for 15 min at high intensity then centrifuged 12 000 × g for 15 min at 4°C and aliquots were stored at −80°C. ChIP assays were performed using Matrix-ChIP on polypropylene plates (8,9). Alternatively, a modified Fast-ChIP protocol was used for the ChIP-Seq libraries (7). Briefly, a volume of chromatin (20–30 μl) with equivalent of 5 × 105 cells was adjusted to 200 μl with immunoprecipitation (IP) buffer [150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, NP-40 (0.5% vol/vol), Triton X-100 (1.0% vol/vol), (pH 7.6)] containing protease and phosphatase inhibitors (Thermo) and incubated with 5 μg of a given antibody for 30 min in ultrasonic bath (Branson, B3510) at 4°C. Next, the chromatin was cleared by centrifugation at 12 000 × g for 10 min at 4°C and transferred to a new tube containing 20 μl of Dynabeads® Protein A (Life Technologies). Tubes were then rotated at 4°C for 60 min on a rotating platform (15 rotations per minute). Beads were separated on a magnetic stand and washed five times with 800 μl of ice cold IP buffer. DNA was recovered from beads by incubating with 50 μl elution buffer [0.5% SDS, 0.1M NaHCO3, 20mg/ml Proteinase K] at 56°C for 30 min with mixing (1000 rpm) in a ThermoMixer (Eppendorf). Samples were separated on a magnetic stand, transferred to a polymerase chain reaction (PCR) tube and then NaCl was added to final concentration of 0.2M followed by overnight reverse crosslinking at 65°C in a thermocycler. For input preparation, the same amount of chromatin as in ChIP was used. After adjusting to 200 μl with TE pH 8.0 buffer containing 20 mg/ml Proteinase K, input samples were incubated overnight at 65°C in a thermocycler. DNA for ChIP and input samples was cleaned up with ChIP DNA Purification Kit (Active motif; 58 002) according to manufacturer's protocol and DNA concentration was determined with Qubit dsDNA HS Assay Kit (Life Technologies) while the respective size distribution was measured with High Sensitivity DNA Analysis Kit on Bioanalyzer 2100 (Agilent). Antibodies used in ChIP studies were as follows: non-specific rabbit IgG (I-1000, Vector Labs), Pol2 (4H8) (Santa cruz; sc-47701), pEGFR (Tyr845) (Cell signaling; 6963), pMek1/2 (Ser217/221) (Cell signaling; 9121), pErk1/2 (Thr202/Tyr204) (Cell signaling; 4370).
ChIP-seq
ChIPed DNA was subjected to a second round of shearing (10) on Covaris S220 AFA ultrasonicator (Covaris Inc.) using the following parameters: Peak Incident Power (W): 175, Duty Factor: 10%, Cycles per Burst: 200, Treatment Time (s): 120. Size distribution was assessed with High Sensitivity DNA Analysis Kit on Bioanalyzer 2100. Sequencing DNA libraries were prepared from up to 20 ng of DNA using TruSeq™ ChIP Sample Prep Kit (Illumina Inc.), according to a standard protocol with slight modifications. Briefly, DNA fragments were end repaired into blunt ends and single ‘A’ nucleotide was added to the 3′ ends. Adenylated fragments were ligated with Illumina's indexed adapters and then amplified during 18-cycles PCR for selective enrichment. Finally, library fragments with insert length of about 120–200 bp were automatically selected using LabChip XT DNA 750 Assay Kit and LabChip XT DNA sizing system. The quality and quantity of all libraries were assessed on 2100 Bioanalyzer using High Sensitivity DNA Kit (Agilent Technologies) and by qPCR with Kapa Library Quantification Kit (KapaBiosystems), respectively.
Samples were sequenced during single 50 bp read run on Illumina HiSeq 2500 system in High Output mode (using TruSeq SR Cluster Kit v3-cBot-HS and TruSeq SBS Kit v3-HS) and Rapid mode (using TruSeq Rapid Duo Sample Loading Kit, TruSeq Rapid SR Cluster Kit and TruSeq Rapid SBS Kit). Base calling, demultiplexing and generation of fastq files was carried on using Illumina RTA and bcl2fastq software.
Post sequencing ChIP-seq data analyses
Sequences were mapped to the human genome assembly (hg19) using Bowtie (11). Binding sites were detected with MACS2 (12). Quality control metrics for ChIP-Seq data including non-redundant fraction (NRF) and PCR Bottleneck Coefficient were computed with jChIP software (13).
Reads counting in gene regions was performed with HTseq (14) using Ensembl gene coordinates (GRCh37, release 75). Number of reads for each antibody and replicate were normalized to equal 5 × 107. Genes with less than 30 reads across all samples for a given antibody were discarded. Genes with more than 4-fold variation between replicates were also discarded.
ChIP-Seq data visualization
Two types of ChIP-seq data visualizations were used. The plot centered on transcription start site (TSS) and a plot depicting an average reads distribution along gene bodies for genes exhibiting ≥2-fold binding induction upon EGF stimulation. The TSS plot was made by taking fixed length (in bp) around the TSS with genes in the orientation 5′ to 3′ open reading frame (Ensembl gene coordinates; GRCh37, release 75). The signal in the frame of 200 bp from all the selected genes was used and then averaged. The second plot was made by normalizing distance from TSS to transcription termination site (TTS) for each gene, where the length of a gene was scaled to fit 2000 bp. The TTS region was estimated as 1000 bp distance from gene end. Additionally, the margins of 1000 bp were included. Each gene was then divided into 100 compartments in which the signal was counted. The profile was then averaged over all the selected genes and presented on a scale showing distance from TSS to TTS as 3000 bp length, so that the normalized frame size was 50 bp. In both visualizations, the plot was smoothed using Running Median algorithm. Both profiles were generated using GenomicTools (15) and visualization was performed in R.
Violin plots were generated for normalized gene length and at the nearest peaks defined with bins spanning range of 200–250 and 2900–2950 bp for TSS and TTS, respectively. Data are presented using log10 scale for Y axis. Values equal to zero were replaced with minimal, non-zero values to fit log scale.
Functional analyses
Peaks for Pol2, pEGFR, pMEK1/2 and pERK1/2 were annotated with cis-regulatory elements from HeLa-S3 combined genome segmentation based on data from ENCODE project (16). Peak annotation was conducted by BEDTools (17). The occurrences of different cis-regulatory elements within peaks were quantified with R-script. To verify whether chromatin feature distribution is different among peaks for studied factors random 622 peaks were selected from HeLa-S3's combined genome segmentation. Distribution/counts of chromatin features in this dataset (i.e. enhancer, promoter, CTCF binding and transcription repression) were compared to counts of features in our ChIP-Seq datasets with chi-square test.
Gene ontology (GO) terms over-representation among selected genes was assessed with GOstats package (18). Selected genes were those with at least 2-fold binding increase upon EGF stimulation. P-values were corrected for multiple hypothesis testing using Benjamini–Hochberg algorithm (19).
Protein IP followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) analyses
Hela-S3 cells (1 × 107) were suspended in 1ml of cold IP buffer containing cocktail of protease and phosphatase inhibitors (Thermo; 78441) and subjected to homogenization at 4°C in a Bioruptor Plus (Diagenode) using a 30 s on-off cycle for 10 min at low intensity followed by centrifugation at 12 000 × g for 15 min in 4°C. IP reaction was performed according to ChIP protocol and proteins were recovered from beads by incubating them with 50 μl 0.1% Trifluoroacetic acid (TFA) for 3 min at room temperature. Samples were transferred to a fresh Eppendorf tube and immediately neutralized by adding 1M NH4HCO3 solution to a final 0.1M concentration and then were reduced, alkylated and trypsin-digested as described earlier (9). Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis of peptides was performed on a LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific) coupled with a nanoAcquity (Waters Corporation) LC system. Acquired MS/MS raw data files were pre-processed with Mascot Distiller (version 2.5.1, Matrix Science) and searched against the SwissProt Homo Sapiens database (release 2015.11).
Endosomal proteins knockdown and western blotting
Cells were transfected with Ambion Silencer Select siRNA (Thermo) specific to dynamin 2 (DNM2) (DNM2_1 - s4212 and DNM2_2- s4213), TSG101 (s14440), VPS28 (s27579) and AP2M1 (AP2M1_1 - s3113 and AP2M1_2 - s3114) or non-complementary (NC, 4390843) siRNA at final concentration of 30 nM using Lipofectamine 3000. Twenty-four hours after transfection cells were transferred to serum free medium and 48 h after quiescence were challenged with EGF (100 ng/ml) and transferrin-Alexa 647 (25 μg/ml, Thermo), and either fixed for immunofluorescence staining or harvested at indicated time points. For western blots cells were lysed then resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene fluoride (PVDF) membrane. Blotted proteins were assessed by western blot analysis using the following antibodies: pEGFR (Tyr845) (Cell signaling; 6963), pMek1/2 (Ser217/221) (Cell signaling; 9121), pErk1/2 (Thr202/Tyr204) (Cell signaling; 4370), DNM2 (BD Biosciences; 610245), TSG101 (Abcam; ab83), VPS28 (Abcam; ab10133), AP2M1 (BD Biosciences; 611350), ACTB (Abcam; ab8226).
Immunofluorescence staining
HeLa-S3 cells were seeded on 12-mm coverslips in a 24-well plate (5 × 104 cells/well). Prior to EGF stimulation, cells were starved for 48 h in serum-free medium and then were stimulated with EGF for 20 min (100 ng/ml), rinsed with PBS and fixed with 3.6% paraformaldehyde in PBS for 15 min at room temperature. Next, cells were permeabilized with 0.1% Triton X-100 and blocked using 10% FBS in PBS for 30 min. Cells were further incubated overnight with primary antibodies against: pEGFR (Tyr845) (Cell signaling; 6963), pMEK1/2 (Ser217/221) (Cell signaling, 9121), pERK1/2 (Thr202/Tyr204) (Cell signaling, 9101) and EEA1 (BD Biosciences; 610457). Next day, cells were incubated with donkey secondary antibodies anti-mouse conjugated with Alexa Fluor 488 and anti-Rabbit conjugated with Alexa Fluor 555 (30 min) in 5% FBS prepared in PBS. Nuclei were visualized by DAPI. Z-stack images were acquired with Zeiss LSM710 microscope using 100×/1.40 oil immersion objective and 1024 × 1024 pixel resolution in ZEN 2009 software. At least ten 12-bit images per experimental condition were captured. Images were imported into MotionTracking/Kalaimoscope (http://motiontracking.mpi-cbg.de) to analyze the integral fluorescence and number of vesicles positive for pEGFR (expressed in arbitrary units, AU) (20,21). Maximum intensity projections of Z-stacks were exported as TIFF and for visual presentation arranged in Adobe Photoshop CS4 Extended software with only linear adjustments.
Statistical analysis
Statistical analyses were performed with a two sided t-test to determine statistical significance between the groups. P-values are represented in given figures as follows: *P-value ≤ 0.05; **P-value ≤ 0.01.
RESULTS
ChIP-Seq analysis of constitutive and EGF-induced genome-wide binding of EGFR and ERK kinase components reveals their colocalization with Pol2 complex
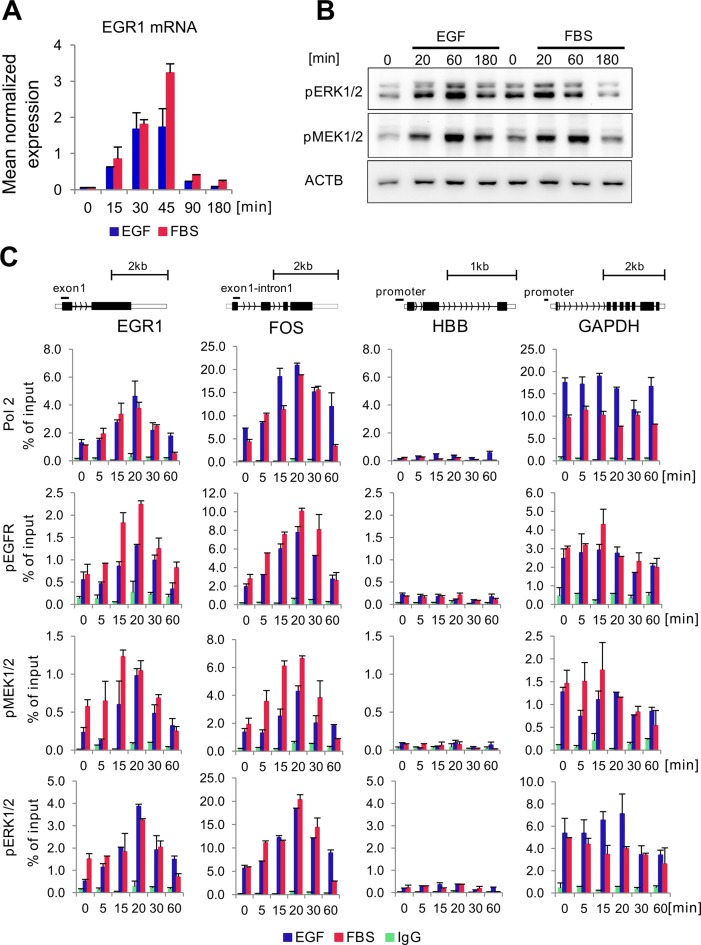
As a model we chose HeLa-S3 since this cell line is the one of the most extensively studied in the ENCODE project (22) allowing to correlate genomic distribution of active EGFR and ERK components with previously established chromatin features in this cell line. In our previous study we used an immediate early (IE) gene, EGR1, as a model locus to study temporal MAPK kinases recruitment in cells stimulated to proliferate with 10% FBS (23). Since proliferation could be triggered by multiple mitogenic ligands present in FBS, we used EGF instead to profile mitogen-induced recruitment of EGFR and ERK kinase components to chromatin. The timing of EGR1 mRNA increase after EGF was similar to FBS response (Figure 1A) and either treatment induced transient phosphorylation of MEK1/2 and ERK1/2 (Figure 1B and Supplementary Figure S1). It has been shown that many of the known cellular inducers of the ERK pathway lead to simultaneous activation of both ERK1 and ERK2 (24), however the antibody we used recognizes both isoforms and we cannot distinguish which of the isoforms is directly tethered to chromatin. Given the 85% identity in sequence for human ERK1 and ERK2 and numerous efforts to establish differences between them, it is thought that the functions of the two isoforms are similar (25), presumably also in the vicinity of chromatin. IP reaction followed by a mass spectrometry (MS) analyses further confirmed the specificity of the anti-phospho antibodies and their ability to pull down the studied factors (Supplementary Table S2). Interestingly, the IP-MS analysis revealed that Pol2 and EGFR co-immunoprecipitated in whole cellular extracts (Supplementary Table S2). Pol2, pEGFR, pMEK1/2 and pERK1/2 binding to EGR1 and FOS, another IE gene, showed their highest recruitment at the first exon following 20 min of EGF treatment (Figure 1C). For all factors patterns of recruitment at EGR1 and FOS in response to EGF was similar to FBS treatment. As expected, these factors were not found at the repressed β-globin (HBB) promoter but were abundantly present at the constitutively highly active glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter (Figure 1C). As 20 min time point revealed highest binding of Pol2 and active EGFR and ERK components at EGR1 (Figure 1C) this time point was chosen to compare their genome wide distribution to that found in untreated cells.
Figure 1.
EGF and FBS-induced EGR1 expression and co-recruitment of Pol2 with active kinases to EGR1, FOS, HBB and GAPDH loci. (A) HeLa S3 cells were treated with EGF (100 ng/ml) or 10% FBS and harvested at indicated time point then RNA extracted with Trizol followed by RT-qPCR measurements. EGR1 expression was normalized to RPLP0 mRNA (n = 3;±SD). (B) Cells were treated as in A and collected at indicated time point. A total of 5 μg of whole cell extract was resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and electrotransferred to PVDF membranes. Western blot analysis was performed using anti-pMEK1/2, anti-pERK1/2 and anti-β-Actin antibodies. (C) HeLa S3 cells were challenged with either EGF (100 ng/ml) or 10% FBS and fixed with 1% formaldehyde at indicated time point. Chromatin was isolated and used in ChIP assays with unspecific IgG, anti-Pol2, anti-pEGFR, anti-pMEK1/2 and anti-pERK1/2 antibody. Isolated DNA was used as the template in qPCR analyses with primers designed to amplify exon1 of EGR1, exon1-intron1 junction of FOS gene and promoters of the β-globin (HBB), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are expressed as a percentage of input chromatin (n = 3;±SD).
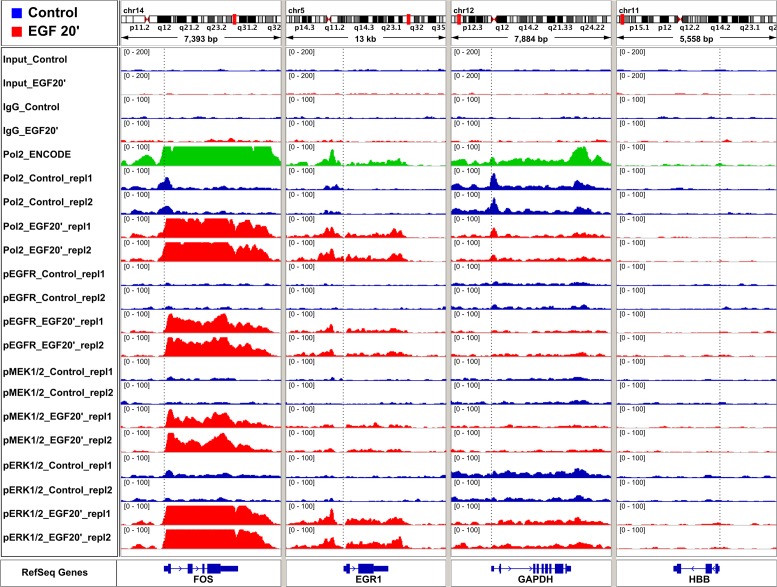
ChIP-Seq experiments were done in biological duplicates. On an average 65 million reads for each factor were mapped to human genome and all libraries for specific antibodies met ENCODE's quality control metrics for ChIP-Seq experiment including NRF and PCR Bottleneck Coefficient (26) (Supplementary Table S3). Inspection of reads in IGV genome browser revealed, as expected, inducible binding of Pol2 and MAPK cascade components to known IE genes including EGR1 and FOS, but no visible induction at GAPDH gene and no binding at the HBB locus. There were also noticeable similarities in Pol2 binding to the above loci between our inducible dataset and Pol2 tracks generated in growing HeLa-S3 cell line in ENCODE project (Figure 2). To further confirm binding specificity in our ChIP-Seq dataset, we evaluated recruitment of these factors to the PHLDA2, SOCS3, TGM2 and DNAJB1 loci that in ChIP-Seq exhibited small-to-moderate level of EGF-induced binding of Pol2, pEGFR, pMEK1/2 and pERK1/2 (Supplementary Figure S2 A). ChIP-qPCR confirmed inducible binding of all factors to selected loci with a peak of recruitment at 20 min upon the EGF challenge (Supplementary Figure S2 B).
Figure 2.
ChIP-seq analysis of Pol2, EGFR and ERK kinases recruitment at selected loci in quiescent and EGF stimulated cells. BigWig input and ChIP-seq files for a given factor and condition were imported to IGV and data range was set for 100 to underscore differences between factors level at given locus. The relative enrichment of each factor at selected sites is inferred from the density of mapped fragments. Pol2 ENCODE ChIP-Seq data for HeLa-S3 proliferating cells (ID: wgEncodeBroadHistoneHelas3Pol2bStdSig) were included to show similarities in transcriptional complex binding between datasets. The direction of transcription is indicated by arrowheads on gene cartoons. A vertical dotted line marks transcription start sites (TSS).
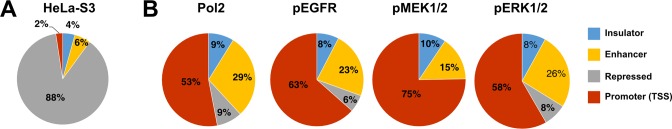
Using MACS we identified 234, 251, 107 and 751 high confidence (FDR < 0.05) differentially occupied peaks for Pol2, pEGFR, pMEK1/2 and pERK1/2, respectively, after treatment with EGF (Supplementary Table S4). These peaks were then aligned with known HeLa-S3 regulatory elements (Figure 3A), described in ENCODE as genome segmentations (16), revealing a preferential Pol2 and ERK kinases co-localization with promoter regions and less frequent interactions with enhancers, insulators and heterochromatin regions (Figure 3B). Differences in regulatory elements counts in comparison with a random HeLa-S3 regulatory elements dataset were statistically significant for all studied factors (maximum P = 2.2 × 10−16; chi-square statistic). Therefore, not unexpectedly, Pol2 and active kinases were found predominantly at regions of active chromatin in EGF stimulated cells.
Figure 3.
Genome-wide functional characterization of Pol2, EGFR and ERK kinases at EGF-responsive binding sites. (A) The percentage frequency of HeLa-S3 regulatory elements described in ENCODE's genome segmentations. (B) Peaks enriched for a given factor in EGF stimulated cells (Supplementary Table S4; FDR ≤ 0.05) were correlated with coordinates of known HeLa-S3 cis-regulatory elements generated within frames of ENCODE project (16).
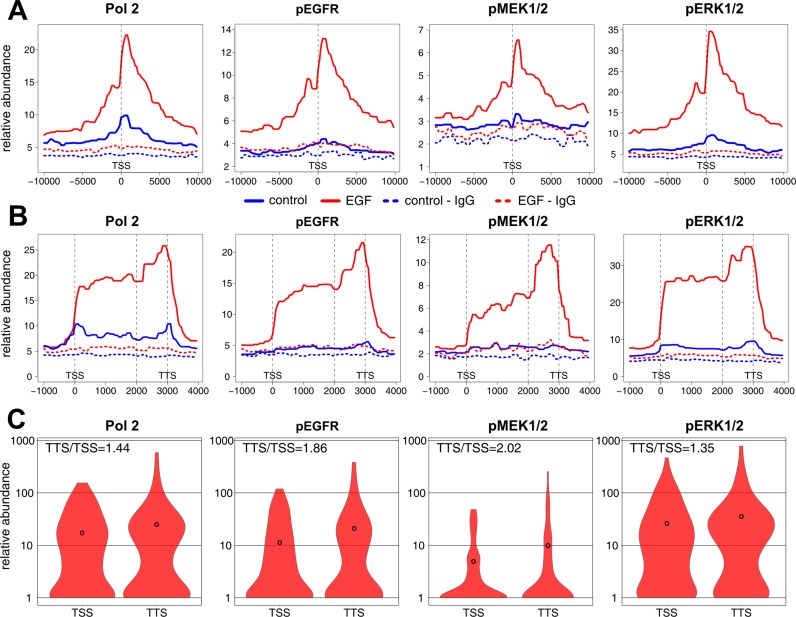
Next, using the HTSeq we estimated differential binding of studied factors to gene regions. Under the ≥2-fold EGF induction threshold we found 127, 96, 69 and 140 genes inducibly occupied by Pol2, pEGFR, pMEK1/2 and pERK1/2, respectively (Supplementary Table S5). Like the known Pol2 binding pattern, EGFR and ERK cascade kinases co-localized at TSS (Figure 4A) with a characteristic bimodal distribution suggestive of divergent transcription at promoters of activated genes (27,28). Further, profiles of MAPK pathway components were significantly correlated with EGF stimulated Pol2 binding at TSS with Pearson correlation coefficient of 0.889, 0.804 and 0.757 for pEGFR, pMEK1/2 and pERK1/2, respectively (Supplementary Figure S3). Average kinase binding profiles along gene bodies as well as at TTS also resembled Pol2 transcriptional complex distribution (Figure 4B). Specifically, levels of a given factor found at TTS/TSS expressed as ratio reached 1.44, 1.86, 2.02 and 1.35 for Pol2, pEGFR, pMEK1/2 and pERK1/2, respectively (Figure 4C). Piling up of Pol2 at the TTS is a well-known phenomenon thought to reflect pausing of terminal transcriptional complex and its disengaging from DNA template (29). Pol2-like binding pattern and relatively higher kinases levels at TTSs suggest EGFR-ERK cascade involvement in EGF-induced processes at 3′ ends of target loci.
Figure 4.
EGF-inducible EGFR, ERK pathway components recruitment to chromatin resembles Pol2 binding at TSS and along gene bodies. (A) Distribution of Pol2, pEGFR, pMEK1/2 and pERK1/2 ChIP-Seq reads near the TSS. Average reads coverage is shown in a bin size of 200 bp for a window 10 kb upstream/downstream from the TSS for a set of genes exhibiting a 2-fold induction in a given factor binding upon EGF stimulation (Supplementary Table S5). The TSS plot was created by taking a fixed length (in bp) around the TSS, with all the selected genes in the same orientation (5′ to 3′ open reading frame), and then averaging the signal across these genes. (B) Distribution of Pol2, pEGFR, pMEK1/2 and pERK1/2 along the genes occupied by MAPK pathway constituents in EGF stimulated HeLa-S3 cells. Genes as in B were used to create an average binding plot spanning the genomic fragment representing a gene body. For each gene in the input a 1 kb upstream and 2 kb downstream sequences were included and then resolution or sampling frequency was set for 100 bp. (C) Violin plots of a given factor binding at TSS and transcription termination site (TTS). The profile was created for the nearest peaks to TSS and TTS. Black circles represent the mean for which the TTS/TSS ratio was computed.
Gene ontology (GO) analysis of loci occupied by EGFR and ERK pathway components identifies cytoskeleton associated genes as targets
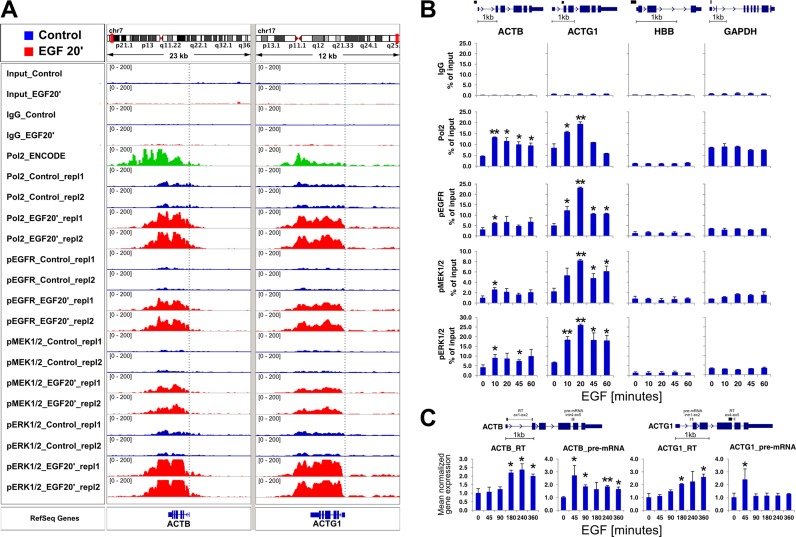
We calculated GO enrichment as way to gain further insight into the biological role of EGFR and ERK cascade components inducibly bound to chromatin encompassing target genes (≥2-fold increase by EGF). This approach identified 38 GO terms with adj. P-value ≤ 0.05 for any factor studied (Supplementary Table S6). We then narrowed down the list to terms that were enriched for at least two factors yielding 14 GO terms (Table 1). The top three out of four GO terms were enriched for all four factors, and were related to DNA binding activities of transcriptional factors. These results are in agreement with EGF induced transcriptional activation of IE genes that encode a functionally related class of TFs (30,31). Unexpectedly, another class of enriched GO terms included ‘actin binding’ genes annotated as interacting with monomeric or multimeric forms of actin (32). Traditionally these genes have been regarded as constitutively active and have been frequently used as control housekeeping genes (33). Examination of individual ChIP-Seq profiles uncovered inducible binding of all factors to β-actin (ACTB) and γ-actin (ACTG1) loci (Figure 5A).
Table 1. Gene ontology (GO) analysis of loci occupied by EGFR and ERK pathway components.
| GO ID | Term | Pol2 | pEGFR | pMek1/2 | pERK1/2 |
|---|---|---|---|---|---|
| GO:0043565 | sequence-specific DNA binding | 0.0033 | 0.0006 | 0.0203 | 0.0126 |
| GO:0003690 | double-stranded DNA binding | 0.0271 | 0.0409 | 0.0203 | 0.0445 |
| GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity | 0.0271 | 0.0114 | 0.0131 | 0.0445 |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 0.0271 | 0.0108 | 0.0390 | 0.0445 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 0.0157 | 0.0013 | 0.0712 | 0.0375 |
| GO:0070412 | R-SMAD binding | 0.0853 | 0.0409 | 0.0203 | 0.0573 |
| GO:0001067 | regulatory region nucleic acid binding | 0.0229 | 0.0042 | 0.0592 | 0.0445 |
| GO:0003707 | steroid hormone receptor activity | 0.0466 | 0.0151 | 0.2169 | 0.0445 |
| GO:0044212 | transcription regulatory region DNA binding | 0.0271 | 0.0162 | 0.5346 | 0.0445 |
| GO:0044822 | poly(A) RNA binding | 0.0271 | 0.0524 | 0.3662 | 0.0082 |
| GO:0003779 | actin binding | 0.0197 | 0.0524 | 0.2613 | 0.0375 |
| GO:0005316 | high-affinity inorganic phosphate:sodium symporter activity | 0.0817 | 0.0409 | 0.0340 | 0.0445 |
| GO:0004586 | ornithine decarboxylase activity | 0.0817 | 0.0409 | NA | 0.0445 |
| GO:0030377 | urokinase plasminogen activator receptor activity | 0.0817 | 0.0409 | NA | 0.0445 |
Adjusted P-values < 0.05 are highlighted. NA: not available.
Figure 5.
EGF-inducible EGFR and ERK pathway components binding at ACTB and ACTG1 loci. (A) ChIP-seq overview of active kinases and Pol2 binding to ACTB and ACTG1 genes depicted in IGV genome browser. Pol2 ENCODE ChIP-Seq data for HeLa-S3 growing cells (ID: wgEncodeBroadHistoneHelas3Pol2bStdSig) were included to show similarities in transcriptional complex binding between datasets. A vertical dotted line marks TSS. (B) Time-course ChIP-qPCR measurements confirm EGFR and ERK pathway components binding to ACTB and ACTG1 following EGF treatment. ChIP analysis of sheared chromatin from a time course of EGF-treated (100 ng/ml EGF for 0, 10, 20, 45 and 60 min) HeLa-S3 cells was done using Matrix-ChIP method in 96-well polypropylene plates as described (8). qPCR was performed using primers (Supplementary Table S1) spanning an area depicted in a cartoon above each gene. ChIP data are expressed as DNA recovered as the percentage (%) of input DNA (n = 3; mean ±SD). Statistical analysis of differences between mean DNA recovery for a given factor in quiescent cells and EGF time points was performed using t-tests. A P-value of ≤ 0.05 (*) was considered significant, **; P-value ≤ 0.01. (C) Time-course RT-qPCR analyses of ACTB and ACTG1 mRNA and pre-mRNA during EGF time course. Cells were harvested at indicated time point, RNA extracted with Trizol, DNAze treated and submitted to RT-qPCR measurements. RNA expression was normalized to RPLP0 mRNA and presented as fold change of expression measured in quiescent cells (n = 3; mean ±SD). Statistical analysis of differences between mean cDNA levels for quiescent cells and given EGF time point was performed using t-tests. A P-value of ≤ 0.05 (*) was considered significant, **; P-value ≤ 0.01. Gene cartoon depicts localization of primers amplifying either mature (RT, spanning exon-exon) or unspliced (pre-mRNA, spanning exon-intron) form of transcript.
To validate the recruitment of MAPK pathway constituents to ACTB and ACTG1 genes, independent ChIP analyses were performed with chromatin from cells challenged with EGF for 10, 20, 45 and 60 min (Figure 5B). These independent measurements showed that active kinases and Pol2 were inducibly recruited to first exons of ACTB and ACTG1 genes, whereas no induction in binding was observed at HBB and GAPDH promoter regions used as negative controls. Notably, while temporal initiation of Pol2 and kinases binding to ACTB and ACTG1 was similar, the pattern at ACTG1 was transient but sustained at ACTB (Figure 5B). Different duration of binding in EGF-treated cells at ACTB versus ACTG1 suggested differences in precursor (pre)-mRNA processing. Therefore, next we measured mRNA and pre-mRNA abundances for these genes in a course of EGF stimulation using RT-qPCR. Mature ACTB and ACTG1 mRNAs increased significantly at 180 min and remained increased up to 360 min (Figure 5C). The pre-mRNA levels of ACTB and ACTG1 reached their highest levels at 45 min. In contrast to ACTG1, the level of ACTB pre-mRNA did not decline to basal level and remained relatively stable from 3 to 6 h with an average FC of 1.7 when compared to quiescent cells. These observations suggest that stable presence of Pol2 and MAPK components at ACTB gene might, in part, reflect differences in pre-mRNA processing mechanisms and factors along these loci in response to EGF treatment (Figure 5A).
Active EGFR, MEK1/2 and ERK1/2 co-localize on endosomes and their recruitment to EGR1 locus depends on the endocytic protein, AP2M1
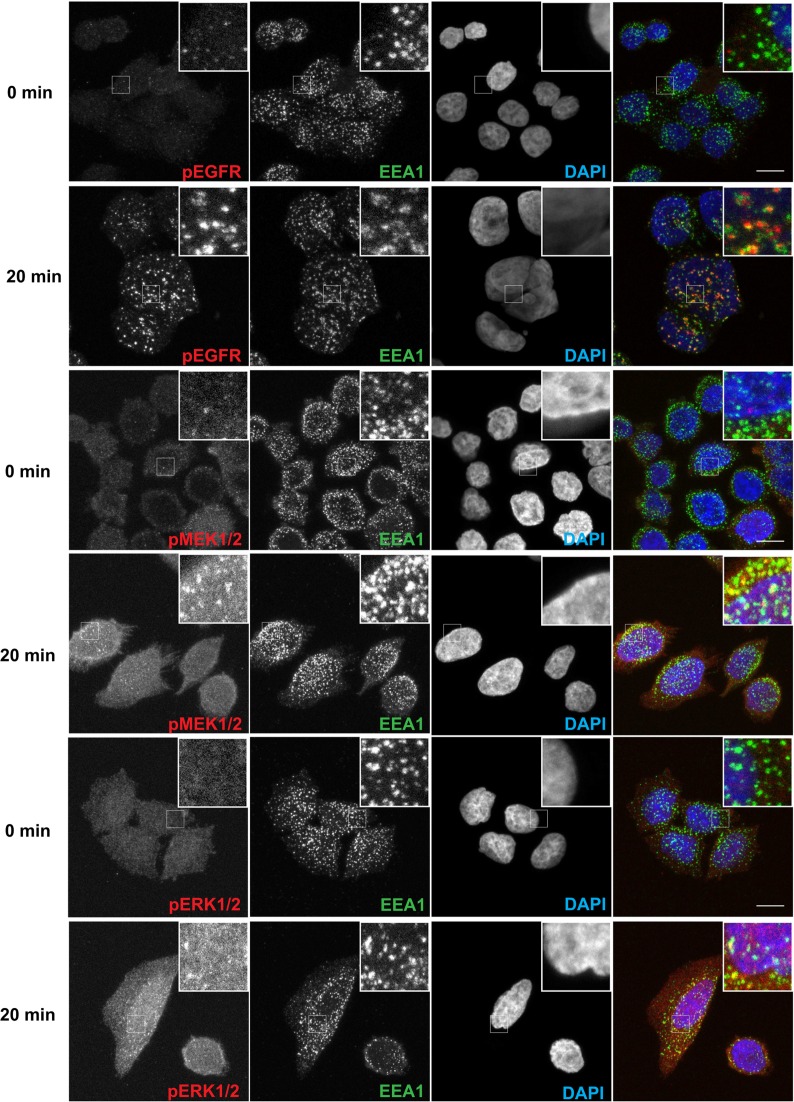
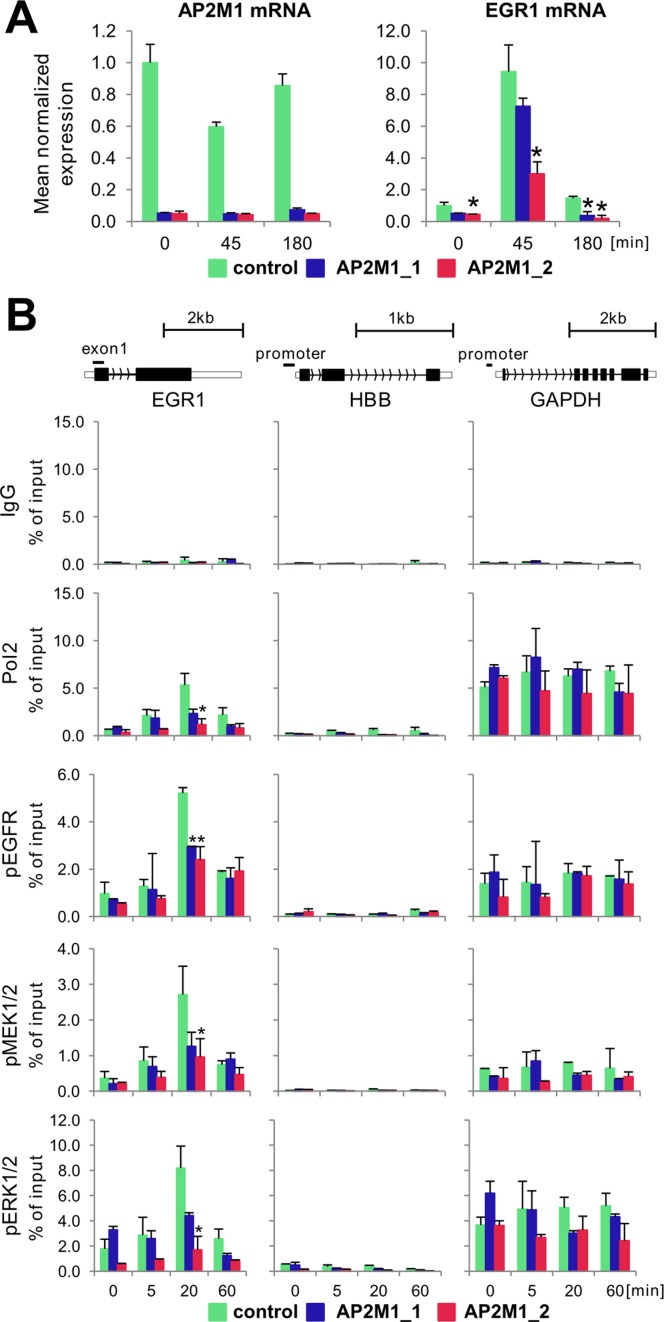
Endosomes appear to play a role in intracellular trafficking of signal transducers (34). The remarkable temporal and spatial similarity of EGFR and ERK kinase components recruitment to target genes suggests that delivery of these proteins is highly coordinated perhaps as a pre-formed signaling complex. Ligand binding triggers internalization of EGFR into endosomes which is a pre-requisite for the nuclear translocation of a fraction of the receptor (6). Using immunofluorescence staining, we confirmed that EGF induces activation of EGFR, visualized by its phosphorylation on Tyr845 with the same antibody as used in ChIP assay (Figure 6). Twenty minutes following EGF treatment, most of the pEGFR signal overlapped with a marker of endosomes, EEA1 protein (35). Importantly, at the same time the signals of active MEK1/2 and ERK1/2 were also clearly present in endosomes, in addition to the expected strong cytosolic staining (Figure 6). We next wished to determine if altering internalization and/or endosomal trafficking could influence the EGF-induced recruitment of EGFR and ERK kinases to chromatin. To this end, we used siRNA to transiently deplete endocytic proteins. To impair internalization, we knocked down a canonical endocytic clathrin adaptor AP2M1 (36) (Supplementary Figure S4) which, as expected, decreased the number as well as the intensity of the pEGFR containing endosomes (Supplementary Figure S5). We attempted to knock down dynamin 2 (DNM2), another mediator of clathrin-dependent endocytosis of EGFR (37). However, we only achieved a partial depletion of DNM2 protein in HeLa cells (Supplementary Figure S4) which was not sufficient to impair internalization of EGFR (data not shown), thus no changes in a transcriptional response to EGF were expected. To perturb endosomal trafficking, we knocked down two components of the ESCRT-I complex, TSG101 and VPS28 which mediate endosomal sorting of EGFR toward lysosomes (38) (Supplementary Figure S4). We then measured EGR1 transcript level as an indicator of the EGF-induced transcriptional program. Among these three endosomal factors only AP2M1 depletion resulted in a significant inhibition of EGR1 transcript expression (Figure 7A). ChIP analysis at EGR1 gene revealed significant inhibition of EGF-stimulated Pol2 and active EGFR and ERK kinases recruitment upon of AP2M1 depletion (Figure 7B).
Figure 6.
EGF-activated EGFR, MEK1/2 and ERK1/2 co-localize in endosomes. HeLa-S3 cells were maintained 48 h in serum-free medium and then stimulated for 20 min with EGF (100 ng/ml). Cells were fixed with paraformaldehyde and subjected to immunofluorescence staining. Endosomes were stained with EEA1 and nuclei are DAPI-labeled. Scale bar, 10 μm.
Figure 7.
AP2M1 depletion reduces EGF-inducible Pol2, EGFR, MEK1/2 and ERK1/2 binding to EGR1 locus. (A) HeLa-S3 cells line were transfected with either one of two AP2M1 siRNAs or non-specific siRNA in a presence of Lipofectamine 3000. Twenty-four hours after transfection, cells were switched to serum free medium, and 48 h after quiescence, cells were treated with EGF (100 ng/ml) for 0, 45 and 180 min. Cells were harvested at indicated time point then RNA extracted with Trizol followed by RT-qPCR measurements. EGR1 and AP2M1 expression was normalized to RPLP0 mRNA (n = 3; ±SD). (B) Cells were prepared as in A and challenged with EGF (100 ng/ml) for 0, 5, 20 and 60 min then fixed, chromatin isolated and sheared. Matrix ChIP assay was done using antibodies to Pol2, pEGFR, pMEK1/2 and pERK1/2. ChIP data are expressed as DNA recovery in percentage (%) of input (means ± S.D., n = 3). Statistical analysis of differences between mean DNA recovery for control and AP2M1-depleted chromatin at given time point was performed using t-tests. ChIP results are shown for PCR product depicted on gene cartoon. A P-value of < 0.05 (*) was considered significant.
These data argue that EGFR–ERK canonical plasma membrane signaling module persists intracellularly, partly on endosomes. If the endosomal pool of this module is diminished by impaired internalization of EGFR this can impact transcription and gene expression. Interestingly, affecting later stages of endosomal trafficking to lysosomes, such as ESCRT-dependent sorting of EGFR toward degradation, has no consequences for transcriptional regulation, as shown before (31). In general, endocytic internalization and localization of EGFR and ERK kinase module to endosomes appear to favor a coordinated delivery of this complex to the nucleus, where it is then synchronously tethered to target genes.
DISCUSSION
Our genome-wide analysis revealed that MAPK pathway constituents including EGFR, MEK1/2 and ERK1/2 are coordinately tethered to clusters of functionally related genomic loci upon EGF mitogen stimulation with a spatiotemporal binding pattern resembling Pol2. Alternation of endosomes formation through AP2M1 depletion in EGF-stimulated cells suggest that these vesicles are in part a repository for organizing nuclear delivery of EGF-responsive signal transducers facilitating their synchronous recruitment to target gene clusters.
Nuclear and chromatin co-localization of EGFR/ERK kinases
Ligand-induced RTKs nuclear localization and recruitment to chromatin, previously unappreciated, is now well recognized non-canonical phenomenon involving EGFR, IGF-1 and insulin (IR) receptors (39–41). Importantly, RTKs contain nuclear localization signals that facilitate their nuclear import (42–44).
Endocytosis plays an essential role in the transport of RTKs from the cell surface to the nucleus (45–47). Although endocytosis typically leads to termination of signaling and degradation of RTKs in lysosomes, in some cases active signaling from endosomes continues after RTK internalization (48). Spatiotemporal distribution of activated EGFR within cell, mediated by endosomal transport, appears to be important for EGF-induced MAPK signaling (49). Thus, endosomes may serve as an accurate and controllable module directing traffic of signaling complexes into various intracellular locations, including the vicinity of nuclear pores and further into the nucleus (50). Our data are consistent with the notion that endocytic internalization may contribute to a transcriptional response to EGF, as endosomes maintain activated EGFR and its associated downstream kinases prior to nuclear import (Figures 6 and 7 and Supplementary Figure S5). Here, EGFR nuclear localization signal (42) could serve as a zip code directing, when needed, pre-assembled signaling kinase modules to the nucleus. Endosome pre-assembled signaling complexes would provide a repository to coordinately induce expression of functionally related gene subsets.
Once in the nucleus, how are components of EGFR/ERK kinase signaling module recruited to target genes? Recent gene-specific and genome-wide studies of ERKs (51), JNKs (52) and other kinases (53,54) have revealed that binding of these proteins to transcribed genes was either directed by a recognition of DNA motifs (53), guided by the presence of specific substrate at promoter (52), directly mediated by Pol2 complex (54) or perhaps in some instances combination of these modes. Consistent with these observations our study showed that most of the Chip-Seq peaks were enriched with kinases upon EGF stimulation within promoter regions of actively transcribed genes and enhancers (Figure 3). Similar inducible genome-wide effect was observed for CK2a, the catalytic subunit of CK2, where kinase was found at promoters, Pol2-transcribed coding genes and active enhancers in testosterone-treated LNCaP prostate line (54). In sum, published work and the current study indicate that genomic kinase co-localization is largely linked to presence of Pol2 and its transcriptional machinery (51,53–55).
Role of chromatin-bound EGFR/ERK kinases in Pol2 transcription
Here, for the first time we show that genome-wide binding of active EGFR matches Pol2 profile. Chromatin-associated RTKs exhibit transcriptional, co-activator and kinase activities (56,57). Compartmentalization of kinase signals, allows spatiotemporal control of substrates activities at specific sites. We speculate that at intracellular sites, including chromatin, RTKs’ role is to coordinate phosphorylation events of their cognate downstream transducers to achieve spatiotemporal specificity (58). For example, compartmentalization of RTKs and downstream kinases allows spatiotemporal control of substrates at target loci ensuring Pol2 productive mRNA synthesis (41).
From yeast to mammals MAPK pathway components are recruited to chromatin where they directly target multiple targets including Pol2 machinery (51,55,59–63). Pol2 phosphorylation within the carboxy terminal domain (CTD), composed of up to 52 heptapeptide repeats (YSPTSPS), regulates transcriptional complex processivity, interactions with auxiliary proteins and transcription associated processes (64). For example, ERK1/2 phosphorylates CTD of Pol2 at serine 5, modification that takes place near TSS (65). MAPK pathway activates the positive elongation factor, P-TEFb, at IE genes upon mitogen stimulation. MEK inhibition by the inhibitor U0126 blocked association of the two components of P-TEFb, cyclin T1 and Cdk9 and decreased phosphorylation of serine 2 of the Pol2 CTD (66). These examples support the notion that the Pol2-like pattern of MAPK binding at TSS and in gene bodies (Figure 4) reflect their activity toward Pol2 CTD and Pol2 associated proteins.
Accumulation of active MAPK constituents at TTSs (Figure 4B and C) suggests their involvement in 3′ end related events at EGF regulated genes including mRNA maturation and/or Pol2 recycling. Eukaryotic pre-mRNAs are processed to their mature forms by a series of coordinated co-transcriptional events including capping, splicing, cleavage and poly(A) addition (67). Importantly, ∼70% of human pre‐mRNAs contain more than one functional polyadenylation signal that allows qualitative as well as quantitative 3′ end adjustments by tuning the type as well as amount of poly(A) mRNA, and ultimately protein synthesis (68–70). For example, serum stimulation of quiescent cells increased ERK mediated phosphorylation and polyadenylation activity of poly(A) polymerase, an enzyme responsible for the synthesis of the poly(A) tails. (71). Thus, our observation that MAPK components accumulate near TTSs suggest their direct involvement in linking external stimuli to 3′ end transcriptional regulation and gene expression. EGFR and ERK kinases have many nuclear partners and targets and thus their role extends beyond direct control of Pol2 activity.
Subsets of genes directly targeted by EGFR/ERK kinases
Analysis of loci genes co-occupied by EGFR/ERK kinases in response to EGF clustered by the means of GO, identified genes encoding TFs involved in mitogenic responses and MAPK phosphatases (Table 1 and Supplementary Table S6) represented by dual-specificity phosphatases (DUSPs). These findings are in line with previous transcriptomic gene expression analyses after EGF challenge in HeLa cells (30). Induction of DUSPs that inactivate ERKs may be part of negative feedback loop (72). For example, recruitment of one of these phosphatases, MKP-1, to insulin-responsive genes follows IR/ERK kinases binding (41). Analysis of our dataset also identified genes belonging to GO term ‘actin binding’ including ACTB and ACTG1 (Table 1 and Figure 5A). Llorens et al. (73) utilizing multiple expression platform, including NGS, also showed their induction after EGF treatment.
Identification of the above gene subsets as EGFR/ERK kinases targets is not surprising given their involvement in mitogenic responses. Thus, the parallel recruitment of EGFR/ERK kinases to these loci provides means to coordinate their expression ensuring effective initiation of cell proliferation. Serum response factor (SRF) and its partner Elk, which are ERK substrates, are known to regulate transcription initiation of some of the genes represented in these clusters (74,75) a fact that could account for their similar kinetics of induction (Figures 1 and 5). But once induced other factors are likely to come into play to regulate the duration of the enhanced transcription as illustrated by the differences between ACTB and ACGT1 (Figure 5). Interestingly, it has recently been shown that b-actin can regulate expression of its own locus and could be one such a factor (76).
In summary, this study provides new details of how mitogen-induced coordinated recruitment of EGFR/ERK kinases to subsets of genes, which appears to involve endosome trafficking, could orchestrate transcriptional events to ensure effective cell proliferation. As such, it serves as a groundwork to define specific molecular mechanisms that mediate their chromatin binding and then drive transcription. Better understanding of the these processes on a genome-wide scale opens up exciting opportunities to identify novel targets to improve clinical outcomes of conditions driven by aberrant mitogenic responses.
Supplementary Material
Acknowledgments
Authors' contributions: M.Mikula., K.B., J.O. conceived the study, analyzed and interpreted the data and drafted the manuscript. M.Mikula., J.K.L., M.Statkiewicz., M.D., U.K., J.K., I.R. cultured cells performed ChIP and RT experiments. M.Skrzypczak., M.G., K.Ginalski. prepared sequencing libraries, performed sequencing and contributed to manuscript drafting. K.Goryca., K.P., M.K. analyzed ChIP-Seq data and contributed to manuscript drafting. M.Miaczynska., K.J. performed and interpreted immunofluorescence data, contributed to manuscript drafting. We thank Daria Zdzalik-Bielecka for technical assistance with immunofluorescence on AP2M1 knockdown cells.
Footnotes
Present address: M. Mikula, Cancer Center-Institute, Roentgena 5, 02-781 Warsaw, Poland.
ACCESSION NUMBERS
Sequencing data was deposited in Gene Expression Omnibus database under entry GSE65323.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Center [2011/01/D/NZ2/05307 to M.Mikula.]; National Science Centre [2015/17/D/NZ2/03711 to M.Skrzypczak.]; Foundation for Polish Science [TEAM], National Science Centre [2011/02/A/NZ2/00014, 2014/15/B/NZ1/03357 to K.Ginalski.]; National Institutes of Health [R01 DK083310, R21 GM111439, R33 CA191135 to K.B.]; Foundation for Polish Science within International PhD Project ‘Studies of nucleic acids and proteins—from basic to applied research’ (to K.J.); European Union—Regional Development Fund (to K.J.). Funding for open access charge: National Science Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1.Turjanski A.G., Vaqué J.P., Gutkind J.S. MAP kinases and the control of nuclear events. Oncogene. 2007;26:3240–3253. doi: 10.1038/sj.onc.1210415. [DOI] [PubMed] [Google Scholar]
- 2.Pearson G., Robinson F., Beers Gibson T., Xu B.E., Karandikar M., Berman K., Cobb M.H. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 3.Yang S.-H., Sharrocks A.D., Whitmarsh A.J. MAP kinase signalling cascades and transcriptional regulation. Gene. 2013;513:1–13. doi: 10.1016/j.gene.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter G., Liao H.-J. Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 2013;5:a008979. doi: 10.1101/cshperspect.a008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M.-K., Hung M.-C. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 2015;282:3693–3721. doi: 10.1111/febs.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikula M., Bomsztyk K., Goryca K., Chojnowski K., Ostrowski J. Heterogeneous nuclear ribonucleoprotein (HnRNP) K genome-wide binding survey reveals its role in regulating 3′-end RNA processing and transcription termination at the early growth response 1 (EGR1) gene through XRN2 exonuclease. J. Biol. Chem. 2013;288:24788–24798. doi: 10.1074/jbc.M113.496679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J., Feng Q., Ruan Y., Komers R., Kiviat N., Bomsztyk K. Microplate-based platform for combined chromatin and DNA methylation immunoprecipitation assays. BMC Mol. Biol. 2011;12:49. doi: 10.1186/1471-2199-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikula M., Rubel T., Karczmarski J., Statkiewicz M., Bomsztyk K., Ostrowski J. Beads-free protein immunoprecipitation for a mass spectrometry-based interactome and posttranslational modifications analysis. Proteome Sci. 2015;13:23. doi: 10.1186/s12953-015-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mokry M., Hatzis P., de Bruijn E., Koster J., Versteeg R., Schuijers J., van de Wetering M., Guryev V., Clevers H., Cuppen E. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS One. 2010;5:e15092. doi: 10.1371/journal.pone.0015092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chojnowski K., Goryca K., Rubel T., Mikula M. jChIP: a graphical environment for exploratory ChIP-Seq data analysis. BMC Res. Notes. 2014;7:676. doi: 10.1186/1756-0500-7-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsirigos A., Haiminen N., Bilal E., Utro F. GenomicTools: a computational platform for developing high-throughput analytics in genomics. Bioinformatics. 2012;28:282–283. doi: 10.1093/bioinformatics/btr646. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E., et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 20.Collinet C., Stöter M., Bradshaw C.R., Samusik N., Rink J.C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M.S., et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 21.Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 22.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikula M., Bomsztyk K. Direct recruitment of ERK cascade components to inducible genes is regulated by heterogeneous nuclear ribonucleoprotein (hnRNP) K. J. Biol. Chem. 2011;286:9763–9775. doi: 10.1074/jbc.M110.213330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefloch R., Pouysségur J., Lenormand P. Total ERK1/2 activity regulates cell proliferation. Cell Cycle. 2009;8:705–711. doi: 10.4161/cc.8.5.7734. [DOI] [PubMed] [Google Scholar]
- 25.Roskoski R. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seila A.C., Core L.J., Lis J.T., Sharp P.A. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 29.Shandilya J., Roberts S.G.E. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim. Biophys. Acta. 2012;1819:391–400. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 31.Brankatschk B., Wichert S.P., Johnson S.D., Schaad O., Rossner M.J., Gruenberg J. Regulation of the EGF transcriptional response by endocytic sorting. Sci. Signal. 2012;5:ra21. doi: 10.1126/scisignal.2002351. [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozera B., Rapacz M. Reference genes in real-time PCR. J. Appl. Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Fiore P.P., von Zastrow M. Endocytosis, signaling, and beyond. Cold Spring Harb. Perspect. Biol. 2014;6:a016865. doi: 10.1101/cshperspect.a016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu F.T., Callaghan J.M., Steele-Mortimer O., Stenmark H., Parton R.G., Campbell P.L., McCluskey J., Yeo J.P., Tock E.P., Toh B.H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine ‘fingers’ and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 36.Rappoport J.Z., Simon S.M. Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J. Cell Sci. 2009;122:1301–1305. doi: 10.1242/jcs.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sousa L.P., Lax I., Shen H., Ferguson S.M., De Camilli P., Schlessinger J. Suppression of EGFR endocytosis by dynamin depletion reveals that EGFR signaling occurs primarily at the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4419–4424. doi: 10.1073/pnas.1200164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams R.L., Urbé S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 39.Hung L.-Y., Tseng J.T., Lee Y.-C., Xia W., Wang Y.-N., Wu M.-L., Chuang Y.-H., Lai C.-H., Chang W.-C. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sehat B., Tofigh A., Lin Y., Trocme E., Liljedahl U., Lagergren J., Larsson O. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 2010;3:ra10. doi: 10.1126/scisignal.2000628. [DOI] [PubMed] [Google Scholar]
- 41.Nelson J.D., LeBoeuf R.C., Bomsztyk K. Direct recruitment of insulin receptor and ERK signaling cascade to insulin-inducible gene loci. Diabetes. 2011;60:127–137. doi: 10.2337/db09-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo H.-W., Ali-Seyed M., Wu Y., Bartholomeusz G., Hsu S.-C., Hung M.-C. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 43.Radulescu R.T. Insulin receptor alpha-subunit: a putative gene regulatory molecule. Med. Hypotheses. 1995;45:107–111. doi: 10.1016/0306-9877(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 44.Kabuta T., Hakuno F., Asano T., Takahashi S.-I. Insulin receptor substrate-3 functions as transcriptional activator in the nucleus. J. Biol. Chem. 2002;277:6846–6851. doi: 10.1074/jbc.M107058200. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y.-N., Yamaguchi H., Huo L., Du Y., Lee H.-J., Lee H.-H., Wang H., Hsu J.-M., Hung M.-C. The translocon sec 61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 2010;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giri D.K., Ali-Seyed M., Li L.-Y., Lee D.-F., Ling P., Bartholomeusz G., Wang S.-C., Hung M.-C. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bild A.H., Turkson J., Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 2002;21:3255–3263. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miaczynska M., Pelkmans L., Zerial M. Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Taub N., Teis D., Ebner H.L., Hess M.W., Huber L.A. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol. Biol. Cell. 2007;18:4698–4710. doi: 10.1091/mbc.E07-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorkin A., von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tee W.-W., Shen S.S., Oksuz O., Narendra V., Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiwari V.K., Stadler M.B., Wirbelauer C., Paro R., Schübeler D., Beisel C. A chromatin-modifying function of JNK during stem cell differentiation. Nat. Genet. 2012;44:94–100. doi: 10.1038/ng.1036. [DOI] [PubMed] [Google Scholar]
- 53.Di Vona C., Bezdan D., Islam A.B.M.M.K., Salichs E., López-Bigas N., Ossowski S., de la Luna S. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol. Cell. 2015;57:506–520. doi: 10.1016/j.molcel.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 54.Basnet H., Su X.B., Tan Y., Meisenhelder J., Merkurjev D., Ohgi K.A., Hunter T., Pillus L., Rosenfeld M.G. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516:267–271. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pokholok D.K., Zeitlinger J., Hannett N.M., Reynolds D.B., Young R.A. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 56.Chou R.-H., Wang Y.-N., Hsieh Y.-H., Li L.-Y., Xia W., Chang W.-C., Chang L.-C., Cheng C.-C., Lai C.-C., Hsu J.L., et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell. 2014;30:224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warsito D., Sjöström S., Andersson S., Larsson O., Sehat B. Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep. 2012;13:244–250. doi: 10.1038/embor.2011.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wortzel I., Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venetianer A., Dubois M.F., Nguyen V.T., Bellier S., Seo S.J., Bensaude O. Phosphorylation state of the RNA polymerase II C-terminal domain (CTD) in heat-shocked cells. Possible involvement of the stress-activated mitogen-activated protein (MAP) kinases. Eur. J. Biochem. 1995;233:83–92. doi: 10.1111/j.1432-1033.1995.083_1.x. [DOI] [PubMed] [Google Scholar]
- 60.Bellier S., Dubois M.F., Nishida E., Almouzni G., Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol. Cell. Biol. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellier S., Chastant S., Adenot P., Vincent M., Renard J.P., Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alepuz P.M., de Nadal E., Zapater M., Ammerer G., Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003;22:2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Proft M., Mas G., de Nadal E., Vendrell A., Noriega N., Struhl K., Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 64.Heidemann M., Hintermair C., Voß K., Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 65.Bonnet F., Vigneron M., Bensaude O., Dubois M.F. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2) Nucleic Acids Res. 1999;27:4399–4404. doi: 10.1093/nar/27.22.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujita T., Ryser S., Piuz I., Schlegel W. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol. Cell. Biol. 2008;28:1630–1643. doi: 10.1128/MCB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perales R., Bentley D. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollerer I., Grund K., Hentze M.W., Kulozik A.E. mRNA 3′end processing: A tale of the tail reaches the clinic. EMBO Mol. Med. 2014;6:16–26. doi: 10.1002/emmm.201303300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Ayoubi A.M., Zheng H., Liu Y., Bai T., Eblen S.T. Mitogen-activated protein kinase phosphorylation of splicing factor 45 (SPF45) regulates SPF45 alternative splicing site utilization, proliferation, and cell adhesion. Mol. Cell. Biol. 2012;32:2880–2893. doi: 10.1128/MCB.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danckwardt S., Gantzert A.-S., Macher-Goeppinger S., Probst H.C., Gentzel M., Wilm M., Gröne H.-J., Schirmacher P., Hentze M.W., Kulozik A.E. p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol. Cell. 2011;41:298–310. doi: 10.1016/j.molcel.2010.12.032. [DOI] [PubMed] [Google Scholar]
- 71.Lee S.-H., Choi H.-S., Kim H., Lee Y. ERK is a novel regulatory kinase for poly(A) polymerase. Nucleic Acids Res. 2008;36:803–813. doi: 10.1093/nar/gkm1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mina M., Magi S., Jurman G., Itoh M., Kawaji H., Lassmann T., Arner E., Forrest A.R.R., Carninci P., Hayashizaki Y., et al. Promoter-level expression clustering identifies time development of transcriptional regulatory cascades initiated by ErbB receptors in breast cancer cells. Sci. Rep. 2015;5:11999. doi: 10.1038/srep11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Llorens F., Hummel M., Pastor X., Ferrer A., Pluvinet R., Vivancos A., Castillo E., Iraola S., Mosquera A.M., González E., et al. Multiple platform assessment of the EGF dependent transcriptome by microarray and deep tag sequencing analysis. BMC Genomics. 2011;12:326. doi: 10.1186/1471-2164-12-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaw P.E., Saxton J. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 2003;35:1210–1226. doi: 10.1016/s1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 75.Odrowaz Z., Sharrocks A.D. ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes. PLoS Genet. 2012;8:e1002694. doi: 10.1371/journal.pgen.1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalo A., Kanter I., Shraga A., Sheinberger J., Tzemach H., Kinor N., Singer R.H., Lionnet T., Shav-Tal Y. Cellular levels of signaling factors are sensed by β-actin alleles to modulate transcriptional pulse intensity. Cell Rep. 2015;11:419–432. doi: 10.1016/j.celrep.2015.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.