Abstract
Gene expression regulation by the stringent response effector, ppGpp, is facilitated by DksA protein; however DksA and ppGpp can play independent roles in transcription. In Escherichia coli, the pArgX promoter which initiates the transcription of four tRNA genes was shown to be inhibited by ppGpp. Our studies on the role of DksA in pArgX regulation revealed that it can stimulate transcription by increasing the binding of RNA polymerase to the promoter and the productive transcription complex formation. However, when DksA is present together with ppGpp a severe down-regulation of promoter activity is observed. Our results indicate that DksA facilitates the effects of ppGpp to drive formation of inactive dead-end complexes formed by RNA polymerase at the ArgX promoter. In vivo, ppGpp-mediated regulation of pArgX transcription is dependent on DksA activity. The potential mechanisms of opposing pArgX regulation by ppGpp and DksA are discussed. pArgX is the first reported example of the promoter stimulated by DksA and inhibited by ppGpp in vitro when an overall inhibition occurs in the presence of both regulators. A dual role is thus proposed for DksA in the regulation of the pArgX promoter activity.
INTRODUCTION
The stringent response is a global regulatory systems in bacteria (1 and refs. therein). Over the years, the study of this control network has yielded numerous data revealing its impact on many bacterial cellular processes. The effectors of the stringent response are the unusual nucleotide, guanosine tetra/pentaphosphate, together referred to as (p)ppGpp, which accumulates rapidly to levels approaching that of ATP upon the onset of various stress conditions, including nutrient deprivation and other environmental stresses. In Escherichia coli two genes are responsible for the synthesis of (p)ppGpp: relA and spoT (1,2). (p)ppGpp was reported to associate with RNA polymerase (RNAP), affecting its activity during transcription (2) and in consequence altering the transcription profile of the entire cell (3,4). In spite of years of intense research, recent reports prove that detailed operation of the stringent response and its mechanisms still remain to be explored (2). The understanding of (p)ppGpp function in the cell was expanded by the discovery of the role of DksA protein in the stringent response. DksA, originally identified as a suppressor of dnaKJ deficiency (5) is a 17-kDa RNAP-associated protein (6). Discovery of the role of DksA was a breakthrough in the understanding of the mechanism of the stringent response and numerous examples described the synergistic role of DksA in ppGpp-mediated transcription regulation (7–10). Contrary to ppGpp, DksA is present at a constant level in the cell during all growth phases (7,11), thus its role has been suggested to sensitize the transcription capacity of RNAP to various ppGpp levels (7,8) by stabilizing the RNAP-ppGpp interaction (6). DksA binds RNAP through the secondary channel imposing structural changes of the enzyme that influence transcription initiation (6,12–14). The main step affected by these alterations is a formation and stability of the promoter-RNAP open complexes and specific effects on a given promoter activity depends on the intrinsic features of transcription initiation intermediates at that promoter (10). For rRNA promoters with particularly unstable open complexes, further destabilization leads to the release of the RNAP from the promoter and inhibition of promoter activity. However, in the case of amino acid biosynthetic promoters stimulated by the stringent response, the destabilization of the stable open complexes facilitate the isomerization step at the formation of open complex and subsequently, effective RNA synthesis.
In a view of the fact that DksA enhances negative and positive effects of ppGpp on promoters’ activities, it was proposed that DksA plays a role as a cofactor of the stringent response (6–8,15). Nevertheless, recent reports indicated that DksA and ppGpp can have independent or opposing effects (16–20) or their effects may very specifically depend on the individual promoters, holoenzymes involved or stress conditions (21). One of the examples is the bacteriophage lambda pR promoter, which was shown to be inhibited by ppGpp and stimulated by DksA (18,22).
The role of ppGpp is to adjust the translational capacity of the cell to reduced levels under unfavorable conditions (e.g. starvation) in order to preserve cellular resources. This is manifested in the inhibition of production of the protein-synthesis system that is translation components: rRNA, r-protein and tRNA (1,2). The promoter initiating the expression of one of the seven rRNA operons in E. coli, rrnB P1, has been extensively studied and is currently one of the best characterized bacterial promoters (23–25). Promoters initiating the synthesis of tRNA (another class of stringently controlled RNA) are far less studied in comparison to rrnB P1; however the mechanism underlying their inhibition during the stringent response was proposed to be similar (26,27). pArgX promoter initiates the transcription of the argX operon containing 4 tRNA genes: argX, hisR, leuT, proM. The activity of this promoter was shown to be inhibited in vivo by the stringent response (28). The upstream sequences were demonstrated to play important roles in pArgX transcription (29). pArgX served as a model for studying the ppGpp interaction with RNAP (30). Further studies revealed that ppGpp leads to the formation of the transcriptionally inactive, dead-end complexes at pArgX leading to promoter inhibition (31). Thus, in this work we aimed to elucidate the role of DksA in pArgX transcription. Surprisingly, we found that pArgX transcription regulation shares many of the features with lambda pR rather than with rrnB P1 in terms of ppGpp and DksA responsiveness. On the other hand, pArgX is unique in the sense that presently it is the only known promoter to be differentially regulated by DksA depending on the presence or absence of ppGpp.
It should be noted that there is some confusion in the databases caused by the presence of argT gene coding for the lysine/arginine/ornithine transporter subunit. Thus, as suggested in the EcoCyc database, the tRNA operon promoter should be named pArgX (according to the first gene of the operon). The previous published data kept to the old nomenclature, using pArgT (31), however, to avoid further confusion, in this work we refer to this promoter as pArgX, indicating that it is a promoter initiating the transcription from argX gene, not argT. We keep this nomenclature even when referring to the work of others, where pArgT name was still in use.
MATERIALS AND METHODS
Bacterial strains and plasmids
The derivatives of Escherichia coli MG1655ΔlacZ strain (30): ppGpp-null (relA spoT) (from M. Cashel), dksA (RK201) (5) and relA spoT dksA (18) were employed in this work. Bacterial cultures were grown in Luria broth (LB) (32) supplemented when needed with ampicillin (100 μg/ml), kanamycin (50 μg/ml). For the supercoiled in vitro transcription template, pTE103 plasmid (33) was used as a vector and the construction was done as follows: the pArgX promoter sequence (400 bp, from −265 to +135) relative to the transcription start site was amplified by polymerase chain reaction (PCR) employing primers argX1 (5′ GTTCCGCTCAATCCTGTTCCCGGGGGCCAATTACG) and argX2 (5′ GCCACCACTACAAAGCTTGTTACGC) which introduced SmaI and HindIII restriction sites at the end of the products. This DNA fragment was then inserted to the corresponding restriction sites of pTE103. The pTE103 contains T7 terminator region and the transcription from pArgX from the pTE103-derived construct leads to the 419 nt transcript. All cloning was done according to standard techniques. The fidelity of PCR-derived DNA and constructs was subsequently verified by DNA sequencing.
Nucleotides and proteins
Nucleotides were purchased from Roche Applied Science, deoxyribonucleotides from Fermentas Bioscience and (α-32P)CTP and (γ-32P)ATP from Hartmann Analytic. Native RNA polymerase from E. coli was purified as described (34) with modifications as in (35). The purification of N-terminal His-tagged DksA was performed according to (17).
RNA extraction, reverse transcription and qPCR analysis
E. coli strains were cultivated at 30°C. Samples were taken at two time points at OD600 = 0.2 and 1.0. Total RNA was extracted using High Pure RNA Isolation Kit (Roche), according to the manufacturer's protocol (which includes DNase I treatment of the samples). The total RNA concentration and purity of samples was assessed using Nanodrop spectrophotometer. Next, reverse transcription was performed using RevertAid H minus Reverse Transcriptase (Thermo Scientific) and specific primers indicated in Table 1, according to manufacturer's protocol. Next, quantification of transcripts in samples was performed by real time RT-qPCR analysis using SYBR Green based method as described previously (36) and it was carry out in LightCycler® 480 instrument (Roche). Briefly, the qPCR reaction (10 μl) contained: 1x SYBR Green I Master Mix (Applied Biosystems), oligonucleotides (final concentration 20 pmol, see Table 1) and 2 μl of cDNA template obtained in a previous step. The qPCR assay was performed with the following amplification program: activation of the enzyme at 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 10 s. Melting curve analysis of the PCR products was performed to ensure specificity of the products. Normalization was done by the amount of total RNA in each RT reaction (100 ng), as well as by the level of expression of a reference gene rrsB. Relative mRNA level in each sample was normalized to wild-type strain sample at OD600 = 0.2.
Table 1. Oligonucleotides used for qRT-PCR.
| Gene name | Product size (bp) | Forward | Reverse |
|---|---|---|---|
| argX | 112 | TAAGCGCCCGTAGCTCAG | ACTACCACCGCAGCTCAAG |
| rrsB | 130 | TGTCGTCAGCTCGTGTTGTG | ATCCCCACCTTCCTCCAGTT |
In vitro transcription reaction
Supercoiled templates for in vitro reactions were obtained by purification of pTE103-derived plasmid (pArg-pTE) from the wild type E. coli strain using ultracentrifugation through CsCl-ethidium bromide density gradients (32). Prior to use, DNA templates were further purified using P-30 desalting columns (BioRad) equilibrated with RNase-free water.
In vitro transcription reactions were performed essentially as described previously (18). The conditions of the transcription reaction were established previously for lambda pR promoter as optimal for assessing the subtle changes in transcription efficiency in the presence of regulators. pArgX as the promoter exhibits similar features and strength, was studied in the same conditions and concentrations of compounds. Single-round reactions were performed in a total volume of 17 μl, in transcription buffer containing 50 mM Tris-HCl pH 8, 10 mM MgCl2, 150 mM KCl, 10 mM β-mercaptoethanol, 10 μg/ml bovine serum albumin, with the final concentration of nucleotides: CTP and GTP of 150 μM, ATP 1 mM, UTP up to 15 μM and (α-32P)UTP up to 10 μCi with heparin (100 μg/ml) for single-round transcription. Template DNA (5 nM) with RNAP (15 nM), KCl (150 mM) and the indicated amounts of the ppGpp and/or DksA protein were incubated for 10 min at 37°C. The reactions were started by the addition of nucleotides (see above) with heparin (100 μg/ml), the samples were incubated at 37°C for 10 min. Reactions were terminated by the addition of 3 μl of the stop buffer (150 mM ethylenediaminetetraacetic acid (EDTA), 1.05 M NaCl, 7 M urea, 10% glycerol, 0.0375% xylene cyanol, 0.0375% bromophenol blue). The samples were separated by electrophoresis in 8% polyacrylamide gel containing 7M urea in TBE buffer at 30 mA. The gel was dried and 419 nucleotides long pArgX-initiated transcript was visualized and quantified using the PhosphorImager system (BioRad) and the ImageQuant program.
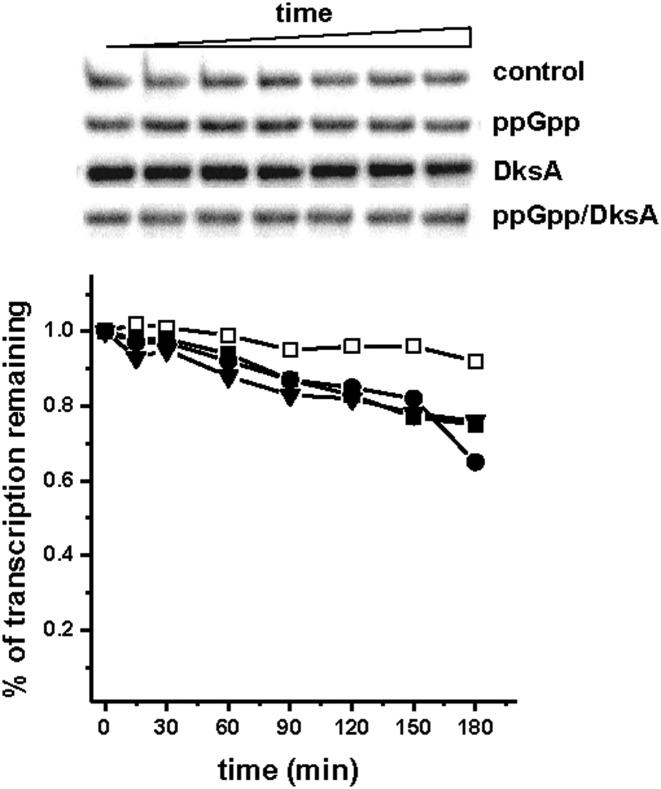
For the analysis of the productive complex formation (Figure 5), the transcription reactions were performed in transcription buffer (see above) supplemented with DNA template (5 nM) with KCl (150 mM) and ppGpp (200 μM) and/or DksA (400 nM) at 20°C for 10 min. The reactions were started by addition of RNAP (15 nM) and mix of nucleotides (see above) with heparin (100 μg/ml). At indicated time-points, samples (17 μl) were withdrawn and after addition of the stop buffer (3 μl) analyzed as described above.
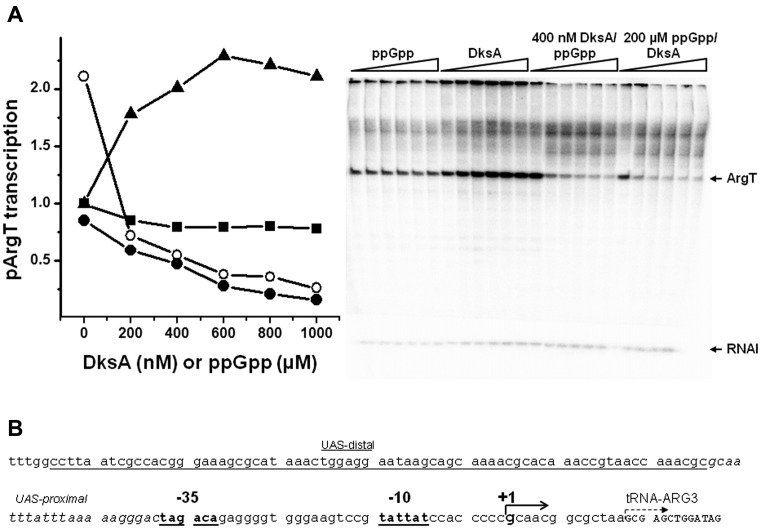
Figure 5.
The effect of ppGpp and DksA on the in vitro productive complex formation at pArgX. The transcription reaction was started by simultaneous addition of RNAP and NTPs to the mixture containing buffer and DNA with no addition (DksA storage buffer, open squares), ppGpp at 200 μM (closed squares), DksA at 400 nM (triangles) or DksA at 400 nM with addition of 200 μM ppGpp (circles). Samples were withdrawn at indicated times and reaction terminated by addition of the stop buffer. (A) Quantification of obtained transcripts. Data were normalized to the level of transcription at 30 min assessed to be 1 for each experimental set-up. The results are from three independent experiments. SD was below 10%. (B) Example of the autoradiogram obtained in the experimental conditions as above.
Open complex stability assay
The experiments for assessment of open complex stability were performed as described in (18). Briefly, RNAP (15 nM), DNA template (5 nM), KCl (150 mM), ppGpp (200 μM) and DksA (400 nM) were incubated for 10 min at 37°C in transcription buffer (as described above). After heparin (100 μg/ml) addition, 15 μl aliquots were removed to a tube containing 2 μl of nucleotides (see above), at indicated times. Reactions were performed for 10 min at 37°C. The reaction was stopped by addition of 3 μl of the stop buffer and analyzed as above.
Electromobility shift assays (EMSA)
The 5′-32P-labeled oligonucleotide argT1FP (5′CAATCCTGTTCCCGGGGGCCAATTACG) and non-labeled argT2FP (5′ GCCACCACTACAAAGCTTGTTACGC) were employed for obtaining of DNA fragment of 420 bp length. Assays were carried out as in (18). Briefly, 15 ng of DNA was incubated in B-buffer (25 mM HEPES, pH 7.5, 0.1 mM EDTA, 5 mM DTT and 10% glycerol) supplemented with 0.1 mg/ml poly(dIdC), 80 mM KCl, 200 μM ppGpp and/or 400 nM DksA with indicated RNAP concentrations (Figure 3) for 20 min in 37°C. The samples were separated on 3.5% Tris–glycine (pH 8.5) polyacrylamide gel at 120 V in 4°C. The DNA bands were visualized and quantified using PhosphorImager system (BioRad).
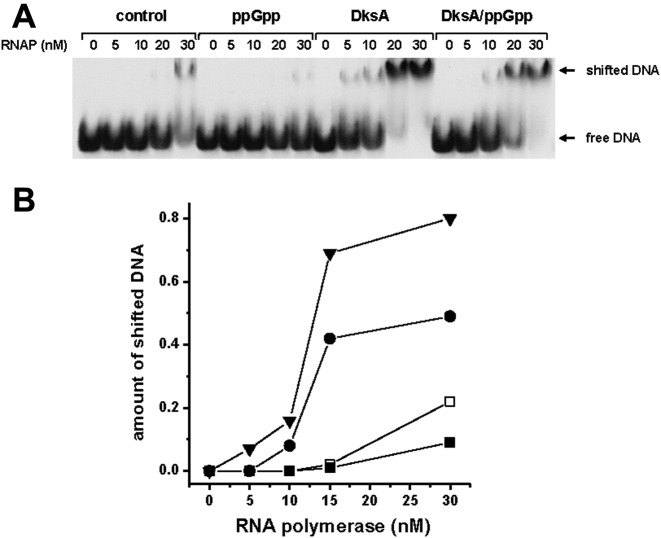
Figure 3.
RNAP binding to the DNA fragment containing pArgX promoter. (A) Electromobility shift assay (EMSA) of 32P-labeled DNA fragment and indicated amounts of RNAP. (B) The quantifications of the percentage of RNAP–DNA complexes (shifted DNA) formed with no addition (DksA storage buffer, open squares), ppGpp at 200 μM (closed squares), DksA at 400 nM (triangles) or DksA at 400 nM with addition of 200 μM ppGpp (circles). The data are from 4 independent experiments with SD below 10%. The example experiment autoradiogram is shown.
DNase I footprinting assay
P32-end-labeled DNA fragments containing pArgX region were obtained by PCR using 5′-labeled (P32) primers: 5′-CCGACAGGATTCGAACCTGAGAC and non-labeled 5′-CTCAGGAGAGCAGCGCTCTATCCAG followed by subsequent purification of the 135 nt DNA fragment. Labeled DNA template was incubated at 37°C for 10 min in transcription buffer supplemented with 25 mM KCl, 150 nM RNAP and/or 400 nM DksA and 600 μM ppGpp, then NTPs (final concentration of each NTP of 1 mM) was added and reaction was carried out at 37°C for 5 min. The final reaction volume was 20 μl. Reaction mixtures were treated with DNase I (Promega Corp.) at 4°C for 15 min, terminated by addition of EDTA (25 mM final), dried and resuspended in 6 μl of loading buffer (95% formamide, 10 mM NaOH, 0.05% bromophenol blue and 0.05% xylene cyanol). The samples were resolved in 7 M urea, 10% polyacrylamide sequencing gels, run in parallel with sequencing reactions obtained by using unlabeled DNA fragment as a template with P32-end labeled primer.
RESULTS
pArgX-initiated transcription is activated by DksA protein and inhibited in the presence of DksA and ppGpp
The transcription from pArgX promoter was reported to be decreased by ppGpp, as well as by an RNA polymerase mutant mimicking the presence of ppGpp (31). These studies, however, did not assess the potential role of DksA on pArgX transcription.
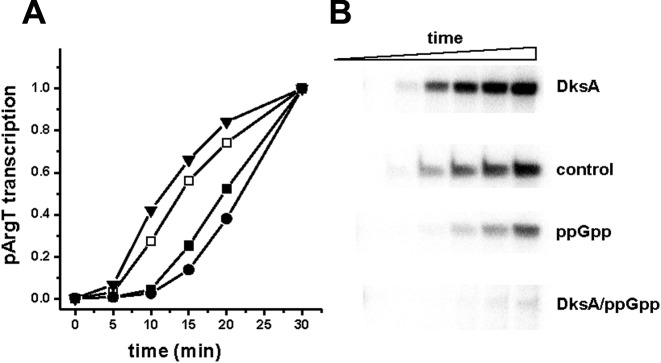
In the recent years, numerous studies have shown DksA, as well as ppGpp, effects on transcription during the stringent response and in further studies, that these regulators can perform independent roles. Among others, the discovery of independent and antagonistic roles of DksA and ppGpp in pR-initiated transcription brought new insight into the understanding of the functions of DksA beyond an auxiliary factor for the stringent response (18). Thus, we asked the question about the possible effect of DksA in pArgX-initiated transcription. We observed that in the in vitro transcription experiment the presence of increasing amounts of DksA activated pArgX transcription with a maximal effect of about 2.5-fold stimulation at 600 nM protein, while ppGpp alone resulted in about 30% inhibition of pArgX activity (Figure 1). This observation was in accordance with the results obtained previously for the pR promoter (18). In our experimental set-up, KCl was present at the concentration of 150 mM throughout the reaction. These in vitro transcription conditions correspond to the experiments performed for pR (18). Fifty percent inhibition of pR transcription was observed only when ppGpp was incubated with RNAP in low salt conditions and at room temperature prior to addition of 150 mM KCl and other components and temperature increase to 37°C (18,22). We tested the ppGpp effect on pArgX transcription in this experimental set-up and found that promoter activity was inhibited by 50%, similarly to pR (data not shown). This observation suggested that pArgX behaves similarly to the lambda pR, in terms of ppGpp and DksA responsiveness. However, the presence of both regulators had a different effect: contrary to pR transcription, DksA and ppGpp present together inhibited dramatically pArgX-initiated transcription (Figure 1). Similar inhibition was observed when DksA was titrated in the presence of 200 μM ppGpp or when increasing ppGpp concentration was added to the reactions containing 400 nM DksA. It should be noted though that in the presence of DksA the start-point of the transcription level is 2-fold higher than transcription with no addition due to stimulatory effect of DksA. The addition of ppGpp even in the lowest concentration (200 μM) transcription was decreased below that observed in the absence of any regulators. Further increase in the DksA concentration up to 1 μM resulted in yet stronger inhibition of promoter activity. The same effect was observed for the increasing concentrations of ppGpp in the presence of 400 nM DksA. In both experiments severe inhibition (down to 10% of promoter activity with no addition) was observed at the highest amounts of regulators: 1 mM ppGpp (with 400 nM DksA) and 1 μM DksA (with 200 μM ppGpp) (Figure 1). These results indicate that DksA is capable of activating pArgX-initiated transcription when present alone, while it acts synergistically with ppGpp resulting in significant transcription inhibition.
Figure 1.
The effects of ppGpp and DksA on the in vitro transcription initiated from pArgX promoter. (A) Transcription was assessed in the presence of increasing concentrations of ppGpp (squares), DksA (triangles), DksA with addition of 200 μM ppGpp (closed circles) or ppGpp with addition of 400 nM DksA (open circles). RNAP was incubated with the template DNA with regulators, where applicable. When two regulators were present, the one present in the constant concentration was added first, then immediately, the indicated amounts of another regulator were added. Then, a single round transcription reaction (10 min at 37°C) was initiated by addition of nucleotides mix with heparin. Transcription in the absence of ppGpp and DksA was set as 1. Data are from three independent experiments with SD below 10%. Autoradiogram shows an example experiment performed as described above. (B) The pArgX promoter region indicating the position of UAS, −35 and −10 sequences as well as a transcription start site (29 and our observations).
The complex regulation of pArgX in vivo (29) may suggest that the regions located upstream of the transcription start site play important role in the regulation of promoter activity, due to, for example, binding of transcription factors, which can affect ppGpp/DksA mediated regulation. Thus, we performed experiments with the construct containing −40 to +5 ArgX promoter region cloned into the same pTE103 vector. The transcription with this minimal promoter region was under very similar regulation of DksA and ppGpp, with mildly increased ppGpp inhibitory effect (data not shown). This indicates that the pArgX promoter regulation by ppGpp and DksA is not dependent on the sequences upstream of the promoter region.
DksA is required for ppGpp-mediated pArgX inhibition in vivo
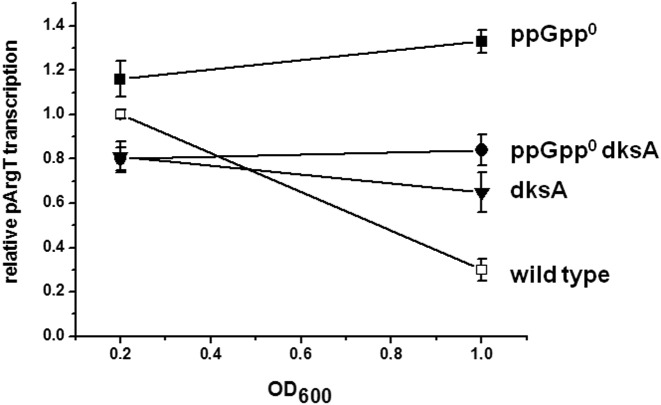
The intriguing results of the in vitro transcription in the presence of ppGpp and DksA led us to investigate pArgX promoter activity in vivo in the genetic context of bacteria lacking either or both these regulators. To address this question, we analyzed level of pArgX-initiation in vivo, employing qRT-PCR in the early exponential and the switch to stationary growth phase. As expected, in the wild-type strain transcription decreased upon the entrance into stationary phase reflecting the increased ppGpp level at this stage of bacterial growth, when in the absence of ppGpp level of transcription was mildly elevated, suggesting that DksA can induce the pArgX transcription. The lack of DksA resulted in slight decrease of pArgX transcription while in the absence of both regulators pArgX activity was not affected by the growth phase (Figure 2). These data indicate that in vivo DksA is necessary for ppGpp-mediated down-regulation of pArgX promoter activity.
Figure 2.
Relative pArgX transcription in vivo assessed by qRT-PCR. The qRT-PCR reaction was performed using the RNA isolated from the samples from bacterial cultures (wild type, open squares, ppGpp0, closed squares, dksA, triangles, dksA ppGpp0, circles as indicated). The results are from 3–6 independent experiments with SD indicated.
DksA facilitates RNAP binding to DNA
It was demonstrated previously that DksA can directly facilitate RNAP binding to a promoter (17,18,37). Based on our previous finding that the mechanism of the DksA-mediated transcription activation of pR promoter was based on this effect, we analyzed the RNAP binding to pArgX promoter region by electromobility shift assay (EMSA). DksA stimulated RNAP association with DNA containing pArgX promoter region as assessed by the amount of the bound (shifted) DNA (Figure 3). ppGpp did not cause any significant decrease in the RNAP binding, however, interestingly, in the presence of ppGpp and DksA the amount of bound DNA was increased comparing to the EMSA with no addition. It may suggest that the process of the RNAP binding to pArgX promoter could be stimulated by DksA irrespective of ppGpp, yet the productive transcription is inhibited by ppGpp (as shown in Figure 1).
DksA and ppGpp do not significantly destabilize open complex at pArgX promoter
Promoters initiating transcription of stable RNA typically form open complexes of relatively short half-life (38). One of the proposed mechanisms of ppGpp/DksA concerted action at stable RNA promoters during the stringent response is destabilization of open complexes hindering the isomerization into productive transcription complexes and leading to the release of RNAP (6,7). In order to dissect the molecular mechanism of the ppGpp and DksA effect on the pArgX promoter, we first assessed the effect of these regulators on the stability of competitor-resistant open complexes in the in vitro transcription reaction conditions as employed for Figure 1. RNAP-promoter complexes were formed on DNA templates in the presence of RNAP pre-treated with DksA and/or ppGpp where indicated. This was followed by the addition of heparin (to prevent re-association of RNAP with DNA), and determination of the relative fraction of active complexes remaining at subsequent times by quantifying the levels of argX- specific transcript. In the competitor control heparin was added prior to the addition of RNAP and as a result no transcripts were observed (data not shown). The decay rate of the open complexes formed at pArgX was relatively slow – after 3 h of heparin challenge the level of active complexes decreased only slightly (Figure 4). It is consistent with previous data (29), which suggested that after binding of RNAP to the promoter region, the distal UAS is looping around polymerase and stabilize the open complex. The presence of DksA and/or ppGpp resulted in a moderate decrease of the open complex stability (25% after 3 h); however, we did not observe significant differences between the effects of these regulators. The complexes formed on linear DNA template were characterized by overall shorter half-lives (3-4 times faster decay), however, the ppGpp/DksA effects were quantitatively similar to those observed for supercoiled DNA (data not shown). In the same experimental conditions, complexes formed at rrnB P1 decayed within minutes in the presence of ppGpp and DksA (data not shown and our previous observations during studying of the pR promoter, 18). Our results confirms that for any given promoter, ppGpp and DksA can destabilize open complexes. However, for the promoters forming stable open complexes, such as pArgX, this effect is unlikely to have a major role in the transcription regulation, what was also suggested for lambda pR promoter (18).
Figure 4.
The pArgX promoter open complex stability in the absence and presence of ppGpp and DksA. The open complexes were generated by 15 nM RNAP and 5 nM DNA. The levels of functional open complexes in the heparin challenge with no addition (DksA storage buffer, open squares), ppGpp at 200 μM (closed squares), DksA at 400 nM (triangles) or DksA at 400 nM with addition of 200 μM ppGpp (circles) were assessed by transcript levels at the different time points. The transcription observed at 20 s after heparin addition was set as 1. The autoradiogram presents an example of the time course experiment performed for pArgX as above.
DksA and ppGpp affect productive complex formation in pArgX transcription initiation
In light of the results obtained from in vitro and in vivo studies, we performed further studies aimed at defining the DksA/ppGpp effect on pArgX transcription initiation. The regulation of transcription from the stable promoters, such as tRNA promoters may involve various steps serving as rate-limiting ones. Thus, we assessed formation of the productive open complexes in the presence of ppGpp/DksA in the single-round transcription. In this assay the reaction was started by simultaneous addition of RNAP and NTPs to promoter DNA in the presence of DksA and ppGpp. We observed that the presence of DksA not only stimulated the transcription similarly as observed in original in vitro reaction (Figures 1 and 5), but also decreased the time necessary for the appearance of the first full-length transcripts (Figure 5A). ppGpp alone decreased the amount of transcripts as compared to the reaction with no addition, while the presence of both regulators resulted in significant inhibition of pArgX transcription (Figure 5B). The delay in the appearance of the first pArgX-specific transcript increased to 15 min in the presence of ppGpp alone and to 20 min when DksA and ppGpp were present together (Figure 5A). It should be noted that in this type of experiment the delay in the formation of full-length transcripts includes the time necessary for RNAP binding to the promoter, formation of closed and subsequently open complexes and finally, RNA polymerization competent complex formation followed by the promoter clearance. The regulators affecting promoter clearance usually lead to the release of the short abortive RNA fragments which influence the overall promoter productivity. Thus, we tested this possibility for pArgX in the in vitro transcription reaction when the level of full-length and abortive transcripts was evaluated in the 20% polyacrylamide gel electrophoresis. We observed that the presence of ppGpp/DksA did not alter the amount of short transcripts produced (data not shown). Therefore, the delay in full-length transcript appearance noted in the presence of DksA/ppGpp, is not likely due to the impaired promoter clearance.
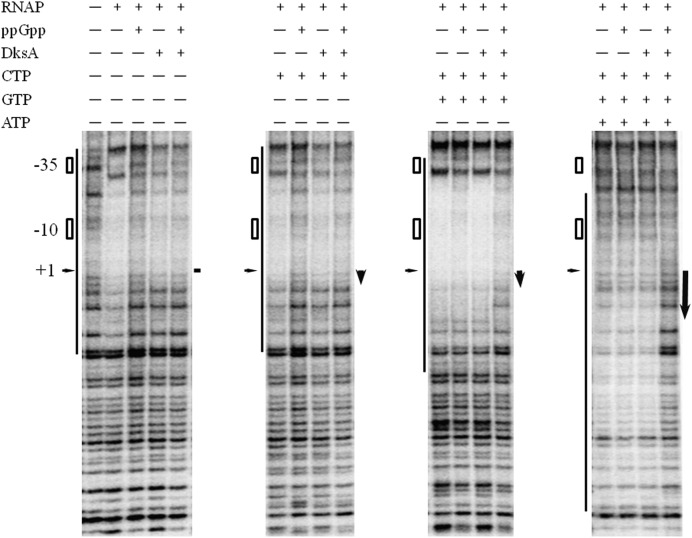
The decreased efficiency of productive complex formation and full length transcript synthesis could be a result of the formation of inactive dead-end complexes which leads to a significant decrease of transcription (31). Thus, we performed DNase I footprinting experiments where the binding of RNAP to the pArgX promoter region was studied under RNA polymerization conditions, in the presence of initiating nucleotides (Figure 6). We observed very effective protection of promoter region by RNAP, with or without the first nucleotide, C. In the same conditions, the addition of DksA caused very similar effects, even with the addition of ppGpp. Whereas, in the presence of ppGpp alone, the footprint was substantially weaker. These data confirmed the results from EMSA experiments (Figure 3), where DksA enhance the RNAP binding to the pArgX promoter region. The discrepancies in the strength of RNAP binding to pArgX in the presence of DksA between footprint and EMSA could be a result of different reaction conditions. In the presence of two initiating nucleotides (CG) the footprint was more prominent and was affected to some extent by ppGpp alone, however, the significant effect of the protection by RNAP was observed when both regulators were present. The presence of two initiating nucleotides plausibly leads to the stabilization of the RNA interaction, resulting in the formation of more stable complex. The addition of three nucleotides (CGA) to the reaction resulted in the shift in the protected region, as RNAP forms a transcription-competent complex. At this stage, the effect of DksA and ppGpp was most pronounced in reducing the protection by RNAP. These data suggest that ppGpp in the presence of DksA strongly affects the formation of binary open promoter complexes at pArgX promoter and the footprint resembles this observed for the closed promoter complex.
Figure 6.
Footprinting analysis of RNAP protection at pArgX promoter in the presence of ppGpp and/or DksA. Reactions contained double stranded template with the 5′-end (P32)-labeled, 150 nM RNAP, 600 mM ppGpp, 400 nM DksA, ATP, CTP and GTP at 1 mM where indicated. The bar represents the region of pArgX promoter protected by RNAP, the positions of −10 and −35 region, as well as +1 is indicated. The arrows indicated length of RNA synthesized at various experimental conditions.
DISCUSSION
The discovery of the role of DksA in the stringent response seemed to resolve the discrepancies in vivo and in vitro results obtained previously for model promoters. DksA was originally found to facilitate ppGpp action at specific promoters, at some promoters this results in inhibition of, at others stimulation of transcription. In addition, DksA has been reported to play an independent or even antagonistic role to ppGpp (16–18). The exact interplay between the various modes of transcription control by these regulators still need to be elucidated. A number of model promoters have been employed to dissect these mechanisms, mainly stable RNA (rRNA and tRNA) promoters, known to be inhibited during the stringent response, which allows the cell to adjust cellular metabolism to match environmental conditions (1,2). The pArgX promoter was already reported to be down-regulated during the stringent response (29) and was employed as a model for studying the effects of ppGpp and RNAP mutants (30,31). In the present work we investigated the role of DksA in pArgX transcription regulation. In the light of the previous data, pArgX was expected to behave similarly to other stable RNA promoters in response to DksA and ppGpp. Surprisingly, our results show that DksA can directly activate pArgX promoter in in vitro transcription. However, in the presence of ppGpp the promoter was significantly down-regulated by DksA. Also, the in vivo control of pArgX transcription by ppGpp requires DksA. This observation led us to examine various steps of transcription initiation to dissect the mechanism of this complex regulation. DksA enhanced the binding of RNAP to the pArgX promoter region (Figure 3), similarly to pR (18) and pfimB (17). The RNA polymerase association was not significantly affected by ppGpp, in the contrary to the remarkable decrease in the promoter activity when relatively low levels of ppGpp were present in addition to DksA (Figure 1). This may indicate that although facilitating RNAP promoter binding is a common feature of DksA in the case of pR and pArgX promoters, the overall role of ppGpp and DksA does overlap only partially. Interestingly, the analysis of open complex stability at pArgX revealed very little effect of DksA and/or ppGpp (Figure 4). However, the open complexes at pArgX are extremely stable, thus this step of transcription initiation is unlikely to play a regulatory role, contrary to rRNA promoters (characterized by very unstable complexes). The effect of DksA extends beyond RNAP association with DNA, since DksA stimulates productive complex formation for pArgX. Interestingly, this effect of DksA can be abolished by ppGpp. The negative transcription regulators acting at the level of promoter escape can exert their function either by the elevated level in the abortive transcription or by formation of stalled dead-end complexes, incapable of promoting transcription (39). For pArgX, the increased abortive transcription as responsible for ppGpp/DksA-mediated inhibition of pArgX was excluded, thus the proposed mechanism would be a formation of dead-end, transcriptionally inactive complexes of RNAP at promoter region in the presence of ppGpp, as already been observed (31) and facilitated by DksA. This hypothesis is supported by the significant delay in the formation of full length transcripts and lower transcription level observed when both regulators were present (Figure 5) as well as by defect in the promoter footprinting in the presence of ppGpp and DksA (Figure 6). The significantly affected RNAP protection pattern was observed in the RNA polymerization conditions, that is when two or three first nucleotides of the transcript were present. The atypical footprint observed in these conditions resembles the pattern occurring for the closed complexes. This is in agreement with the previously reported observation about the ppGpp effect on pArgX transcription (31). Also, as was reported by Hsu et al. (29), RNA polymerase at the pArgX promoter did not recycle normally under high NTP concentration.
Studies on the pArgX promoter transcription driven by mutant RNAP, which could mimic the effect of ppGpp, led to the model where the inhibition of the promoter function by ppGpp was due to accumulation of unproductive dead-end RNAP-DNA complexes (31). DksA-promoted allosteric modification in RNAP was proposed to affect the transition between transcription initiation intermediates at rrnB P1 promoter (13). For pArgX, DksA considerably facilitate the binding of RNAP to the promoter region, however, when ppGpp is present, the net effect is inhibitory (Figures 2 and 5), plausibly by the isomerization of the complexes into unproductive closed ones. The mechanism underlying DksA-mediated stimulation of pArgX transcription could involve different steps in transcription initiation than inhibition exerted by ppGpp together with DksA. The apparent facilitation of the ppGpp effect by DksA could be a result of a higher number of RNAP molecules bound to the DNA due to DksA effect on this step. However, DksA could increase sensitization of RNAP to ppGpp to enhance its effect on formation of unproductive complexes (7,8).
pArgX shares some features with pR, rather than with the rrnB P1 promoter, in terms of the stability of the open complexes and DksA activation by enhancing the RNAP binding to DNA. However, there are important differences originating from different context of these promoters and their physiological roles. Namely, pArgX is down-regulated by the concerted action of ppGpp and DksA, while DksA can activate pR in the presence of ppGpp. DksA deficiency in vivo leads to moderate decrease in the pArgX transcription, and notably to impairment of ppGpp-mediated down-regulation of the promoter activity while the net transcription was not affected, while the lack of both ppGpp and DksA resulted in low level of transcription from pR. Our results indicate that promoters with similar kinetics properties, like lambda pR and E. coli pArgX could be differentially affected by stringent response factors ppGpp and DksA. In light of the different physiological requirements of pR and pArgX control, the differential effects of ppGpp and DksA on the promoter activity might be of advantage. The role of pR is to start the efficient transcription of lambda genes leading to lytic development, thus during host stress, signaled by accumulation of ppGpp, the viral prophage may implement an exit strategy cumulating in lytic development. pArgX promoter initiates transcription of four genes encoding for tRNAs (29), thus the appropriate host response to elevated ppGpp level associated with nutrient starvation is inhibition of pArgX activity (reported already in 28).
It should be noted that in vivo there are more players in the regulation of RNAP activity. For example, the transcription factors, GreA and GreB, which share structure similarities with DksA, can associate with the RNA polymerase secondary channel (40). Their effect as well as their cellular concentration is a subject of the complex regulation and the net effect of the transcription at a given promoter could be a result of the balance or cooperation of various factors (20), including DNA supercoiling, Fis and H-NS (41,42). The in vitro results lead to the conclusion that DksA stimulates pArgX transcription in the absence of ppGpp. The physiological role of this stimulation could be hypothesized as a part of the complex interplay between factors associating with RNAP that could compete for the same binding site. Moreover, during rapid growth, ppGpp level is very low, estimated to be in the range of few μM, (43,44), the transcription from pArgX could be stimulated by DksA, promoting the synthesis of tRNAs needed during intense cell growth. In concert with this observation, in the absence of ppGpp, DksA slightly increases promoter activity (Figure 2), by increasing the binding of RNAP to the promoter region, as shown in EMSA experiments (Figure 3). When bacteria enter stationary phase, or when nutrient deficiency conditions occur, ppGpp level increases dramatically affecting pArgX-initiated transcription. Nevertheless, the ppGpp-mediated in vivo effect requires DksA (Figure 2). The observed effect of down-regulation of pArgX activity upon amino acid starvation (28) corresponds to the situation in vitro when both factors are present. DksA level in the cell is constant (7,11) while ppGpp concentration varies with growth phases and conditions. Thus, DksA/ppGpp concerted action inhibits pArgX promoter in unfavorable growth conditions, however, in rapidly growing cells, when ppGpp level is extremely low and high pArgX activity is required, the promoter takes advantage of DksA-mediated stimulation.
Thus, our studies on pArgX transcription regulation show new aspects of opposite roles of ppGpp and DksA which includes DksA-mediated transcription activation in the absence of ppGpp and facilitating ppGpp-mediated inhibition at high ppGpp levels. Such an intriguing dual function of DksA can shed new light on the function of this protein in bacteria and give us new insight into the dissection of the molecular mechanisms of the function of DksA and ppGpp. To the best of our knowledge, pArgX transcription regulation is the first example for condition-dependent opposing function of DksA. Whether this could be a general mechanism for DksA effects on other promoters (e.g. tRNA promoters), remains yet to be elucidated.
Acknowledgments
The authors thank Dr Grzegorz Węgrzyn and Dr Katarzyna Potrykus for encouragement and critical reading of the manuscript and Dr Michael Cashel for discussion. The authors are grateful to anonymous reviewers for comments that led to significant improvement of the manuscript.
Dedication: This work is dedicated to the memory of Jim Hernandez.
FUNDING
Polish Ministry of Science and Higher Education [N N301 161635 to A.S.-P.]; National Science Centre, Poland [2013/09/D/NZ1/03347 to R.Ł.]; Foundation for Polish Science, programme ‘START’ (to R.Ł); Educators for the elite-integrated training program for PhD students, post-docs and professors as academic teachers at University of Gdańsk, program founded by the European Social Found, framework - Human Capital Operational Programme, Action 4.1.1 - Improving the quality of educational offer of tertiary education institutions [to R.Ł] Funding for open access charge: National Science Centre, Poland [2013/09/D/NZ1/03347 to R.Ł.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Cashel M., Gentry D., Hernandez V.J., Vinella D. Escherichia coli and Salmonella: Cellular and Molecular Biology. I. Washington DC: American Society for Microbiology; 1996. The stringent response; pp. 1458–1496. [Google Scholar]
- 2.Potrykus K., Cashel M. ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 3.Durfee T., Hansen A.M., Zhi H., Blattner F.R., Jin D.J. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 2007;90:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traxler M.F., Zacharia V.M., Marquardt S., Summers S.M., Nguyen H.T., Stark S.E., Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol. Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang P.J., Craig E.A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perederina A., Svetlov V., Vassylyeva M.N., Tahirov T.H., Yokoyama S., Artsimovitch I., Vassylyev D.G. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 7.Paul B.J., Barker M.M., Ross W., Schneider D.A., Webb C., Foster J.W., Gourse R.L. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Paul B.J., Berkmen M.B., Gourse R.L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemke J.J., Durfee T., Gourse R.L. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haugen S.P., Ross W., Gourse R.L. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrangsu P., Lemke J.J., Gourse R.L. The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol. Microbiol. 2011;80:1337–1348. doi: 10.1111/j.1365-2958.2011.07649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blankschien M.D., Lee J.H., Grace E.D., Lennon C.W., Halliday J.A., Ross W., Gourse R.L., Herman C. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009;28:1720–1731. doi: 10.1038/emboj.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford S.T., Villers C.L., Lee J.H., Ross W., Gourse R.L. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lennon C.W., Gaal T., Ross W., Gourse R.L. Escherichia coli DksA binds to free RNA polymerase with higher affinity than to RNA polymerase in an open complex. J. Bacteriol. 2009;191:5854–5858. doi: 10.1128/JB.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts J.W. Promoter-specific control of E. coli RNA polymerase by ppGpp and a general transcription factor. Genes Dev. 2009;23:143–146. doi: 10.1101/gad.1770509. [DOI] [PubMed] [Google Scholar]
- 16.Magnusson L.U, Gummesson B., Joksimović P., Farewell A., Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aberg A., Shingler V., Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- 18.Łyżeń R., Kochanowska M., Węgrzyn G., Szalewska-Pałasz A. Transcription from bacteriophage λ pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 2009;37:6655–6664. doi: 10.1093/nar/gkp676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrikh H., Ferrazzoli A.E., Lovett S.T. Growth phase and ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J. Bacteriol. 2009;191:7436–7446. doi: 10.1128/JB.00412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinella D., Potrykus K., Murphy H., Cashel M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol. 2012;194:261–273. doi: 10.1128/JB.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopalkrishnan S., Nicoloff H., Ades S.E. Co-ordinated regulation of the extracytoplasmic stress factor, sigmaE, with other Escherichia coli sigma factors by ppGpp and DksA may be achieved by specific regulation of individual holoenzymes. Mol. Microbiol. 2014;93:479–493. doi: 10.1111/mmi.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potrykus K., Wegrzyn G., Hernandez V.J. Multiple mechanisms of transcription inhibition by ppGpp at the lambda p(R) promoter. J. Biol. Chem. 2002;277:43785–43791. doi: 10.1074/jbc.M208768200. [DOI] [PubMed] [Google Scholar]
- 23.Gralla J.D. Escherichia coli ribosomal RNA transcription: Regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol. Microbiol. 2005;55:973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- 24.Paul B.J., Ross W., Gaal T., Gourse R.L. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 25.Kolmsee T., Delic D., Agyenim T., Calles C., Wagner R. Differential stringent control of Escherichia coli rRNA promoters: effects of ppGpp, DksA and the initiating nucleotides. Microbiology. 2011;157:2871–2879. doi: 10.1099/mic.0.052357-0. [DOI] [PubMed] [Google Scholar]
- 26.Figueroa-Bossi N., Guérin M., Rahmouni R., Leng M., Bossi L. The supercoiling sensitivity of a bacterial tRNA promoter parallels its responsiveness to stringent control. EMBO J. 1998;17:2359–2367. doi: 10.1093/emboj/17.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pemberton I.K., Muskhelishvili G., Travers A.A., Buckle M. The G+C-rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. J. Mol. Biol. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- 28.Rowley K.B., Elford R.M., Roberts I., Holmes W.M. In vivo regulatory responses of four Escherichia coli operons which encode leucyl-tRNAs. J. Bacteriol. 1993;175:1309–1315. doi: 10.1128/jb.175.5.1309-1315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu L.M., Giannini J.K., Leung T.W., Crosthwaite J.C. Upstream sequence activation of Escherichia coli argT promoter in vivo and in vitro. Biochemistry. 1991;30:813–822. doi: 10.1021/bi00217a035. [DOI] [PubMed] [Google Scholar]
- 30.Toulokhonov I.I., Shulgina I., Hernandez V.J. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta'-subunit. J. Biol. Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- 31.Maitra A., Shulgina I., Hernandez V.J. Conversion of active promoter-RNA polymerase complexes into inactive promoter bound complexes in E. coli by the transcription effector, ppGpp. Mol. Cell. 2005;17:817–829. doi: 10.1016/j.molcel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd edn. NY: Cold Spring Harbor Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 33.Elliott T., Geiduschek E.P. Defining a bacteriophage T4 late promoter: absence of a ‘‘-35’’ region. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- 34.Burgess R.R., Jendrisak J.J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 35.Hager D.A., Jin D.J., Burgess R.R. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 36.Guantes R., Cayrol B., Busi F., Arluison V. Positive regulatory dynamics by a small noncoding RNA: Speeding up responses under temperature stress. Mol. BioSyst. 2012;8:1707–1715. doi: 10.1039/c2mb05479e. [DOI] [PubMed] [Google Scholar]
- 37.Perron K., Comte R., van Delden C. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 2005;56:1087–1102. doi: 10.1111/j.1365-2958.2005.04597.x. [DOI] [PubMed] [Google Scholar]
- 38.Gourse R.L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu LM. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 40.Laptenko O., Lee J., Lomakin I., Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pokholok D.K., Redlak M., Turnbough C.L., Jr, Dylla S., Holmes W.M. Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli. J. Bacteriol. 1999;181:5771–5782. doi: 10.1128/jb.181.18.5771-5782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opel M.L., Aeling K.A., Holmes W.M., Johnson R.C., Benham C.J., Hatfield G.W. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 2004;53:665–674. doi: 10.1111/j.1365-2958.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- 43.Neubauer P., Ahman M., Tornkvist M., Larsson G., Enfors S.O. Response of guanosine tetraphosphate to glucose fluctuations in fed-batch cultivations of Escherichia coli. J. Biotechnol. 1995;43:195–204. doi: 10.1016/0168-1656(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 44.Słomińska M., Neubauer P., Węgrzyn G. Regulation of Bacteriophage Lambda Development by Guanosine 5′-Diphosphate-3′diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]