Abstract
Casposons are a recently discovered group of large DNA transposons present in diverse bacterial and archaeal genomes. For integration into the host chromosome, casposons employ an endonuclease that is homologous to the Cas1 protein involved in protospacer integration by the CRISPR-Cas adaptive immune system. Here we describe the site-preference of integration by the Cas1 integrase (casposase) encoded by the casposon of the archaeon Aciduliprofundum boonei. Oligonucleotide duplexes derived from the terminal inverted repeats (TIR) of the A. boonei casposon as well as mini-casposons flanked by the TIR inserted preferentially at a site reconstituting the original A. boonei target site. As in the A. boonei genome, the insertion was accompanied by a 15-bp direct target site duplication (TSD). The minimal functional target consisted of the 15-bp TSD segment and the adjacent 18-bp sequence which comprises the 3′ end of the tRNA-Pro gene corresponding to the TΨC loop. The functional casposase target site bears clear resemblance to the leader sequence-repeat junction which is the target for protospacer integration catalyzed by the Cas1–Cas2 adaptation module of CRISPR-Cas. These findings reinforce the mechanistic similarities and evolutionary connection between the casposons and the adaptation module of the prokaryotic adaptive immunity systems.
INTRODUCTION
Most of the archaea and many bacteria possess an adaptive immune system, the CRISPR (Clustered Regularly Interspaced Palindromic Repeats)-Cas (CRISPR-associated proteins) system, whose role is to neutralize invading foreign DNA of viral or plasmid origin (1,2). This is achieved by maintaining an archive of fragments that are derived from previously encountered foreign genomes (3). These fragments are subsequently transcribed into an RNA molecule that, after processing, ends up as a guide for a distinct complex of Cas proteins which then recognizes and degrades further invading DNAs bearing the same (or in some case, closely related) sequence (for reviews see refs. (1,4–9)).
The library of foreign DNA fragments is located in the genome at the CRISPR locus, in which the fragments are integrated as spacers between identical, often palindromic, repeats of about 25–50 bp each (CRISPR) (9,10). Integration of new fragments is carried out by a complex that consists of the two most conserved Cas proteins, Cas1 and Cas2 (2,11) and in some cases, includes additional Cas proteins as well (12–15). Spacers are integrated at the 5′ end of the CRISPR array, resulting in duplication of the corresponding repeat (for recent reviews see refs (16,17)). The integrase activity of Escherichia coli Cas1 has been characterized in vitro (18). Cas1 has been shown to promote the integration of a synthetic, double-stranded spacer into a target plasmid. This reaction is strongly stimulated by Cas2 and occurs only with supercoiled target DNA unless integration host factor (IHF) is additionally supplied (19,20). When the target plasmid contained a CRISPR locus, the spacer was inserted preferentially at the border of the CRISPR array although some integration was also observed in the control plasmid pUC19 not bearing such repeats. In the latter case, however, the preferred site of integration was located in the vicinity of an inverted repeat with a potential to form a cruciform structure, suggesting that, as with bona fide CRISPR, such structures are important for spacer integration (18,21). It has been further shown that Cas1 alone displays sequence-specific activity, with a clear preference for the nucleotides flanking the integration site at the leader-repeat 1 boundary of the CRISPR locus, suggesting that the inherent sequence specificity of Cas1 is a major determinant of the adaptation process (11,22,23). The Cas1–Cas2 machinery is at the heart of the adaptation process and is considered to be the hallmark of the prokaryotic CRISPR-Cas immunity (1,2). However, how the adaptation machinery and the specificity of protospacer incorporation at the leader-proximal repeat boundary have evolved remains largely unknown.
Most of the cas1 genes identified in sequenced genomes belong to CRISPR-Cas systems; however, some Cas1 homologs are encoded outside of the CRISPR loci and are not associated with other cas genes (24,25). Instead, these genes appear to be part of clusters that always additionally include genes for family B DNA polymerases and a variable complement of other genes, including, in some cases, Cas4-like nucleases, another widespread component of the CRISPR-Cas systems (1,25), as well as other nucleases (24). These clusters are delimited by terminal inverted repeats (TIR) which themselves are flanked by direct repeats typical of the target site duplications (TSD) resulting from fill-in repair of the single-strand gaps generated upon staggered nick-mediated integration of various transposons (26–28). Consequently, we hypothesized that these gene clusters represent a novel group of transposons, dubbed casposons to emphasize the conservation of the Cas1 homologs and their likely involvement in transposition (24). Notably, the TIRs of some casposons show sequence and secondary structure similarity (i.e. palindromic organization) with certain CRISPR repeats. Based on these observations and phylogenetic analysis of Cas1 proteins, it has been proposed that the adaptation module of the CRISPR-Cas systems evolved from casposons (29). Although less common than many other families of transposons, casposons are present in both bacterial and archaeal genomes and are classified into four families based on the gene content and casposase phylogeny (30).
The integrase activity of the Cas1 protein encoded in the casposon of Aciduliprofundum boonei, hereinafter called casposase, has been demonstrated in vitro (31). The recombinant casposase was able to incorporate a 5′-labeled fluorescent oligonucleotide duplex consisting of the 26 terminal nucleotides of the TIR into the pUC19 plasmid. Integration proceeded by transesterification leading to the formation of a new phosphodiester bond between the 3′ end of the incoming duplex and the target strand of the plasmid, and to nicking of the plasmid. As a result of the nicking, the supercoiled form of the plasmid was converted to the relaxed form. A second nicking and integration event occurring on the opposite strand in the close proximity of the first one then linearized the plasmid (Supplementary Figure S1) (31).
The reaction displayed a strong specificity for the distal part of the TIR borne by the incoming DNA. Such specificity contrasts the case of CRISPR system, in which the only documented preference concerning the protospacer sequence in the subtype IE CRISPR-Cas system of E. coli appears to involve the 3′ terminal cytidine that is complementary to the G of the AAG protospacer adjacent motif (PAM) that is required both for protospacer acquisition and for targeting the cognate foreign DNA (18,32). Even more prominent difference between the A. boonei casposase and CRISPR Cas1 was that, in contrast to protospacer integration by CRISPR Cas1, casposon integration by the casposase seemed to display little specificity with respect to the site of insertion of the incoming casposon. However, this lack of specificity was deduced from the results of integration experiments performed with pUC19, a plasmid that carries no similarity to the sequences that flank the original casposon in the A. boonei genome. In contrast, recent comparative genomic analysis of a large collection of Methanosarcina mazei strains provided the first evidence of intra-genomic mobility of these elements and indicated that casposon integration is sequence-specific in vivo (33). These observations prompted us to re-examine the insertion specificity of the A. boonei casposase. To this end, we used a target plasmid carrying the regions flanking the casposon in the A. boonei genome and identified the site of integration of two substrates corresponding to the pre-cleaved casposon ends. These substrates were, respectively, a fluorescent oligonucleotide duplex matching the distal end of the casposon TIR and an artificial casposon consisting of a kanamycin resistance gene flanked by TIRs on both sides. The target site of the casposase identified through this approach resembles the target for protospacer integration catalyzed by the Cas1–Cas2 adaptation module of CRISPR-Cas, thus reinforcing the mechanistic similarity between the casposons and CRISPR-Cas which presumably stems from the evolutionary link between the two.
MATERIALS AND METHODS
Strains and plasmids
The lists of strains and plasmids used in this study are summarized in Supplementary Tables S1 and 2. A codon-optimized version of the sequence encoding the casposase of A. boonei was ordered from GeneArt™ (a subsidiary of Thermo Fisher Scientific). The gene, including the stop codon, was recloned between the Klenow-filled NcoI and the XhoI sites of the pETM-11, generating the pETM-11-Cas1 plasmid. The plasmid was then introduced into SoluBL21(DE3)™ (amsbio) bearing the pDIA17 plasmid (34).
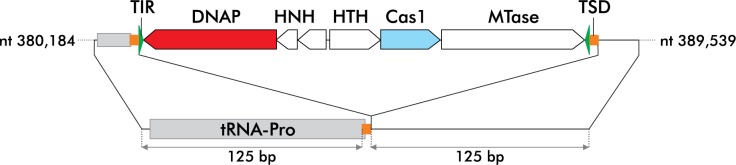
The pMA-Target plasmid was ordered from GeneArt™. It comprises the region extending from nt 380 184 to nt 380 323 fused with the region extending from nt 389 415 to nt 389 539 of the genome sequence of A. boonei T469 (accession n°CP001941.1). Thus, the synthesized segment includes a single copy of the duplicated target site CCCCACTACGAGGAG flanked by the adjacent 125 nt on either side (Figure 1). This DNA segment is inserted between the SacI and KpnI sites of the pMA plasmid (map available from the Thermofisher web site: https://www.thermofisher.com/content/dam/LifeTech/Documents/geneart/geneart-vector-map.pdf).
Figure 1.
Structure of the insert borne by pMA-Target. TIR: terminal inverted repeat; DNAP; DNAP: PolB-like DNA polymerase; HNH: HNH nuclease; HTH: helix-turn-helix protein; MTase: methyl transferase; TSD: TSD segment. The TIR, TSD and the tRNA-Pro gene are not drawn to scale.
The pMA-ΔTarget plasmid was constructed by digesting pMA-Target with NcoI and KpnI, followed by polishing the ends with Klenow DNA polymerase and recircularizing the vector, thus removing most of the A. boonei sequence. Other plasmids carrying deletions of pMA-Target were constructed by polymerase chain reaction (PCR) amplification of pMA-Target using divergent primers flanking the region to be deleted and carrying either SacI or KpnI extensions. The amplified segments were then gel-purified, digested with the corresponding enzymes plus DpnI to destroy the original template, and recircularized. The pMA-Target plasmid and its derivatives were introduced into TOP10 chemically competent cells (Invitrogen).
The pMA-Abooneicasp-kana plasmid was designed by inserting the kanamycin resistance gene of pET28, preceded by its promoter and followed by a T7 terminator, between the 36-bp left and right TIR of the A. boonei casposon. The plasmid then served as a template for the PCR amplification of the minicasposon encoding kanamycin resistance.
Production and purification of the casposase
A 1-L culture of SoluBL21(DE3)(pDIA17)(pETM-11-Cas1) was started at 30°C in 2YT medium containing 30 μg/ml kanamycin and 30 μg/ml chloramphenicol. The growth of the culture was followed by monitoring the OD600. When the OD600 reached 0.9–1.1, the culture was chilled for 5 min in a 13°C water bath, followed by induction with IPTG at a final concentration of 1 mM. After further incubating the culture overnight with shaking at 14°C, cells were centrifuged and the pellets kept at −80°C until processing.
The frozen pellet from a 1-L culture was resuspended in 20 ml buffer 1 (50 mM NaHPO4 pH 7.8 containing 1.2 M NaCl, 20 mM imidazole (neutralized to pH 8) and two pellets of protease inhibitor cocktail (Roche cOmplete Ultra mini®). Cells were ruptured by two passages with a French press set at 100 MPa and the crude extract was centrifuged for 30 min at 35 000 g. The supernatant was loaded on a column containing 1.5 ml Ni-NTA (Qiagen) equilibrated with the same buffer (but without protease inhibitors) and washed with several volumes of the same buffer. Remaining nucleic acids retained by the casposase were washed at room temperature with 5 column volumes of buffer 2 (50 mM Tris–HCl buffer pH 8 containing 4.75 M NaCl), followed by three volumes of buffer 1 and four volumes of buffer 3 (same as buffer 1 except 35 mM imidazole). The purified protein was eluted with 50 mM NaHPO4 pH 7.8 containing 1.2 M NaCl and 300 mM imidazole. Eluted fractions containing the protein were dialyzed against 50 mM Tris–HCl pH 8 containing 0.8 M NaCl and concentrated by ultrafiltration. In order to remove the N-terminal His-tag, which consisted of the sequence MKHHHHHHPMSDYDIPTTENLYFQ, part of the preparation (2 mg) was treated over night at 25°C with 1.6 mg His-tagged Tev protease in 4 ml 50 mM Tris-HCl pH 8 containing 0.5 M NaCl, 1 mM dithiothreitol and 0.25 mM ethylenediaminetetraacetic acid (EDTA). The de-tagged protein, which carried the two residues GA upstream of the original initiation methionine of Cas1, was separated from the tagged polypeptides (cleaved His-tag, tagged Tev protease and remaining His-tagged casposase) by passage over a 1.5 ml Ni-NTA column equilibrated with buffer 1. Unbound fractions were collected, concentrated by ultrafiltration, buffer was exchanged against 25 mM Tris–HCl pH 7.5 containing 0.5 M NaCl and 15% (v/v) glycerol and the preparation stored at −20°C.
Integration of a fluorescent oligonucleotide duplex
The integration of a fluorescent oligonucleotide duplex was performed as described in (31). Briefly, 200 nM of ds oligonucleotide formed by hybridizing LE26 and 6-FAM-labeled LE26r (Supplementary Table S3) was incubated for 1 h at 37°C in 100 μl Tris–HCl 25 mM containing 150 mM KCl, 5 mM MnCl2, 50 μg/ml bovine serum albumin (BSA), 1.5 μg/ml plasmid DNA and various concentrations of A. boonei casposase carrying or not an N-terminal His tag. The reaction was stopped by adding 25 mM EDTA (final), and the reaction was digested for 1 h with 60 μg proteinase K 30 u/mg (Eurobio, Paris). Nucleic acids were precipitated with ethanol in the presence of glycogen and 0.3 M sodium acetate, pH 5, washed with 70% ethanol, resuspended in 10 mM Tris–HCl, 1 mM EDTA, pH 8, heated at 65°C for 5 min and run on a 1% agarose gel in 1 × TBE (Tris/Borate/EDTA). Electrophoresis equipment was soaked in 2.6% hypochlorite to destroy any remaining ethidium bromide. The integration of the fluorescent duplex into the relaxed and linearized form of the plasmids was visualized on a GE Typhoon FLA 9500 imager.
Integration of a mini-casposon encoding kanamycin resistance
A mini-casposon containing a kanamycin resistance gene flanked by the 37-bp TIR from A. boonei was generated by PCR using primers LE41 and RE41 and pMA-Casp-kana as a template. The product was treated with DpnI and gel-purified to eliminate the template. Integration reactions were performed as described by Hickman and Dyda (31). Four μg/ml mini-casposon was incubated over night at 37°C in 100 μl Tris–HCl 25 mM containing 150 mM KCl, 5 mM MnCl2, 50 μg/ml BSA, 1.5 μg/ml plasmid DNA and 75 nM A. boonei casposase carrying or not an N-terminal His tag. Reaction products were column purified using the Macherey Nagel Gel and PCR purification kit, precipitated with ethanol and resuspended in 8 μl water. Two microliter were used to electroporate 20 μl of ElectroMax E. coli DH10B cells (ThermoFisher) as directed by the supplier. Cells were plated on carbenicillin plus kanamycin plates. The sites of integration of the casposon were determined by isolating the plasmids borne by the transformants and sequencing casposon boundaries with the divergent primers 533r and Seq3.
RESULTS
Insertion of the TIR-derived oligonucleotide duplex into pMA-Target and pMA-Δtarget
To investigate the specificity of casposon integration, we compared the integration of the 6-FAM-labeled LE26 duplex, which matches the distal part of the left TIR of the A. boonei casposon, into plasmids pMA-Target and pMA-ΔTarget. The pMA-Target plasmid carries a synthetic DNA segment that reconstitutes the target region of integration of the A. boonei casposon, with a single copy of the TSD segment flanked on each side by 125 bp of the original A. boonei chromosomal sequence. This segment was deleted in the pMA-ΔTarget plasmid. A. boonei casposase had been shown to perform both single and double-ended integration of the LE26 duplex into pUC19 in vitro, generating fluorescently labeled relaxed and linear forms of the plasmid, respectively (31).
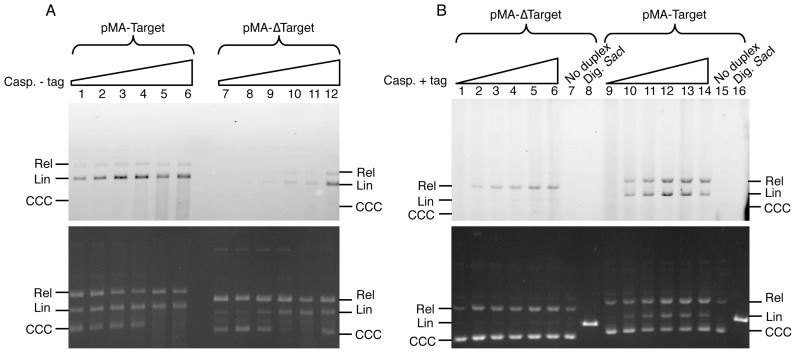
Figure 2A shows that, in agreement with previous findings (31), the de-tagged enzyme is able to generate the tandem integration of the duplex regardless of the presence or absence of the target sequence. In these reactions, labeled linearized forms of both pMA-Target and pMA-ΔTarget were generated. However, in the absence of the target sequence, (lanes 7–12), integration of the 6-FAM-tagged duplex was considerably less efficient.
Figure 2.
Integration by the Aciduliprofundum boonei casposase of the 6-FAM-labeled LE26 duplex into pMA-ΔTarget and pMA-Target. (A) Integration by de-tagged casposase. Integration of the 6-FAM-labeled LE26 duplex (200 nM) was monitored as described in ‘Materials and Methods’ section using 1.5 μg/ml pMA-Target (lanes 1–6) or pMA-ΔTarget (lanes 7–12) and increasing concentrations of de-tagged Cas1: 15 nM (lanes 1 and 7), 20 nM (lanes 2 and 8), 30 nM (lanes 3 and 9), 40 nM (lanes 4 and 10), 60 nM (lanes 5 and 11) and 75 nM (lanes 6 and 12). The gel was scanned to visualize integration of the fluorescent duplex (top panel) followed by staining with ethidium bromide (bottom panel). Rel, Lin and CCC: relaxed, linear and covalently closed, circular forms of the plasmids, respectively. (B) Integration by His-tagged casposase. Integration of the 6-FAM-labeled LE26 duplex (200 nM) was monitored as described in ‘Materials and Methods’ section using 1.5 μg/ml pMA-ΔTarget (lanes 1–6) or pMA-Target (lanes 9–14) and increasing concentrations of His-tagged Cas1: 0 nM (lanes 1 and 9), 15 nM (lanes 2 and 10), 30 nM (lanes 3 and 11), 45 nM (lanes 4 and 12), 70 nM (lanes 5 and 13) and 100 nM (lanes 6 and 14). In lanes 7 and 15, each plasmid was incubated with 100 nM Cas1, but no duplex. pMA-ΔTarget and pMA-Target linearized with SacI were loaded in lanes 8 and 16, respectively. After visualizing the 6-FAM fluorescence of the integrated duplex (top panel), the gel was stained with ethidium bromide (bottom panel). Similar results were obtained in two independent experiments.
Strikingly, when the reaction was performed with the His-tagged enzyme (Figure 2B), the tandem integration of the labeled LE26 duplex occurred exclusively in the presence of the A. boonei target site, as in pMA-Target. In contrast, integration in pMA-ΔTarget was less efficient and led almost exclusively to the relaxed form, indicating either single integration events or, possibly, double-ended integrations occurring at distantly located sites.
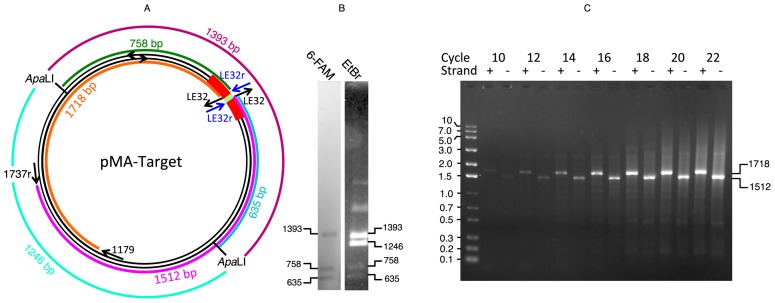
In order to locate the site of double-ended integration on pMA-Target, we performed the integration reaction with His-tagged casposase and pMA-Target DNA that had been pre-digested with ApaLI. ApaLI generates a 1393 nt fragment that carries the target site and a 1246 nt fragment that does not, thus single integration at the target site is expected to label the 1393 fragment, while tandem integration should yield two further fragments of 758 and 635 nt. Figure 3B confirms these predictions. Furthermore, it demonstrates that A. boonei casposase is able to perform integration on linear DNA, unlike the E. coli CRISPR Cas1–Cas2 complex (18).
Figure 3.
Determination of the site and strand of integration of the LE32 duplex. (A) Map of the pMA-Target plasmid and of expected PCR fragments (see panel C). The target region in shown in red, with the TSD segment in green; the LE32 and LE32r oligonucleotides are shown in black and blue, respectively; the PCR fragments generated in reactions 1 and 2 are in brown and purple, respectively. (B) Integration by His-tagged casposase of 6-FAM labeled LE-26 duplex into ApaLI-digested pMA-Target. Fifteen ng/μl ApaLI-digested pMA-Target was incubated for 1 h at 37°C in a total volume of 100 μl reaction buffer with 200 nM 6-FAM labeled LE-26 duplex and 75 nM His-tagged casposase. The reaction mixture was treated with proteinase K, precipitated and analyzed as described in ‘Materials and Methods’ section. Similar results were obtained for four independent incubations. The two unlabeled bands with slower migration seen in the ethidium bromide stain correspond to undigested closed circular and relaxed DNA molecules that did not undergo integration of the 6-FAM duplex. (C) Identification the strand bearing the integrated duplex upon single integration events. A preparative integration reaction was set up by incubating 450 ng of pMA-Target together with 200 nM LE32 duplex and 320 nM de-tagged Cas1 in a total volume of 300 μl reaction buffer for 95 min at 37°C. The relaxed form of pMA-Target was gel-purified and subjected to two PCR reactions with different pairs of primers: reaction 1 was performed with primers LE32r and 1179 and reaction 2 with primers LE32r and 1737r. Samples were withdrawn at the end of the elongation step at defined cycles of amplification from either PCR reaction and subjected to agarose gel electrophoresis. Integration revealed by amplification in reactions 1 and 2 were defined as occurring in strand + and −, respectively. The expected sizes of amplified fragments for integration at the TSD site are 1718 and 1512 bp for reactions 1 and 2, respectively (panel A).
We then sought to determine whether single integration of an oligonucleotide duplex by casposase proceeds with a preference for a specific strand of the plasmid. For this, we incubated pMA-Target with the de-tagged casposase in the presence of the LE32 duplex which matches the distal end of the left TIR. We then purified the relaxed form, corresponding to single-site integration, and subjected it to PCR amplification using the primers LE32r + 1179 or LE32r + 1737r (Supplementary Table S3). Samples were taken at different cycles of amplification and analyzed by agarose gel electrophoresis. Given that 1179 and 1737r hybridize to opposite strands of pMA-Target (Figure 3), the appearance of a defined fragment in either PCR implies that a single integration of the LE32 duplex has occurred at a specific site on the opposite strand.
Figure 3 shows two salient findings. First, there is clearly a preferential, but not exclusive integration site that yields defined fragments of about 1700 bp in the case of the PCR performed with LE32r + 1179 and 1500 bp in the case of the PCR performed with LE32r + 1737r. The length of these fragments corresponds to the length expected for integration of the duplex at the target site, i.e. 1718 and 1512 bp, respectively. Second, Figure 3 also shows that the two fragments appear and increase in the amount concomitantly and with the same intensity at each increasing number of PCR cycles. This result indicates that a single integration of the duplex preferentially occurs at the defined target site on both strands with little or no strand preference.
Insertion of an artificial casposon encoding kanamycin resistance
In their recent study, Hickman and Dyda (31) described an artificial mini-casposon with a kanamycin resistance gene flanked by the 15 or 30 distal bp of the left and right TIR of the A. boonei casposon. These mini-casposons were successfully integrated by the casposase into the pUC19 plasmid. Mapping and sequencing of the integration sites has shown that integration consistently involved duplication of a 14- or 15-bp sequence from the target site but occurred essentially randomly, with neither sequence signature nor orientation preference (31).
Given the specificity of TIR-derived duplex integration into the native target site, we tested the insertion specificity of the artificial mini-casposon consisting of a kanamycin resistance gene flanked by the full-length (37 bp) left and right TIRs from the genome of A. boonei. A summary of the results is presented in Tables 1 and 2, and the details of each clone can be found in Supplementary Tables S4 and 5.
Table 1. Modes of integration of a mini-casposon encoding kanamycin resistance into pMA-Target catalyzed by tagged and de-tagged Cas1.
| Enzyme | De-tagged Cas1 | Tagged Cas1 | ||
|---|---|---|---|---|
| Number of clones recovered | 129 ± 34a (2)b | 59 ± 19a (4)b | ||
| Total number of clones sequenced | 9 | 28 | ||
| Site of insertion | Insertion at the A. boonei target site | Insertion outside of the A. boonei target site | Insertion at the A. boonei TSD | Insertion outside of the A. boonei target site |
| Number of clones sequenced | 4 clones | 5 clones | 26 clones | 2 clones |
| Size of TSD at insertion site | 15 bp (2 clones) 4 bp (2 clones) | 16 bp (1 clone) 15 bp (1 clone) Unstable (1 clone) Deletion (2 clones) | 15 bp (26 clones) | 22 bp (1 clone) deletion (1 clone) |
anumber of clones ± standard deviation recovered per independent experiment.
bnumber of independent experiments (insertion + transformation).
Following transformation and selection for resistance to kanamycin, transformants were picked randomly and the regions flanking the left and right TIR were sequenced to determine the site of integration and the extent of sequence duplication occurring at the target site.
Table 2. Modes of integration catalyzed by tagged and de-tagged Cas1 into pMA-ΔTarget of a mini-casposon encoding kanamycin resistance.
| Enzyme | De-tagged Cas1 | Tagged Cas1 |
|---|---|---|
| Number of clones recovered | 80 ± 22a (3)b | 2 ± 1.8a (4)b |
| Number of clones sequenced | 9 clones | 8 clones |
| Size of TSD at insertion site | 13 bp (1 clone) 14 bp (3 clones) 15 bp (3 clones) 155 bp (1 clone) 1421 bp (1 clone) | 3 bp (1 clone) 14 bp (2 clones) 15 bp (3 clones) 546 bp (1 clone) 1802 bp (1 clone) |
anumber of clones ± standard deviation recovered per independent experiment.
bnumber of independent experiments (insertion + transformation).
Following transformation and selection for resistance to kanamycin, transformants were picked randomly and the regions flanking the left and right TIR were sequenced to determine the site of integration and the extent of sequence duplication occurring at the target site.
When performing the reaction with de-tagged casposase and pMA-Target, in four of the nine tested clones, insertion occurred at the A. boonei native target site with a duplication of the adjacent 15-bp segment, hereafter the TSD segment (Table 1 and Supplementary Table S4). In the remaining five clones, integration occurred randomly at sites remote from the A. boonei target site. This finding is consistent with the result of the LE32 duplex integration: Figure 3C shows a defined band arising from integration at the target site, which is surrounded by a background likely reflecting random integration at other sites.
When the His-tagged casposase was used to integrate the mini-casposon into pMA-Target, in 26 of the 28 clones, the mini-casposon was found inserted at the original target site reconstituted from the A. boonei genome, with a duplication of the 15-bp TSD segment. At variance with the original A. boonei sequence, an additional T was found in most cases between the upstream TSD segment and the beginning of the casposon TIR, and an A was found between the other end of the casposon and the downstream TSD (Supplementary Table S4). Most likely, the presence of these extra nucleotides is an artefact of PCR amplification with the Taq DNA polymerase, which is known to add a 3′-terminal A via its nucleotidyltransferase activity (35). Two clones were also identified in which casposon insertion did not result in duplication of the TSD. In one of these, the casposon was inserted at a different site and was flanked by a 22-bp repeat; in the other one, one end of the casposon was flanked by the A. boonei TSD segment, whereas the other end was inserted at a position located 288 bp upstream from the TSD segment (Table 1 and Supplementary Table S4).
The de-tagged casposase was then used to catalyze the integration of the mini-casposon into pMA-ΔTarget. The recovery of recombinant clones was about 50–60% of that obtained with pMA-Target. There was no obvious preference regarding the site of insertion, which seemed to occur randomly, consistent with the previous results (31). The majority (seven out of nine) of the clones tested displayed a reiteration of 13–15 bp flanking the casposon; however, two casposons were flanked by much larger reiterations of 155 and 1421 bp, respectively (Table 2 and Supplementary Table S5).
When the His-tagged casposase was used to integrate the mini-casposon into pMA-ΔTarget, the efficiency of insertion declined by an order of magnitude. The integration sites were located randomly; in addition, although five of the eight sequenced clones contained 14–15 bp repeats flanking either side of the casposon, three clones were flanked by repeats ranging from 3 to 1802 bp (Table 2 and Supplementary Table S5), indicating a less stringent application of the ‘15 bp yardstick’. No transformants were obtained when the reaction was performed in the absence of the enzyme, excluding the possibility that the observed integrations were the result of illegitimate recombination events taking place in E. coli.
Regardless of the substrate plasmid or the casposase preparation, in 52 of the 58 sequenced recombinant plasmids, insertion occurred such as to yield the same orientation for the kanamycin resistance gene borne by the mini-casposon and the ampicillin resistance gene borne by the plasmid. This uniformity is likely due to the selection by the host, which was plated with both antibiotics, and to the ability of the casposon to switch orientation by recombination between the TIRs. Indeed, PCR with appropriate primers demonstrated that single colonies contained both orientations but in such different proportions that sequencing nonetheless yielded a non-ambiguous result.
Determination of the target sequence required for recognition by the casposase
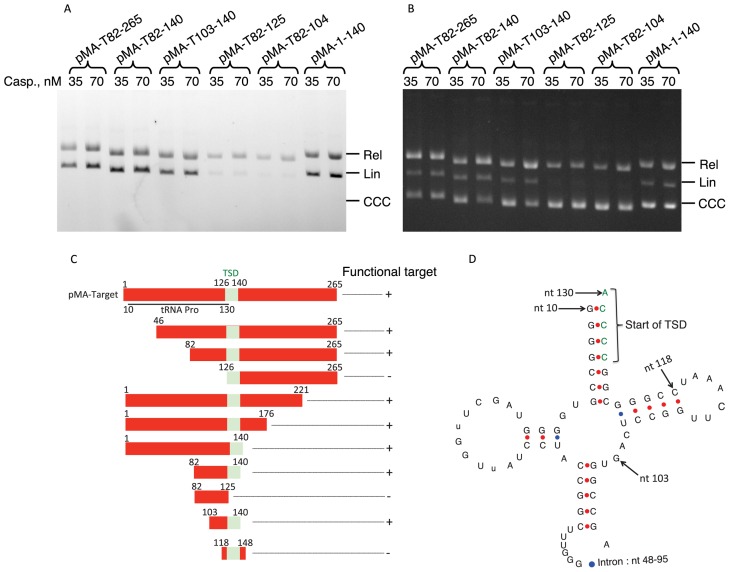
In order to delimit the minimum segment recognized by the casposase, we made use of the fact that the target sequence is essential for double-ended integration of the fluorescent LE26 duplex by His-tagged casposase, which generates the labeled, linear form of the plasmid. We thus generated a set of deletions in the pMA-Target and tested the incorporation of the duplex, as shown in Figure 4 A and B. The results are summarized in Figure 4C. The 15-bp TSD segment, where the two DNA strands are nicked, was insufficient to support tandem integration of LE26. Whereas the sequence downstream of the TSD segment was dispensable, the upstream region was strictly essential for integration. The shortest fragment tested that still bears the recognition sequence includes the 15-bp TSD segment and an 18-bp adjacent segment.
Figure 4.
Functional mapping of the region required for tandem integration of the LE26 duplex by His-taged Cas1. (A and B) Integration of the 6-FAM-labeled LE26 duplex (200 nM) was monitored as described in ‘Materials and Methods’ section with either 35 or 70 nM His-tagged casposase and 1.5 μg/ml of plasmids harboring various deletions of the target segment borne by pMA-Target. The concentration of the casposase and the borders of each target site are indicated above each lane.A: gel scanned for 6-FAM fluorescence; B, same as A, after ethidium bromide staining. (C) Diagram summarizing the results obtained for the tandem integration of the 6-FAM-labeled LE26 duplex using the plasmids mentioned in Table 2. The TSD segment is shown in green. (D) Structure of the tRNA encoded by the region upstream of the Aciduliprofundum boonei TSD segment. Arrows indicate the position of the tRNA gene and its intron relative to the insert of pMA-Target and of features such as the beginning of the TSD or the beginning of the insert borne by the pMA-T103-140 plasmid. Numbering starts at the beginning of the insert borne by pMA-Target. The Figure was drawn using the software available online and described in ref. (45).
An interesting feature of the A. boonei TSD segment is that it overlaps with the 3′ end of a gene encoding tRNA-Pro by 5-bp (Figure 4D). The 103–140 segment (Figure 4C) essential for casposon integration includes the segment corresponding to the TΨC loop of the tRNA.
DISCUSSION
The previous study on the A. boonei casposase has shown that the protein catalyzes non-sequence-specific integration of mini-casposons and generates TSD in the process (31). As confirmed in the present work, this is indeed what happens when a plasmid lacking the specific target site is used as a substrate for integration reactions. More specifically, we detected tandem integration into plasmids of a fluorescent oligonucleotide duplex corresponding to the distal part of the TIR; likewise, an artificial casposon containing a kanamycin resistance gene flanked by TIRs was integrated with the concomitant generation, in most cases, of a 15 ± 2 nt duplication at the integration site. When performed with a plasmid that lacked the original target region from A. boonei, these reactions displayed no obvious preference regarding the site of integration. Importantly, however, here we show that when the original A. boonei target site is present, this site is strongly preferred for integration by the casposase. In addition, the version of the enzyme containing a His-tag was almost incapable of performing tandem integration in the absence of the target site, thus displaying, somewhat surprisingly, an enhanced target specificity compared to the de-tagged, native form. The nature of this effect of the His-tag has not been directly addressed; conceivably, the concentration of the de-tagged casposase used in the experiments was sufficient to oversaturate the genuine, high affinity target site and lead to binding and integration into other, low-affinity random sites. Decreasing the affinity of the casposase would prevent oversaturation of the specific site and occupation of the non-specific sites. The present demonstration that the casposase shows considerable specificity toward the native integration sites is fully consistent with the comparative genomic findings which indicate that casposon integration in methanogenic archaea occurs in a sequence-specific manner (33). Interestingly, virtually all known site-specific recombinases belong to one of only two superfamilies, the tyrosine recombinases and the serine recombinases (36,37). Casposases and CRISPR Cas1 belong to neither of these groups and thus can be considered to represent the third superfamily of recombinases with preferred integration sites.
Integration by A. boonei casposase typically generates a duplication of the 15-bp TSD segment (plus or minus one or two nucleotides). However, in some cases, mini-casposon integration resulted from staggered transesterifications located at sites separated by distances widely differing from the 15-bp ‘yardstick’. These events were detected only when integration occurred outside of the canonical target site. A scenario compatible with these observations could be that the casposase binds to DNA and, while bound to a particular site, performs concerted transesterifications at sites separated by 15 bp. The A. boonei casposase has been shown to behave as a dimer in solution (31), as also is the case for CRISPR Cas1 in the absence of Cas2 (38,39). The dimer configuration provides a pair of catalytic sites poised for concerted reactions at a specific distance. However, if the interaction between casposase and DNA is weakened, the enzyme could dissociate before both reactions are completed. This would prevent linearization of the plasmid; furthermore, in cases when the complex binds again randomly to a second site, integrations would lead to duplications of random sizes. In this scenario, two factors would influence the stability of the functional interaction between the casposase and DNA. One is the target sequence: the enzyme displays a high, specific affinity for the segment reconstituted from the A. boonei integration site, making it a strongly preferred target for integration. Another one would be the presence of the N-terminal His-tag, which would destabilize the interaction. This would explain why the tagged enzyme is nearly incapable of performing the linearization of the plasmid unless the A. boonei target sequence is present; it would also account for the random size duplications observed when integration occurs outside of the canonical target site. The same effects likely would have been observed if the presence of the target sequence or the His-tag were to increase or decrease, respectively, the rates of both concerted integration reactions.
As a site-preferring integrase, A. boonei casposase resembles CRISPR Cas1, which also shows a strong preference for sites adjacent to CRISPR palindromic repeats (18). In the case of the A. boonei casposase, the target site includes the palindromic repeat formed by the sequence encoding the TΨC loop of tRNA-Pro. However, the presence of a palindrome potentially able to form a cruciform structure is unlikely to be the only factor that determines the site of integration. The segment 82–125, which includes the TΨC loop, but not the 15-bp region corresponding to the TSD, is not a functional target (Figure 4), and nor is the segment 126–265, which includes another palindrome, AGTCCCCCTTTGGGACT, that potentially could form a hairpin loop of greater stability than that of the TΨC loop (40).
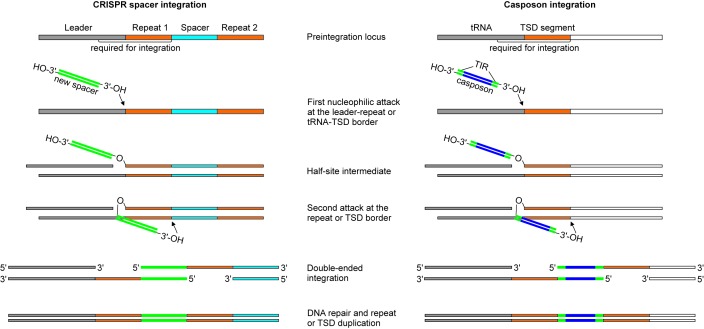
Notably, the sequence features of the casposon target site are functionally similar to those required for directional insertion of new protospacers into CRISPR arrays (Figure 5). In both systems, the functional target site consists of two components: (i) a sequence which gets duplicated upon integration of the incoming DNA duplex (i.e. the TSD segment in the case of casposon and a CRISPR unit during protospacer integration) and (ii) the upstream region which further determines the exact location of the integration (i.e. the leader sequence located upstream of the CRISPR array (11,22,23) and the 18-bp segment encoding the TΨC loop of tRNA-Pro in A. boonei). Directional integration of the protospacers at the leader-repeat 1 boarder is an important property of the CRISPR-Cas systems. The catalog of CRISPR repeats provides a sequential historical account of prior encounters between the host and mobile genetic elements (41). Similarly, the inherent features of the target site would in principle allow multiple casposon integrations in tandem, each new casposon being inserted upstream of the previous one, and every casposon being separated from its neighbors by a TSD segment. Such tandem casposon arrays have been indeed identified in the genomes of certain Methanosarcina species (33). Thus, the insertion of casposons appears to be deeply similar to the insertion of CRISPR spacers, which further emphasizes the evolutionary relationships between the two systems. However, in spite of clear structural and functional similarities, they also display notable differences. In particular, the spacers integrated by the CRISPR Cas1–Cas2 complex have a defined size and are generally short, ranging between 26 and 72 bp (9), whereas the casposase was able to integrate a 1024-bp artificial casposon whose only similarity to the original 9.1 kb casposon of A. boonei included the two TIRs. Furthermore, integration of casposons is strictly dependent on the sequence of the distal ends of the TIRs (31), whereas in the E. coli CRISPR-Cas system, the specificity of incorporation of new spacers into CRISPR loci is limited to single nucleotide originating from the adjacent PAM motif (18,42). In this respect, A. boonei casposase resembles other transposases, which also recognize TIR (reviewed in ref (26)). Another difference is that integration by the A. boonei casposase does not depend on the DNA conformation and can proceed with linear DNA (Figure 3 and ref. (31)), whereas the CRISPR Cas1–Cas2 complex requires a supercoiled target plasmid, unless bending of the target DNA is effected by the IHF (19). Finally, no homolog of CRISPR Cas2 has yet been identified within the casposons described so far. Investigating the crystal structure of A. boonei casposase, particularly in complex with its substrates, will enable detailed comparison with the CRISPR-Cas structures (20,43,44) and shed light on the differences and further similarities between the two systems. Such comparison will also provide crucial information on the features that determine the recognition specificity of the casposon TIR and of the integration target site. It is to be noted that for both systems, only the integration reaction so far has been amenable to successful in vitro dissection whereas the excision of CRISPR spacers or of casposons from their original locations still awaits a similar development.
Figure 5.
Comparison of models of CRISPR spacer (left) and casposon (right) integration. The first nucleophilic attack occurs at the leader-repeat or the tRNA-TSD segment border by the 3′-OH end of the spacer or the casposon, respectively. After the formation of the half-site intermediate, the second nucleophilic attack occurs on the opposite strand at the repeat-spacer border (for CRISPR spacer) or at the junction between the TSD segment and the flanking DNA, distal to the tRNA gene (for casposon). The resulting single-stranded DNA gaps are repaired by uncharacterized mechanisms. The left panel has been modified from ref (19).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) [UE 340440 to P.F.]; US Department of Health and Human Services Intramural Funds (to the National Library of Medicine) (to E.V.K.). Funding for open access charge: ERC [UE 340440 to P.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 4.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Attar S., Westra E.R., van der Oost J., Brouns S.J. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 2011;392:277–289. doi: 10.1515/BC.2011.042. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 8.Charpentier E., Richter H., van der Oost J., White M.F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorek R., Kunin V., Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 10.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y., Terns R.M., Terns M.P. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorontsova D., Datsenko K.A., Medvedeva S., Bondy-Denomy J., Savitskaya E.E., Pougach K., Logacheva M., Wiedenheft B., Davidson A.R., Severinov K., et al. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015;43:10848–10860. doi: 10.1093/nar/gkv1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amitai G., Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 17.Sternberg S.H., Richter H., Charpentier E., Qimron U. Adaptation in CRISPR-Cas systems. Mol. Cell. 2016;61:797–808. doi: 10.1016/j.molcel.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Nuñez J.K., Lee A.S., Engelman A., Doudna J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuñez J.K., Bai L., Harrington L.B., Hinder T.L., Doudna J.A. CRISPR immunological memory requires a host factor for specificity. Mol. Cell. 2016;62:824–833. doi: 10.1016/j.molcel.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Nuñez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature. 2015;527:535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arslan Z., Hermanns V., Wurm R., Wagner R., Pul U. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 2014;42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rollie C., Schneider S., Brinkmann A.S., Bolt E.L., White M.F. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife. 2015;4:e08716. doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Y., Chesne M.T., Terns R.M., Terns M.P. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015;43:1749–1758. doi: 10.1093/nar/gku1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krupovic M., Makarova K.S., Forterre P., Prangishvili D., Koonin E.V. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova K.S., Wolf Y.I., Koonin E.V. The basic building blocks and evolution of CRISPR-CAS systems. Biochem. Soc. Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickman A.B., Dyda F. Mechanisms of DNA Transposition. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0034-2014. doi:10.1128/microbiolspec.MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siguier P., Gourbeyre E., Varani A., Ton-Hoang B., Chandler M. Everyman's guide to bacterial insertion sequences. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0030-2014. doi:10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 28.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin E.V., Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat. Rev. Genet. 2015;16:184–192. doi: 10.1038/nrg3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krupovic M., Koonin E.V. Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems. Curr. Opin. Microbiol. 2016;31:25–33. doi: 10.1016/j.mib.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickman A.B., Dyda F. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 2015;43:10576–10587. doi: 10.1093/nar/gkv1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 33.Krupovic M., Shmakov S., Makarova K.S., Forterre P., Koonin E.V. Recent mobility of casposons, self-synthesizing transposons at the origin of the CRISPR-Cas immunity. Genome Biol. Evol. 2016;8:375–386. doi: 10.1093/gbe/evw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munier H., Gilles A.M., Glaser P., Krin E., Danchin A., Sarfati R., Barzu O. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 35.Marchuk D., Drumm M., Saulino A., Collins F.S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grindley N.D., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 37.Rice P.A. Resolving integral questions in site-specific recombination. Nat. Struct. Mol. Biol. 2005;12:641–643. doi: 10.1038/nsmb0805-641. [DOI] [PubMed] [Google Scholar]
- 38.Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., Gagarinova A., Pogoutse O., Brown G., Binkowski A., et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiedenheft B., Zhou K., Jinek M., Coyle S.M., Ma W., Doudna J.A. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fineran P.C., Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Mojica F.J., Diez-Villaseñor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 43.Nuñez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.